Abstract
Presenilins have been implicated in the genesis of Alzheimer’s disease and in facilitating LIN-12/Notch activity during development. All presenilins have multiple hydrophobic regions that could theoretically span a membrane, and a description of the membrane topology is a crucial step toward deducing the mechanism of presenilin function. Previously, we proposed an eight-transmembrane-domain model for presenilin, based on studies of the Caenorhabditis elegans SEL-12 presenilin. Here, we describe experiments that support the view that two of the hydrophobic regions of SEL-12 function as the seventh and eighth transmembrane domains. Furthermore, we have shown that human presenilin 1 behaves like SEL-12 presenilin when analyzed by our methods. Our results provide additional experimental support for the eight-transmembrane-domain model of presenilin topology.
Alzheimer’s disease (AD) is the most common cause of dementia among elderly people. The etiology and pathogenesis of AD are currently unknown. AD is characterized by the presence of amyloid deposits and neurofibrillary tangles in the brains of affected individuals. The majority of cases are sporadic, but mutations in three genes have been found to cause early-onset familial Alzheimer’s disease (FAD). The first identified genetic lesions were found in the gene encoding the β-amyloid precursor protein (β-APP) gene; mutations in β-APP account for a relatively small percentage (5–7%) of early-onset FAD cases (1). Mutations in the other two genes, encoding the proteins presenilin 1 (PS1) and presenilin 2 (PS2), are responsible for a significant portion (up to 60%) of early-onset FAD cases (2). An understanding of how presenilin proteins normally function, and how mutant proteins malfunction, will no doubt provide clues to the genesis of AD.
Proteins that are highly similar to the presenilins have been found in Caenorhabditis elegans (3, 4), Drosophila melanogaster (5, 6), Xenopus laevis (7), and mice (8, 9). Functional data exist for the SEL-12 and HOP-1 presenilins of C. elegans, and for PS1 of mice. The sel-12 gene was found by a genetic screen for mutations that affect the activity of a transmembrane receptor of the LIN-12/Notch family (3). The SEL-12 protein appears to be a bona fide presenilin, because human PS1 or PS2 can substitute for SEL-12 in C. elegans (10). The HOP-1 protein was identified by searching a genomic sequence database. HOP-1 also appears to be a bona fide presenilin, because it can substitute for SEL-12 and appears to be functionally redundant with SEL-12 in facilitating the activity of LIN-12/Notch proteins (4). The intimate connection between presenilin activity and LIN-12/Notch signaling also has been observed in mice, where the phenotype of mutants lacking PS1 has been shown to resemble the phenotype of mutants deficient in Notch1 (11, 12).
The mechanism of presenilin function is unknown. All presenilins have multiple hydrophobic regions that could theoretically span a membrane. On the basis of a study of SEL-12∷LacZ protein fusions, we previously provided evidence for an eight-transmembrane-domain topological model for presenilin proteins (13). Conventional biochemical studies of human PS1 placed the amino and carboxyl terminus and hydrophilic loop in the cytosol (14, 15), consistent with our eight-transmembrane-domain model. However, there have been challenges to this model. In particular, a six-transmembrane-domain model (16) as well as a seven-transmembrane-domain model (17) have been proposed.
It is crucial to resolve these differences and achieve an understanding of presenilin topology. A knowledge of presenilin topology has a direct bearing on issues of presenilin function. For example, the seven-transmembrane-domain topological model has been used to promote the hypothesis that presenilins are serpentine receptors activated by β-amyloid precursor protein binding (18). Furthermore, a knowledge of the correct topology is also important for interpreting experiments involving physical interactions. For example, it has been suggested that PS1 physically interacts with δ-catenin, a cytosolic protein (19); in evaluating the possible functional relevance of this interaction, it is important to know if this interaction involves a cytosolic region of PS1.
Here, we provide strong additional support for our original eight-transmembrane-domain model (13). In the design of these experiments, we have sought to bolster the key claims we made in our original study, specifically by providing additional evidence that the hydrophobic regions that we assigned as the seventh and eighth transmembrane domains actually function as transmembrane domains, to counter assertions that one or both of these hydrophobic regions do not function as transmembrane domains (16, 17). Furthermore, we show that human PS1 behaves like SEL-12 presenilin when analyzed by our methods, to counter the argument that C. elegans and human presenilins have different topologies (16).
MATERIALS AND METHODS
Genetic Methods and Strains Used.
Methods for handling and culturing C. elegans have been described by Brenner (20). The wild-type parent for all strains used was C. elegans var. Bristol strain N2 (20). Topology experiments examined transgenes in an N2 genetic background; transgenic lines were grown at 25°C to maximize β-galactosidase activity. Rescue experiments examined transgenes in a sel-12(ar171) unc-1(e538) background (3) and were performed at 20°C.
Plasmids.
The expression vector pLSX (13) was employed for rescue experiments and topological analysis. pLSX contains 2.8 kb of sel-12 5′-flanking region, a polylinker, and 755 bp of 3′ noncoding region including a polyadenylation sequence from the unc-54 gene (21). This vector leads to good expression in a subset of cells that express sel-12 (X.L., D. Levitan, and I.G., unpublished data) and sufficient expression to enable rescue of sel-12(ar171) by an inserted sel-12(+) cDNA (4). However, pLSX does not lead to as much expression as is seen when other segments of the sel-12 coding region are included (10, 22).
LacZ or TM∷LacZ cDNAs were fused in-frame to portions of a sel-12 or PS1 cDNA at the carboxyl terminus. Hybrid cDNAs were placed into pLSX. The in-frame fusions or truncations were generated by PCR-mediated cloning. The integrity of all PCR products was confirmed by sequencing.
To generate SEL-12ΔHR8–9 cDNA, the carboxyl-terminal portion of sel-12 cDNA was amplified by using PCR with one primer SEL12d2TM-5′-CTGCTAGCAGCACTCCCGGCTCTGC harboring an NheI site and the sequence encoding amino acids 408–412, ALPAL, and the other primer IST3–5′-ACTAAAGGGAACAAAAGCTGG, which is complementary to a region in the vector pBluescript. This amplified cDNA product was joined to the NheI site of the amino-terminal portion of sel-12 cDNA at amino acid L336. This fusion resulted in the deletion of SEL-12 from amino acid A338 to amino acid R407, including HR8 and HR9.
To generate SEL-12ΔHR8, the C-terminal portion of sel-12 cDNA was amplified by using PCR with one primer SEL12mut1F-5′-GAAGCTAGCCCCTGACTGGAACACGACTATCG harboring an NheI site and amino acids 380–385, DWNTTI, and the other primer IST3. Similarly this amplified cDNA was linked to the NheI site of sel-12 cDNA at amino acid L336. This fusion resulted in the deletion of SEL-12 from amino acid A338 to amino acid F379, including HR8.
To generate SEL-12ΔHR6–8 for LacZ fusions, the amino-terminal portion of sel-12 cDNA was amplified by using PCR with one primer SEL12TM5R-5′-CGTCTAGAGGTAGGTACTTGATAAAGACC, which is complementary to amino acids 205–211, VFIKYLP, and contains an XbaI site at the 5′ end, and the other primer IST7–5′-ACTCACTATAGGGCGAATTGG, which contains a sequence in the vector pBluescript. The carboxyl-terminal portion of sel-12 cDNA was amplified by using PCR with one primer SEL12del2–5′-CTAGCTAGCTTGGAACACGACTATCGCTTG harboring an NheI site and amino acids 381–386, WNTTIA, and the other primer SEL12TM8R-5′-GCGGATCCGGGAGTGCTCGTTTGAAG, which is complementary to the sequence encoding amino acids 405–410, FKRALP, and contains a BamHI site at the 5′ end. These two portions were linked through the XbaI site of the amino-terminal product and the NheI site of the carboxyl-terminal product. This fusion resulted in the deletion of SEL-12 from amino acid E212 to amino acid D380, including HR6, HR7, and HR8.
To generate SEL-12HR8*, the amino-terminal portion of sel-12 cDNA was amplified by using PCR with one primer SEL12mut1R-5′-CGTCTAGAACTACAGCCACGCCGAGGAGAACAGAGTAG, which is complementary to the sequence encoding amino acids YSVLLGVAVVL and contains an XbaI site at the 5′ end, and the other primer IST7. The carboxyl-terminal portion of sel-12 cDNA was amplified by using PCR with one primer SEL12mut1F harboring the sequence encoding amino acids PDWNTTI and an NheI site at the 5′ end and the other primer IST3. These two portions of sel-12 cDNA were joined through the XbaI site of the amino-terminal product and the NheI site of the carboxyl-terminal product. This fusion resulted in the replacement of amino acids 374–379, KASSYF, in HR8 of SEL-12 by amino acids VAVVLAP.
To generate PS1HR7 and PS1HR9 for LacZ fusions, the portions of PS1 cDNA were amplified by using PCR with the forward primer PS1CF-5′-CGATCGATCAGCAAAAATGACAGAGTTACCTGCACCGT, which is complementary to the sequence encoding amino acids 1–7, MTELPAP, and the reverse primer PS1LRX for PS1HR7 and the reverse primer PS1TM8RB for PS1HR9. PS1LRX is 5′-CGTCTAGAAGTTTTACTCCCCTTTCCTCTGG, which is complementary to the sequence encoding amino acids 370–377, PEERGVKL, and contains an XbaI site at the 5′end. PS1TM8RB is 5′-GCGGATCCGGCAATGCTTTCTTGAAAATGGC, which is complementary to the sequence encoding amino acids 422–429, AIFKKALP, and contains a BamHI site at the 5′ end.
To generate PS1ΔHR8 for LacZ fusions, the carboxyl-terminal portion of PS1 cDNA was amplified by using PCR with one primer PS1TM8FX-5′-CGTCTAGACTGGAACACAACCATAG harboring the sequence encoding amino acids 399–404, DWNTTI, and an XbaI site at the 5′ end and the other primer PS1TM8RB. This portion of the cDNA product was linked to the previously generated PS1HR7 cDNA product through XbaI site ligation. This fusion resulted in the deletion of PS1 from amino acids G378 to G398, including HR8.
Transgenic Lines.
Transgenic lines were established by microinjection of plasmid mixtures into the hermaphrodite germ line to create extrachromosomal arrays (23). For the rescue experiments, plasmids were injected at a concentration of 20 μg/ml into recipient sel-12(ar171) unc-1(e538) hermaphrodites along with 100 μg/ml pRF4. pRF4 is a plasmid containing the cloned dominant rol-6(su1006) gene, which has been widely used as a cotransformation marker in C. elegans (23). F1 Roller progeny were picked and F2 Roller progeny were used to establish stable lines. For the topology experiments, plasmids were injected at a concentration of 20 μg/ml into recipient N2 hermaphrodites along with 100 μg/ml pRF4. F1 Roller progeny were picked and F2 Roller progeny were used to establish stable lines.
Rescue Assays.
Twenty to 61 L4 Roller progeny from each independent transgenic line were picked individually to assess the ability of the altered sel-12 cDNAs to rescue the egg-laying defect of sel-12(ar171) worms. Because sel-12(ar171) never lay eggs (3), transgenic animals were scored daily for the ability to lay eggs. Animals were scored as Egl+ if they displayed robust egg-laying for 2 days as adults, characteristic of wild-type hermaphrodites.
β-Galactosidase Staining.
Transgenic lines expressing hybrid LacZ proteins were grown at 25°C and fixed with an acetone fixation protocol (24). For transgenes encoding SEL-12∷LacZ and SEL-12∷TM∷LacZ, fixed animals were stained overnight at room temperature for β-galactosidase activity. For transgenes encoding PS1∷LacZ and PS1∷TM∷LacZ, fixed animals were stained for 2 days at room temperature for β-galactosidase activity.
RESULTS
General Strategies and Issues.
Throughout this paper, we will use the term hydrophobic region (HR) for a segment of a protein with the potential to span the membrane [as assessed by hydropathy analysis (25)] and the term transmembrane domain (TM) for an HR that our data indicate actually spans a membrane (see below).
Hydropathy analysis suggests that C. elegans SEL-12 presenilin, human PS1, and human PS2 each have 10 HRs (see Fig. 1). We previously proposed that eight of these hydrophobic regions in SEL-12 function as transmembrane domains in vivo. HR1–6 correspond to TM1–6, HR8 corresponds to TM7, and HR9 corresponds to TM8; HR7 and HR10 do not function as transmembrane domains (13). Another C. elegans presenilin, HOP-1, lacks HR7 and has only nine hydrophobic regions, and available topological data are consistent with an eight-transmembrane-domain topology for HOP-1 presenilin (4).
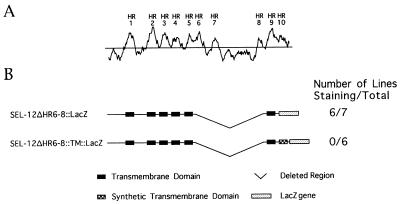
Figure 1.
Hydrophobicity plot of SEL-12 and schematic representation of SEL-12 constructs. (A) Hydrophobicity plot of SEL-12 constructed by using the Kyte–Doolittle algorithm (25) with a window size of 15. HRs are numbered as in ref. 13. (B) SEL-12 constructs used in the LacZ fusion approach and the rescue assay. SEL-12ΔHR6–8∷LacZ, LacZ placed after the HR9 of SEL-12 with the region of HR6 through HR8 deleted; SEL-12ΔHR6–8∷TM∷LacZ, a construct similar to SEL-12ΔHR6–8∷LacZ but including a synthetic transmembrane domain (21) at the fusion junction (amino acid P410 of SEL-12) between sel-12 and lacZ cDNAs.
One approach we have used in this study, as in our previous study (13), has been to use β-galactosidase activity to probe topology. This approach relies on the observation that β-galactosidase is active within the cytoplasm of cells but not in the extracytosolic compartment. When β-galactosidase is fused at different points in a membrane protein, its location in the cytosol or in an extracytosolic compartment, and hence the topology of the membrane protein, can be deduced by its activity (26–30). Thus, we constructed transgenes encoding hybrid SEL-12∷LacZ (or PS1∷LacZ) proteins, in which lacZ was fused in-frame to portions of sel-12 (or PS1) cDNA fragments at the carboxyl terminus. We also constructed analogous transgenes encoding hybrid SEL-12∷TM∷LacZ (or PS1∷TM∷LacZ) proteins, which include a synthetic transmembrane domain at the fusion junction. Transgenic C. elegans animals expressing hybrid proteins in which the LacZ moiety is located in the cytosol should consistently display β-galactosidase activity, whereas animals expressing hybrid proteins in which the LacZ moiety is located in the extracytosolic compartments should not display β-galactosidase activity. For a pair of hybrid proteins with the same fusion point but differing only in the presence or absence of a synthetic transmembrane domain, we would expect that one hybrid would have β-galactosidase activity but the other would not.
We have also used a presenilin functional assay to assess the activity of engineered SEL-12 or PS1 mutant proteins. Mutations that reduce or eliminate the activity of sel-12 cause a distinctive egg-laying defective (Egl) phenotype. The activity of normal and mutant presenilins can be assessed by creating transgenic C. elegans lines that express cDNAs under the control of lin-12 (10) or sel-12 regulatory sequences (4, 22) (see also Fig. 2) and assaying hermaphrodites for their egg-laying ability.
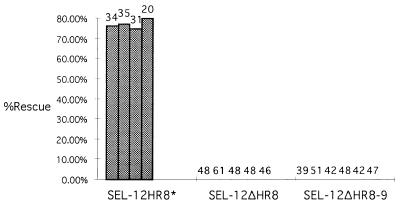
Figure 2.
Rescue of the sel-12(ar171) Egl and abnormal vulva phenotype by the SEL-12 constructs. The data shown for transgenic lines are generated by injecting a construct that places a modified sel-12 cDNA (encoding SEL-12HR8*, in which the hydrophobicity of HR8 has been increased; see Fig. 3) or a truncated sel-12 cDNA (encoding SEL-12ΔHR8 or SEL-12ΔHR8–9) under the control of sel-12 5′ flanking sequence. Each line in the histogram represents data for an independent transgenic line and the number of hermaphrodites scored is shown above each line. The altered protein encoded by the transgene is indicated on the horizontal axis. The percentage of rescued hermaphrodites is indicated on the vertical axis. Transgenic animals expressing the pLSX vector do not exhibit any rescuing activity (4).
All of the proposed models agree that the first six hydrophobic regions function as transmembrane domains. The major differences among the models are as follows: (i) According to the six-transmembrane-domain model, only HR1–HR6 function as transmembrane domains; both the amino and carboxyl termini are located in the cytosol (16). (ii) According to the seven-transmembrane-domain model, HR1–HR6 function as transmembrane domains; the seventh transmembrane domain corresponds to HR9, and the amino terminus is located extracytosolically (17). (iii) According to the eight-transmembrane-domain model, both the amino and carboxyl termini are located in the cytosol, and HR8 and HR9 function as transmembrane domains in addition to HR1–HR6 (13). We have focused on these differences in the experiments described below.
We note that the high degree of amino acid sequence conservation of HR8 and HR9 in C. elegans and mammalian presenilins (see ref. 4) is a strong suggestion that these regions are functionally relevant. Studies of mutant SEL-12 proteins support this contention. Proteins in which HR8 is deleted singly (SEL-12ΔHR8) or along with HR9 (SEL-12ΔHR8–9) do not appear to have presenilin activity in our rescue assay (Fig. 2). In addition, sel-12(ar133), which has a stop codon mutation in the sel-12 gene before HR9, is a strong loss-of-function mutant (3), even though truncation of SEL-12 just after HR9 does not appear to affect rescuing activity (10). Thus, the issue of the membrane topology of this region of the protein is relevant to understanding the function of the presenilins.
The Ninth Hydrophobic Region of SEL-12 Can Function as a Transmembrane Domain.
We previously showed that a SEL-12∷LacZ hybrid protein in which the fusion junction is located after HR9 has β-galactosidase activity, but the corresponding SEL-12∷TM∷LacZ hybrid protein does not. When HR8 was deleted from these proteins, the apparent position of the LacZ moiety “switched”; that is, SEL-12∷ΔHR8∷HR9∷LacZ did not display β-galactosidase activity, but the corresponding SEL-12∷ΔHR8∷HR9∷TM∷LacZ hybrid protein did. These results led us to suggest that HR8 and HR9 function as transmembrane domains (13).
To provide additional experimental evidence for these proposals, we engineered a deletion mutant that specifically tests the ability of HR9 to function as a TM. We deleted the region encompassing HR6, HR7, the loop, and HR8 of SEL-12∷LacZ and SEL-12∷TM∷LacZ proteins in which LacZ was placed after the HR9 of SEL-12 (Fig. 1). According to our eight-transmembrane-domain topology model, this deletion should remove two transmembrane domains, TM6 (HR6) and TM7 (HR8). According to the six- or seven-transmembrane-domain models, this deletion should remove only one transmembrane domain, TM6 (HR6). In principle, removal of an even number of transmembrane domains will not affect the location of the LacZ moiety; however, removal of an odd number of transmembrane domains will switch the location of the LacZ moiety. Thus, if our model is correct, then the apparent position of the LacZ moiety of the SEL-12ΔHR6–8∷LacZ protein should not change. However, if the six- or seven-transmembrane-domain models are correct, then the apparent position of the LacZ moiety of the SEL-12ΔHR6–8∷LacZ protein will change, from cytosolic to extracytosolic.
The results we obtained fulfill the prediction of the eight-transmembrane-domain model: transgenic lines expressing the hybrid SEL-12ΔHR6–8∷LacZ protein in which LacZ was placed after HR9 consistently display staining (Fig. 1), as is the case with the original SEL-12∷LacZ hybrid protein in which LacZ was placed after HR9 (13). Similarly, transgenic lines expressing the corresponding SEL-12ΔHR6–8∷TM∷LacZ hybrid protein do not display staining (Fig. 1), as is the case with the original SEL-12∷TM∷LacZ hybrid protein in which LacZ was placed after HR9 (13). These results strongly support our original suggestion that HR9 functions as a transmembrane domain.
Increasing the Hydrophobicity of HR8/TM7 of SEL-12 Does Not Affect Its Apparent Topology or Its Presenilin Activity.
One argument that has been advanced in support of six- and seven-transmembrane-domain models is the fact that HR8 from SEL-12, PS1, and PS2 is relatively short and not very hydrophobic. In our previous study (13), the main evidence in favor of HR8 function as a transmembrane domain was the switch in apparent topology when HR8 was deleted from SEL-12∷LacZ and SEL-12∷TM∷LacZ hybrid protein in which the fusion junction is located after HR9. However, we could not obtain evidence in favor of HR8 function as a transmembrane domain by comparing the activity of SEL-12∷LacZ and SEL-12∷TM∷LacZ proteins in which the fusion junction is located after HR8 directly because neither of these proteins gave reproducible β-galactosidase activity.
To explore the function of HR8 further we constructed a modified SEL-12 protein, in which HR8 is made considerably more hydrophobic (Fig. 3). The rationale behind this experiment is that, if HR8 does not normally function as a transmembrane domain, then increasing its hydrophobicity so that it demonstrably functions as a transmembrane domain will disrupt its function. We replaced six amino acids of HR8 (KASSYF, amino acids 374–379) by seven amino acids, VAVVLAP. The six amino acids that were changed are less conserved among presenilins, whereas the remainder of HR8 is highly conserved (see ref. 4). These amino acid changes make HR8 of the modified SEL-12 protein (SEL-12HR8*) highly hydrophobic (Fig. 3). We found that HR8* can function as a transmembrane domain, since most transgenic lines expressing the SEL-12HR8*∷LacZ hybrid protein do not display staining, whereas most transgenic lines expressing the corresponding SEL-12HR8*∷TM∷LacZ hybrid protein display strong staining (Fig. 3). Because HR8* forms a transmembrane domain in the context of the LacZ fusions, we expect that it also would form a transmembrane domain in the context of full-length SEL-12.
Figure 3.
Hydrophobicity plots of SEL-12 and SEL-12HR8*. The algorithm of Kyte and Doolittle (25) was used with a window size of 15 residues. (A) Hydrophobicity plot of SEL-12. (B) Hydrophobicity plot of SEL-12HR8*, a modified SEL-12 protein, in which the six amino acids of HR8 (KASSYF, amino acids 374–379) are replaced by the seven amino acids VAVVLAP. This modification makes the new HR8 (indicated by * in the hydrophobicity plot) of the modified SEL-12 protein highly hydrophobic. The arrow indicates the fusion point of SEL-12HR8*∷LacZ and SEL-12HR8*∷TM∷LacZ at the newly introduced L378.
We also constructed a SEL-12 protein containing the modified HR8* region, and we assessed its function as a presenilin. We found that SEL-12HR8* has presenilin activity, because expression of this modified protein can efficiently rescue the egg-laying defect of a strong sel-12 loss-of-function allele (Fig. 2). The topology and rescue results with HR8* taken together strongly suggest that HR8 normally functions as a transmembrane domain.
Human PS1 and C. elegans SEL-12 Have Similar Membrane Topologies.
Human PS1 and C. elegans SEL-12 display a high degree of amino acid sequence similarity, particularly in the hydrophobic regions (3, 13). Furthermore, human PS1 can substitute for SEL-12 in C. elegans (10). Nevertheless, an objection has been made to inferring features of human PS1 topology based on studies of SEL-12 topology (16). We therefore tested the topology of human PS1 by using the LacZ hybrid protein approach.
Transgenic lines expressing PS1∷LacZ hybrid proteins in which LacZ was located in the loop region (PS1HR7∷LacZ) or after the ninth hydrophobic region (PS1HR9∷LacZ) display staining, whereas transgenic lines expressing corresponding PS1∷TM∷LacZ hybrid proteins (PS1HR7∷TM∷LacZ and PS1HR9∷TM∷LacZ) did not display staining (Fig. 4). These results suggest that the loop and the region after HR9 are located in the cytosol. Furthermore, these results are consistent to what we have observed for the corresponding LacZ hybrid proteins with C. elegans SEL-12 and HOP-1 presenilins (4, 13) and what has been observed by conventional biochemical methods for human PS1 (14, 15).
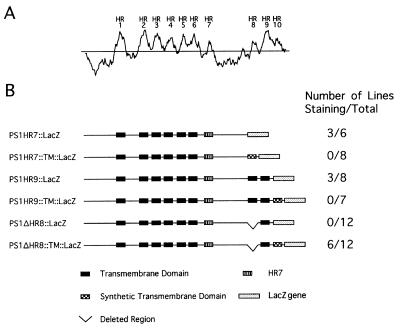
Figure 4.
Hydrophobicity plot of human PS1 and schematic representations of PS1∷LacZ and PS1∷TM∷LacZ constructs. (A) Hydrophobicity plot of PS1, using a window size of 15 (ref. 25). The hydrophobic regions are numbered as in ref. 13. (B) PS1 constructs used in the LacZ fusion approach for topological studies. PS1HR7∷LacZ, LacZ placed after the HR7 of PS1; PS1HR7∷TM∷LacZ, a fusion similar to PS1HR7∷LacZ with a synthetic transmembrane domain at the fusion junction (amino acid L377 of PS1); PS1HR9∷LacZ, LacZ placed after the HR9 of PS1; PS1HR9∷TM∷LacZ, a fusion similar to PS1HR9∷LacZ with a synthetic transmembrane at the fusion junction (amino acid P429 of PS1); PS1ΔHR8∷LacZ, LacZ placed after the HR9 of PS1 with the HR8 deleted; PS1ΔHR8∷TM∷LacZ, a fusion similar to PS1ΔHR8∷LacZ with a synthetic transmembrane domain at the fusion junction (amino acid P429 of PS1).
To determine whether HR8 and HR9 of PS1 function as transmembrane domains, we constructed a pair of hybrid proteins (PS1ΔHR8∷LacZ and PS1ΔHR8∷TM∷LacZ), in which LacZ or TM∷LacZ is fused after the ninth hydrophobic region of PS1 with the eighth hydrophobic region deleted. We observed β-galactosidase activity of PS1ΔHR8∷TM∷LacZ hybrid but not PS1ΔHR8∷LacZ hybrid. Thus, deletion of HR8 switches the location of the LacZ moiety of PS1∷LacZ hybrid proteins with a fusion junction after the ninth hydrophobic region, suggesting that HR8 and HR9 of PS1 function as transmembrane domains. The results with PS1 are entirely consistent with what we have observed before with SEL-12 presenilin (13).
DISCUSSION
Our results with the LacZ hybrid protein approach for both SEL-12 and human PS1 lead to an eight-transmembrane-domain model for presenilin proteins. The inferred topology is illustrated in Fig. 5. Conventional biochemical studies of human PS1 provide data in support of this model: the amino and carboxyl termini and loop region have been shown to be cytosolic (14, 15, 31).
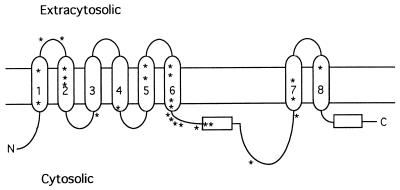
Figure 5.
Inferred topology of presenilin proteins. Arabic numerals indicate the transmembrane domains deduced from this study of SEL-12 and PS1 and our previous study of SEL-12 (13). Horizontal rectangles indicate the seventh or the tenth hydrophobic region in presenilins, which do not appear to span the membrane. Asterisks indicate the positions of mutations found in PS1 that are associated with early-onset familial Alzheimer’s disease (ref. 36 and references therein).
The biochemical data for PS1 (14, 15, 31) would also be consistent with a six-transmembrane-domain model. Lehmann et al. (16) have proposed a six-transmembrane-domain model based on a study using the glycosylation of PS1 hybrid proteins to probe for membrane topology. A glycosylation tag was placed after each of the 10 hydrophobic regions; thus, the experimental design is conceptually similar to ours. When the proteins were expressed in reticulocyte lysates or COS cells, the authors reported that glycosylation occurred only when the tag was placed after HR1, HR3, and HR5. They therefore proposed that all regions of the protein after HR6 are located in the cytosol.
The failure of Lehmann et al. (16) to detect glycosylation of a tag placed after HR8 is reminiscent of the situation we encountered in our original study, where we could not unambiguously interpret the results of LacZ hybrid proteins with a fusion junction after HR8 (13) (see above). HR8 is relatively short and of relatively low hydrophobicity compared with many transmembrane domains. Indeed, the summed hydrophobicities of uncharged 9-amino acid stretches in HR8/TM7 of all presenilins are close to -20 kcal/mol (data not shown), which is at the lower end of transition range for a transmembrane segment to export a reporter in prokaryotes (32). If this “rule” applies to eukaryotic proteins, then it might account for the failure to detect export of tags placed after HR8/TM7.
However, in the intact presenilin protein, HR8/TM7 might be stabilized, for example by interactions with other transmembrane domains. Thus, another test of the function of HR8 is to examine the effects of deletions on the inferred topology of tagged proteins: deletion of single transmembrane domain (HR8 in this case) located upstream of HR9 would be expected to switch the location of a reporter placed after HR9 from the cytosolic to the extracytosolic compartment, whereas deletion of two transmembrane domains located upstream of HR9 would not switch the location of a reporter placed after HR9. When we performed such tests, here and in our previous study (13), we found consistent evidence that HR8/TM7 functions as a transmembrane domain.
We have now further addressed the possible function of HR8 as a transmembrane domain by increasing its hydrophobicity (“HR8*”). We found that the significant increase in hydrophobicity of HR8* does not compromise the function of SEL-12 in vivo and leads to a clear function as a transmembrane domain when LacZ and TM∷LacZ reporters are placed after HR8*. These results strongly support our interpretation that HR8 normally functions as a transmembrane domain. Furthermore, deletion analysis demonstrates that HR9 can function as a transmembrane domain. Thus, in this study we have provided additional experimental evidence that HR8 functions as TM7 and HR9 functions as TM8.
Dewji and Singer (17) have proposed a seven-transmembrane-domain topology in which the amino terminus and loop regions are extracytosolic, as they are in G-protein-coupled receptors. This topology was primarily based on the observation that antibodies to the amino terminus and loop region of PS1 can stain living cells transfected with PS1. However, it is not clear if the antibodies used are specific for PS1, because they recognize proteins in extracts from untransfected cells and were selected on the basis of their staining properties (33).
The proposed seven-transmembrane-domain topology is inconsistent with two observations. First, several biochemical studies have placed the amino terminus and loop region in the cytosol (14, 15, 31) and have suggested that there is intracellular proteolysis of PS1 in the loop region (31, 34). Dewji et al. (35) have argued that in those studies the intracellular proteolysis of PS1 is an artifact of the way protein is extracted from tissue. However, we have made an observation in vivo that is consistent with an intracellular (cytosolic) proteolytic cleavage event in the loop: when we stain transgenic worms expressing a LacZ protein containing a nuclear localization signal fused in the loop, we see localization of the protein in the nucleus, consistent with a cleavage event after TM6 (13). Second, the seven-transmembrane-domain model predicts that HR8 is part of an extracytosolic loop. This prediction is difficult to reconcile with our observation that increasing the hydrophobicity of HR8 enables it to function as a transmembrane domain and does not appreciably compromise its function as a presenilin.
In summary, we feel that our detailed analyses of SEL-12 and PS1 topology in vivo using the LacZ hybrid protein approach (ref. 13; this work), coupled with corroborating biochemical characterization of the amino-terminal, carboxyl-terminal, and loop regions of PS1 (14, 15), strongly supports an eight-transmembrane-domain topology for presenilins. This topological model will be increasingly useful as additional structural and functional data on presenilins are generated, and it will inform models as to how presenilins normally function and how mutant presenilins malfunction to cause Alzheimer’s disease.
Acknowledgments
We thank David Hirsh and Barth Grant, and Diane Levitan and other members of our laboratory, for helpful comments on this manuscript. This work was supported by National Institutes of Health Grant NS35556 to I.G. I.G. is an Associate Investigator of the Howard Hughes Medical Institute.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: PS1 and PS2, presenilins 1 and 2; HR, hydrophobic region; TM, transmembrane domain.
References
- 1.Tanzi R, Kovacs D, Kim T, Moir R, Guenette S, Wasco W. Neurobiol Dis. 1996;3:159–168. doi: 10.1006/nbdi.1996.0016. [DOI] [PubMed] [Google Scholar]
- 2.Lamb B. Nat Med. 1997;3:28–29. doi: 10.1038/nm0197-28. [DOI] [PubMed] [Google Scholar]
- 3.Levitan D, Greenwald I. Nature (London) 1995;377:351–354. doi: 10.1038/377351a0. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Greenwald I. Proc Natl Acad Sci USA. 1997;94:12204–12209. doi: 10.1073/pnas.94.22.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong C S, Koo E H. NeuroReport. 1997;8:665–668. doi: 10.1097/00001756-199702100-00017. [DOI] [PubMed] [Google Scholar]
- 6.Boulianne G L, Livne-Bar I, Humphreys J M, Liang Y, Lin C, Rogaev E, St. George-Hyslop P. NeuroReport. 1997;8:1025–1029. doi: 10.1097/00001756-199703030-00041. [DOI] [PubMed] [Google Scholar]
- 7.Tsujimura A, Yasojima K, Hashimoto-Gotoh T. Biochem Biophys Res Commun. 1997;231:392–6. doi: 10.1006/bbrc.1996.6043. [DOI] [PubMed] [Google Scholar]
- 8.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Nature (London) 1995;375:754–760. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 9.Vito P, Wolozin B, Ganjei J K, Iwasaki K, Lacana E, D’Adamio L. J Biol Chem. 1996;271:31025–31028. doi: 10.1074/jbc.271.49.31025. [DOI] [PubMed] [Google Scholar]
- 10.Levitan D, Doyle T G, Brousseau D, Lee M K, Thinakaran G, Slunt H H, Sisodia S S, Greenwald I. Proc Natl Acad Sci USA. 1996;93:14940–14944. doi: 10.1073/pnas.93.25.14940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen J, Bronson R T, Chen D F, Xia W, Selkoe D J, Tonegawa S. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 12.Wong P C, Zheng H, Chen H, Becher M W, Sirinathsinghji D J S, Trumbauer M W, Chen H Y, Price D L, Van der Ploeg L H T, Sisodia S S. Nature (London) 1997;387:288–292. doi: 10.1038/387288a0. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Greenwald I. Neuron. 1996;17:1015–1021. doi: 10.1016/s0896-6273(00)80231-7. [DOI] [PubMed] [Google Scholar]
- 14.Doan A, Thinakaran G, Borchelt D R, Slunt H H, Ratovitsky T, Podlisny M, Selkoe D J, Seeger M, Gandy S E, Price D L, Sisodia S S. Neuron. 1996;17:1023–1030. doi: 10.1016/s0896-6273(00)80232-9. [DOI] [PubMed] [Google Scholar]
- 15.De Strooper B, Beullens M, Contreras B, Levesque L, Craessaerts K, Cordell B, Moechars D, Bollen M, Fraser P, St. George-Hyslop P, Leuven F V. J Biol Chem. 1997;272:3590–3598. doi: 10.1074/jbc.272.6.3590. [DOI] [PubMed] [Google Scholar]
- 16.Lehmann S, Chiesa R, Harris D A. J Biol Chem. 1997;272:12047–12051. doi: 10.1074/jbc.272.18.12047. [DOI] [PubMed] [Google Scholar]
- 17.Dewji N N, Singer S J. Proc Natl Acad Sci USA. 1997;94:14025–14030. doi: 10.1073/pnas.94.25.14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dewji N N, Singer S J. Science. 1996;271:159–160. doi: 10.1126/science.271.5246.159. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J, Liyanage U, Medina M, Ho C, Simmons A D, Lovett M, Kosik K S. NeuroReport. 1997;8:2085–2090. doi: 10.1097/00001756-199705260-00054. [DOI] [PubMed] [Google Scholar]
- 20.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fire A, Harrison S W, Dixon D. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- 22.Baumeister R, Leimer U, Zweckbronner I, Jakubek C, Grunberg J, Haass C. Genes Funct. 1997;1:149–159. doi: 10.1046/j.1365-4624.1997.00012.x. [DOI] [PubMed] [Google Scholar]
- 23.Mello C C, Kramer J M, Stinchcomb D T, Ambros V A. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fire A. Genet Anal Tech Appl. 1993;9:151–158. doi: 10.1016/1050-3862(92)90042-4. [DOI] [PubMed] [Google Scholar]
- 25.Kyte J, Doolittle R. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 26.Silhavy T J, Beckwith J R. Microbiol Rev. 1985;49:398–418. doi: 10.1128/mr.49.4.398-418.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Froshauer S, Green G N, Boyd D, McGovern K, Beckwith J. J Mol Biol. 1988;200:501–511. doi: 10.1016/0022-2836(88)90539-6. [DOI] [PubMed] [Google Scholar]
- 28.Traxler B, Boyd D, Beckwith J. J Membr Biol. 1993;132:1–11. doi: 10.1007/BF00233047. [DOI] [PubMed] [Google Scholar]
- 29.Henn D K, Baumann A, Kaupp U B. Proc Natl Acad Sci USA. 1995;92:7425–7429. doi: 10.1073/pnas.92.16.7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai C C, Hong K, Kinnel M, Chalfie M, Driscoll M. J Cell Biol. 1996;133:1071–1081. doi: 10.1083/jcb.133.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thinakaran G, Borchelt D R, Lee M K, Slunt H H, Spitzer L, Kim G, Ratovitsky T, Davenport F, Nordstedt C, Seeger M, et al. Neuron. 1996;17:181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 32.Lee E, Manoil C. J Biol Chem. 1994;269:28822–28828. [PubMed] [Google Scholar]
- 33.Dewji N N, Singer S J. Proc Natl Acad Sci USA. 1997;94:9926–9931. doi: 10.1073/pnas.94.18.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim T-W, Pettingell W H, Jung Y K, Kovacs D M, Tanzi R E. Science. 1997;277:373–376. doi: 10.1126/science.277.5324.373. [DOI] [PubMed] [Google Scholar]
- 35.Dewji N N, Do C, Singer S J. Proc Natl Acad Sci USA. 1997;94:14031–14036. doi: 10.1073/pnas.94.25.14031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohan de Silva H A, Patel A J. NeuroReport. 1997;8:i–xii. [PubMed] [Google Scholar]