Abstract
An important question in memory development is understanding the differences between effector CD8 T cells that die versus effector cells that survive and give rise to memory cells. In this study, we provide a comprehensive phenotypic, functional, and genomic profiling of terminal effectors and memory precursors. Using killer cell lectin-like receptor G1 as a marker to distinguish these effector subsets, we found that despite their diverse cell fates, both subsets possessed remarkably similar gene expression profiles and functioned as equally potent killer cells. However, only the memory precursors were capable of making interleukin (IL) 2, thus defining a novel effector cell that was cytotoxic, expressed granzyme B, and produced inflammatory cytokines in addition to IL-2. This effector population then differentiated into long-lived protective memory T cells capable of self-renewal and rapid recall responses. Experiments to understand the signals that regulate the generation of terminal effectors versus memory precursors showed that cells that continued to receive antigenic stimulation during the later stages of infection were more likely to become terminal effectors. Importantly, curtailing antigenic stimulation toward the tail end of the acute infection enhanced the generation of memory cells. These studies support the decreasing potential model of memory differentiation and show that the duration of antigenic stimulation is a critical regulator of memory formation.
The potential to rapidly proliferate, mount accelerated effector functions, and thereby blunt the severity of a second infection with the same pathogen makes the generation of memory CD8 T cells an important vaccination goal. In the past several years there have been major advances in our understanding of memory CD8 T cell differentiation and maintenance (1–9). During an acute infection, antigenic stimulation leads to expansion and differentiation of naive CD8 T cells into effector cells (10–14). Elaboration of effector functions via cytotoxic molecules (perforin and granzymes) and effector cytokines (IFN-γ and TNF-α) results in effective pathogen control, after which, ∼5–10% of the antigen-specific cells present at the peak of the effector response survive and differentiate into long-lived memory cells.
Differentiation of memory CD8 T cells is a progressive process in which key genotypic, phenotypic, and functional properties are acquired over several weeks after antigen clearance (15–17). CD8 T cells that are fated to differentiate into long-lived memory cells (memory precursors) are believed to exist within the effector pool, and cell-surface markers such as IL-7Rα have been proposed to distinguish them from terminal effector cells that are destined to die during the contraction phase (6, 18–23). It has also been shown that a brief period of antigen encounter can commit naive cells to a program of effector and memory differentiation (10–14). However, it is important to consider that the duration of stimulation for most acute infections is prolonged until the virus is cleared at the peak of the effector response. During this primary course of an acute infection, how repetitive stimulations shape CD8 T cell lineage decisions remains largely undefined. Thus, signals accumulated during the expansion phase of acute infections and their role in regulating the generation of memory precursors and terminal effectors remain areas of active investigation (5, 24).
Moreover, whether memory cells pass through an effector phase is a central question in memory differentiation that continues to be debated (2, 5, 25, 26). Although several lines of evidence support the notion that memory cells are direct descendants of effector cells (15, 27, 28), recent data in both mouse and human infection models suggest that memory cells do not pass through a fully differentiated effector phase (29–36). In this case, memory cells are proposed to directly differentiate from naive cells by virtue of qualitative and quantitative differences in signals perceived (antigen, cytokine, co-stimulation, CD4 T cell help, etc.), putatively because of a differential timing of recruitment into the immune response or localization. In particular, early inflammatory cytokine signals (IL-12, type-I IFNs, IFN-γ, IL-2, IL-21, etc.), now referred to as the “third signal,” have been shown to be intimately involved in the promotion of effector T cell responses (37–42). Recently, asymmetry in cell division has also been invoked in support of the divergent model of memory differentiation (43), whereby unequal partitioning of proteins during the first division leads to the generation of two distinct daughter cells: one cell is fated toward an effector lineage (i.e., it expresses IFN-γR, t-bet, granzyme B, and CD43), and the other one has attributes of a memory cell (i.e., lower expression of granzyme B, t-bet, and CD43, and higher IL-7Rα and CD62L expression).
In this study, we sought to enhance our understanding of the mechanisms regulating the development of memory CD8 T cells and gain further insights into their developmental path. Our current experiments demonstrate the differentiation of naive CD8 T cells into potent effector cells early during the immune response, and it is from these effector cells that memory cells eventually arise after antigen clearance. We also delineate an as of yet undescribed novel effector subset (IL-2–producing, IFN-γ+, TNF-α+, granzyme B+ killer cells), distinguishable by lower expression of killer cell lectin-like receptor G 1 (KLRG-1), which is distinct from non–IL-2–producing KLRG-1Hi terminal effector cells that are also identifiable in the expanding IL-7RαLo antigen-specific population when antigen is still around. Finally, gene profiling and mechanistic experiments examining the generation of terminal effector cells versus memory precursors demonstrate that terminal effectors are driven further down the effector differentiation pathway by continued proliferation and stimulation toward the tail end of antigen clearance, thereby making them metabolically and functionally unfit for memory lineage.
RESULTS
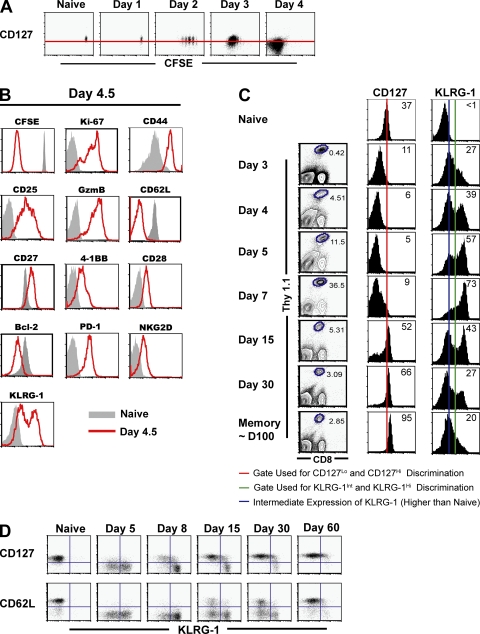
Effector CD8 T cells uniformly down-regulate CD127 but can be distinguished on the basis of KLRG-1 expression
Selective expression of IL-7Rα on a subset of antigen-specific cells correlates with an increased potential for the generation of long-lived memory cells and is functionally necessary, albeit not sufficient, for memory development (18, 19, 44–46). However, it is unclear whether (a) IL-7Rα expression is down-regulated uniformly on all antigen-specific cells after activation, such that memory precursors subsequently reexpress IL-7Rα after antigen clearance, or (b) whether memory precursors retain IL-7Rα expression through the course of infection. Thus, we performed a detailed analysis of IL-7Rα expression on antigen-specific CD8 T cells during expansion (days 1–7) after lymphocytic choriomeningitis virus (LCMV) infection. We observed that IL-7Rα was down-regulated in a uniform manner on early antigen-specific cells independent of the extent of cell division (during the first 5–6 rounds; Fig. 1 A), and it continued to decline progressively until ∼4–5 d postinfection (p.i.), when the antigen-specific cells had undergone at least 8–10 rounds of cell division (Fig. 1, A and C). Consistent with previous reports (18, 19, 47), a fraction of the effector cells subsequently reexpressed CD127 by day 7 p.i. Thereafter (days >7 p.i.), the relative frequency of CD127+ cells as well as the expression levels of CD127 progressively increased. These data demonstrate that after activation, CD127 expression is uniformly down-regulated on antigen-specific cells and is selectively regained on a subset of cells after viral clearance. This selective reexpression of IL-7Rα by a subset of previously CD127Lo cells suggests differential programming of memory precursors and terminal effector cells during expansion.
Figure 1.
Effector CD8 T cells uniformly down-regulate CD127 but can be distinguished on the basis of KLRG-1 expression. (A) Cell-surface expression of IL-7Rα with respect to cell division at days 1–4 after LCMV infection. 106 naive CFSE-labeled P14 CD8 T cells were adoptively transferred into naive mice that were subsequently infected with LCMV. All plots are gated on CD8+ Thy1.1+ P14 splenocytes directly stained ex vivo for IL-7Rα expression. An uninfected naive control is also shown, and the red horizontal line indicates the naive level of expression. (B) Phenotypic properties of CD127Lo antigen-specific cells at day 4.5 p.i. B6 mice containing ∼105 naive Thy1.1+ P14 cells were infected with LCMV, and expression of the indicated cell-surface and intracellular markers on CD8+ Thy1.1+ splenocytes was assessed 4.5 d later (red line histograms). Gray histograms represent naive cells from uninfected control mice. (C) Longitudinal analysis of cell-surface IL-7Rα and KLRG-1 expression. B6 mice containing ∼105 naive Thy1.1+ P14 cells were infected with LCMV, and the cell-surface expression of IL-7Rα and KLRG-1 was analyzed at the indicated time points. Histograms depict the MFI of IL-7Rα expression and percentages of KLRG-1Hi cells gated on CD8+ Thy1.1+ P14 cells. (D) Inverse association of KLRG-1 expression with the memory markers CD127 and CD62L. B6 mice containing 105 CD8+ Thy1.1+ P14 cells were infected with LCMV, and KLRG-1 expression with respect to CD127 or CD62L was analyzed at the indicated time points.
Therefore, we hypothesized that heterogeneous expression of other markers within the homogenously CD127Lo pool of antigen-specific cells may delineate such differential programming. Hence a detailed marker analysis of day 4.5 CD8 T cells was performed. For the most part, there was no clear heterogeneity in the expression of ∼30 phenotypic markers that were evaluated (Fig. 1 B and not depicted). The day 4.5 CD127Lo antigen-specific cells demonstrated the typical characteristics of effector cells: they produced high levels of granzyme B, uniformly down-regulated lymph node–homing molecule CD62L to aid in peripheral homing, and underwent at least 8–10 rounds of division (CFSE dilution). Expression of IL-2Rα (CD25) was higher on responding cells than naive cells, indicating that cells were responding to antigen in the environment. Co-stimulatory receptors (CD28, CD27, and 4-1BB), survival and apoptotic molecules like Bcl-2 and caspase-3, and cytokine receptor components (IL-15Rα, CD132, and CD122) were all uniformly expressed. The effector cells also uniformly up-regulated the expression of a variety of inhibitory molecules, including programmed death 1 and NK cell inhibitory receptors (NKG2A, NKG2D, 2B4, etc.). Despite this evident uniformity in most markers, the CD127Lo antigen-specific cells exhibited a striking heterogeneity in the cell-surface expression of the NK cell receptor KLRG-1 (Fig. 1, B and C) (48). Although naive cells expressed low levels of KLRG-1 (KLRG-1Lo mean fluorescence intensity [MFI] = 16–20), two distinct subpopulations of activated CD8 T cells with increased KLRG-1 expression were distinguishable as early as days 4–5 p.i. (when CD127 expression is uniformly low): KLRG-1Int (MFI = 82–90) and KLRG-1Hi (MFI = 350–495; Fig. 1 C).
An analysis of KLRG-1 expression with respect to cell-surface markers classically associated with long-lived memory cells is likely to provide us with clues about which effector subset (KLRG-1Int or KLRG-1Hi) may give rise to the long-lived memory lineage. Memory cells express higher levels of CD127, and increased CD62L expression is typically associated with the long-lived central memory lineage (5). A longitudinal analysis of KLRG-1 expression with respect to these markers revealed that even though heterogeneous KLRG-1 expression subdivided the CD127LoCD62LLo early effector population at days 4–5 p.i., KLRG-1 expression was inversely associated with CD127 and CD62L later in the immune response (Fig. 1 D).
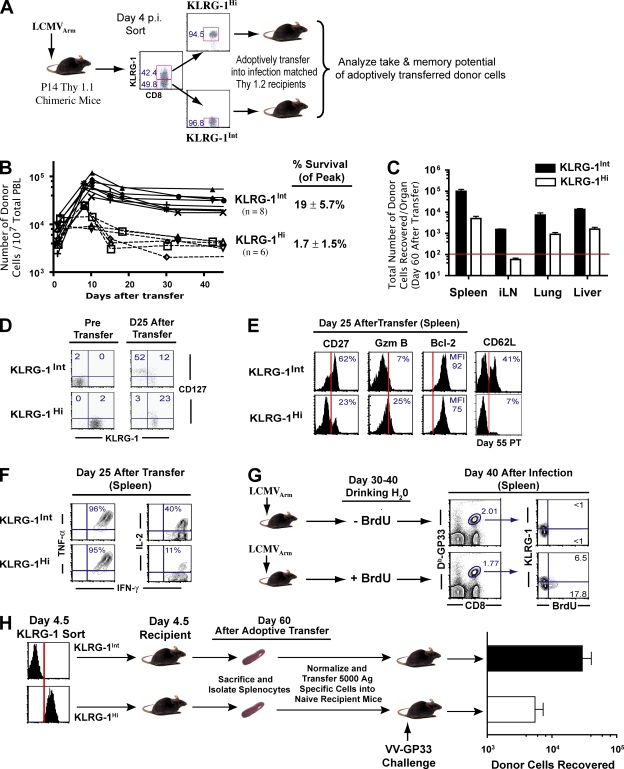
Heterogeneity in KLRG-1 expression identifies effector CD8 T cells with distinct memory lineage fates
The inverse expression pattern of KLRG-1 with respect to CD127 and CD62L suggested that after antigen clearance CD127Hi CD62LHi memory CD8 T cells were differentiating from the day 4–5 KLRG-1Int effector cells. However, it may also be possible that this simply represented a conversion of KLRG-1Hi cells into KLRG-1Int memory cells after antigen clearance. To distinguish between these two possibilities, we analyzed the memory potential of day 4–5 KLRG-1Int and KLRG-1Hi CD8 T cells by FACS purifying the two populations and adoptively transferring equal numbers into infection-matched recipient mice (Fig. 2 A). We chose infection-matched recipients instead of naive recipients for two reasons: (a) this allowed us to follow the differentiation of sorted donor cells in the same environment as the donor mice, and (b) this bypassed potential problems of virus transfer from day 4–5–infected donor mice into naive mice and initiating a new round of infection. To evaluate the possibility of differential engraftment of KLRG-1Int and KLRG-1Hi cells initially, which may lead to different levels of memory cells arising from these two effector subsets, we first analyzed the “take” of donor cells in recipient mice within 8 h of adoptive transfer (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20071641/DC1). Similar numbers of KLRG-1Int and KLRG-1Hi cells were engrafted into lymphoid and nonlymphoid tissues (spleen, lymph node, liver, i.p. cavity, and blood). However, strikingly different memory outcomes were observed: KLRG-1Hi donor cells largely died during contraction (Fig. 2, A and B), whereas 7–10-fold higher levels of KLRG-1Int donor cells were detectable in blood after contraction. At day 60 after adoptive transfer, KLRG-1Int donor cells gave rise to 5–12-fold higher total numbers of surviving memory cells, in both lymphoid and nonlymphoid tissues, compared with KLRG-1Hi donor cells (Fig. 2, B and C).
Figure 2.
Heterogeneity in KLRG-1 expression identifies effector CD8 T cells with distinct memory lineage fates. KLRG-1Int effector cells preferentially give rise to long-lived memory cells. (A) KLRG-1Int and KLRG-1Hi cells were sorted from B6 mice containing 106 Thy1.1+ P14 cells 4 d after LCMV infection. Equal numbers (∼106) of sorted cells were transferred into infection-matched Thy1.2 recipients. The numbers indicate percentages of corresponding gated populations. (B) Expansion and contraction of adoptively transferred KLRG-1Int and KLRG-1Hi cells was longitudinally assessed in the blood of recipient mice by staining for the Thy1.1+ marker. (C) Long-term survival of the sorted donor KLRG-1Int and KLRG-1Hi effector cells was determined by enumerating the total number of donor cells in the spleen, lymph node, lung, liver, and blood ∼60 d after adoptive transfer. The horizontal red line indicates the lower limit of detection for absolute numbers of antigen-specific cells. Mean data from six to eight mice are presented, and error bars represent SEM. (D and E) Detailed phenotypic characterization of surviving KLRG-1Int and KLRG-1Hi donor cells in spleens at memory. Percentages of donor cells expressing the indicated markers are depicted. The MFI of Bcl-2 expression is presented. Vertical red lines indicate the negative expression gate for the respective markers. (F) Cytokine production by donor cells at day 25 after transfer. Production of intracellular cytokines (IFN-γ, TNF-α, and IL-2) in Thy1.1+ donor cells was evaluated after 5 h of stimulation with GP33-41 peptide in the presence of BFA in vitro. Percentages of donor cells producing IFN-γ, TNF-α, and IL-2 are depicted in the respective histograms. (G) Homeostatic proliferation of KLRG-1Int and KLRG-1Hi memory cells was assessed in LCMV-infected B6 mice. Immune mice were fed BrdU in their drinking water between days 30 and 40 p.i. Subsequently, BrdU incorporation in KLRG-1Int and KLRG-1Hi splenic DbGP33–specific memory cells was evaluated by flow cytometry. The numbers indicate percentages of corresponding gated populations. (H) Recall proliferation and boosting ability of KLRG-1Int and KLRG-1Hi memory cells. KLRG-1Int and KLRG-1Hi P14 cells were adoptively transferred into infection-matched mice; 30 d later, equal numbers of surviving memory cells (Thy 1.1+) from each group were transferred into Thy1.2+ naive mice, which were infected i.v. with 2 × 106 PFU VV-GP33. Donor cells recovered from the spleen at day 60 after challenge are plotted. Mean data from six mice are presented, and error bars represent SEM.
Consistent with their enhanced survival, KLRG-1Int cells preferentially reexpressed CD127 compared with KLRG-1Hi cells (Fig. 2 D), indicating that the decision to up-regulate CD127 after antigen clearance was selectively imprinted in the KLRG-1Int cells early during expansion. It is noteworthy that although a fraction of KLRG-1Int donor cells up-regulated KLRG-1 expression (Fig. 2 D), KLRG-1Hi donor cells did not convert back to KLRG-1Int cells. In addition to CD127 expression, a fraction of the KLRG-1Int donor cells also progressively acquired CD62L expression after antigen clearance (Fig. 2 E) and exhibited preferential localization in lymph nodes compared with peripheral lung and liver tissues (not depicted). Thus, KLRG-1Int donor cells gave rise to both central and effector memory cells. In contrast, the few surviving KLRG-1Hi donor cells (1–2% of peak donors) did not up-regulate CD62L and largely exhibited effector memory properties (higher granzyme B and lower Bcl-2 and CD27 expression [Fig. 2 E], and preferential distribution in peripheral lung and liver tissues [not depicted]). Functionally, memory cells arising from KLRG-1Int donor cells and the small fraction of surviving KLRG-1Hi donor cells both similarly produced IFN-γ and TNF-α, but KLRG-1Int donor cells exhibited enhanced IL-2 production (Fig. 2 F), a key central memory property (16).
We next examined functional memory properties (homeostatic proliferation and recall proliferation to antigen) associated with KLRG-1Int and KLRG-1Hi cells. Long-lived memory CD8 T cells exhibit antigen-independent longevity by virtue of slow homeostatic proliferation in response to the cytokines IL-15 and IL-7 (5, 8, 49). To compare the homeostatic proliferation potential of KLRG-1Int and KLRG-1Hi cells, we administered BrdU in the drinking water of B6 mice that were infected with LCMV 30 d earlier. Selective BrdU incorporation by KLRG-1Int memory cells indicated that their longevity could be attributed to enhanced antigen-independent homeostatic proliferation (Fig. 2 G).
To rigorously test the recall proliferation and boosting of memory cells generated from KLRG-1Int and KLRG-1Hi day 4.5 donor cells, we adoptively transferred equal numbers of surviving memory cells from each group into naive mice and challenged them with recombinant vaccinia virus (VV) expressing the GP33-41 epitope (VV-GP33). We found that memory cells derived from KLRG-1Int donor cells boosted 5–10–fold better than those from KLRG-1Hi cells (Fig. 2 H). Thus, KLRG-1Int donor cells composed the memory precursor subset that preferentially differentiated into fully functional memory cells capable of self-renewal and superior recall proliferation. In contrast, KLRG-1Hi donor cells represented terminal effector cells that largely died during contraction. Collectively, these data indicate that the lineage commitment to long-lived lymphoid and nonlymphoid memory subsets is imprinted on antigen-specific cells early on during the expansion phase of the primary CD8 T cell response.
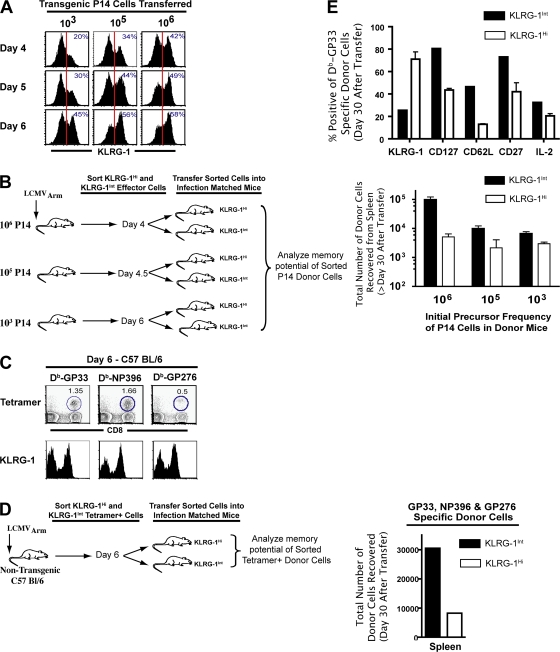
Diverse cell fates associated with KLRG-1Int and KLRG-1Hi effector CD8 T cells are independent of initial precursor frequencies of antigen-specific cells and are seen with both transgenic (Tg) T cells and endogenous cells in normal B6 mice
To detect cells at early time points (days 1–5), we used DbGP33-specific TCR Tg P14 cells at a high precursor frequency in the experiments shown in Figs. 1 and Figs.2. It has been recently proposed that the higher than physiological initial precursor frequency of naive TCR Tg cells could lead to altered differentiation of memory CD8 T cells compared with endogenous cells (22, 50). Thus, to address this issue, we evaluated KLRG-1 expression on P14 cells at various input numbers (low, intermediate, and high), as well as on tetramer-specific endogenous cells in B6 mice. As shown in Fig. 3, the evident heterogeneity in KLRG-1 expression was observed irrespective of the dose of Tg P14 cells transferred (103, 105, and 106 cells; Fig. 3 A). Notably, at P14 doses (103 cells) representing endogenous naive precursor frequencies of CD8 T cells (50, 51), heterogeneity in the expression of KLRG-1 was also observed as early as day 4–5 p.i. (Fig. 3 A). Likewise, endogenous GP33-, NP396-, and GP276-specific CD8 T cells also exhibited similar heterogeneity in KLRG-1 expression during expansion (Fig. 3 C). More importantly, irrespective of the naive precursor frequency of Tg cells used in donor mice, KLRG-1Int donor cells exhibited a preferential capacity to survive and differentiate into long-lived memory cells compared with KLRG-1Hi donor cells (Fig. 3 B). In the most stringent of tests, when we sorted KLRG-1Int and KLRG-1Hi cells from B6 mice infected 6 d earlier and adoptively transferred them into infection-matched recipients, we again observed a preferential generation of memory cells from KLRG-1Int cells (Fig. 3 D). Moreover, we observed that endogenous tetramer+ KLRG-1Int cells gave rise to both CD62LHi and CD62LLo memory cells. And, similar to our data with Tg cells, KLRG-1Int cells preferentially differentiated into both lymphoid and nonlymphoid memory cells (not depicted) that expressed higher levels of CD127, CD62L, CD27, and IL-2 (Fig. 3 E), in contrast to KLRG-1Hi cells. Thus, both Tg and endogenous CD8 T cells exhibit similar heterogeneity in the cell-surface expression of KLRG-1, which is associated with their diverse memory fates.
Figure 3.
Diverse cell fates associated with KLRG-1Int and KLRG-1Hi effector CD8 T cells are independent of initial precursor frequencies of antigen-specific cells and are seen with both Tg T cells and endogenous cells in normal B6 mice. (A) B6 mice adoptively transferred with low (103) or high (105 or 106) doses of naive P14 cells were infected with LCMV. CD8+ Thy1.1+ donor cells in the spleen were analyzed for cell-surface KLRG-1 expression at days 4, 5, and 6 p.i. Vertical red lines indicate the gate for intermediate KLRG-1 expression. (B) KLRG-1Int and KLRG-1Hi effector cells were sorted from B6 mice containing 106, 105, or 103 naive P14 cells at days 4, 4.5, and 6 p.i., respectively. KLRG-1Int and KLRG-1Hi donors from each group were transferred into infection-matched recipients, and total numbers of memory cells in the spleen were enumerated at days 40–60 after transfer. Mean data are plotted, and error bars represent SEM. (C) Non-Tg B6 mice were infected with LCMV, and 6-d p.i. cell-surface expression of KLRG-1 on DbGP33-, DbNP396-, and DbGP276-specific CD8 T cells was assessed using anti-CD8α, anti–KLRG-1, and the respective MHC class I tetramers. Numbers show the frequency of tetramer-positive cells. (D) Tetramer-specific cells were sorted into KLRG-1Int and KLRG-1Hi populations from Thy1.2+ B6 mice infected with LCMV 6 d earlier. A total of 1.5 × 106 KLRG-1Hi– and KLRG-1Int–sorted DbGP33+ DbNP396+ DbGP276+ cells were transferred into infection-matched Thy1.1 recipients. Total numbers of DbGP33+ DbNP396+ DbGP276+ donor cells in the spleen are shown at 20 d after transfer. (E) Surface markers were analyzed on DbGP33-specific CD8 T cells by direct ex vivo staining of splenocytes, and production of IL-2 by memory cells was determined by ex vivo stimulation with GP33 peptide for 5 h. Bar graphs represent the fraction of donor cells expressing the indicated markers. Mean data are plotted, and error bars represent SEM.
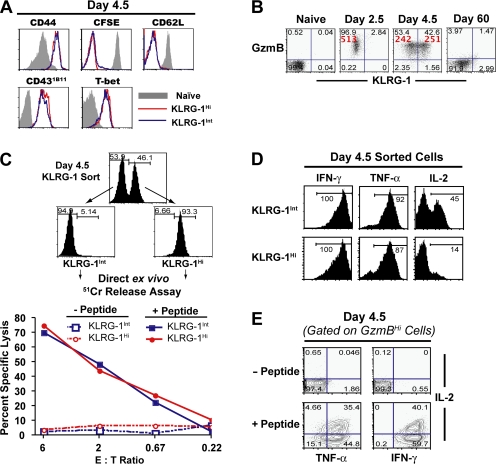
Memory cells differentiate from a novel IL-2–producing effector CD8 T cell subset capable of granzyme B production and direct ex vivo killing
The differentiation path followed by memory cells is much debated, and it has been proposed that memory cells do not arise from effector cells but compose an independent lineage (2, 5). Thus, we investigated whether the KLRG-1Int and KLRG-1Hi CD8 T cell subsets exhibited effector function. Typically, effector CD8 T cells are classified based on their ability to migrate to peripheral sites of infection and to produce effector cytokines (IFN-γ and TNF-α) and effector molecules (granzyme B and perforin). More importantly, effector cytotoxic T lymphocytes, as implied by their name, are capable of direct ex vivo killing. A comparison of these effector properties between memory precursors and terminal effectors demonstrated that both subsets expressed similar levels of effector cell markers such as granzyme B and CD43 (Fig. 4, A and B). Similar down-regulation of lymphoid-homing molecules like CD62L on KLRG-1Int and KLRG-1Hi cells also attested to their differentiation into effector cells. Notably, both subsets expressed similarly high levels of transcription factor T-bet at day 4–5 p.i. (Fig. 4 A), which has been functionally implicated in CD8 T cell effector differentiation (52, 53). To further assess the effector differentiation of memory precursors and terminal effectors, we performed a conventional in vitro Cr51 release assay with day 4.5 KLRG-1Hi– and KLRG-1Int–sorted cells. Also in terms of this important CTL function of direct ex vivo killing, both subsets exhibited equally potent cytotoxicity (Fig. 4 C), even at the lowest effector/target ratios evaluated. And consistent with their rapid direct ex vivo killing potential, purified KLRG-1Int and KLRG-1Hi subsets also produced similarly high levels of the effector cytokines IFN-γ and TNF-α (Fig. 4 D), indicating that both subsets are potent effector cells.
Figure 4.
Memory cells differentiate from a novel IL-2–producing effector CD8 T cell subset capable of granzyme B production and direct ex vivo killing. (A) Phenotypic characterization of KLRG-1Int (blue lines) and KLRG-1Hi (red lines) subsets at day 4.5 p.i. B6 mice containing ∼105 naive P14 cells were infected with LCMV, and the cell-surface expression of the indicated phenotypic markers on the donor cells was analyzed with respect to KLRG-1 expression at day 4.5 p.i. All plots are gated on CD8+ Thy1.1+ donor cells. Gray histograms represent naive controls. For analysis of cell proliferation, naive donor cells were labeled with CFSE before adoptive transfer and infection. (B) Direct ex vivo granzyme B production by naive P14 cells and P14 cells at days 2.5, 4.5, and 60 p.i. Granzyme B expression with respect to KLRG-1 expression in Thy1.1+ splenocytes is presented as dot plots, and quadrant frequencies are indicated. The MFI of granzyme B expression is given in red. (C) Direct ex vivo cytolytic activity of KLRG-1Int– and KLRG-1Hi–sorted cells during expansion. A standard 5-h Cr release assay was performed to assess direct ex vivo cytolytic activity of the KLRG-1Int and KRG-1Hi effector cells at the indicated effector/target cell ratios. (D) Production of IFN-γ, TNF-α, and IL-2 by FACS-purified KLRG-1Int and KLRG-1Hi effector cells at day 4.5 p.i. was evaluated after 5 h of in vitro stimulation with GP33-41 peptide in the presence of BFA. Percentages of cytokine-producing cells are indicated. (E) IFN-γ, TNF-α, and IL-2 production by day 4.5 granzyme B–expressing effector cells is shown. Plots are gated on CD8+ Thy1.1+ granzyme B+ cells, and quadrant frequencies are indicated.
One aspect that distinguished KLRG-1Int from KLRG-1Hi cells was their IL-2 production (Fig. 4 D). Although both early subsets were capable of producing similar levels of IFN-γ and TNF-α, the ability to produce IL-2 was exclusively limited to memory precursors. Strong IL-2 production is not usually associated with effector cells (3, 54, 55) but is instead a property typically ascribed to naive and central memory cells (16). To be sure that IL-2 was being produced by effector CTL, we gated on granzyme B+ cells and checked their ability to produce IL-2. As shown in Fig. 4 E, a subset of granzyme B+, IFN-γ+, TNF-α+ effector cells was clearly capable of IL-2 production. It is also noteworthy that the association of IL-2 production with KLRG-1Int cells was consistent at every time point investigated (days 5, 8, 15, 30, and 60 p.i.; unpublished data), suggesting that IL-2 production is a selective property of long-lived antigen-specific CD8 T cells throughout the course of CD8 T cell memory differentiation. Combined with the observation that all antigen-specific cells pass through a uniformly granzyme BHi effector state (day 2.5 and 4.5) before differentiating into granzyme BLo memory cells (Fig. 4 B), similar effector properties of memory precursors and terminal effectors strongly support the notion that memory cells directly differentiate from a novel subset of effector cytotoxic T cells capable of IL-2 production.
Genome-wide microarray analysis of memory precursors and terminal effectors
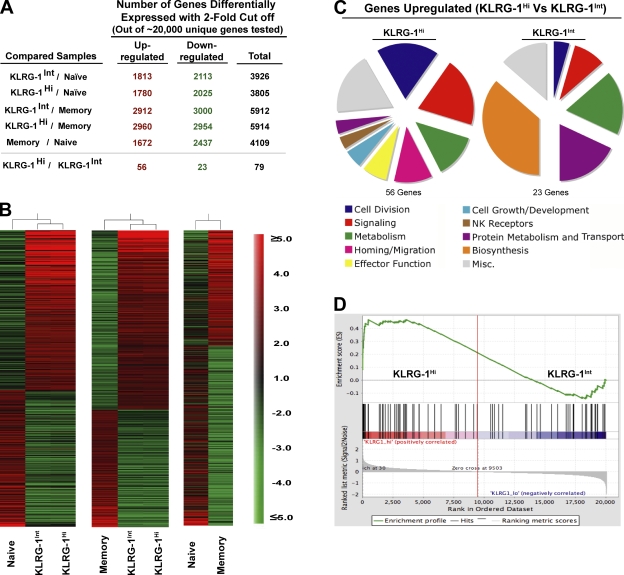
To further characterize the two effector subsets, we performed a genome-wide microarray analysis of KLRG-1Int and KLRG-1Hi subsets at days 4–5 p.i. Heat map (Fig. 5 B) and scatter plot analyses (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20071641/DC1) of gene expression of the naive, memory, KLRG-1Int, and KLRG-1Hi subsets demonstrated that both effector populations were strikingly different from either naive or memory CD8 T cells in terms of their global gene expression profiles (Fig. 5 and Fig. S2). They differed in ∼4,000 genes from naive CD8 T cells and in ∼5,000 genes from memory cells, with fold differences for several genes as high as 10–100 (Fig. 5, A and B; Tables S1 and S2; and Fig. S2). Both subsets exhibited gene expression patterns characteristic of activated effector cells (up-regulated IL-2Rα; granzyme A, B, and K; IFN-γ; T-bet; and down-regulated IL-7Rα and L-selectin). And, consistent with their actively cycling nature, both subsets exhibited increased transcription of genes involved in DNA replication, repair, and cell division relative to naive and memory cells (ribonucleotide reductase, RAD51, thymidine kinase 1, and cyclin B1 and B2, with fold changes in the range of 10–100; Tables S1 and S2). Thus, KLRG-1Int and KLRG-1Hi cells had differentiated from their naive counterparts into potent effector cells but had not yet acquired the characteristic memory gene expression profile.
Figure 5.
Genome-wide microarray analysis of memory precursors and terminal effectors. Expression levels of mRNA were measured by microarray analysis of sorted P14 CD44Lo naive cells, KLRG-1Int and KLRG-1Hi effector cells at day 4.5 p.i., and P14 memory cells at day 60 p.i. Gene ratios (fold change) represent the mean value of three to four independent hybridizations to Affymetrix microarrays. (A) Total number of transcripts differentially expressed (up- or down-regulated; twofold cutoff) between the indicated comparison groups is shown. (B) Relative intensities of all genes with a 2.5-fold cutoff in KLRG-1Int and KLRG-1Hi cells with respect to naive and memory populations are plotted as heat maps to depict the relationship between various populations. (C) All genes differentially expressed (twofold cutoff) between KLRG-1Int and KLRG-1Hi cells are functionally classified under broad categories based on information found in the Gene Ontology and Ingenuity Pathways databases and plotted as pie charts. (D) Enrichment profile of effector signature genes in KLRG-1Hi versus KLRG-1Int effector cells. GSEA of a set of genes corresponding to differentiated effectors (D8) in the rank-ordered list of genes differentially expressed between KLRG-1Int versus KLRG-1Hi effector cells is shown.
A closer comparison of gene expression profiles of memory precursors and terminal effectors demonstrated that the two effector subsets were quite closely related; a scatter plot analysis of KLRG-1Int and KLRG-1Hi gene expression levels revealed a distinctive linear correlation characteristic of strikingly similar populations (Fig. S2). Out of ∼22,000 unique genes investigated, only 79 genes were differentially expressed between the two subsets and even that was with a highest fold difference of only ∼4, as opposed to 10–100-fold differences observed with naive or memory cells (Figs. 5 A; Tables S1 and S2; and Fig. S2). The list of 56 genes that were up-regulated and 23 genes that were down-regulated in terminal effectors compared with memory precursors is presented in Table S3 (available at http://www.jem.org/cgi/content/full/jem.20071641/DC1) and summarized in Fig. 5 C. We observed that although memory precursors exhibited increased expression of genes signifying metabolic fitness (e.g., galactokinase 1 and spermidine synthase), terminal effector cells appeared more “effector-like” and expressed modestly higher levels of transcripts encoding certain effector molecules (granzyme A, lysozyme, and Fas ligand), genes involved in cell proliferation (Ki-67, cdca1, PRC1, etc.), and cell adhesion and migration (CCR2 and 5; Fig. 5 C; and Table S3). To rigorously test whether KLRG-1Hi terminal effector cells represent a comparatively more differentiated effector subpopulation than KLRG-1Int memory precursor cells, we next used gene set enrichment analysis (GSEA; Fig. 5 D). For this, a rank-ordered list of “effector signature” genes was generated. We compared profiles of effector cells at day 8 p.i. with naive cells and identified the top ∼200 genes increased in expression in effector cells. We next validated this set of genes enriched in an independent dataset of samples from day 8 effector and naive cells (15). Using GSEA, we identified those genes most enriched in effector cells versus naive cells in both effector datasets. This validated effector signature from day 8 p.i. was then used to interrogate the genes differentially expressed by KLRG-1Hi versus KRLG-1Int cells. If KLRG-1Hi cells represented a more differentiated effector population, one would predict the day 8 effector signature to be more enriched in this population than in KLRG-1Int cells. Using GSEA, we found highly significant (P < 0.001) enrichment of the differentiated effector signature in KLRG-1Hi compared with KLRG-1Int effector cells (Fig. 5 D), confirming our hypothesis that terminal effector cells are relatively more differentiated effectors than memory precursors.
Effector CD8 T cells that continue to proliferate toward the tail end of antigen clearance contribute less to the long-lived memory lineage
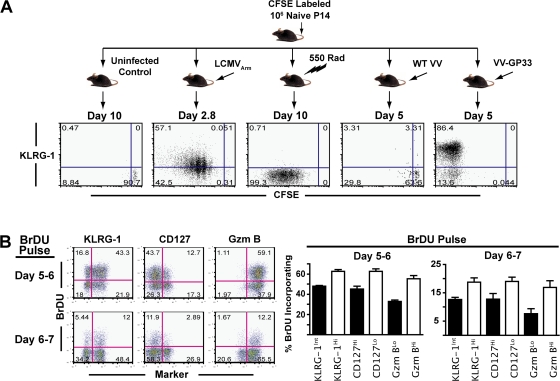
Our initial experiments (Figs. 1 and Figs.4) showed that both KLRG-1Int and KLRG-1Hi cells had undergone >8–10 rounds of cell division at days 4–5 p.i., as measured by CFSE dilution. Significantly higher expression of cell-cycle genes in KLRG-1Hi compared with KLRG-1Int cells by microarray analysis (Fig. 5 C) suggested that there may be differences in cell proliferation between the two subsets. Thus, we evaluated the extent of KLRG-1 expression with respect to cell proliferation at early (days 0–3) and late (days 5–7) time points of CD8 T cell expansion after infection. Analysis of KLRG-1 expression versus CFSE dilution between days 0–3 p.i. demonstrated that KLRG-1 expression increased after TCR-induced cell division (Fig. 6 A). Moderately higher KLRG-1 expression on cells in higher rounds of cell division indicated that KLRG-1Hi cells represented a relatively “more divided” subset compared with KLRG-1Int cells. These data are consistent with previous reports that KLRG-1 expression is associated with extensive proliferation (56). Nevertheless, it should be noted that lymphopenia-induced proliferation alone, or inflammatory stimuli alone in the absence of TCR stimulation (as delivered by VV infection of mice containing LCMV GP33-specific naive TCR Tg cells; Fig. 6 A), did not lead to up-regulation of KLRG-1 expression.
Figure 6.
Effector cells that continue to proliferate toward the tail end of antigen clearance contribute less to the long-lived memory lineage. (A) Cell-surface expression of KLRG-1 with respect to cell division at the indicated days after infection with LCMV, WT-VV, VV-GP33, or γ irradiation was evaluated using106 CFSE-labeled P14 CD8 T cells. All plots are gated on CD8+ Thy1.1+ P14 splenocytes stained directly ex vivo, and quadrant frequencies are indicated. (B) B6 mice containing 106 Thy1.1+ P14 cells were administered 1 mg BrdU i.p. per mouse at day 5 or 6 p.i. BrdU incorporation in CD8+ Thy1.1+ cells was assessed at the end of the pulse with respect to KLRG-1, CD127, and granzyme B expression. Percentages of BrdU+ cells were calculated within effector subsets expressing higher or lower levels of KLRG-1, CD127, and granzyme B and are summarized as bar graphs with mean and SEM.
To further monitor the cell proliferation of memory precursor and terminal effector subsets around the tail end of antigen clearance and peak of CD8 T cell responses, we used BrdU to mark dividing cells (Fig. 6 B). Mice were pulsed with BrdU between days 5–6 and 6–7 after LCMV infection, and the phenotype of cells dividing during this phase was evaluated. We found that between days 5–6 and 6–7, terminal effectors that are not committed to the memory lineage (KLRG-1Hi, CD127Lo cells) incorporated relatively more BrdU compared with KLRG-1Int, CD127Hi memory precursor cells. Furthermore, the KLRG-1Hi cells that continued to proliferate toward the tail end of antigen presentation also expressed more granzyme B, suggesting that terminal effector cells were “late leavers” that continued to perceive stimulation and divide longer than memory precursors. Based on these observations, we speculated that curtailing antigenic stimulation and, hence, proliferation toward the tail end of viral clearance during expansion could limit the generation of terminally differentiated KLRG-1Hi cells.
Curtailing stimulation toward later stages of infection enhances memory generation potential of effector cells
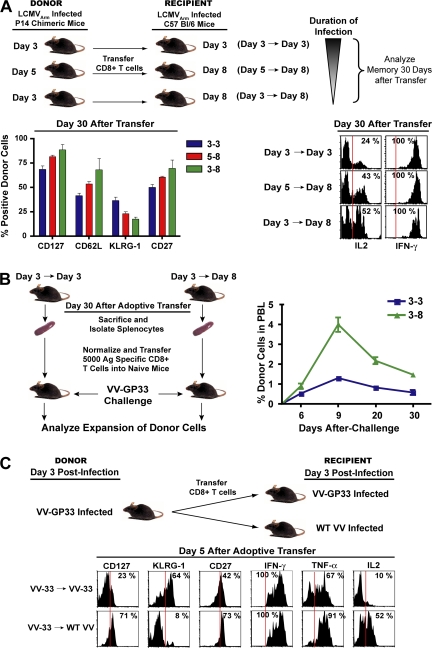
The observations that terminal effectors continue to proliferate during late stages of antigen clearance suggested that cells that are stimulated longer during expansion contribute less to the long-lived memory pool. Thus, we examined the influence of CD8 T cell stimulation during the late stages of infection on memory CD8 T cell generation. We controlled the duration of infection by isolating antigen-specific CD8 T cells at days 3 and 5 p.i. and adoptively transferring them into day 8 recipient mice that had controlled the infection and contained minimum to no antigen (Fig. 7). These adoptive transfer techniques curtailed the duration of stimulation to ∼3 and 5 d, respectively. Day 3 cells were also transferred back into day 3 mice; in this case, effector CD8 T cells encountered the complete duration of infection. In this set of experiments, we found that decreasing the duration of stimulation resulted in enhanced acquisition of memory properties (Fig. 7 A). Cells exposed to the shortest duration of infection (day 3 to day 8 transfer) exhibited the lowest levels of KLRG-1 and the highest levels of memory markers (CD127, CD62L, and CD27; Fig. 7 A) 30 d after adoptive transfer. They also produced the highest levels of IL-2 (Fig. 7 A) and exhibited preferential lymphoid tissue localization consistent with higher levels of CD62L expression (not depicted). Likewise, cells exposed to an intermediate duration of stimulation (day 5 to day 8 transfer) exhibited intermediate levels of KLRG-1, CD127, CD62L, CD27, and IL-2 compared with cells that perceived the shortest (day 3 to day 8 transfer) or longest (day 3 to day 3 transfer) duration of stimulation. Consistent with their phenotype, we found that preventing cells from perceiving stimulation toward the later stages of infection resulted in memory cells with superior recall proliferation compared with cells that were exposed to the entire duration of infection (Fig. 7 B). Because of the larger extent of donor cell proliferation in the day 3→day 3 group, the overall size of memory was two- to threefold larger than the day 3→day 8 group, but on a per cell basis shortening the duration of infection resulted in the generation of memory cells with superior recall proliferative potential (Fig. 7 B).
Figure 7.
Curtailing antigenic stimulation toward later stages of infection enhances memory generation potential of effector cells. (A) B6 mice containing 106 Thy1.1+ P14 cells were infected with LCMV and CD8+ splenocytes were isolated at days 3 and 5 p.i. and adoptively transferred into day 8 LCMV-infected recipients. Day 3 cells were also transferred into infection-matched day 3 recipients as controls. Donor cells were analyzed for cell-surface expression of CD62L, CD127, KLRG-1, and CD27 30 d after transfer. Bar graphs indicate mean with SEM. Production of IL-2 and IFN-γ by donor cells are plotted as histograms. (B) Recall proliferation potential of donor cells isolated from spleens 30 d after transfer was analyzed. Equal numbers of memory cells from each group were transferred into Thy1.2 naive mice. 1 d later, recipient mice were infected i.p. with 2 × 106 PFU VV-GP33. The expansion of donor cells was longitudinally assessed in blood by staining for cell-surface CD8 and Thy1.1. Data are plotted as the mean ± SEM. (C) C57BL/6 mice containing 106 Thy1.1+ P14 cells were infected with VV-GP33. At day 3 after infection, CD8+ splenocytes were isolated and adoptively transferred into either into VV-GP33–infected or WT-VV–infected recipient mice. Donor cells were analyzed for cell-surface expression of CD127, KLRG-1, and CD27 5 d after transfer. Production of ex vivo IFN-γ, TNF-α, and IL-2 cytokines by donor cells was also analyzed after stimulation with GP33-41 peptide. Vertical red lines in B and D indicate the negative expression gate for the respective markers.
To dissect the contribution of environmental and TCR stimuli in the generation of terminally differentiated effector cells, we adoptively transferred KLRG-1Int effector cells from mice infected with VV-GP33 into VV-GP33–infected or wild-type VV (WT-VV)–infected recipient mice (Fig. 7 C). Upon transfer into WT-VV–infected recipients, the donor cells were exposed to the same environment as that of VV-GP33 recipients but did not receive any further TCR stimuli. This shortening of TCR stimulation resulted in lower expression of KLRG-1, higher expression of CD127 and CD27, and superior IL-2 production on a per cell basis (Fig. 7 C). These studies underscore the contribution of antigenic stimulation during the tail end of immune response in dictating the lineage fate of effector CD8 T cells. In summary, these data demonstrate that after optimal stimulation, curtailing prolonged CD8 T cell stimulation and excessive proliferation toward the tail end of antigen clearance decreases terminal effector differentiation and favors the generation of long-lived memory cells. These data provide further evidence in favor of the decreasing potential model of memory differentiation, which proposes that prolonged stimulation of CD8 T cells during an acute infection progressively decreases the potential of effector cells to give rise to long-lived memory cells.
DISCUSSION
How the generation of memory precursors and terminal effector cells is regulated, and the precise differentiation path followed by memory cells is not completely understood. In this study, we performed a comprehensive phenotypic, functional, and genomic profiling of memory precursors and terminal effector cells. Such an analysis revealed that both subsets pass through a potent effector phase. However, cells that are destined to die continue to be stimulated and proliferate longer than memory precursors during the course of an acute infection. Consistent with this, genome-wide microarray analyses further demonstrated that terminal effectors are more differentiated and less metabolically fit compared with memory precursors. These data support the decreasing potential model of memory differentiation (5, 16, 24, 57–61) and extend it further to propose that cumulative stimuli accrued after optimal stimulation result in a spectrum of fully functional effector cells (62) that vary in their ability to differentiate into long-lived memory cells. Thus, cells that continue to perceive stimulation during later stages of infection (late leavers) are more terminally differentiated and largely die after antigen clearance.
These studies identify KLRG-1 as a marker that distinguishes terminal effector cells from memory precursors during an ongoing acute infection when both subsets have down-regulated IL-7Rα, a marker that identifies memory precursors later, after the infection has been resolved. KLRG-1 is an inhibitory C-type lectin expressed on NK cells and activated CD8 T cells (48) and has been implicated previously as a senescence marker (56, 63). Likewise, we find that during primary expansion KLRG-1 up-regulation marks more terminally differentiated cells that are compromised in their recall proliferation (Fig. 2). Our studies further show that terminal differentiation, as marked by KLRG-1 up-regulation, is related to the collective stimulation and proliferative history of a cell and not solely the extent of cell division (Fig. 6).
The effects of antigen dose, duration of antigenic stimulation, and inflammatory stimuli on terminal differentiation of effector CD8 T cells are not completely understood and are an active area of investigation. Recent studies have shown that inflammation can drive terminal differentiation of effector CD8 T cells (21, 64, 65). In these studies the role of inflammation was examined in the presence of antigen. We wanted to know whether inflammation alone could regulate memory T cell differentiation. Our experiments to address the role of infection (antigen plus inflammation) versus inflammation alone showed that inflammation by itself does not drive terminal differentiation of effector CD8 T cells (Fig. 6 A). Furthermore, we found that specifically shortening the duration of antigenic stimulation during an ongoing infection (Fig. 7 C) with an unchanged virally induced inflammatory milieu decreased terminal differentiation of effector CD8 T cells. In a recent study, Joshi et al. (65) examined the effect of dose of antigen (epitope density) on KLRG-1 expression on effector T cells using VV recombinants that express varying amounts of antigen. They found that these various VV recombinants, despite expressing different levels of antigen, induced a similar effector differentiation program. It should be noted that in this elegant experiment the amount of antigen varied but the duration of antigenic stimulation was the same for the different VV recombinants. In our study, we have shown that antigenic duration is a key determinant of effector and memory CD8 T cell differentiation. Collectively, these data suggest that memory differentiation is regulated by both the duration of antigenic stimulation and inflammation.
Why one subset of effector cells (KLRG-1Hi) perceives antigen longer and is more terminally differentiated than the other is an intriguing question. One can speculate that increased expression of KLRG-1 on a subset of effector cells during primary immune response may be a stochastic event whereby cells recruited earlier into the immune response enter the proliferative cycle first and continue to perceive antigen longer (Figs. 6 and Figs.7). This further implies that these late leaver cells that continue to encounter antigen longer are likely to kill a higher number of times, thereby compromising their metabolic fitness and memory potential. Alternatively, memory precursors and terminal effectors may also arise as a result of differential localization in specialized niches with different microenvironments (antigenic and/or inflammatory stimuli). Until recently KLRG-1 was an orphan receptor, but two separate reports now show that it binds cadherins (66, 67), which are ubiquitously expressed in vertebrates and mediate cell–cell adhesion. Around the peak of CD8 T cell expansion, KLRG-1Hi cells predominate in peripheral tissues (unpublished data). Thus, it will be interesting to see if interactions between KLRG-1 and cadherins exert a functional role in maintaining KLRG-1Hi cells in the periphery.
The lineage of memory cells is a key issue that is much debated (2, 5). Recently, a tantalizing notion that divergent memory and effector cell fates are decided after the first cell division has been presented (43). Considering the fact that the initial antigenic encounter programs CD8 T cells to enter a mode of extensive proliferation, it is prudent to predict that in the setting of an ongoing acute infection, events transpiring beyond the first round of cell division are likely to play a major role in cell fate decisions. Indeed, in the biologically relevant setting of an acute primary infection, our studies provide evidence that potent effector properties are acquired by antigen-specific CD8 T cells when they divide multiple times (nearly 100% of antigen-specific CD8 T cells express granzyme B at days 2–3 p.i.; Fig. 4). Moreover, by delineating memory precursors in the presence of antigen, we show that memory precursors pass through a potent effector phase and are remarkably similar to terminal effectors in their effector phenotype, function, and gene profile. Both subsets elaborated similar levels of direct ex vivo cytotoxicity and produced similarly high levels of effector molecules, including IFN-γ, TNF-α, and granzyme B (∼97–98% of KLRG-1Int and KLRG-1Hi donor cells expressed similarly high levels of granzyme B; Fig. 4 B). Assuming a similar rate of expansion of granzyme B Hi and Lo cells within the KLRG-1Int and KLRG-1Hi effector subsets, it is unlikely that the minor 2–3% of KLRG-1Int effector cells expressing lower granzyme B could expand to compose the 20% surviving population at memory (Fig. 2). Collectively, these data strongly argue in favor of the notion that memory cells pass through a fully functional effector stage in the presence of antigen.
Our studies define KLRG-1Int cells as a novel effector subset that has differentiated away from naive cells and not yet acquired memory properties, but has clearly retained the naive and memory properties of IL-2 production (Fig. 4). Because some markers are evidently shared among various differentiation states, superior IL-2 production by the KLRG-1Int effector subset reiterates the importance of considering all functional and phenotypic properties of a cell in totality before its classification as a naive, effector, or memory cell. Increased expression of Blimp-1 has been shown to inhibit IL-2 production by CD4 and CD8 T cells (55). Consistent with this, our microarray analysis also demonstrated slightly increased levels of Blimp-1 in non–IL-2–producing terminal effectors compared with memory precursors, although the differences were <1.5-fold. Recently, the importance of IL-2 signals in programming secondary expansion of memory cells has been demonstrated (68). Thus, it is interesting to speculate that IL-2 production by memory precursors may be functionally important for memory development. However, during expansion IL-2 signals are abundantly available from other sources (e.g., CD4 T cells), and it is possible that IL-2 production by self may not exert a functional role. It will also be interesting to evaluate whether epigenetic changes at the IL-2 locus correlate with memory potential.
While providing important insights into the mechanisms regulating the development of memory cells, we hope that these early phenotypic, functional, and genomic profiling studies of CD8 T cell differentiation will provide further leads for dissecting the precise signaling events that regulate CD8 T cell memory generation, thus facilitating rational vaccine design.
MATERIALS AND METHODS
Mice, virus, and infections.
C57BL/6 mice (Thy1.2+ or Thy1.1+) were purchased from the Jackson Laboratory. Thy1.1+ P14 mice bearing the DbGP33-specific TCR were fully backcrossed to C57BL/6 and maintained in our animal colony. LCMV Armstrong (LCMVArm), WT-VV, recombinant VV that expresses the GP33-41 epitope of LCMV (VV-GP33), and recombinant Listeria monocytogenes strain XFL203 that expresses the GP33-41 epitope of LCMV (LM-GP33) were propagated, titered, and used as previously described (15, 16). B6 mice were directly infected with 2 × 105 PFU LCMVArm i.p., with 2 × 106 PFU VV-GP33 or WT-VV i.v., or with 5 × 104 CFU LM-GP33 i.v. P14 chimeric mice were generated by adoptively transferring 103, 105, or 106 naive TCR Tg T cells into naive nonirradiated B6 mice followed by LCMVArm infection (referred to as P14 chimeras). Infection of P14 Tg chimeras with LCMVArm was similar to the infection of B6 mice. All mice were used in accordance with National Institutes of Health and Emory University Institutional Animal Care and Use Committee guidelines.
Antibodies, flow cytometry, and intracellular cytokine staining.
All antibodies were purchased from BD Biosciences, except for the antibody to mouse KLRG-1 (SouthernBiotech), anti–mouse CD127 (eBioscience), and anti–human granzyme B (Caltag). MHC class I peptide tetramers were made and used as previously described (69). Cells were stained for surface or intracellular proteins and cytokines as previously described (15, 16). For the analysis of intracellular cytokines, 106 lymphocytes per well were stimulated with 0.2 μg/ml GP33-41 peptide in the presence of brefeldin A (BFA) for 5 h, followed by surface staining for CD8 and intracellular staining for IFN-γ, TNF-α, or IL-2.
Isolation of T cells, adoptive transfers, CTL assays, and CFSE labeling.
Effector Thy1.1+ P14 CD8 T cells were isolated from the spleens of mice infected 3, 4–5, or 8 d earlier using anti-CD8 magnetic beads (Miltenyi Biotec), according to the manufacturer's instructions. To further purify KLRG-1Int and KLRG-1Hi cells, purified CD8 T cells were stained with anti-CD8α, anti-Thy1.2, and anti–KLRG-1 on ice in PBS containing 1% FCS, and were FACS sorted based on high or low expression of KLRG-1 (FACSAria; Becton Dickinson). The purity of FACS-sorted samples was ∼99%, and the purity of cells isolated by magnetic beads ranged from 95 to 99%. The number of Thy1.1+ P14 cells was calculated in each population, and equal numbers (∼0.5–2 × 106) were transferred i.v. into infection-matched C57BL/6 mice. Donor cells were distinguished using Thy1.1 mAb, and donor cells in the spleen, lymph node, liver, lung, and blood were isolated as previously described (16). Cells were labeled with CFSE for proliferation analyses (Invitrogen) as described previously (15, 16). Homeostatic proliferation of memory cells was also assessed by administering 0.8 mg/ml BrdU in drinking water for ∼10 d, after which BrdU incorporation in CD8 T cells was measured by staining for intracellular BrdU (BD Biosciences), according to the manufacturer's instructions. GP33-41–specific CTL activity was determined by a 5-h 51Cr release with purified KLRG-1Int and KLRG-1Hi effector populations, as previously described (15). For analysis of KLRG-1 expression with respect to cell division, naive Thy1.1+ P14 cells were labeled with CFSE according to manufacturer's instructions (Invitrogen), and 106 cells were adoptively transferred i.v. into naive B6 mice, which were subsequently infected with LCMVArm.
Microarray hybridization and analysis.
Naive, memory, and day 4 KLRG-1Int and KLRG-1Hi Thy1.1+ P14 CD8 T cells were FACS sorted, and RNA was isolated from cells in TRIzol (Invitrogen) according to manufacturer's protocol. cDNA was synthesized using the SuperScript Choice cDNA synthesis kit (Invitrogen) and an oligo (dT) primer containing a T7 promoter. The MEGAscript T7 kit (Ambion) was used to amplify cRNA from the cDNA. The cRNA was reverse transcribed with biotinylated nucleotides using the Enzo BioArray High Yield RNA transcript labeling kit in the second round of cRNA synthesis, fragmented, and hybridized on mouse 430.2 microarray chips (Affymetrix) at the Vanderbilt University Microarray Shared Resource, according to the manufacturer's protocols. Raw data were normalized using the RMA package built within Genespring 7.0 (Agilent Technologies), and fold-change comparisons between the groups indicated in the figures were subsequently made. Microarray datasets generated in this study have been made publicly available through the National Center for Biotechnology Information Gene Expression Omnibus database under accession nos. GSM257826, GSM257827, GSM257828, GSM257829, GSM257830, GSM257831, GSM257832, GSM257833, GSM257834, GSM257835, GSM257836, and GSM257837.
To test for the enrichment of effector genes in KLRG-1Int and KLRG-1Hi cells, GSEA was performed as previously described (70). In brief, GSEA is a method for determining whether a rank-ordered list of genes for a particular comparison of interest (e.g., effector CD8 T cells isolated 8 d after LCMV infection compared with naive cells) is enriched in genes derived from an independently generated gene set (e.g., KLRG-1Int vs. KLRG-1Hi effector cells). GSEA provides an enrichment score (ES) that measures the degree of enrichment of a given gene set at the top (highly correlated) or bottom (anticorrelated) of the second, rank-ordered dataset. A nominal p-value is used to assess the significance of the ES.
Online supplemental material.
Fig. S1 shows the engraftment of donor cells in recipients. Fig. S2 depicts scatter plot comparisons of naive, memory, and KLRG-1Int and KLRG-1Hi effector subset gene expression profiles. Table S1 provides a list of transcripts that are differentially expressed by KLRG-1Int and KLRG-1Hi effector subsets compared with naive cells. Table S2 provides a list of transcripts that are differentially expressed by KLRG-1Int and KLRG-1Hi effector subsets compared with memory cells. Table S3 provides a list of transcripts that are differentially expressed between KLRG-1Int and KLRG-1Hi effector subsets. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20071641/DC1.
Supplemental Material
Acknowledgments
We wish to thank Robert Karaffa and Michael Hulsey for their excellent assistance with FACS.
The authors wish to acknowledge support from National Institutes of Health grant AI30048 (to R. Ahmed), Bill and Melinda Gates Foundation Grant CAVD 38645 (to R. Ahmed), and Elizabeth Glaser Pediatric AIDS Foundation Scholar awards (to S. Sarkar and V. Kalia).
The authors have no conflicting financial interests.
Abbreviations used: BFA, brefeldin A; GSEA, gene set enrichment analysis; KLRG-1, killer cell lectin-like receptor G 1; LCMV, lymphocytic choriomeningitis virus; MFI, mean fluorescence intensity; p.i., postinfection; Tg, transgenic; VV, vaccinia virus; WT-VV, wild-type VV.
S. Sarkar and V. Kalia contributed equally to this study.
References
- 1.Welsh, R.M., L.K. Selin, and E. Szomolanyi-Tsuda. 2004. Immunological memory to viral infections. Annu. Rev. Immunol. 22:711–743. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, R., and B.T. Rouse. 2006. Immunological Memory. Immunol. Rev. 211:5–7. [Google Scholar]
- 3.Badovinac, V.P., and J.T. Harty. 2006. Programming, demarcating, and manipulating CD8+ T-cell memory. Immunol. Rev. 211:67–80. [DOI] [PubMed] [Google Scholar]
- 4.Fearon, D.T., J.M. Carr, A. Telaranta, M.J. Carrasco, and J.E. Thaventhiran. 2006. The rationale for the IL-2-independent generation of the self-renewing central memory CD8+ T cells. Immunol. Rev. 211:104–118. [DOI] [PubMed] [Google Scholar]
- 5.Kalia, V., S. Sarkar, T.S. Gourley, B.T. Rouse, and R. Ahmed. 2006. Differentiation of memory B and T cells. Curr. Opin. Immunol. 18:255–264. [DOI] [PubMed] [Google Scholar]
- 6.Lefrancois, L. 2006. Development, trafficking, and function of memory T-cell subsets. Immunol. Rev. 211:93–103. [DOI] [PubMed] [Google Scholar]
- 7.Pearce, E.L., and H. Shen. 2006. Making sense of inflammation, epigenetics, and memory CD8+ T-cell differentiation in the context of infection. Immunol. Rev. 211:197–202. [DOI] [PubMed] [Google Scholar]
- 8.Surh, C.D., O. Boyman, J.F. Purton, and J. Sprent. 2006. Homeostasis of memory T cells. Immunol. Rev. 211:154–163. [DOI] [PubMed] [Google Scholar]
- 9.Williams, M.A., B.J. Holmes, J.C. Sun, and M.J. Bevan. 2006. Developing and maintaining protective CD8+ memory T cells. Immunol. Rev. 211:146–153. [DOI] [PubMed] [Google Scholar]
- 10.van Stipdonk, M.J., E.E. Lemmens, and S.P. Schoenberger. 2001. Naive CTLs require a single brief period of antigenic stimulation for clonal expansion and differentiation. Nat. Immunol. 2:423–429. [DOI] [PubMed] [Google Scholar]
- 11.Kaech, S.M., and R. Ahmed. 2001. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nat. Immunol. 2:415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong, P., and E.G. Pamer. 2001. Cutting edge: antigen-independent CD8 T cell proliferation. J. Immunol. 166:5864–5868. [DOI] [PubMed] [Google Scholar]
- 13.van Stipdonk, M.J., G. Hardenberg, M.S. Bijker, E.E. Lemmens, N.M. Droin, D.R. Green, and S.P. Schoenberger. 2003. Dynamic programming of CD8+ T lymphocyte responses. Nat. Immunol. 4:361–365. [DOI] [PubMed] [Google Scholar]
- 14.Carrio, R., O.F. Bathe, and T.R. Malek. 2004. Initial antigen encounter programs CD8+ T cells competent to develop into memory cells that are activated in an antigen-free, IL-7- and IL-15-rich environment. J. Immunol. 172:7315–7323. [DOI] [PubMed] [Google Scholar]
- 15.Kaech, S.M., S. Hemby, E. Kersh, and R. Ahmed. 2002. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 111:837–851. [DOI] [PubMed] [Google Scholar]
- 16.Wherry, E.J., V. Teichgraber, T.C. Becker, D. Masopust, S.M. Kaech, R. Antia, U.H. von Andrian, and R. Ahmed. 2003. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 4:225–234. [DOI] [PubMed] [Google Scholar]
- 17.Roberts, A.D., K.H. Ely, and D.L. Woodland. 2005. Differential contributions of central and effector memory T cells to recall responses. J. Exp. Med. 202:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaech, S.M., J.T. Tan, E.J. Wherry, B.T. Konieczny, C.D. Surh, and R. Ahmed. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4:1191–1198. [DOI] [PubMed] [Google Scholar]
- 19.Huster, K.M., V. Busch, M. Schiemann, K. Linkemann, K.M. Kerksiek, H. Wagner, and D.H. Busch. 2004. Selective expression of IL-7 receptor on memory T cells identifies early CD40L-dependent generation of distinct CD8+ memory T cell subsets. Proc. Natl. Acad. Sci. USA. 101:5610–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madakamutil, L.T., U. Christen, C.J. Lena, Y. Wang-Zhu, A. Attinger, M. Sundarrajan, W. Ellmeier, M.G. von Herrath, P. Jensen, D.R. Littman, and H. Cheroutre. 2004. CD8alphaalpha-mediated survival and differentiation of CD8 memory T cell precursors. Science. 304:590–593. [DOI] [PubMed] [Google Scholar]
- 21.Badovinac, V.P., B.B. Porter, and J.T. Harty. 2004. CD8+ T cell contraction is controlled by early inflammation. Nat. Immunol. 5:809–817. [DOI] [PubMed] [Google Scholar]
- 22.Marzo, A.L., K.D. Klonowski, A. Le Bon, P. Borrow, D.F. Tough, and L. Lefrancois. 2005. Initial T cell frequency dictates memory CD8+ T cell lineage commitment. Nat. Immunol. 6:793–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachmann, M.F., P. Wolint, K. Schwarz, P. Jager, and A. Oxenius. 2005. Functional properties and lineage relationship of CD8+ T cell subsets identified by expression of IL-7 receptor alpha and CD62L. J. Immunol. 175:4686–4696. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed, R., and D. Gray. 1996. Immunological memory and protective immunity: understanding their relation. Science. 272:54–60. [DOI] [PubMed] [Google Scholar]
- 25.Foulds, K.E., C.Y. Wu, and R.A. Seder. 2006. Th1 memory: implications for vaccine development. Immunol. Rev. 211:58–66. [DOI] [PubMed] [Google Scholar]
- 26.Swain, S.L., J.N. Agrewala, D.M. Brown, D.M. Jelley-Gibbs, S. Golech, G. Huston, S.C. Jones, C. Kamperschroer, W.H. Lee, K.K. McKinstry, et al. 2006. CD4+ T-cell memory: generation and multi-faceted roles for CD4+ T cells in protective immunity to influenza. Immunol. Rev. 211:8–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacob, J., and D. Baltimore. 1999. Modelling T-cell memory by genetic marking of memory T cells in vivo. Nature. 399:593–597. [DOI] [PubMed] [Google Scholar]
- 28.Opferman, J.T., B.T. Ober, and P.G. Ashton-Rickardt. 1999. Linear differentiation of cytotoxic effectors into memory T lymphocytes. Science. 283:1745–1748. [DOI] [PubMed] [Google Scholar]
- 29.Lanzavecchia, A., and F. Sallusto. 2000. Dynamics of T lymphocyte responses: intermediates, effectors, and memory cells. Science. 290:92–97. [DOI] [PubMed] [Google Scholar]
- 30.Lauvau, G., S. Vijh, P. Kong, T. Horng, K. Kerksiek, N. Serbina, R.A. Tuma, and E.G. Pamer. 2001. Priming of memory but not effector CD8 T cells by a killed bacterial vaccine. Science. 294:1735–1739. [DOI] [PubMed] [Google Scholar]
- 31.Manjunath, N., P. Shankar, J. Wan, W. Weninger, M.A. Crowley, K. Hieshima, T.A. Springer, X. Fan, H. Shen, J. Lieberman, and U.H. von Andrian. 2001. Effector differentiation is not prerequisite for generation of memory cytotoxic T lymphocytes. J. Clin. Invest. 108:871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fearon, D.T., P. Manders, and S.D. Wagner. 2001. Arrested differentiation, the self-renewing memory lymphocyte, and vaccination. Science. 293:248–250. [DOI] [PubMed] [Google Scholar]
- 33.Wong, P., M. Lara-Tejero, A. Ploss, I. Leiner, and E.G. Pamer. 2004. Rapid development of T cell memory. J. Immunol. 172:7239–7245. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Y., G. Joe, E. Hexner, J. Zhu, and S.G. Emerson. 2005. Host-reactive CD8+ memory stem cells in graft-versus-host disease. Nat. Med. 11:1299–1305. [DOI] [PubMed] [Google Scholar]
- 35.van Leeuwen, E.M., G.J. de Bree, I.J. ten Berge, and R.A. van Lier. 2006. Human virus-specific CD8+ T cells: diversity specialists. Immunol. Rev. 211:225–235. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, M., S. Byrne, N. Liu, Y. Wang, A. Oxenius, and P.G. Ashton-Rickardt. 2007. Differential survival of cytotoxic T cells and memory cell precursors. J. Immunol. 178:3483–3491. [DOI] [PubMed] [Google Scholar]
- 37.Curtsinger, J.M., C.M. Johnson, and M.F. Mescher. 2003. CD8 T cell clonal expansion and development of effector function require prolonged exposure to antigen, costimulation, and signal 3 cytokine. J. Immunol. 171:5165–5171. [DOI] [PubMed] [Google Scholar]
- 38.Zeng, R., R. Spolski, S.E. Finkelstein, S. Oh, P.E. Kovanen, C.S. Hinrichs, C.A. Pise-Masison, M.F. Radonovich, J.N. Brady, N.P. Restifo, et al. 2005. Synergy of IL-21 and IL-15 in regulating CD8+ T cell expansion and function. J. Exp. Med. 201:139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitmire, J.K., J.T. Tan, and J.L. Whitton. 2005. Interferon-γ acts directly on CD8+ T cells to increase their abundance during virus infection. J. Exp. Med. 201:1053–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curtsinger, J.M., J.O. Valenzuela, P. Agarwal, D. Lins, and M.F. Mescher. 2005. Type I IFNs provide a third signal to CD8 T cells to stimulate clonal expansion and differentiation. J. Immunol. 174:4465–4469. [DOI] [PubMed] [Google Scholar]
- 41.Kolumam, G.A., S. Thomas, L.J. Thompson, J. Sprent, and K. Murali-Krishna. 2005. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J. Exp. Med. 202:637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mescher, M.F., J.M. Curtsinger, P. Agarwal, K.A. Casey, M. Gerner, C.D. Hammerbeck, F. Popescu, and Z. Xiao. 2006. Signals required for programming effector and memory development by CD8+ T cells. Immunol. Rev. 211:81–92. [DOI] [PubMed] [Google Scholar]
- 43.Chang, J.T., V.R. Palanivel, I. Kinjyo, F. Schambach, A.M. Intlekofer, A. Banerjee, S.A. Longworth, K.E. Vinup, P. Mrass, J. Oliaro, et al. 2007. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 315:1687–1691. [DOI] [PubMed] [Google Scholar]
- 44.Klonowski, K.D., K.J. Williams, A.L. Marzo, and L. Lefrancois. 2006. Cutting edge: IL-7-independent regulation of IL-7 receptor alpha expression and memory CD8 T cell development. J. Immunol. 177:4247–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun, J.C., S.M. Lehar, and M.J. Bevan. 2006. Augmented IL-7 signaling during viral infection drives greater expansion of effector T cells but does not enhance memory. J. Immunol. 177:4458–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hand, T.W., M. Morre, and S.M. Kaech. 2007. Expression of IL-7 receptor alpha is necessary but not sufficient for the formation of memory CD8 T cells during viral infection. Proc. Natl. Acad. Sci. USA. 104:11730–11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schluns, K.S., W.C. Kieper, S.C. Jameson, and L. Lefrancois. 2000. Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat. Immunol. 1:426–432. [DOI] [PubMed] [Google Scholar]
- 48.McMahon, C.W., A.J. Zajac, A.M. Jamieson, L. Corral, G.E. Hammer, R. Ahmed, and D.H. Raulet. 2002. Viral and bacterial infections induce expression of multiple NK cell receptors in responding CD8(+) T cells. J. Immunol. 169:1444–1452. [DOI] [PubMed] [Google Scholar]
- 49.Purton, J.F., J.T. Tan, M.P. Rubinstein, D.M. Kim, J. Sprent, and C.D. Surh. 2007. Antiviral CD4+ memory T cells are IL-15 dependent. J. Exp. Med. 204:951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Badovinac, V.P., J.S. Haring, and J.T. Harty. 2007. Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8(+) T cell response to infection. Immunity. 26:827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blattman, J.N., R. Antia, D.J. Sourdive, X. Wang, S.M. Kaech, K. Murali-Krishna, J.D. Altman, and R. Ahmed. 2002. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J. Exp. Med. 195:657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Intlekofer, A.M., N. Takemoto, E.J. Wherry, S.A. Longworth, J.T. Northrup, V.R. Palanivel, A.C. Mullen, C.R. Gasink, S.M. Kaech, J.D. Miller, et al. 2005. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6:1236–1244. [DOI] [PubMed] [Google Scholar]
- 53.Takemoto, N., A.M. Intlekofer, J.T. Northrup, E.J. Wherry, and S.L. Reiner. 2006. Cutting Edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J. Immunol. 177:7515–7519. [DOI] [PubMed] [Google Scholar]
- 54.Fuller, M.J., D.A. Hildeman, S. Sabbaj, D.E. Gaddis, A.E. Tebo, L. Shang, P.A. Goepfert, and A.J. Zajac. 2005. Cutting edge: emergence of CD127high functionally competent memory T cells is compromised by high viral loads and inadequate T cell help. J. Immunol. 174:5926–5930. [DOI] [PubMed] [Google Scholar]
- 55.Gong, D., and T.R. Malek. 2007. Cytokine-dependent Blimp-1 expression in activated T cells inhibits IL-2 production. J. Immunol. 178:242–252. [DOI] [PubMed] [Google Scholar]
- 56.Voehringer, D., C. Blaser, P. Brawand, D.H. Raulet, T. Hanke, and H. Pircher. 2001. Viral infections induce abundant numbers of senescent CD8 T cells. J. Immunol. 167:4838–4843. [DOI] [PubMed] [Google Scholar]
- 57.Williams, M.A., and M.J. Bevan. 2004. Shortening the infectious period does not alter expansion of CD8 T cells but diminishes their capacity to differentiate into memory cells. J. Immunol. 173:6694–6702. [DOI] [PubMed] [Google Scholar]
- 58.van Faassen, H., M. Saldanha, D. Gilbertson, R. Dudani, L. Krishnan, and S. Sad. 2005. Reducing the stimulation of CD8+ T cells during infection with intracellular bacteria promotes differentiation primarily into a central (CD62LhighCD44high) subset. J. Immunol. 174:5341–5350. [DOI] [PubMed] [Google Scholar]
- 59.Jelley-Gibbs, D.M., J.P. Dibble, S. Filipson, L. Haynes, R.A. Kemp, and S.L. Swain. 2005. Repeated stimulation of CD4 effector T cells can limit their protective function. J. Exp. Med. 201:1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.D'Souza, W.N., and S.M. Hedrick. 2006. Cutting edge: latecomer CD8 T cells are imprinted with a unique differentiation program. J. Immunol. 177:777–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jelley-Gibbs, D.M., J.P. Dibble, D.M. Brown, T.M. Strutt, K.K. McKinstry, and S.L. Swain. 2007. Persistent depots of influenza antigen fail to induce a cytotoxic CD8 T cell response. J. Immunol. 178:7563–7570. [DOI] [PubMed] [Google Scholar]
- 62.Lanzavecchia, A., and F. Sallusto. 2005. Understanding the generation and function of memory T cell subsets. Curr. Opin. Immunol. 17:326–332. [DOI] [PubMed] [Google Scholar]
- 63.Thimme, R., V. Appay, M. Koschella, E. Panther, E. Roth, A.D. Hislop, A.B. Rickinson, S.L. Rowland-Jones, H.E. Blum, and H. Pircher. 2005. Increased expression of the NK cell receptor KLRG1 by virus-specific CD8 T cells during persistent antigen stimulation. J. Virol. 79:12112–12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Badovinac, V.P., B.B. Porter, and J.T. Harty. 2002. Programmed contraction of CD8(+) T cells after infection. Nat. Immunol. 3:619–626. [DOI] [PubMed] [Google Scholar]
- 65.Joshi, N.S., W. Cui, A. Chandele, H.K. Lee, D.R. Urso, J. Hagman, L. Gapin, and S.M. Kaech. 2007. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 27:281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grundemann, C., M. Bauer, O. Schweier, N. von Oppen, U. Lassing, P. Saudan, K.F. Becker, K. Karp, T. Hanke, M.F. Bachmann, and H. Pircher. 2006. Cutting edge: identification of E-cadherin as a ligand for the murine killer cell lectin-like receptor G1. J. Immunol. 176:1311–1315. [DOI] [PubMed] [Google Scholar]
- 67.Ito, M., T. Maruyama, N. Saito, S. Koganei, K. Yamamoto, and N. Matsumoto. 2006. Killer cell lectin-like receptor G1 binds three members of the classical cadherin family to inhibit NK cell cytotoxicity. J. Exp. Med. 203:289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams, M.A., A.J. Tyznik, and M.J. Bevan. 2006. Interleukin-2 signals during priming are required for secondary expansion of CD8+ memory T cells. Nature. 441:890–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murali-Krishna, K., J.D. Altman, M. Suresh, D.J. Sourdive, A.J. Zajac, J.D. Miller, J. Slansky, and R. Ahmed. 1998. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 8:177–187. [DOI] [PubMed] [Google Scholar]
- 70.Subramanian, A., P. Tamayo, V.K. Mootha, S. Mukherjee, B.L. Ebert, M.A. Gillette, A. Paulovich, S.L. Pomeroy, T.R. Golub, E.S. Lander, and J.P. Mesirov. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.