Abstract
Embryonic stem (ES) cells have a unique regulatory circuitry, largely controlled by the transcription factors Oct4, Sox2, and Nanog, which generates a gene expression program necessary for pluripotency and self-renewal. How external signals connect to this regulatory circuitry to influence ES cell fate is not known. We report here that a terminal component of the canonical Wnt pathway in ES cells, the transcription factor T-cell factor-3 (Tcf3), co-occupies promoters throughout the genome in association with the pluripotency regulators Oct4 and Nanog. Thus, Tcf3 is an integral component of the core regulatory circuitry of ES cells, which includes an autoregulatory loop involving the pluripotency regulators. Both Tcf3 depletion and Wnt pathway activation cause increased expression of Oct4, Nanog, and other pluripotency factors and produce ES cells that are refractory to differentiation. Our results suggest that the Wnt pathway, through Tcf3, brings developmental signals directly to the core regulatory circuitry of ES cells to influence the balance between pluripotency and differentiation.
[Keywords: Wnt pathway, embryonic stem cells, Tcf3, regulatory circuitry]
Embryonic stem (ES) cells provide a unique opportunity to study early development and hold great promise for regenerative medicine (Thomson et al. 1998; Reubinoff et al. 2000; Pera and Trounson 2004). ES cells are derived from the inner cell mass of the developing blastocyst, and can be propagated in culture in an undifferentiated state while maintaining the capacity to generate any cell type in the body. Discovering how signaling pathways and transcriptional regulatory circuitry contribute to self-renewal and pluripotency is essential for understanding early development and realizing the therapeutic potential of ES cells.
A model for the core transcriptional regulatory circuitry of ES cells has emerged from studying the target genes of the ES cell transcription factors Oct4, Sox2, and Nanog (Boyer et al. 2005; Loh et al. 2006). These master regulators occupy the promoters of active genes encoding transcription factors, signal transduction components, and chromatin-modifying enzymes that promote ES cell self-renewal. They also occupy the promoters of a large set of developmental transcription factors that are silent in ES cells, but whose expression is associated with lineage commitment and cellular differentiation. Polycomb Repressive Complexes co-occupy the genes encoding these developmental transcription factors to help maintain a silent transcriptional state in ES cells (Boyer et al. 2006; Lee et al. 2006; Wilkinson et al. 2006; Rajasekhar and Begemann 2007; Stock et al. 2007).
External signals can promote ES cell pluripotency or cause these cells to differentiate, but precisely how these pathways are connected to the ES cell regulatory network has not been determined. These signals are produced by the stem cell niche in the developing blastocyst or, for cultured ES cells, can be produced by added factors or serum to maintain stem cell identity or promote differentiation. Recent studies have demonstrated the importance of several signaling pathways in maintaining or modifying ES cell state, including the Activin/Nodal, Notch, BMP4, and Wnt pathways (Rao 2004; Boiani and Scholer 2005; Friel et al. 2005; Kristensen et al. 2005; Valdimarsdottir and Mummery 2005; Dreesen and Brivanlou 2007; Pan and Thomson 2007). By understanding how these signaling pathways influence the gene expression program of ES cells, it should be possible to discover how they contribute to ES cell identity or promote specific differentiation programs.
The Wnt/β-catenin signaling pathway has multiple roles in ES cell biology, development, and disease (Logan and Nusse 2004; Reya and Clevers 2005; Clevers 2006). Several studies have shown that activation of the Wnt pathway can cause ES cells to remain pluripotent under conditions that induce differentiation (Kielman et al. 2002; Sato et al. 2004; Hao et al. 2006; Ogawa et al. 2006; Singla et al. 2006; Miyabayashi et al. 2007; Takao et al. 2007), while other studies have shown that the Wnt pathway has an important role in directing differentiation of ES cells (Otero et al. 2004; Lindsley et al. 2006). Recent studies have shown that T-Cell Factor-3 (Tcf3), a terminal component of the Wnt pathway, acts to repress the Nanog gene in ES cells (Pereira et al. 2006), providing an important clue for at least one mechanism by which the Wnt pathway regulates stem cell state. Nonetheless, we have an incomplete understanding of how the pathway exerts its effects, in part because few target genes have been identified for its terminal components in ES cells.
Stimulation of the canonical Wnt signaling pathway causes the transcriptional coactivator β-catenin to translocate to the nucleus, where it interacts with constitutively DNA-bound Tcf/Lef proteins to activate target genes (Behrens et al. 1996; Brantjes et al. 2001; Cadigan 2002). Tcf3, a member of the Tcf/Lef family, is highly expressed in murine ES (mES) cells, and is critical for early embryonic development (Korinek et al. 1998; Merrill et al. 2004; Pereira et al. 2006). To determine how the Wnt pathway is connected to the gene expression program of ES cells, we determined the genome-wide binding profile of Tcf3 and examined how perturbations of the pathway affect the gene expression program. Remarkably, the genome-wide data reveal that Tcf3 co-occupies the ES cell genome with the pluripotency transcription factors Oct4 and Nanog. These and other results reveal that the Wnt pathway brings developmental signals directly to the core regulatory circuitry of ES cells, which consists of the pluripotency transcription factors and Tcf3, together with their mutual target genes.
Results
Identification of Tcf3-binding sites genome-wide
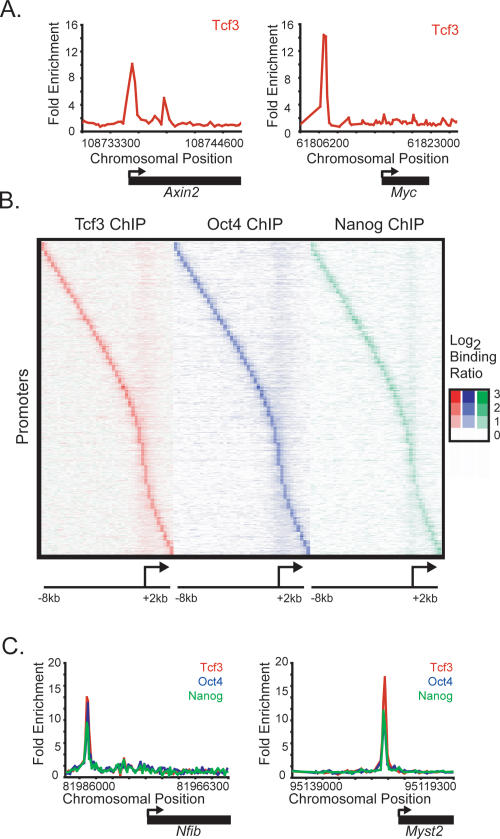
To determine how the Wnt pathway regulates the gene expression program of mES cells, we first identified genes occupied by Tcf3. mES cells were grown under standard conditions (Supplemental Fig. S1) and DNA sequences occupied by Tcf3 were identified using chromatin immunoprecipitation (ChIP) combined with DNA microarrays (ChIP–Chip). For this purpose, DNA microarrays were designed with 60-mer oligonucleotide probes tiling the entire nonrepeat portion of the mouse genome. The results revealed that Tcf3 occupies >1000 murine promoters (Supplemental Table S1), including those of the known Wnt targets Axin2 and Myc (Fig. 1A; He et al. 1998; Yan et al. 2001; Jho et al. 2002).
Figure 1.
Tcf3, Oct4, and Nanog co-occupy the genome in mouse ES cells. (A) Tcf3 binds to known target genes. Examples of previously known Tcf3-bound regions are displayed as unprocessed ChIP-enrichment ratios for all probes within the chromosomal region indicated below the plot. The gene is depicted below the plot, and the TSS and direction are denoted by an arrow. (B) Tcf3, Oct4, and Nanog display nearly identical binding profiles. Analysis of ChIP–chip data from genes bound by Tcf3, Oct4, or Nanog reveals that the three factors bind to similar genomic regions at all promoters. Regions from −8 kb to +2 kb around each TSS were divided into bins of 250 bp. The raw enrichment ratio for the probe closest to the center of the bin was used. If there was no probe within 250 bp of the bin center then no value was assigned. For genes with multiple promoters, each promoter was used for analysis. The analysis was performed on 3764 genes, which represents 4086 promoters. Promoters are organized according to the distance between the maximum Tcf3-binding ratio and the TSS. (C) Tcf3, Oct4, and Nanog bind in close proximity at target genes. Plots display unprocessed ChIP-enrichment ratios for all probes within the chromosomal region indicated below the plot. The gene is depicted below the plot, and the TSS and direction are denoted by an arrow.
Tcf3 Co-occupies the genome with ES cell master regulators
Inspection of the genes occupied by Tcf3 revealed a large set that were previously shown to be bound by the homeodomain transcription factor Oct4 (Boyer et al. 2005; Loh et al. 2006), which is an essential regulator of early development and ES cell identity (Nichols et al. 1998; Hay et al. 2004). To examine the overlap of gene targets more precisely, we carried out ChIP–Chip experiments with antibodies directed against Oct4 in mES cells and used the same genome-wide microarray platform employed in the Tcf3 experiment. Remarkably, the binding profiles of Tcf3 and Oct4 revealed that they bind the same genomic regions and display identical spatial distribution patterns with regard to transcription start sites (TSSes) (Fig. 1B; Supplemental Fig. S2). These results identified a set of 1224 genes that are co-occupied by Tcf3 and Oct4 at high confidence (Supplemental Table S1) and suggested that the Wnt pathway connects directly to genes regulated by Oct4 through Tcf3.
Previous studies in human ES cells have shown that Oct4 shares target genes with the transcription factors Nanog and Sox2 (Boyer et al. 2005), suggesting that Tcf3-occupied genes in mES cells should also be occupied by Nanog and Sox2. Additional genome-wide ChIP–Chip experiments with antibodies directed against Nanog revealed that it does indeed bind the same sites occupied by Oct4 and Tcf3 (Figs. 1B,C, 2; Supplemental Fig. S2). The fact that Oct4 and Sox2 form heterodimers in ES cells (Dailey and Basilico 2001; Okumura-Nakanishi et al. 2005) and frequently co-occupy promoters in human ES cells (Boyer et al. 2005) makes it likely that Tcf3 co-occupies much of the genome with Oct4, Nanog, and Sox2.
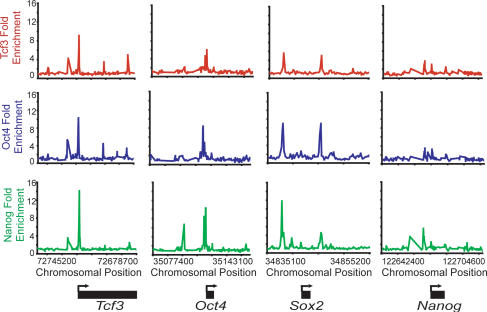
Figure 2.
Tcf3, Oct4, and Nanog bind the promoters of Tcf3, Oct4, Sox2, and Nanog. Plots display unprocessed ChIP-enrichment ratios for all probes within the chromosomal region indicated below the plot. The gene is depicted below the plot, and the TSS and direction are denoted by an arrow.
The observation that Tcf3 co-occupies much of the genome with the ES cell pluripotency transcription factors has a number of implications for the regulatory circuitry of these cells. Tcf3 binds its own promoter as well as the promoters of genes encoding Oct4, Sox2, and Nanog (Fig. 2). Thus, Tcf3 is an integral component of an interconnected autoregulatory loop, where all four transcription factors together occupy each of their own promoters (Fig. 3A). This feature of ES cell regulatory circuitry was described previously for Oct4, Sox2, and Nanog alone (Boyer et al. 2005), and has been postulated to be a common regulatory motif for master regulators of cell state (Chambers et al. 2003; Odom et al. 2004, 2006; Okumura-Nakanishi et al. 2005; Rodda et al. 2005). Autoregulation is thought to provide several advantages to the control of cell state, including reduced response time to environmental stimuli and increased stability of gene expression (McAdams and Arkin 1997; Thieffry et al. 1998; Rosenfeld et al. 2002; Shen-Orr et al. 2002). It is also notable that Tcf3 and the pluripotency transcription factors together occupy genes encoding many Wnt pathway components (Supplemental Fig. S3), suggesting that this transcription factor regulates much of its own signaling pathway apparatus together with the pluripotency factors.
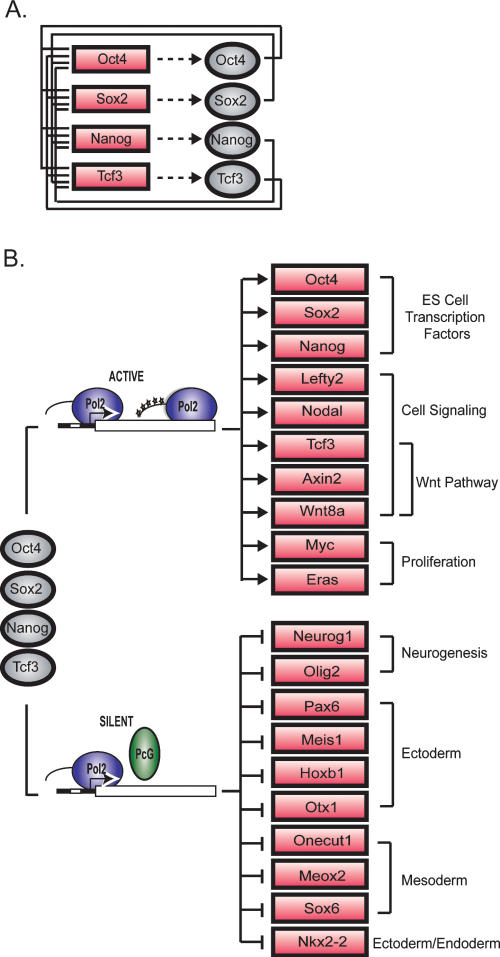
Figure 3.
Tcf3 is an integral component of the core regulatory circuitry of ES cells. (A) Tcf3 forms an interconnected autoregulatory loop with Oct4, Sox2, and Nanog. Proteins are represented by ovals and genes are indicated by rectangles. (B) Model showing a key portion of the regulatory circuitry of mES cells where Oct4, Sox2, Nanog, and Tcf3 occupy both active and silent genes. The evidence that Oct4, Nanog, and Tcf3 occupy these genes is described here; Sox2 occupancy is inferred from previous studies in human ES cells (Boyer et al. 2005). Evidence that the transcriptionally silent genes are occupied by Polycomb Repressive Complexes is from Boyer et al. (2006), and unpublished data and that these genes have stalled RNA polymerases is from Guenther et al. (2007) and Stock et al. (2007). Proteins are represented by ovals and genes are indicated by rectangles.
A model for the core regulatory circuitry of ES cells has been proposed in which the genes bound by the master regulators Oct4, Sox2, and Nanog fall into two classes: transcriptionally active genes encoding transcription factors, signaling components, and other products that support the stem cell state; and transcriptionally inactive genes, consisting mostly of developmental regulators, where Polycomb is bound and RNA polymerase II is recruited, but transcription is stalled (Boyer et al. 2005, 2006; Lee et al. 2006; Guenther et al. 2007; Stock et al. 2007; Zeitlinger et al. 2007). Our results reveal that Tcf3, together with the pluripotency regulators, is associated with both classes of genes, and thus provide a modified model of the core regulatory circuitry of ES cells (Fig. 3B). The association of Tcf3 with the set of genes encoding key transcription factors, signaling pathway components, and developmental regulators suggests that the Wnt signaling pathway contributes to the regulation of these genes, thereby impacting ES pluripotency and self-renewal.
Expression analysis of Tcf3 knockdown in mES cells
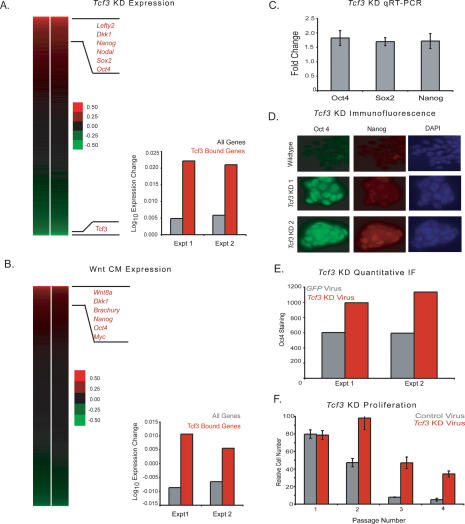
Genes bound by Tcf/Lef proteins are thought to be repressed in the absence of Wnt/β-catenin signaling and to be activated upon Wnt pathway stimulation (Behrens et al. 1996; Cavallo et al. 1998; Brantjes et al. 2001; Daniels and Weiss 2005; Miyabayashi et al. 2007). mES cells have low endogenous Wnt activity in standard culture conditions, and the Wnt pathway can be further stimulated in culture (Supplemental Fig. S4; Dravid et al. 2005; Yamaguchi et al. 2005; Lindsley et al. 2006; Ogawa et al. 2006; Anton et al. 2007; Takao et al. 2007). Thus, it is unclear whether Tcf3-occupied genes are being repressed or activated at the low level of Wnt activity characteristic of standard ES cell culture conditions. To investigate whether the effect of Tcf3 occupancy is to repress or to activate genes, RNAi constructs were used to deplete Tcf3 mRNA in mES cells in two independent experiments (Supplemental Fig. S5) and changes in global mRNA levels were assayed with DNA microarrays (Fig. 4A). The ∼3.5% of mouse genes whose mRNA levels changed by at least twofold were significantly enriched for Tcf3 targets relative to genes whose expression was unaltered by the Tcf3 knockdown (P-value < 2 × 10−10) (Supplemental Fig. S6; Supplemental Table S2). The genes whose expression increased upon loss of Tcf3 included those encoding the master regulators Oct4, Sox2, and Nanog, other genes involved in pluripotency such as Lefty2 and Nodal, and the Wnt pathway component Dkk1 (Fig. 4A). The fact that up-regulated genes are strongly enriched for Tcf3 binding suggests that Tcf3 mainly acts to repress genes. Upon loss of Tcf3, target genes are no longer repressed and can now be activated by other factors (such as Oct4, Sox2, and Nanog) present at their promoters.
Figure 4.
Knockdown of Tcf3 and activation of the Wnt pathway in mES cells reveal a role for Tcf3 in repression of target genes and a role in regulating pluripotency. (A) Tcf3 knockdown results in up-regulation of target genes. The effect of Tcf3 knockdown on gene expression was measured by hybridization of labeled RNA prepared from Tcf3 knockdown cells against RNA prepared from cells infected with nonsilencing control lentivirus at 48 h post-infection. A heat map of biological replicate data sets of expression changes was generated where genes are ordered according to average expression change. Tcf3 target genes have a higher average expression change than the average for all genes upon knockdown of Tcf3. (B) Wnt CM results in up-regulation of Tcf3 target genes. The effect of Wnt activation on gene expression was measured by hybridization of labeled RNA prepared from mES cells grown in Wnt CM against RNA prepared from cells grown in mock CM. A heat map of biological replicate data sets of expression change upon addition of Wnt CM where genes are ordered according to average expression change of replicates. Tcf3 target genes have a higher average expression change than the average for all genes upon addition of Wnt CM. (C) Tcf3 knockdown results in increased expression of Oct4, Sox2, and Nanog. Real-time PCR demonstrates that Oct4, Sox, and Nanog have increased expression upon knockdown of Tcf3. Values are normalized to Gapdh transcript levels, and fold change is relative to cells transfected with a nonsilencing hairpin. (D) Tcf3 knockdown results in increased staining for Oct4 and Nanog. Immunofluorecence was performed on mES cells grown one passage off of feeders that were either infected with Tcf3 knockdown lentivirus or infected with nonsilencing control lentivirus. Cells were fixed with 4% paraformaldehyde 96 h post-infection. Cells were stained with Oct4, Nanog, and DAPI. Images for Oct4 and Nanog staining were taken at 40× magnification and an exposure time of 300 msec. Tcf3 KD 1 and KD 2 represent different knockdown hairpin constructs. Tcf3 KD 2 is the virus also used in A, C, E, and F. (E) Tcf3 knockdown results in a significant increase of Oct4 staining. Quantification of Oct4 staining was performed in cells infected with Tcf3 or Gfp knockdown virus. (F) Tcf3 knockdown cells proliferate over more passages in the absence of LIF. Relative cell numbers of ES cells transfected with Tcf3 or control virus through multiple passages off of feeders in the presence or absence of LIF. Identical cell numbers were initially plated, and cells were split 1:12 every 2–3 d. Cells were counted at each passage and values for cells grown in the absence of LIF were normalized to cells grown in the presence of LIF.
While expression of Tcf3 target genes was often up-regulated upon loss of Tcf3, the expression of a substantial number of Tcf3-bound genes remained unchanged, and a relatively small number of Tcf3-bound genes showed reduced expression (Fig. 4A). Nearly half of the genes occupied by Tcf3, Oct4, and Nanog are co-occupied by Polycomb Repressive Complexes (Boyer et al. 2006; Lee et al. 2006; Wilkinson et al. 2006; Rajasekhar and Begemann 2007), and their transcriptional state would not be expected to change as Polycomb would prevent elongation of transcripts at these genes (Stock et al. 2007). Indeed, we find that expression of genes occupied by Tcf3 and Polycomb do not show a significant expression change upon loss of Tcf3 (P-value > 0.4). There were some Tcf3 target genes whose expression was down-regulated upon loss of Tcf3; because mES cells have a low level of Wnt pathway activation, it is possible that sufficient β-catenin enters the nucleus in order to associate with and activate this subset of genes. Indeed, we found that some amount of β-catenin does associate with Tcf3 and Oct4, as β-catenin can be detected in cross-linked chromatin extracts immunoprecipitated for either Tcf3 or Oct4 (Supplemental Fig. S7). It is also possible that the loss of expression of this set of Tcf3 target genes is a secondary consequence of the knockdown. The repressive activity of Tcf3 appears to be its dominant function for most genes under these conditions, as the set of Tcf3-bound genes were found to have a significantly higher increase in expression upon knockdown compared with all genes (P-value < 7 × 10−5) (Fig. 4A).
Expression analysis of Wnt pathway activation in mES cells
We next studied the effect of increased stimulation of the Wnt pathway on Tcf3 target genes in mES cells. Cells were treated with Wnt3a conditioned media in (CM) two independent experiments, and changes in global mRNA levels were assayed with DNA microarrays (Fig. 4B). The <1% of mouse genes whose mRNA levels changed by at least twofold in the Wnt-treated cells were significantly enriched for Tcf3 targets relative to genes whose expression was unaltered by the addition of Wnt (P-value < 1.5 × 10−5) (Supplemental Fig. S8; Supplemental Table S3). The genes whose expression most increased encode the pluripotency factors Oct4 and Nanog, Wnt pathway components such as Wnt8a and Dkk1, and known Wnt targets such as Brachury (Fig. 4B). These results are consistent with a model where Tcf3 acts to partially repress many of its target genes under standard mES cell culture conditions, yet contributes to increased expression of its target genes under conditions of increased Wnt stimulation. We would therefore expect a correlation between genes up-regulated upon loss of Tcf3 and genes up-regulated upon Wnt stimulation. Indeed, we do find these gene sets to be significantly correlated (P-value < 1 × 10−8) (Supplemental Fig. S9). Although a significant portion of Tcf3 target genes undergo expression changes upon Wnt stimulation, it is possible that a second class of Tcf3 target genes are regulated independently of Wnt signaling, and therefore are uninfluenced by changes in pathway activation (Yi and Merrill 2007). In fact, several studies have shown a β-catenin-independent role for Tcf3 (Kim et al. 2000; Merrill et al. 2001; Roël et al. 2002). It should also be noted that ES cells express other mammalian Tcf/Lef proteins and that these factors may also mediate the functional consequences of Wnt signaling (Pereira et al. 2006).
Influence of Tcf3 and Wnt on pluripotency regulators and ES cell state
Evidence that Tcf3 is an integral component of the core transcriptional circuitry of ES cells that functions to partially repress transcription of pluripotency genes led us to examine whether Tcf3 knockdown enhances features of ES cells associated with pluripotency and self-renewal. Quantitative real-time PCR analysis demonstrated that Tcf3 knockdown in mES cells results in higher transcript levels for the pluripotency genes Oct4, Sox2, and Nanog (Fig. 4C). Up-regulation of Nanog upon Tcf3 depletion confirms a previous report that Tcf3 acts to repress this gene under normal ES cell growth conditions (Pereira et al. 2006). Thus, the results of the Tcf3 knockdown experiment indicate that under normal conditions Tcf3 functions to reduce expression of the three pluripotency regulators.
We next measured the levels of Oct4 and Nanog proteins in ES cells subjected to Tcf3 knockdown. The results of immunofluorescence experiments show that there are substantial increases in the levels of Oct4 and Nanog transcription factors in the nucleus of such cells (Fig. 4D). There is a significant increase of Oct4 in Tcf3 knockdown cells compared with control cells based on quantitative measurements of staining intensity using Cellomics software (Fig. 4E). Remarkably, Tcf3 knockdown mES cells display enhanced proliferation and Oct4 staining in the absence of feeders and leukemia inhibitory factor (LIF) compared with control cells, supporting previous results (Fig. 4F; Supplemental Fig. S10; Pereira et al. 2006). Previous studies have demonstrated that activation of the Wnt/β-catenin pathway can have similar effects on ES cell pluripotency (Sato et al. 2004; Hao et al. 2006; Singla et al. 2006), and we also found that cells treated with Wnt CM show increased staining of Oct4 (Supplemental Fig. S11). The observation that Tcf3 knockdown and Wnt stimulation have similar functional consequences is consistent with the expression data described above for ES cells subjected to Tcf3 knockdown and ES cells treated with Wnt3a CM. These studies demonstrate the functional importance of Tcf3 occupancy and Wnt pathway activation for a subset of target genes that includes the pluripotency regulators.
Discussion
It is fundamentally important to determine how signaling pathways control ES cell pluripotency and differentiation, and how these pathways connect to discrete sets of target genes to affect such states. We found that a terminal component of the Wnt signaling pathway, the transcription factor Tcf3, is physically associated with the same genomic sites as the pluripotency regulators Oct4 and Nanog in mES cells. This result reveals that the Wnt pathway is physically connected to the core regulatory circuitry of these cells. This core circuitry consists of two key features: an interconnected autoregulatory loop, and the set of target genes that are mutually bound by the pluripotency transcription factors and Tcf3.
The genome-wide data sets we report here enhance our knowledge of the targets of Oct4, Nanog, and Tcf3. These new data sets were generated using the same protocols and genome-wide tiling microarrays in ES cells grown under identical conditions, allowing more reliable conclusions about the overlap of these factors throughout the genome; previous data sets for these factors came from different mES cells grown in different settings, using different ChIP analysis platforms, and these data were not always genome-wide (Boyer et al. 2005, 2006; Loh et al. 2006). The new data reveal, for example, the remarkable extent to which Oct4 and Nanog binding overlap throughout the ES cell genome and the striking association of Tcf3 with those sites (Fig. 1B). The new data also provide a revised model for the core regulatory circuitry of mES cells, which incorporates Tcf3 and high-confidence target genes of key ES cell regulators (Fig. 3).
The revised model of core regulatory circuitry extends our knowledge of how extracellular signals from the Wnt pathway contribute to stem cell state. Pereira et al. (2006) demonstrated that Tcf3 binds the Nanog promoter and represses its mRNA expression in mES cells. Our data confirm Tcf3 binding and function at Nanog and extend our knowledge of Tcf3 targets to the other well-characterized pluripotency regulators Oct4 and Sox2, as well as most of their target genes. Pereira et al. (2006) proposed a model wherein Tcf3-mediated control of Nanog levels allows stem cells to balance the creation of lineage-committed and undifferentiated cells. Our results also support this model, but argue that Tcf3 contributes to the balance through its functions in the core regulatory circuitry described here.
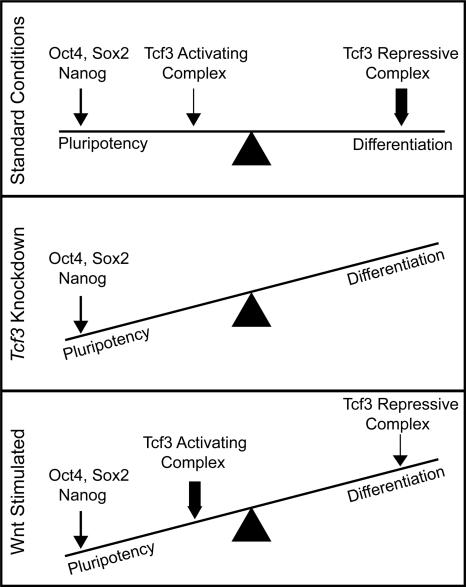
Our results suggest that the Wnt pathway, through Tcf3, influences the balance between pluripotency and differentiation in ES cells, as modeled in Figure 5. Under standard culture conditions, where there is a low-level of Wnt activation, ES cells are poised between the pluripotent state and any of a number of differentiated states. It is well established that Oct4, Sox2, and Nanog act to promote the pluripotent state, as depicted in the model where the influence of these factors is shown by an arrow. Under standard culture conditions, Tcf3 may exist in an activating or repressive complex, but is predominantly in a repressive complex promoting differentiation. The loss of Tcf3 in Tcf3 knockdown cells, would, in this model, favor pluripotency. Wnt stimulation converts the repressive complex to an activating complex and thus promotes pluripotency. Our results suggest that the Wnt pathway, through Tcf3, influences the balance between pluripotency and differentiation by bringing developmental signals directly to the core regulatory circuitry of ES cells. The observation that the Wnt pathway can be manipulated to affect the balance between pluripotency and differentiation suggests that perturbation of this pathway may impact the efficiency of reprogramming somatic cells into pluripotent stem cells.
Figure 5.
Model depicting the influence of Wnt pathway components on pluripotency and differentiation in ES cells. ES cells are poised between the pluripotent state and any of a number of differentiated states. Oct4, Sox2, and Nanog act to promote the pluripotent state (depicted by an arrow). Tcf3 can exist in an activating complex with β-catenin or a repressive complex with Groucho (Reya and Clevers 2005). Under standard growth conditions, the Wnt pathway is only active at low levels (Supplemental Fig. S4; Dravid et al. 2005; Yamaguchi et al. 2005; Lindsley et al. 2006; Ogawa et al. 2006; Anton et al. 2007; Takao et al. 2007). Therefore, Tcf3 is mainly in a repressive complex promoting differentiation (depicted by a thick arrow), although some Tcf3 associates with β-catenin to activate target genes and promote pluripotency (depicted by a thin arrow). In Tcf3 knockdown cells, there is no influence from Tcf3 on cell state. Thus, the balance is tipped toward maintaining pluripotency. Upon Wnt stimulation, the balance again tips toward maintaining pluripotency as more Tcf3 associates with β-catenin in an activating complex (depicted by a thick arrow). This model is not meant to imply that Wnt or Tcf3 are themselves pluripotency factors, but rather that they can influence cell state in the presence of other pluripotency factors, such as Oct4, Sox2, and Nanog.
Materials and methods
mES cell culture conditions
V6.5 mES cells were grown on irradiated murine embryonic fibroblasts (MEFs) unless otherwise stated. Cells were grown under mES cell conditions as described previously in Boyer et al. (2005). Briefly, cells were grown on 0.2% gelatinized tissue culture plates in DMEM-KO (Invitrogen, 10829-018) supplemented with 15% fetal bovine serum (Hyclone, characterized SH3007103), 1000 U/mL LIF (ESGRO, ESG1106), 100 μM nonessential amino acids (Invitrogen, 11140-050), 2 mM L-glutamine (Invitrogen, 25030-081), 100 U/mL pennicillin, 100 μg/mL streptomycin (Invitrogen, 15140-122), and 8 nL/mL 2-mercaptoethanol (Sigma, M7522).
Genome-wide location analysis
ChIP protocol
Protocols describing ChIP methods were downloaded from http://jura.wi.mit.edu/young_public/hESregulation/ChIP.html with slight modifications. Briefly, 108 mES cells were grown for one passage off of feeders and then cross-linked using formaldehyde. Cells were resuspended, lysed in lysis buffer, and sonicated to solubilize and sheer cross-linked DNA. Triton X-100 and SDS were added to the lysate after sonication to final concentrations of 1% and 0.1%, respectively. The resulting whole-cell extract was incubated overnight at 4°C with 100 μL of Dynal Protein G magnetic beads that had been preincubated with 10 μg of the appropriate antibody overnight. After 16–18 h, beads were washed with the following four buffers for 4 min per buffer: low-salt buffer (20 mM Tris at pH 8.1, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS), high-salt buffer (20 mM Tris at pH 8.1, 500 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS), LiCl buffer (10 mM Tris at pH 8.1, 250 mM LiCl, 1 mM EDTA, 1% deoxycholate, 1% NP-40), and TE+ 50 mM NaCl. Bound complexes were eluted from the beads in elution buffer by heating at 65°C with occasional vortexing, and cross-links were reversed by overnight incubation at 65°C.
ChIP antibodies
Cell extracts were immunoprecipitated using antibodies against Tcf3 (Santa Cruz Biotechnology, sc-8635), Oct4 (Santa Cruz Biotechnology, sc-8628), or Nanog (Bethyl Laboratories, bl1662).
Array design
The murine 244k whole-genome array was purchased from Agilent Technology (http://www.agilent.com). The array consists of 25 slides each containing ∼244,000 60-mer oligos (slide ID 15310-3, 15317, 15319-21, 15323, 15325, 15327-30, 15332-7, 15339-41, and 15343-44) covering the entire nonrepeat portion of the mouse genome at a density of about one oligo per 250 base pairs (bp).
Data normalization and analysis
Data normalization and analyses were performed as described previously in Boyer et al. (2005).
Tcf3 knockdown
Lentiviral production
Lentivirus was produced according to Open Biosystems Trans-lentiviral shRNA Packaging System (TLP4614). The shRNA constructs targeting murine Tcf3 were designed using an siRNA rules-based algorithm consisting of sequence, specificity, and position scoring for optimal hairpins that consist of a 21-base stem and a 6-base loop (RMM4534-NM-009332). Five hairpin constructs were used to produce virus targeting Tcf3. A negative control virus was made from the pLKO.1 empty vector (RHS4080).
Lentiviral infection of mES cells
V6.5 mES cells were plated at ∼30% confluence on the day of infection. Cells were seeded in 2× mES media with 6 μg/mL polybrene (Sigma, H9268-10G) and Tcf3 knockdown or control (pLKO.1) virus was immediately added. After 24 h, infection media was removed and replaced with mES media with 2 μg/mL Puromycin (Sigma, P8833). RNA was harvested at 48 h after infection.
Knockdown efficiency
Knockdown efficiency was measured using real-time PCR to measure levels of Tcf3 mRNA (Supplemental Fig. S5).
RNA isolation, real-time PCR, and analysis of transcript levels
To determine transcript levels by RT–PCR, RNA was isolated from ∼106 to 107 mES cells using TRIzol reagent following the protocol for cells grown in monolayer (Invitrogen, 15596-026). Samples were treated with DNase I (Invitrogen, 18068-015) and cDNA was prepared using SuperScript III RT kit (Invitrogen, 180808-051) using oligo dT primed first-strand synthesis. Real-time PCR was carried out on the 7000 ABI Detection System using TaqMan probes for the housekeeping gene Gapdh (Applied Biosystems, Mm99999915_g1) as a control and genes of interest (Applied Biosystems: Tcf3, Mm00493456_m1; Oct4, Mm00658129_gH; Sox2, Mm00488369_s1; Nanog, Mm02384862_g1).
Expression arrays
Genomic expression analysis was measured using Agilent Whole-Mouse Genome Microarrays (Agilent, G4122F). Two micrograms of RNA were labeled for each sample using the Two-color Low RNA Input Linear Amplification Kit PLUS (Agilent, 5188-5340). RNA from the treated sample (either Tcf3 KD cells or cells treated with Wnt3a CM) were labeled with Cy5, and RNA from control cells (infected with empty vector virus or a mock CM control, respectively) were labeled with Cy3. Labeled cRNA was hybridized overnight at 65°C. Slides were washed according to the Agilent protocol and scanned on an Agilent DNA microarray scanner BA. Data were analyzed using Agilent Feature Extraction version 9.5.3 with default settings recommended by Agilent. Flagged and low-intensity spots were then removed and spots representing a single gene were averaged.
Wnt pathway activation
Wnt pathway activity in mES cells was stimulated using Wnt3a CM (American Type Culture Collection [ATCC], CRL-2647) and mock CM (ATCC, CRL-2648) was used as a control. Preparation of CM was performed as per protocol provided with the cells. CM was diluted with mES media at a ratio of 1:1.
Immunohistochemical analysis
Mouse ES cells were cross-linked for 10 min at room temperature with 4% paraformaldehyde. Cells were permeabilized with 0.2% Triton X-100 for 10 min and stained for Oct4 (Santa Cruz Biotechnology, sc-5279; 1:200 dilution), Nanog (Abcam, ab1603; 1:250 dilution), and DAPI Nucleic Acid Stain (Invitrogen D1306; 1:10000 dilution) overnight at 4°C. After several washes, cells were incubated for 2 h at room temperature with goat anti-mouse-conjugated Alexa Fluor 488 (Invitrogen; 1:200 dilution) or goat anti-rabbit-conjugated Alexa Fluor 568 (Invitrogen; 1:200 dilution).
Quantitative image acquisition and data analysis
Image acquisition and data analysis were performed essentially as described in Moffat et al. (2006). Five days post-infection, cells were fixed and stained with Oct4 and Hoechst 33342 (1:1000 dilution). Stained cells were imaged on an Arrayscan HCS Reader (Cellomics) using the standard acquisition camera mode (10× objective, nine fields). Hoechst was used as the focus channel and intrawell focusing was done every three fields. The Apotome feature was applied to acquire all images. Objects selected for analysis were identified based on the Hoechst staining intensity using the Target Activation Protocol and the Isodata Threshold method. Parameters were established requiring that individual objects pass an intensity and size threshold. The Object Segmentation Assay Parameter was adjusted for maximal resolution. Following object selection, the average Oct4 intensity was determined and then a mean value for each well was calculated. All wells used for subsequent analysis contained at least 5000 selected objects.
Accession numbers
All microarray data from this study are available at ArrayExpress at the EBI (http://www.ebi.ac.uk/arrayexpress) under the accession designation E-TABM-409.
Acknowledgments
We thank Stuart Levine, Alex Marson, Martin Aryee, and Sumeet Gupta for experimental and analytical support; Warren Whyte for the Gfp lentivirus vector; Roshan Kumar for knockdown and microarray advice; Jennifer Love for microarray advice; Laurie Boyer and Mathias Pawlak for cell culture advice; and Tony Lee, Scott McCuine, Brett Chevalier, and Rudolph Jaenisch for helpful discussions. Images for immunofluorescence were collected using the W.M. Keck Foundation Biological Imagining Facility at the Whitehead Institute and Whitehead-MIT Bioimaging Center. The SSEA-1 monoclonal antibody developed by D. Solter and B.B. Knowles was obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICGH, and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA. This work was supported by grants from the NIH and The Whitehead Institute. S.J. was supported by an NSF Predoctoral Training Fellowship, and M.K. was supported by an NIH NIGMS Post-doctural Fellowship.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1642408.
References
- Anton R., Kestler H.A., Kuhl M. β-Catenin signaling contributes to stemness and regulates early differentiation in murine embryonic stem cells. FEBS Lett. 2007;581:5247–5254. doi: 10.1016/j.febslet.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Behrens J., von Kries J.P., Kuhl M., Bruhn L., Wedlich D., Grosschedl R., Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Boiani M., Scholer H.R. Regulatory networks in embryo-derived pluripotent stem cells. Nat. Rev. Mol. Cell Biol. 2005;6:872–884. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brantjes H., Roose J., De van Wetering M., Clevers H. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 2001;29:1410–1419. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan K.M. Wnt signaling—20 years and counting. Trends Genet. 2002;18:340–342. doi: 10.1016/s0168-9525(02)02707-5. [DOI] [PubMed] [Google Scholar]
- Cavallo R.A., Cox R.T., Moline M.M., Roose J., Polevoy G.A., Clevers H., Peifer M., Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Dailey L., Basilico C. Coevolution of HMG domains and homeodomains and the generation of transcriptional regulation by Sox/POU complexes. J. Cell. Physiol. 2001;186:315–328. doi: 10.1002/1097-4652(2001)9999:9999<000::AID-JCP1046>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Daniels D.L., Weis W.I. β-Catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 2005;12:364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- Dravid G., Ye Z., Hammond H., Chen G., Pyle A., Donovan P., Yu X., Cheng L. Defining the role of Wnt/β-catenin signaling in the survival, proliferation, and self-renewal of human embryonic stem cells. Stem Cells. 2005;23:1489–1501. doi: 10.1634/stemcells.2005-0034. [DOI] [PubMed] [Google Scholar]
- Dreesen O., Brivanlou A.H. Signaling pathways in cancer and embryonic stem cells. Stem Cell Rev. 2007;3:7–17. doi: 10.1007/s12015-007-0004-8. [DOI] [PubMed] [Google Scholar]
- Friel R., van der Sar S., Mee P.J. Embryonic stem cells: Understanding their history, cell biology and signalling. Adv. Drug Deliv. Rev. 2005;57:1894–1903. doi: 10.1016/j.addr.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J., Li T.G., Qi X., Zhao D.F., Zhao G.Q. WNT/β-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Dev. Biol. 2006;290:81–91. doi: 10.1016/j.ydbio.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Hay D.C., Sutherland L., Clark J., Burdon T. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells. 2004;22:225–235. doi: 10.1634/stemcells.22-2-225. [DOI] [PubMed] [Google Scholar]
- He T.C., Sparks A.B., Rago C., Hermeking H., Zawel L., da Costa L.T., Morin P.J., Vogelstein B., Kinzler K.W. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Jho E.H., Zhang T., Domon C., Joo C.K., Freund J.N., Costantini F. Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielman M.F., Rindapaa M., Gaspar C., van Poppel N., Breukel C., van Leeuwen S., Taketo M.M., Roberts S., Smits R., Fodde R. Apc modulates embryonic stem-cell differentiation by controlling the dosage of β-catenin signaling. Nat. Genet. 2002;32:594–605. doi: 10.1038/ng1045. [DOI] [PubMed] [Google Scholar]
- Kim C.H., Oda T., Itoh M., Jiang D., Artinger K.B., Chandrasekharappa S.C., Driever W., Chitnis A.B. Repressor activity of Headless/Tcf3 is essential for vertebrate head formation. Nature. 2000;407:913–916. doi: 10.1038/35038097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V., Barker N., Willert K., Molenaar M., Roose J., Wagenaar G., Markman M., Lamers W., Destree O., Clevers H. Two members of the Tcf family implicated in Wnt/β-catenin signaling during embryogenesis in the mouse. Mol. Cell. Biol. 1998;18:1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen D.M., Kalisz M., Nielsen J.H. Cytokine signalling in embryonic stem cells. APMIS. 2005;113:756–772. doi: 10.1111/j.1600-0463.2005.apm_391.x. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley R.C., Gill J.G., Kyba M., Murphy T.L., Murphy K.M. Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development. 2006;133:3787–3796. doi: 10.1242/dev.02551. [DOI] [PubMed] [Google Scholar]
- Logan C.Y., Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- McAdams H.H., Arkin A. Stochastic mechanisms in gene expression. Proc. Natl. Acad. Sci. 1997;94:814–819. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill B.J., Gat U., DasGupta R., Fuchs E.2001Lef-1 and Tcf-3 transcription factors mediate tissue specific Wnt signaling during Xenopus development Genes & Dev. 151688–1705.11445543 [Google Scholar]
- Merrill B.J., Pasolli H.A., Polak L., Rendl M., Garcia-Garcia M.J., Anderson K.V., Fuchs E. Tcf3: A transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263–274. doi: 10.1242/dev.00935. [DOI] [PubMed] [Google Scholar]
- Miyabayashi T., Teo J.L., Yamamoto M., McMillan M., Nguyen C., Kahn M. Wnt/β-catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. 2007;104:5668–5673. doi: 10.1073/pnas.0701331104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J., Grueneberg D.A., Yang X., Kim S.Y., Kloepfer A.M., Hinkle G., Piqani B., Eisenhaure T.M., Luo B., Grenier J.K., et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Scholer H., Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Odom D.T., Zizlsperger N., Gordon D.B., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom D.T., Dowell R.D., Jacobsen E.S., Nekludova L., Rolfe P.A., Danford T.W., Gifford D.K., Fraenkel E., Bell G.I., Young R.A. Core transcriptional regulatory circuitry in human hepatocytes. Mol. Syst. Biol. 2006;2:2006.0017. doi: 10.1038/msb4100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K., Nishinakamura R., Iwamatsu Y., Shimosato D., Niwa H. Synergistic action of Wnt and LIF in maintaining pluripotency of mouse ES cells. Biochem. Biophys. Res. Commun. 2006;343:159–166. doi: 10.1016/j.bbrc.2006.02.127. [DOI] [PubMed] [Google Scholar]
- Okumura-Nakanishi S., Saito M., Niwa H., Ishikawa F. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J. Biol. Chem. 2005;280:5307–5317. doi: 10.1074/jbc.M410015200. [DOI] [PubMed] [Google Scholar]
- Otero J.J., Fu W., Kan L., Cuadra A.E., Kessler J.A. β-Catenin signaling is required for neural differentiation of embryonic stem cells. Development. 2004;131:3545–3557. doi: 10.1242/dev.01218. [DOI] [PubMed] [Google Scholar]
- Pan G., Thomson J.A. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- Pera M.F., Trounson A.O. Human embryonic stem cells: Prospects for development. Development. 2004;131:5515–5525. doi: 10.1242/dev.01451. [DOI] [PubMed] [Google Scholar]
- Pereira L., Yi F., Merrill B.J. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol. Cell. Biol. 2006;26:7479–7491. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekhar V.K., Begemann M. Concise review: Roles of polycomb group proteins in development and disease: A stem cell perspective. Stem Cells. 2007;25:2498–2510. doi: 10.1634/stemcells.2006-0608. [DOI] [PubMed] [Google Scholar]
- Rao M. Conserved and divergent paths that regulate self-renewal in mouse and human embryonic stem cells. Dev. Biol. 2004;275:269–286. doi: 10.1016/j.ydbio.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Reubinoff B.E., Pera M.F., Fong C.Y., Trounson A., Bongso A. Embryonic stem cell lines from human blastocysts: Somatic differentiation in vitro. Nat. Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- Reya T., Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Rodda D.J., Chew J.L., Lim L.H., Loh Y.H., Wang B., Ng H.H., Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- Roël G., Hamilton F.S., Gent Y., Bain A.A., Destrée O., Hoppler S. Lef-1 and Tcf-3 transcription factors mediate tissue-specific Wnt signaling during Xenopus development. Curr. Biol. 2002;12:1941–1945. doi: 10.1016/s0960-9822(02)01280-0. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N., Elowitz M.B., Alon U. Negative autoregulation speeds the response times of transcription networks. J. Mol. Biol. 2002;323:785–793. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- Sato N., Meijer L., Skaltsounis L., Greengard P., Brivanlou A.H. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Shen-Orr S.S., Milo R., Mangan S., Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- Singla D.K., Schneider D.J., LeWinter M.M., Sobel B.E. wnt3a but not wnt11 supports self-renewal of embryonic stem cells. Biochem. Biophys. Res. Commun. 2006;345:789–795. doi: 10.1016/j.bbrc.2006.04.125. [DOI] [PubMed] [Google Scholar]
- Stock J.K., Giadrossi S., Casanova M., Brookes E., Vidal M., Koseki H., Brockdorff N., Fisher A.G., Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Takao Y., Yokota T., Koide H. β-Catenin up-regulates Nanog expression through interaction with Oct-3/4 in embryonic stem cells. Biochem. Biophys. Res. Commun. 2007;353:699–705. doi: 10.1016/j.bbrc.2006.12.072. [DOI] [PubMed] [Google Scholar]
- Thieffry D., Salgado H., Huerta A.M., Collado-Vides J. Prediction of transcriptional regulatory sites in the complete genome sequence of Escherichia coli K-12. Bioinformatics. 1998;14:391–400. doi: 10.1093/bioinformatics/14.5.391. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Valdimarsdottir G., Mummery C. Functions of the TGFβ superfamily in human embryonic stem cells. APMIS. 2005;113:773–789. doi: 10.1111/j.1600-0463.2005.apm_3181.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson F.H., Park K., Atchison M.L. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc. Natl. Acad. Sci. 2006;103:19296–19301. doi: 10.1073/pnas.0603564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Ogura S., Ishida M., Karasawa M., Takada S. Gene trap screening as an effective approach for identifcation of Wnt-responsive genes in the mouse embryo. Dev. Dyn. 2005;233:484–495. doi: 10.1002/dvdy.20348. [DOI] [PubMed] [Google Scholar]
- Yan D., Wiesmann M., Rohan M., Chan V., Jefferson A.B., Guo L., Sakamoto D., Caothien R.H., Fuller J.H., Reinhard C., et al. Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/β-catenin signaling is activated in human colon tumors. Proc. Natl. Acad. Sci. 2001;98:14973–14978. doi: 10.1073/pnas.261574498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F., Merrill B.J. Stem cells and TCF proteins: A role for β-catenin-independent functions. Stem Cell Rev. 2007;3:39–48. doi: 10.1007/s12015-007-0003-9. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J., Stark A., Kellis M., Hong J.W., Nechaev S., Adelman K., Levine M., Young R.A. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]