Abstract
C→U plant organellar RNA editing is required for the translation of evolutionarily conserved and functional proteins. 28 different C targets of RNA editing have been identified in maize chloroplasts, and hundreds of Cs are edited in mitochondria. Mutant analysis in Arabidopsis has indicated that absence of a single site-specific recognition protein can result in loss of editing of a single C target, raising the possibility that each C target requires a recognition protein. Here we show that transcripts encompassing two editing sites, ZMrpoB C467 and ZMrps14 C80, can compete editing activity from each other in vitro despite limited sequence similarity. The signal causing competition overlaps a 5′-cis element required for editing efficiency. A single five-nucleotide mutation spanning the region from –20 to –16 relative to the edited C of rpoB C467 is sufficient to eliminate its substrate editing as well as its ability to compete editing activity from rps14 C80 substrates. A corresponding mutation in an rps14 C80 competitor likewise eliminated its ability to compete editing activity from rpoB C467 substrates. Taken together, our results indicate that the RNA sequences mediating both editing efficiency and cross-competition are highly similar and that a common protein is involved in their editing. Sharing of trans-factors can facilitate editing of the large number of different C targets in plant organelles so that a different protein factor would not be required for every editing site.
Post-transcriptional modification of plant organellar mRNAs by RNA editing is required for maintenance of functional protein sequences (1–3) and also for the introduction of translation initiation codons in particular transcripts (4, 5). Typically, chloroplast genomes of higher land plants have on the order of 30–40 editing sites, whereas mitochondria generally have greater than 400 (6–12). To date, 28 cytidine-to-uridine editing sites have been identified in 15 chloroplast transcripts in maize (12, 13), all of which alter the encoded amino acid, except one site in the 5′-untranslated region of ndhG.
It is currently believed that the plant organellar RNA editing machinery consists of two distinct components: the cis-element, which uniquely identifies a given editing site by its sequence and structure within the transcript itself, and the trans-acting factors, which are likely to be proteins that recognize the cis-element and catalyze the editing reaction (14). The sequences surrounding all editing sites in a given organism do not show obvious similarity to each other either by direct sequence alignment or by secondary structure prediction. However, transplastomic tobacco that overexpress a fragment of maize rpoB or tobacco ndhF transcripts spanning the rpoB C467 or ndhF C290 editing sites, respectively, showed reduced editing at the cognate tobacco sites, as well as at additional sites (15), indicating that at least some cis-elements are related. These three editing sites therefore form a “cluster” affected by overexpression of transcripts carrying only one C target (Fig. 1). Furthermore, three 2–3-nt2 regions of sequence identity exist between the sites of the rpoB C473 cluster within 20 nt 5′ of the edited C, when gaps are introduced in the sequences (15).
FIGURE 1.
Sequence identity in members of the NTrpoB C473 editing site cluster. A, alignment of sequences from tobacco editing sites affected by overexpression of ZMrpoB C467 transgene, as reported in Ref. 15. Shaded boxes indicate regions of sequence identity when the alignment allows gaps. B, orthologous maize editing sites aligned using the tobacco model.
Two nuclear-encoded protein factors have been identified that are believed to be responsible for sequence recognition of editing sites in the ndhD transcript of Arabidopsis: CRR4, which is critical for ATndhD C2 editing, and CRR21, which is required for ATndhD C383 editing (16, 17). Both of these proteins are members of the pentatricopeptide repeat class of proteins, which consists of >450 members in Arabidopsis that are largely targeted to chloroplasts and/or mitochondria (18). Pentatricopeptide repeat motif-containing proteins have been implicated in additional organellar RNA processing or maturation events as well as RNA editing (19–21). As crr4 or crr21 knockouts specifically affect editing at one C target only, they are believed to be sequence recognition factors for the sites they affect. Although CRR4 and CRR21 evidently do not affect editing of multiple C targets, in vivo competition data regarding other editing sites as well as our data from in vitro analysis presented here suggest that trans-factors required for editing of multiple C targets will be described in the future. Possibly genes encoding such trans-factors rarely emerge in mutant screens because their loss of function would often have lethal consequences.
An in vitro editing assay has been developed to study cis-elements near C targets of editing. Editing of RNA substrates, transcribed in vitro, occurs in extracts prepared from isolated chloroplasts (22, 23). A major benefit of this strategy for studying the editing machinery is that many mutant substrates can be studied for editing efficiency at the same time, under controlled conditions. Alternatively, cis-elements can be studied in vivo in transgenic plants; however, this technique is very limited in terms of the number of substrates that can be tested by the amount of time and expense required to generate such transgenic plants, as well as variability due to transformation and regeneration. The in vitro strategy has previously been used in our laboratory to identify cis-elements of the tobacco rpoB C473 and psbE C214 editing sites (23–25).
We have previously studied the tobacco rpoB C473 site extensively both in vivo and in vitro. A transgene containing the sequence from 20 nt 5′ to 6 nt 3′ (–20/+6) of the NTrpoB C473 site is sufficient for tobacco editing in vivo (26), and a –31/+22 transgene is edited more efficiently in vivo (24). Furthermore, a synthetic substrate containing the –31/+22 region of rpoB C473 was shown to be sufficient for in vitro editing in both tobacco and maize extracts (24). Here we report our findings on the relationship of the cis-element of maize rpoB C467 to that of its cluster member, rps14 C80. The existence of a five-member rpoB C473 cluster has been functionally proven in tobacco using an in vivo approach (15); however, in the putative orthologous maize cluster, editing occurs only at rpoB C467 and rps14 C80 because the remaining three members of the maize cluster have a genomically encoded U at the position of the C target in tobacco (Fig. 1). We have found that synthetic maize rpoB C467 and rps14 C80 RNAs can both be edited by maize chloroplast extracts and that both are capable of reducing the editing extent of transcripts carrying either the rpoB C467 or the rps14 C80 sites. We have taken advantage of our competition assay to localize the cis-elements of each editing site that are responsible for the competition effect. Previously, sequences responsible for cross-competition in vivo could not be studied by in vitro editing assays because substrates representing two or more cluster members could not be edited in vitro. We observe that the cis-elements causing the competition effect co-localize with those that determine editing efficiency, indicating that the same trans-factor is likely to mediate both responses.
EXPERIMENTAL PROCEDURES
Synthesis of Editing Substrates in Vitro—DNA templates for RNA substrates were made by PCR amplification (Taq MasterMix kit, Qiagen, Valencia, CA) from maize genomic DNA using gene-specific primers (Integrated DNA Technologies, Coralville, IA) containing overhanging bacterial fragments SK and KS (supplemental Table S1). The T7 promoter sequence was then added by a subsequent PCR step to the 5′ end of the templates. Mutant templates were made by incorporation of mismatches in primers used for PCR. RNA substrates were then transcribed in vitro from their DNA templates and purified as described previously (25).
Preparation of Editing Competent Maize Extracts—Maize chloroplast extracts were prepared from leaves of 7–10-day-old plants as described previously (24). Leaf tissue was homogenized, and intact chloroplasts were isolated by gradient sedimentation using Percoll (Amersham Biosciences). Buffers and conditions for chloroplast isolation, extraction, and dialysis were as described previously (23). Extracts contained 2–4 μg/μl protein.
In Vitro Editing Reaction—Editing reactions for substrate editing without competitor RNAs were as described previously (23), using 0.1 fmol of RNA and 4 μl of extract. For competition experiments, competitor RNAs were added to the editing reaction mixture prior to the addition of 10 fmol of RNA substrate. Following incubation to allow editing, 1 μl of the editing reaction mixture was used for cDNA synthesis and subsequent PCR amplification as in Ref. 23. All substrates, with the exception of the two competition substrates, used the KS primer for RT and SK and KS primers for PCR. For competition experiments, RT of the substrate RNA used the SK(-s) primer, and the subsequent PCR used KS(-s) and SK(-s) primers. The pTri competitor is a 128-nt control transcript containing a fragment of conserved human 18 S rRNA sequence unrelated to any known editing site and is transcribed from a template included with the T7MEGAshortscript kit (Ambion).
Poisoned Primer Extension—To determine the editing efficiency in a given reaction, poisoned primer extension was performed as described previously (23). Different oligonucleotides were used for extension of substrates from each site, depending on the presence of mutations either 5′ or 3′ of the edited C, and are listed in supplemental Table S1.
RESULTS
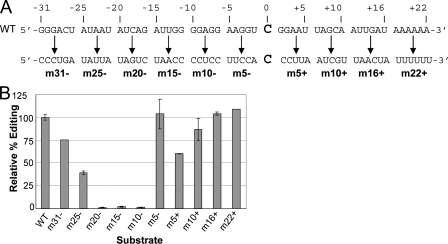
A 54-nt ZMrpoB C4673 substrate, containing 31 nt upstream and 22 nt downstream of the target C and flanked by SK and KS sequences on the 5′ and 3′ ends, respectively, was found to edit 60 ± 4% in replicate experiments. Substrates were made with blocks of 5 or 6 nts at a time mutated to the complementary nucleotides along the length of the sequence (Fig. 2A) to evaluate the significance of each sequence block for C467 editing (Fig. 2B). Six of the mutant substrates had reduced editing efficiency relative to the wild-type substrate, and three of these six were virtually unedited using standard assay conditions. The three 5-nt blocks that contain these critical sequences were m20–, m15–, and m10–, which collectively span the region from –20 to –6 (Fig. 2).
FIGURE 2.
Editing efficiency in vitro of ZMrpoB C467 substrates containing multiple mutations. A, each substrate consisted of WT sequence (upper line), with the exception of the 5–6 nts indicated in the lower line. Substrates also contained SK and KS sequences on the 5′ and 3′ ends, respectively, for RT-PCR amplification. B, relative editing efficiencies of the 10 substrates indicated in A, as compared with a WT substrate, which was 63% edited under the reaction conditions. Error bars represent 1 S.D. from the mean in replicate samples.
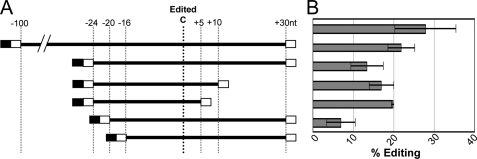
A 131-nt ZMrps14 C80 substrate (100 nt upstream and 30 nt downstream) was edited in vitro to 28 ± 5%. Substrates containing 5′ and 3′ truncations of the 131-nt substrate were assayed for editing efficiency to determine the minimal sequence required for editing to proceed (Fig. 3). Truncation substrates containing at least 20-nt 5′ and at least 5-nts 3′ retained editing efficiency, but substrates with less than 20-nt 5′ had a marked decrease in editing efficiency.
FIGURE 3.
Editing efficiency in vitro of rps14 C80 substrates with varying amounts of sequence around the edited C. A, substrate lengths tested. Open boxes indicate sequences used for universal amplification of substrates during RT-PCR, SK on 5′ ends of substrates, and KS on 3′ ends. Black boxes refer to the T7 promoter sequence for in vitro transcription. B, editing efficiency of each substrate shown in A. Error bars represent 1 S.D. from the mean in replicate samples.
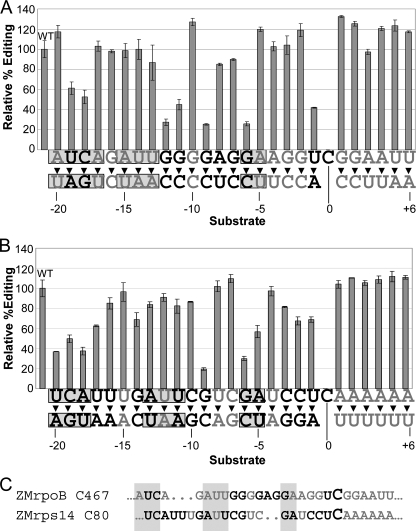
As determined above, the critical cis-elements directing editing of both rpoB C467 and rps14 C80 lie within 20-nt 5′ of the target C, and some sequence further 5′ or on the 3′ side of the target may also be involved in recognition. To further explore the sequence requirements for editing of each substrate, we made single nucleotide mutations at each position within the –20/+6 windows for each site by altering the wild-type sequence to the complementary nucleotide, and each was assayed for editability (Fig. 4). The relative editing efficiency for each substrate was calculated. For rpoB C467, 35% of the mutated substrates had reduced editing efficiency, 35% had enhanced editing efficiency, and 30% had no effect. Of the rps14 C80 mutated substrates, 58% had reduced editing efficiency, 8% had enhanced editing efficiency, and 35% had no effect.
FIGURE 4.
Effect of single nucleotide mutations within the critical editing windows of ZMrpoB C467 and ZMrps14 C80 on in vitro editing efficiency. A, relative editing efficiencies of rpoB C467 54-nt substrate containing mutations to the complementary base at the specified positions, as compared with WT substrate, which was edited 60–74% in replicate experiments. Shaded boxes indicate regions of sequence identity between the two sites, and critical nucleotides (positions that, when mutated, cause a reduction of editing efficiency below WT level) are shown in bold. Error bars represent 1 S.D. from the mean. B, same as A, except for the rps14 C80 131-nt substrate. Actual editing efficiency of WT substrate was 47–61% under the reaction conditions. C, alignment as in Fig. 1, with critical single nucleotides from A and B shown in bold; sequence identity in the cluster is marked by shaded boxes.
The raw (unaligned) comparison between the two critical windows reveals six common positions that negatively affect editing when mutated, and only three of these are the same nucleotide in both sequences. Using gaps to align the rpoB C467 and rps14 C80 editing sites, as reported by Ref. 15, reveals six positions in common that negatively affect editing when mutated, and five of these six are the same nucleotide in both sequences (Fig. 4C).
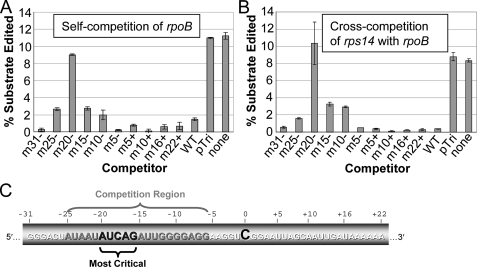
To further investigate cis-elements affecting editing and the similarity between the rpoB C467 and rps14 C80 sites, we performed in vitro competition experiments. First, we established that 100-fold self-competitor was sufficient to reduce editing of rpoB C467 substrate to virtually undetectable levels, whereas inclusion of pTri 18 S RNA, the 128-nt control transcript of unrelated sequence, did not reduce editing of C467. Cross-competition was observed; a 100-fold amount of rpoB C467 substrate reduced rps14 C80 substrate editing to a similarly low level as self-competitor, and rps14 C80 reciprocally reduced rpoB C467 editing.
Fig. 5A shows the results of self-competition experiments using rpoB C467 substrate and rpoB C467 wild-type and multiple-nt mutated competitors. Three 5-nt mutated competitors had reduced competition relative to wild-type, and in the case of competition with the m20– sequence, editing efficiency of the substrate was >80% relative to the no-competitor and pTri controls. In the case of rpoB cross-competition of rps14 C80 substrate editing, four of the 5-nt mutated competitors exhibited reduced competition relative to wild-type competitors (Fig. 5B). Three of these also showed reduced competition of the rpoB C467 substrate (Fig. 5A). The same 5-nt mutation had the largest effect on competition and completely eliminated the ability of the rpoB C467 competitor to reduce editing of rps14 C80. Furthermore, when the –20 to –16 region of the rps14 C80 competitor was mutated, cross-competition was likewise eliminated (Fig. 6).
FIGURE 5.
Effect of ZMrpoB substrate mutations on in vitro self- and cross-competition. A, actual percentage edited of WT ZMrpoB C467 54-nt substrate when competitors were added to in vitro editing reactions at a ratio of 100:1. Substrates m31– to m22+ refer to those indicated in Fig. 2. Error bars represent 1 S.D. from the mean from replicate reactions. B, same asA except for testing rpoB competitors for cross-competition with the rps14 131-nt substrate.
FIGURE 6.
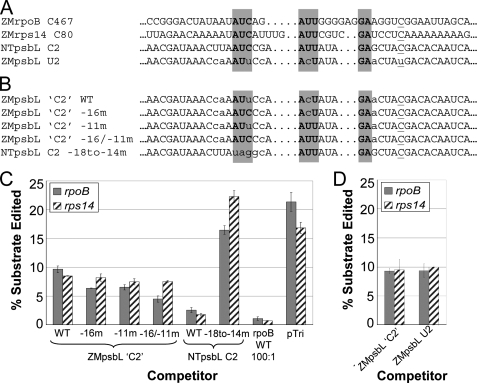
Effect of rps14 competitor mutations on in vitro cross-competition with rpoB C467 substrate. A, sequence alignments of WT and mutant rpoB C467 and rps14 C80 competitor RNAs. Boxed regions show the 5 nts altered in the mutated competitors, and shaded positions represent sequence identity from Fig. 1. B, editing efficiency of rpoB C467 substrate under competition from the RNAs indicated.
In tobacco, overexpression of the maize rpoB C467 site in vivo reduces editing of psbL C2 as well as rps14 C80. The nucleotide at the editing site in the maize psbL transcript is a genomically encoded U, and thus drift could have occurred in a cis-element that might have been present in a progenitor of maize in which psbL C2 was edited. However, we noted that some of the nucleotides found to be critical for editing of maize rpoB C467 and rps14 C80 are present at comparable positions in maize upstream of psbL U2. We therefore wondered whether a maize psbL RNA might reduce editing of rpoB C467 and rps14 C80. Indeed, we found that maize psbL RNA was able to reduce editing of rpoB C467 and rsp14 C80 substrates if a higher competitor-to-substrate ratio was used. Although rpoB C467 and rps14 C80 could compete editing activity at 100-fold competitor-to-substrate, maize psbL U2 competitor or an artificial psbL C2 competitor did not cause an appreciable decrease in substrate editing efficiency unless a 1000-fold competitor-to-substrate ratio was used (Fig. 7).
FIGURE 7.
Effect of psbL C2 competitors on editing efficiency of rpoB C467 and rps14 C80 substrates. A, rpoB C467 editing site cluster as described previously (15), showing nucleotide differences between the tobacco and maize sequences. Conserved elements are shown in shaded boxes, and divergence of the maize gene from the tobacco gene is shown by the use of lowercase letters. B, psbL C2 RNAs used for in vitro competition experiments, including conserved element restoration mutants. U2 in the WT sequence was mutated to a C in all competitor RNAs to correspond to the tobacco psbL C2 editing site. C, shaded bars indicate the editing extent of the rpoB C467 substrate, and dashed bars indicate the editing extent of the rps14 C80 substrate. The ratio of competitor to substrate was 1000 to 1, except in the rpoB 100:1 column, in which the ratio was 100:1. D, competition as in C, except using ZMpsbL C2 and ZMpsbL U2 competitor RNAs.
Comparison of the psbL transcripts from maize and tobacco reveals several sequence differences upstream of the editing site in addition to the C/U editing site difference itself (Fig. 7A). At the 1000-fold competitor-to-substrate ratio needed to observe competition by psbL substrates, a competitor containing the tobacco sequence was able to reduce editing of rpoB C467 and rps14 C80 substrates more efficiently than one containing the maize sequence (Fig. 7C). Two of the differences between the tobacco and maize psbL sequences, at the –16 and –11 positions of the maize transcript, relative to the editing site, are changes in the conserved sequence elements identified in the tobacco editing site cluster. Restoration of these conserved positions in maize psbL C2 competitors (Fig. 7B) enhanced rpoB C467 competition slightly but not to the level of competition exerted by the tobacco psbL C2 competitor (Fig. 7C). The improvement in competition of by the mutated maize competitors was higher for rpoB C467 substrate than for rps14 C80 substrate. Mutating the tobacco psbL competitor in the region corresponding to the rpoB m20– mutant competitor likewise eliminated its ability to compete editing activity from either rpoB C467 or rps14 C80 substrates (Fig. 7C).
Competitors carrying the genomically encoded U at the editing site of maize psbL RNAs did not compete differently from those with a C at this position. When these maize psbL U2 and psbL C2 competitors were added to reactions containing rpoB C467 substrate, the editing efficiencies were reduced to 9%. There also was no significant difference between the psbL U2 and psbL C2 competitors on rps14 C80 substrate editing; with either competitor, the editing efficiencies were reduced to 10%.
DISCUSSION
Self-competition of editing of endogenous psbL transcript with psbL transgene transcripts was first observed in vivo by Chaudhuri et al. (27) and provided the initial indirect evidence for the existence of site-specific recognition factors for chloroplast RNA editing. Subsequently, when Hirose and Sugiura (28) developed editing-competent tobacco chloroplast extracts, they observed that oligoribonucleotides carrying sequences 5′ to psbL C2 or a ndhB C target of editing were effective self-competitors for editing but that no cross-competition occurred when either psbL or ndhB RNAs were added to radiolabeled oligoribonucleotide substrates specified by the other gene. Likewise, Miyamoto et al. (22) showed that psbE and petB editing substrates will undergo self-competition but not cross-competition in vitro. Mutations in either psbE or petB RNAs between –5 and –1 upstream of the C target of editing did not affect self-competition extent (29). These prior reports utilized a thin-layer chromatography separation of radiolabeled C and U to assay editing and competition extent, which is not easily quantified. We have used a sensitive and precise poisoned primer extension assay (25) to quantify the effect of mutation of substrates and competitors on the extent of editing in maize chloroplast extracts. We show that swapping the SK/KS flanking sequences of substrate and competitor RNAs is a convenient method to assay self-competition in vitro.
Our report is the first to demonstrate cross-competition between transcripts of two different editing sites in vitro. The cross-competition of maize rpoB C467 and rps14 C80 substrates in vitro is consistent with our prior finding of cross-competition of the orthologous editing sites in vivo by overexpression of rpoB transgene transcripts in tobacco transplastomic plants (15).
We have demonstrated that psbL competitors carrying either a C or a U at the location of the edited nucleotide are equally effective in reducing editing of rpoB C467 and rps14 C 80 in maize extracts in vitro. The effectiveness of either C-containing or U-containing competitors was not unexpected given that ndhF transgene transcripts carrying either C or U at the editing site were effective competitors of endogenous ndhF transcript editing in transplastomic tobacco plants in vivo (30). Evidently, both unedited and edited transcripts can be recognized by trans-factors.
Cross-competition between editing sites in vivo and in vitro reveals similarities in cis-elements between sequences surrounding different C targets of editing. The simplest explanation for cross-competition would be the existence of a single trans-factor that recognizes these similar editing sites. This hypothesis is an attractive explanation for the capability of the plant to recognize hundreds of different Cs with a high degree of specificity. Imperfect selectivity of a trans-factor in recognition of C targets could explain how a new T → C mutation could be corrected at the transcript level in the plant in which it first arises, leading to evolutionary improvement of editing efficiency at new targets through modification of trans-factors to recognize multiple targets. Alternatively, there could be different factors, perhaps evolutionarily related, that recognize each editing site, perhaps with different efficiencies. Transcripts carrying one editing site could possibly bind both factors, resulting in reduced editing extent of two different C targets when the competitor transcript is in great excess.
A study of an Arabidopsis editing mutant has shown that the lack of a single trans-factor, CRR4, can prevent editing of the ATndhD C2 target (16) and that CRR4 specifically binds to an RNA fragment containing ndhD C2 (17). In tobacco, editing of ndhD C2 is affected by overexpression of an ndhF transgene, and sequences 5′ to both C targets exhibit some similarity, suggesting that the two sites may share the same or related trans-factors. The Arabidopsis ndhF C290 and ndhD C2 editing sites also exhibit 5′ sequence similarity, but no effect on ndhF C290 editing in the ndhD C2 editing-deficient mutant was detected (16). Several hypotheses can be created to explain these apparently contradictory findings. First, possibly an ortholog of CRR4 in tobacco can bind both to ndhD C2 and to the ndhF C290 site, and thus ndhF C290 editing would be reduced in the presence of excess ndhF transcript. Second, there may be a trans-factor that is shared between ndhF C290 and ndhF C290 editing sites that is not an ortholog of CRR4 but is a factor that remains to be discovered. If such a factor is in limiting quantities and is bound by ndhF transcript, editing of ndhD C2 could be reduced even if a CRR4 ortholog only binds to ndhD transcript. Future identification of all of the components of editing complexes that act on C targets with related 5′ sequence should reveal the nature of the relationships between the editing complexes responsible for converting different plant organelle Cs to Us.
Supplementary Material
This work was supported by National Science Foundation MCB-0344007 and MCB-0716888 (to M. R. H.) and National Institutes of Health graduate traineeships (to W. P. H. and M. L. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental table.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: nt, nucleotide; RT-PCR, reverse transcription-PCR; WT, wild-type.
ZMrpoB C467, C at nt 467 from A of ATG of the Zea mays rpoB gene, nomenclature for editing sites as proposed by Hayes et al. (24).
References
- 1.Bock, R., Kossel, H., and Maliga, P. (1994) EMBO J. 13 4623–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sasaki, Y., Kozaki, A., Ohmori, A., Iguchi, H., and Nagano, Y. (2001) J. Biol. Chem. 276 3937–3940 [DOI] [PubMed] [Google Scholar]
- 3.Zito, F., Kuras, R., Choquet, Y., Kossel, H., and Wollman, F. A. (1997) Plant Mol. Biol. 33 79–86 [DOI] [PubMed] [Google Scholar]
- 4.Hoch, B., Maier, R. M., Appel, K., Igloi, G. L., and Kossel, H. (1991) Nature 353 178–180 [DOI] [PubMed] [Google Scholar]
- 5.Kudla, J., Igloi, G. L., Metzlaff, M., Hagemann, R., and Kossel, H. (1992) EMBO J. 11 1099–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corneille, S., Lutz, K., and Maliga, P. (2000) Mol. Gen. Genet. 264 419–424 [DOI] [PubMed] [Google Scholar]
- 7.Giege, P., and Brennicke, A. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 15324–15329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Handa, H. (2003) Nucleic Acids Res. 31 5907–5916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inada, M., Sasaki, T., Yukawa, M., Tsudzuki, T., and Sugiura, M. (2004) Plant Cell Physiol 45 1615–1622 [DOI] [PubMed] [Google Scholar]
- 10.Notsu, Y., Masood, S., Nishikawa, T., Kubo, N., Akiduki, G., Nakazono, M., Hirai, A., and Kadowaki, K. (2002) Mol. Genet. Genomics 268 434–445 [DOI] [PubMed] [Google Scholar]
- 11.Tillich, M., Funk, H. T., Schmitz-Linneweber, C., Poltnigg, P., Sabater, B., Martin, M., and Maier, R. M. (2005) Plant J. 43 708–715 [DOI] [PubMed] [Google Scholar]
- 12.Tsudzuki, T., Wakasugi, T., and Sugiura, M. (2001) J. Mol. Evol. 53 327–332 [DOI] [PubMed] [Google Scholar]
- 13.Tillich, M., Schmitz-Linneweber, C., Herrmann, R. G., and Maier, R. M. (2001) Maize Genetics Cooperation Newsletter 75 42–44 [Google Scholar]
- 14.Shikanai, T. (2006) CMLS Cell. Mol. Life Sci. 63 698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chateigner-Boutin, A. L., and Hanson, M. R. (2002) Mol. Cell. Biol. 22 8448–8456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotera, E., Tasaka, M., and Shikanai, T. (2005) Nature 433 326–330 [DOI] [PubMed] [Google Scholar]
- 17.Okuda, K., Myouga, F., Motohashi, R., Shinozaki, K., and Shikanai, T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 8178–8183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lurin, C., Andres, C., Aubourg, S., Bellaoui, M., Bitton, F., Bruyere, C., Caboche, M., Debast, C., Gualberto, J., Hoffmann, B., Lecharny, A., Le Ret, M., Martin-Magniette, M. L., Mireau, H., Peeters, N., Renou, J. P., Szurek, B., Taconnat, L., and Small, I. (2004) Plant Cell 16 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barkan, A., Walker, M., Nolasco, M., and Johnson, D. (1994) EMBO J. 13 3170–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashimoto, M., Endo, T., Peltier, G., Tasaka, M., and Shikanai, T. (2003) Plant J. 36 541–549 [DOI] [PubMed] [Google Scholar]
- 21.Meierhoff, K., Felder, S., Nakamura, T., Bechtold, N., and Schuster, G. (2003) Plant Cell 15 1480–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyamoto, T., Obokata, J., and Sugiura, M. (2002) Mol. Cell. Biol. 22 6726–6734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegeman, C. E., Hayes, M. L., and Hanson, M. R. (2005) Plant J. 42 124–132 [DOI] [PubMed] [Google Scholar]
- 24.Hayes, M. L., Reed, M. L., Hegeman, C. E., and Hanson, M. R. (2006) Nucleic Acids Res. 34 3742–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayes, M. L., and Hanson, M. R. (2007) RNA (Cold Spring Harbor) 13 281–288 [Google Scholar]
- 26.Reed, M. L., Peeters, N. M., and Hanson, M. R. (2001) Nucleic Acids Res. 29 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaudhuri, S., Carrer, H., and Maliga, P. (1995) EMBO J. 14 2951–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirose, T., and Sugiura, M. (2001) EMBO J. 20 1144–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamoto, T., Obokata, J., and Sugiura, M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 48–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed, M. L., Lyi, S. M., and Hanson, M. R. (2001) Gene (Amst.) 272 165–171 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.