Abstract
Determining the composition of aggregated superoxide dismutase 1 (SOD1) species associated with amyotrophic lateral sclerosis (ALS), especially with respect to co-aggregated proteins and post-translational modifications, could identify cellular or biochemical factors involved in the formation of these aggregates and explain their apparent neurotoxicity. The results of mass spectrometric and shotgun-proteomic analyses of SOD1-containing aggregates isolated from spinal cords of symptomatic transgenic ALS mice using two different isolation strategies are presented, including 1) resistance to detergent extraction and 2) size exclusion-coupled anti-SOD1 immunoaffinity chromatography. Forty-eight spinal cords from three different ALS-SOD1 mutant mice were analyzed, namely G93A, G37R, and the unnatural double mutant H46R/H48Q. The analysis consistently revealed that the most abundant proteins recovered from aggregate species were full-length unmodified SOD1 polypeptides. Although aggregates from some spinal cord samples contained trace levels of highly abundant proteins, such as vimentin and neurofilament-3, no proteins were consistently found to co-purify with mutant SOD1 in stoichiometric quantities. The results demonstrate that the principal protein in the high molecular mass aggregates whose appearance correlates with symptoms of the disease is the unmodified, full-length SOD1 polypeptide.
Mutations in the gene encoding superoxide dismutase 1 (SOD1)7 induce familial amyotrophic lateral sclerosis (ALS), and aggregation of the mutant SOD1 protein is hypothesized to cause pathogenesis (1). Support for the aggregation hypothesis includes pathological, cell-culture, and biophysical data such as: 1) the appearance of fibrillar, spherical or irregularly shaped SOD1-containing aggregates in murine and human ALS spinal cord (2–8); 2) the formation of detergent-insoluble SOD1 aggregates with the onset of motor neuron degeneration in ALS mice (2, 9–12); 3) the observation that cultured neural cells that form SOD1 aggregates die faster than cells that do not (13); and 4) various biophysical and structural perturbations to ALS variant SOD1 proteins (14–18) that accelerate aggregation in vitro (1).
Although the aggregation of SOD1 is a conspicuous signature of SOD1-linked ALS, the protein composition of SOD1 aggregates remains unclear. Identifying proteins that might be co-aggregated with SOD1 could help explain how the aggregates are formed and why they are apparently toxic. For example, the expression of ALS-SOD1 variants in mice and cell cultures can induce: the slowing of axonal transport (19), glutamate excitotoxicity (20, 21), and proteasomal inhibition (22, 23). Data also suggest that variant SOD1 proteins aggregate with other proteins and lower the concentration of these proteins below a threshold for normal viability (24–27). Therefore, the identification of binding interactions or co-aggregation between SOD1 and specific proteins involved in any of these processes could provide clues about the cellular processes involved in ALS pathogenesis.
Another unanswered question pertains to whether the SOD1 polypeptide is pathogenic as it is expressed or if post-translational modifications to SOD1 trigger the polypeptide's pathogenicity. Immunohistochemical analysis of SOD1 inclusions in mouse spinal cord using antibodies that are specific for oxidized proteins have suggested that aggregated SOD1 proteins are extensively modified (e.g. carboxymethylation of lysine residues (4)). Likewise, the self-oxidation of metal coordinating histidine residues by SOD1 has been shown to inactivate SOD1 in vitro (28–31). Such inactivation would be expected to cause metal ion loss and structural destabilization of SOD1, which could promote SOD1 aggregation.
Current information regarding the protein composition of aggregated SOD1 and the post-translational state of aggregated SOD1 is based upon immunohistochemical and Western blot analysis of tissue inclusions and detergent-resistant complexes that form in murine ALS spinal cord. For example, in addition to containing SOD1 (3, 6), the inclusions that are visible by light microscopy in ALS-affected spinal cord tissue are recognized by antibodies to neurofilament components (3, 12), ubiquitin (3, 6, 32), vimentin (9), heat shock proteins (33), Dorfin (34), and NEDL1 (35). The immunostaining of detergent-insoluble SOD1 aggregates from the spinal cord of ALS mice has suggested that many of the same proteins detected in visible SOD1 inclusions are also contained in detergent-insoluble aggregates (11, 32). Consequently, hypotheses have emerged regarding the role of some of these proteins in ALS pathogenesis. Other reports, however, suggest that the co-localization of some of these proteins with SOD1 in inclusion bodies is minimal, inconsistent, and rare, i.e. ubiquitin (9, 32) and heat shock proteins (36).
Immunohistochemical analyses have provided invaluable information on the composition of various types of SOD1 aggregates; however, the limitations to immuno-based analyses involve: 1) the possibility of cross-reactivity or nonspecific binding that produces a false positive identification of proteins; 2) a protein bias; that is, the researcher must know which protein or chemical modification to assay; and 3) the inability to discern if proteins that co-sediment or are co-localized with SOD1 are in fact aggregated with SOD1.
In an attempt to directly characterize the SOD1 proteins that aggregate in ALS spinal cord and to broadly identify proteins that might be co-aggregated with SOD1, we have isolated aggregate forms of SOD1 from the spinal cords of transgenic ALS mice and analyzed these aggregate species with mass spectrometry, a technique used recently to analyze SOD1 aggregates formed in vitro (37). Two different methods were used to isolate SOD1-containing aggregates from mouse spinal cord. One method involves detergent extraction and differential centrifugation and has been previously described (11, 38). This method isolates aggregates based upon their high molecular mass and resistance to various detergents. It is very possible, however, that a detergent extraction could dissociate meta-stable oligomeric species that might play a role in ALS pathogenesis (39, 40). Therefore we also isolated SOD1-aggregate species from mouse spinal cord using a less harsh, detergent-free method that employs size exclusion liquid chromatography (SE-LC) coupled with anti-SOD1 immunoaffinity liquid chromatography (IA-LC).
Analysis of the isolated aggregate species with electrospray ionization mass spectrometry (ESI-MS) and matrix-assisted laser desorption time-of-flight (MALDI-TOF) mass spectrometry has revealed that the detergent resistant SOD1 aggregates are composed predominantly of the full-length, non-ubiquitinated SOD1 polypeptide that is bearing no covalent or oxidative modifications. With the exception of trace amounts of highly abundant proteins, there were no other proteins that were found to be consistently co-aggregated with SOD1.
EXPERIMENTAL PROCEDURES
Transgenic Mice—Transgenic mice expressing hWT, G93A, and G37R SOD1 and the double mutant mouse, expressing H46R/H48Q ALS SOD1 mutations were sacrificed, and their spinal cords were harvested as previously described (2, 11). In short, mice were sacrificed at the end stage of motor neuron degeneration when hind limb paralysis was pronounced. Non-transgenic mice and hWT SOD1 transgenic mice were age-matched with ALS mice when sacrificed. A total of 75 murine spinal cords were analyzed in this study: 12 G93A, 27 H46R/H48Q, 18 G37R, 9 hWT, and 9 non-transgenic.
Detergent Extraction and Sedimentation of Aggregates from ALS Murine Spinal Cord—Detergent extraction and differential centrifugation of high molecular mass, detergent-stable complexes was performed as previously described (11, 38). Spinal cords were sonicated in 10 volumes of TEN buffer (10 mm Tris, pH 7.5, 1 mm EDTA, pH 8.0, 100 mm NaCl), and the homogenate was centrifuged at 800 × g for 10 min. This initial pellet (denoted P1) was discarded, and the remaining supernatant was treated with 1/10 volume of TEN buffer containing 10% of Nonidet P-40. This solution was mixed well, sonicated, and centrifuged at 100,000 × g. The resulting supernatant from this preparation is referred to as S1, and the pellet is referred to as P2. An additional, harsher detergent extraction was performed on the P2 pellet by resuspension of the P2 pellet in 1 ml of TEN buffer with 0.5% Nonidet P-40, 0.25% SDS, and 0.5% deoxycholate. This homogenate was sonicated and centrifuged at 100,000 × g for 30 min. The resulting pellet is denoted as the P3 pellet. The P3 pellet was also subjected to an additional washing step; the P3 pellet was resuspended in 1 ml of TEN with 0.5% Nonidet P-40, 0.25% SDS, and 0.5% deoxycholate, and then sonicated at centrifuged at 100,000 × g for 30 min. The supernatant was discarded. For storage and analysis, the washed P3 pellet was resuspended in 200 μl of water and kept at –80 °C. The number of spinal cords analyzed from mice expressing G93A, H46R/H48Q, G37R, and hWT SOD1 were 12, 9, 3, and 3, respectively.
Dissolution of Detergent-resistant Pellets and Analysis with LC-ESI MS—Frozen detergent-resistant pellet (DRP) samples were thawed, and 100-μl aliquots were mixed with 400 μl of a denaturing solution consisting of 3 m guanidine thiocyanate and 0.5 m DTT. Frozen S1 samples were also thawed and 1/10 volume (∼100 μl) was mixed with 400 μl of 3 m guanidine thiocyanate and 0.5 m DTT. These mixtures were incubated at room temperature for 30 min. Two 200-μl aliquots were removed and placed in separate Eppendorf tubes and to each tube methanol (600 μl), chloroform (200 μl), and water (400 μl) was added. The tubes were mixed vigorously for 2 min, yielding a white suspension. The tubes were then centrifuged for 1 min, yielding a biphasic mixture with a white precipitate at the aqueous/organic interface. The upper methanol/aqueous layer was removed, and 600 μl of methanol was added to each tube. The tubes were then inverted gently to yield one organic phase. The tubes were centrifuged again for 1 min. The organic layer was removed, and the white solid was allowed to dry for 2 min at room temperature and pressure (41, 42).
The white solid was dissolved in 88% formic acid (total volume: 90 μl), and the entire volume was immediately injected onto the HPLC column and separated using a two-solvent gradient. The solvents used were 0.1% trifluoroacetic acid in water (solvent “A”) and 49.9% isopropyl alcohol, 49.9% acetonitrile, 0.05% trifluoroacetic acid (solvent “B”). A compound linear gradient was ramped from 5% B at 5 min to 40% B at 30 min and to 100% B at 150 min. The chromatography was conducted at 40 °C using an PLRPS column, with a flow rate of 100 μl/min (43). The output from the HPLC was outfitted with a splitter affording an equal flow of analyte to the mass spectrometer and to the fraction collector. The fraction collector collected 1-min (50 μl) fractions. The mass spectrometer (API III+, Sciex, Concord, Canada) was tuned and calibrated as described previously (44) and scanned from 600 to 2300 m/z with a step size of 0.3 and a dwell of 1 ms.
MS/MS Identification of Proteins Contained in DRPs—LC-ESI-MS provides a survey of the number of proteins present in the pellet, but mass alone is insufficient for an unambiguous identification of proteins. To identify proteins detected with LC-ESI-MS, they must be trypsinized and sequenced with tandem mass spectrometry (LC-ESI-MS/MS). Therefore, the collected fractions (∼30 in number) from the LC-ESI-MS separation were reduced, alkylated, and treated with trypsin (Promega, sequencing grade) for analysis with LC-ESI-MS/MS. ESI-LC-MS/MS analysis of DRPs was performed on a Q-STAR XL mass spectrometer (Applied Biosystems, Foster City, CA). See supplemental information for additional experimental details of the trypsinization of DRP samples and analysis with ESI-LC-MS/MS.
Size Exclusion and Anti-hSOD1 Immunoaffinity Isolation of Soluble and Aggregated SOD1—Spinal cord tissue was weighed and homogenized in 10 volumes of chilled phosphate-buffered saline, pH 7.4, using an ultrasonic disintegrator. During sonication, the homogenate was kept on ice. The homogenate was then centrifuged (800 × g, 10 min). This low speed centrifugation only pellets the major cellular debris and does not pellet smaller structures such as mitochondria or aggregates. To obtain SOD1 quantities sufficient for detection with mass spectrometry, it was determined that the SE/IA procedure had to be scaled up to include three spinal cords. The supernatants from three individual spinal cords were combined and then loaded onto the G-75 SE column. The numbers of spinal cords analyzed from mice expressing H46R/H48Q, G37R, and hWT SOD1 were 18, 9, and 6, respectively.
The size exclusion/immunoaffinity method used here involved the separation of SOD1-containing complexes first with size exclusion chromatography followed by anti-SOD1 immunoaffinity chromatography. Sephadex G-75 medium was packed into glass columns (1.0 cm × 50 cm and 1.5 cm × 75 cm, respectively). The packed SE columns were connected to a Bio-Logic fraction collector (Bio-Rad) programmed to collect 10-min fractions. The running buffer was phosphate-buffered saline (the chromatography methods did not use detergent). Typically, all of these columns flowed at 200–300 μl/min. To identify SOD1-positive fractions, a 10-μl aliquot from each collected fraction was analyzed with SDS-PAGE and anti-hSOD1 Western blotting. The collected SOD1-positive fractions corresponding to the void volume were combined, concentrated with Centricon centrifugal filtration devices (molecular mass allowance of 3000 Da, Millipore), and subjected to an immunoaffinity (IA) purification using the anti-SOD1 antibody-Sepharose matrix. The resolving fractions were also combined, concentrated with 3-kDa centrifugation filtration devices, and subjected to the anti-SOD1 IA column.
SOD1-positive fractions that eluted from the SE column were incubated with the anti-SOD1-Sepharose beads overnight at 4 °C. The bound species were eluted using 0.1 m glycine/HCl buffer, pH 2.8, according to the same procedure used for the purification of anti-SOD1 antibodies from the SOD1-Sepharose media (see supplemental information for experimental details). The fractions were then washed with 10 mm Tris (pH 8.0) using Centricon centrifugal filtration devices (molecular mass allowance of 3000 Da) to prepare the sample for digestion with trypsin. In short, samples were concentrated to ∼200 μl, followed by the addition of 1.8 ml 10 mm Tris, pH 8.0. This procedure was repeated three to four times with the samples being finally concentrated to ∼200 μl.
Mass Spectrometry of Soluble and Aggregated SOD1 Isolated with Size Exclusion/Anti-SOD1 Immunoaffinity Chromatography—SE/IA purified samples were analyzed directly with MALDI-TOF mass spectrometry. To dissociate aggregated SOD1, the samples were heated at ∼80 °C for 2 min in the presence of 100 mm DTT. Standard protocols were followed for MALDI-TOF-MS analysis (42). In short, 1 μl of matrix solution (1.0 mg of sinapic acid dissolved into 100 μl of 70% acetonitrile/0.1% trifluoroacetic acid) was spotted onto the MALDI plate along with 0.5 μl of protein sample and 0.5 μl of myoglobin (1 nmol/100 μl of H2O; added as an internal calibrant). Spots were allowed to dry at room temperature, and data were collected on a Voyager-DE STR instrument (Applied Biosystems) and analyzed with Data Explorer software (Version 5.10).
MS/MS Identification of Proteins Co-eluting with SOD1 in Chromatographic Fractions—SE/IA samples were concentrated to a volume between 150 and 200 μl using centrifugal filtration devices (Millipore, molecular mass allowance of 3000 Da) and subjected to digestion with trypsin. An aqueous solution of DTT was added to the digest so that the final concentration was 1 mm. Tryptic digests were analyzed by μLC-MS/MS with data-dependent acquisition using an LCQ-DECA ion trapquadrupole ESI-MS (ThermoFinnigan, San Jose, CA). See supplemental information for additional experimental details on the trypsinization of samples and analysis with LC-ESI-MS/MS.
Preparation of Oxidatively Modified ALS Variant SOD1—The ALS variant H48Q SOD1 was expressed in yeast and purified as previously described (45). Purified H48Q SOD1 (1.5 mg/ml) was reacted with 5 mm hydrogen peroxide (5 mm phosphate buffer, pH 7.4) for 1 h at room temperature. The resulting protein solution was analyzed with LC-ESI-MS using the same gradient used for analysis of the DRP samples.
RESULTS
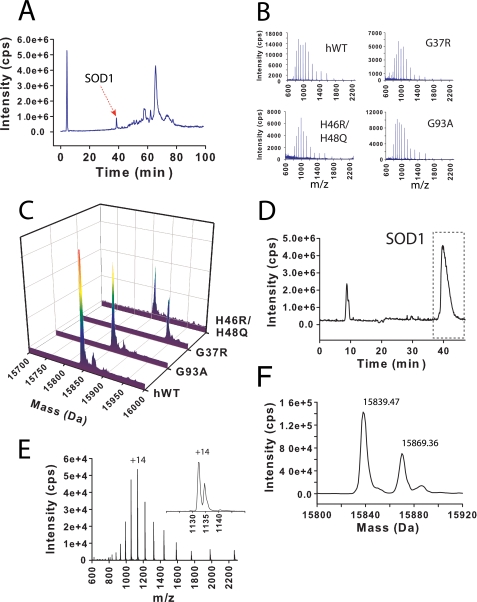
Mass Spectral Profile of Soluble SOD1—Detergent-soluble fractions (i.e. the supernatant of spinal cord homogenate referred to as S1 and prepared as described under “Experimental Procedures”) were analyzed with LC-MS. Both hWT and ALS variant SOD1 proteins eluted from the HPLC column several minutes before most other molecular species. The retention time for hWT or ALS variant SOD1 was between 39 and 41 min (Fig. 1A). This elution profile allowed the collection of easily interpretable mass spectra (Fig. 1, B and C). A deconvolution of the raw mass spectra (m/z) produced a molecular mass reconstruct (m, Da) wherein each spectrum had one predominant signal that was consistent with the molecular mass of each SOD1 variant (Fig. 1C). The calculated average masses for N-terminally acetylated, Met1 removed hWT, G37R, H46R/H48Q, and G93A SOD1 were 15846.62, 15945.75, 15856.66, and 15860.65 Da, respectively. The experimentally measured masses were 15847.4, 15946.6, 15857.2, and 15861.7 Da, respectively (Table 1). These values are within experimental error consistent with normal Met-1 removal and N-terminal acetylation (Table 1). As summarized in Fig. 1, the mass distribution of both hWT and ALS variant SOD1 proteins is predominantly unimodal. This peak shape indicates that soluble hWT and ALS variant SOD1 proteins are largely unmodified. There are, however, low levels of modified SOD1 species that can be observed in each de-convoluted mass spectrum. The mass adducts were +25 Da (hWT), +13 Da (G37R), +26 Da (G93A), and +24 Da (H46R/H48Q) and are most probably due to oxidation (+16 Da) or formylation (+28 Da) as a consequence of brief exposure to 90% formic acid during solubilization. Sodium (+22 Da) and potassium (+38 Da) adducts are rarely seen under the conditions used. It is reasonably assumed that modest adduction does not change the ionization efficiency of the protein, and based on this assumption we conclude that at least 80% of the SOD1 protein is unmodified. It is further noted that a high concentration of DTT was added to the pellet at the beginning of the methanol/chloroform extraction (see “Experimental Procedures”), and thus any modifications easily reversible with DTT, such as O on Cys (+16 Da) may not be seen. The reader is reminded that there are no Met residues in the mature SOD1 sequence.
FIGURE 1.
LC-ESI-MS of soluble hWT and ALS variant SOD1 from ALS-mouse spinal cord. A, total ion current LC chromatogram from LC-ESI-MS analysis of supernatant solution (referred to as “S1”). ALS variant and hWT SOD1 elute at ∼39–41 min (peak corresponding to hSOD1 marked with red arrow). B, raw mass spectra of SOD1 species eluting between 38 and 41 min from hWT, G37R, H46R/H48Q, and G93A mice. C, reconstructed mass spectra that were deconvoluted from the corresponding raw spectra in B (see Table 1 for mass values). D–F, oxidative modifications to SOD1 do not significantly alter retention time on HPLC column. D, LC-ESI-MS total ion current chromatogram of recombinantly purified H48Q SOD1 that has been partially oxidized in vitro with H2O2. Peak at 39–41 min corresponds to H48Q SOD1 (peak at ∼9 min is eluted salt from sample injection). E and F, raw mass spectrum (E) and deconvoluted mass spectrum (F) of H48Q SOD1 species eluting in selected region of chromatogram in D. Two SOD1 species are present and have co-eluted, demonstrating that oxidatively modified SOD1 in mouse spinal cord would have eluted with unmodified SOD1 at ∼39–41 min. Species with mass of 15839.47 Da corresponds to unmodified H48Q SOD1; species with 15869.36 Da corresponds to oxidatively modified H48Q SOD1.
TABLE 1.
Molecular mass values for aggregated and soluble SOD1 proteins isolated from spinal cord of symptomatic mice expressing ALS mutant SOD1 proteins (and from non-symptomatic mice expressing human WT SOD1)
|
hSOD1 variant
|
Molecular
massa
|
No. of spinal cords analyzed
|
||||
|---|---|---|---|---|---|---|
| Calculated | Soluble | Aggregate | ||||
| Da | ||||||
| H46R/H48Q | 15856.7 | 15857.2 | 15857.0 | 9 | ||
| G37R | 15945.8 | 15946.6 | 15945.0 | 9 | ||
| G93A | 15860.7 | 15861.7 | 15862.5 | 12 | ||
| WT | 15846.7 | 15847.4 | -b | 3 | ||
Mass values for “soluble” and “aggregated” SOD1 refers to SOD1 proteins detected in supernatant of spinal cord homogenate and in detergent-insoluble aggregates (P2 preparations), respectively.
-, denotes that no SOD1 protein was detected in detergent insoluble aggregates. Theoretical molecular mass values account for N terminus methionine cleavage and N terminus acetylation.
It is possible that oxidized SOD1 would elute from the HPLC column separately from unmodified SOD1, as can be observed for some proteins upon their oxidation (46). To determine if oxidative modifications to SOD1 result in a substantial change in the HPLC retention time, we prepared an oxidatively modified ALS variant SOD1 protein by incubating a recombinantly purified SOD1 protein (H48Q SOD1) with hydrogen peroxide. This reaction did not result in the appearance of an additional peak in the HPLC total ion current chromatogram (Fig. 1D); however, the mass spectra of the SOD1 protein did exhibit a +32 Da modification (Fig. 1, E and F; note: a similarly prepared +32 Da modified SOD1 protein has been previously shown to involve the oxidation of copper-coordinating histidine residues (47)). Therefore, oxidatively modified SOD1 proteins that might exist in ALS spinal cord can be expected to elute with the unmodified SOD1 protein.
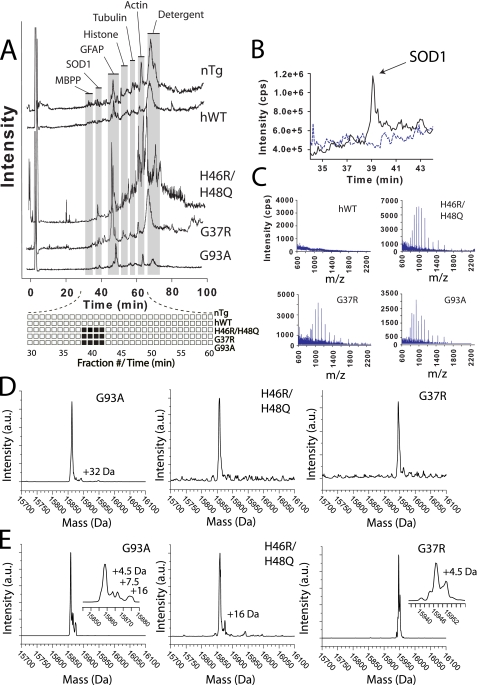
Mass Spectral Profile of SOD1 Proteins Recovered from DRPs—ALS-variant proteins were extracted from detergent-resistant pellets after reduction using methanol/chloroform and then analyzed with LC-MS (Fig. 2). All three ALS variant SOD1 proteins recovered from the DRPs (G37R, G93A, and H46R/H48Q) had identical elution times compared with the soluble SOD1 proteins from supernatant (39–41 min, Fig. 2A). The most noticeable difference between the LC-MS results for SOD1 proteins from the S1 supernatant (Fig. 1) compared with proteins from the DRPs is that no signal was detected for the hWT SOD1 protein in the DRP samples (Fig. 2 and Table 1). This absence of hWT SOD1 in DRPs suggests that hWT SOD1 does not accumulate into detergent-insoluble and sedimentable complexes (Fig. 2C). This negative result is corroborated by previous Western blot analyses of DRPs from ALS mouse spinal cord (48) and supports the key observation that SOD1 only accumulates in DRPs when animals begin to show symptoms of the disease.
FIGURE 2.
LC-ESI mass spectra of ALS variant SOD1 from DRPs prepared from symptomatic ALS mouse spinal cord. DRPs that were resistant to 10% Nonidet P-40 are denoted “P2”; pellets resistant to 10% Nonidet P-40 and an additional exposure to 0.5% Nonidet P-40, 0.25% SDS, and 0.5% deoxycholate are denoted “P3.” A, total ion current chromatograms of P2 DRPs (solubilized for LC-MS analysis with methanol/chloroform) from non-transgenic (nTg) and hWT, H46R/H48Q, G37R, and G93A mice are overlaid. B, expanded and overlaid view of SOD1 elution peak at 39–41 min shows that hWT SOD1 is not present in DRPs from transgenic mice expressing hWT SOD1; dashed and solid lines represent hWT and H46R/H48Q samples, respectively. C, raw mass spectra of ALS variant SOD1 recovered from P2 DRP. D and E, reconstructed mass spectra of ALS variant SOD1 deconvoluted from raw mass spectra from P2 (D) and P3 (E) DRPs, respectively. See Table 1 for mass values of SOD1.
The ALS variant SOD1 proteins that form DRPs had mass values consistent with full-length, unmodified SOD1 (similar to the soluble SOD1 proteins from supernatant). The G93A, G37R, and H46R/H48Q SOD1 proteins were found to have mass values of 15862.5, 15945.0, and 15857.0 Da, respectively (Table 1). Raw mass spectra of ALS variant SOD1 proteins from P2 pellets are shown in Fig. 2C, and the de-convoluted mass spectra for both P2 and P3 pellets are shown in Fig. 2 (D and E), respectively. (See “Experimental Procedures” or Fig. 2 for a description of the different detergent mixtures used to generate P2 and P3 pellets.) The mass distributions for all three SOD1 variants are predominantly unimodal, and only low intensity mass adducts were detected with values of +4.5, 7.5, 16, and 32 Da. It should be noted that the +16 and +32 Da species that are observable with some SOD1 proteins from DRPs (Fig. 2, E and D) are also routinely observed to form over time, in vitro, with recombinantly purified hWT and ALS variant SOD1 proteins during purification, storage, and analysis (15). These +16 and +32 Da adducts are likely due to the air oxidation of free cysteine residues present in human SOD1 (i.e. Cys-6 and/or Cys-111) (14, 15, 49, 50) that occurs during detergent extraction and organic solvent extraction. Previous MS/MS analysis of modified hWT SOD1 showed that a +32 Da modification was located on cysteine 111 (49). Trypsinization and MS/MS analysis of our detergent-extracted SOD1 proteins, however, did not yield MS/MS spectra consistent with modified SOD1 peptides, and this is probably due to the low abundance of these modified proteins in the detergent-extracted protein samples.
The oxidation of Trp-32 in SOD1 has also been observed, in vitro with purified SOD1 solutions containing hydrogen peroxide and bicarbonate (51) and with ALS mutant SOD1 purified from the blood of transgenic mice (52). It should also be pointed out that the +4.5-Da modifications to G93A and G37R SOD1 in the P3 samples (Fig. 2E) could be due to the oxidation of tryptophan to kynurenine, which results in a +3.9-Da modification. These 4.5-Da adducts were only observed in P3 samples and not P2 samples, and could have formed during the additional detergent extraction step. Regardless, these species do not represent the majority of SOD1 in the DRP samples and are low in abundance relative to the unmodified SOD1 protein.
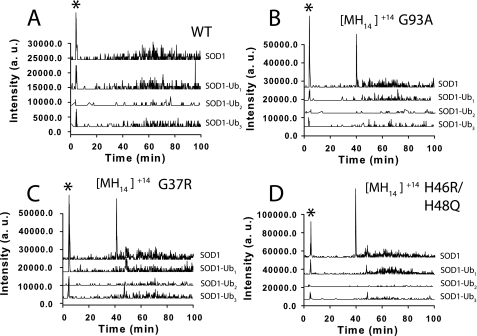
It is very likely that SOD1 proteins bearing single or multiple 8-kDa ubiquitin modifications would have altered elution times compared with non-ubiquitinated SOD1 proteins. Therefore, we used Analyst software to search the entire LC-MS chromatogram of P2 samples from hWT and ALS mutant samples for ions corresponding to mono-, di-, and tri-ubiquitinated SOD1 proteins. No distributions of ions were detected that corresponded to hWT or ALS variant SOD1 having 1–3 conjugated ubiquitin polypeptides. Fig. 3 shows the results of this ion extraction for the +20 molecular ion of mono and di-ubiquitinated SOD1 (denoted SOD1-Ub1 and SOD1-Ub2) and the +30 ion for tri-ubiquitinated SOD1 (SOD1-Ub3). Although only the +20 or +30 ions are shown in Fig. 3, we performed an ion extraction for a complete ion series (+20 to +40) of SOD1-Ubn with no significant signal detected (data not shown). All four plots (A–D) in Fig. 3 show, as a reference, an ion extract chromatogram for the +14 ion of unmodified SOD1. As can be seen from Fig. 3 (A–D), the +14 ion is extremely abundant at times of 39–41 min, where unmodified SOD1 was shown to elute. It should be noted that the ion extract for unmodified hWT SOD1 does not exhibit a strong signal, consistent with no hWT SOD1 being present in the DRPs. It should also be noted that in Fig. 3 (C and D), a small signal can be seen for SOD1-Ub1 and SOD1-Ub2 at ∼45 min. This signal is also present in the SOD1 extracts (e.g. SOD1-Ub0), and these ions were found to be members of an ion distribution for glial fibrillary acidic protein and not SOD1 or ubiquitinated SOD1.
FIGURE 3.
Ubiquitinated-SOD1 (SOD1-Ub) ion extract of LC-ESI-MS chromatogram for DRP from ALS variant and hWT SOD1 mouse spinal cord. G93A, G37R, H46R/H48Q, and hWT SOD1 ion extracts for +20 molecular ion are shown for SOD1-Ub1 and SOD1-Ub2; +30 ion shown for SOD1-Ub3. The ion extract for the +14 molecular ion of unmodified SOD1 is shown for each variant, with strong signals appearing between 39 and 41 min, where SOD1 elutes (note: signal marked with asterisk in each plot corresponds to injection peak and is due to salt clusters across a broad m/z range).
It should be remembered that aggregated proteins that could not be dissociated during the DRP dissolution would be difficult to detect with ESI-MS. We believe, however, that we achieved complete dissociation and recovery of aggregated proteins based upon the lack of an observable pellet after dissolution and the absence of an increase in pressure after each sample was loaded onto the HPLC column (data not shown).
To approximate the amounts of SOD1 proteins present in S1, P2, and P3 samples, we integrated the mass spectral signals for each variant and determined the quantity of SOD1 using an SOD1 calibration curve (see supplemental information for additional experimental details). Calibration curves were prepared using recombinantly purified hWT and ALS variant SOD1 (e.g. H48Q) of known concentration. The results are summarized in supplemental Fig. S2. The results show that ∼90–96% of the SOD1 from all three ALS mutant samples was present in the supernatant with ∼4–10% contained in the P2 pellet (supplemental Fig. S2C).
MS/MS Identification of Proteins Co-sedimented with SOD1 in Detergent-resistant Pellets—The organic extraction of proteins from DRPs and the subsequent analysis with LC-MS generated chromatograms wherein the predominant signals were found to correspond to molecular species with masses consistent with human SOD1, glial fibrillary acidic protein, myelin basic protein precursor, tubulin, and actin (Fig. 2A). Trypsinization of collected fractions and sequence analysis with MS/MS confirmed the identity of each protein (see supplemental Table S1 and tryptic map of SOD1 in the supplemental information). All of these proteins, except for SOD1, were also found in the nTg and hWT SOD1 controls, indicating that their detergent-insoluble, high molecular mass nature is a non-pathogenic occurrence or artifact of the DRP preparation. No other proteins were detected in the DRPs with LC-MS. However, the trypsinization and LC-MS/MS analysis of LC fractions collected from the DRP LC-MS analysis did result in the detection and identification of additional proteins that were present along with SOD1, glial fibrillary acidic protein, actin, myelin basic protein precursor, and tubulin in the DRPs (see supplemental Table S1 in supplemental information for a list of these proteins). None of these proteins, however, were consistently detected in aggregates samples from spinal cords of all three ALS mutant mice except for vimentin, a highly abundant cytoskeletal protein. Therefore, the only proteins that were consistently detected with ESI-MS/MS to be present in detergent-resistant samples from only ALS mutant spinal cords were SOD1, and vimentin.
The detection of additional proteins such as vimentin in the trypsinization and LC-MS/MS experiments (but not in the LC-MS experiments) is likely due to a combination of factors, including: 1) the greater relative sensitivity of the LC-MS/MS instrumentation, compared with the instrumentation used for the LC-MS (75-μm versus 2-mm column; see “Experimental Procedures”); 2) the additional concentration step included in the trypsinization protocol (see “Experimental Procedures”); and 3) the increased ionization efficiency of tryptic peptides, relative to unproteolyzed, full-length proteins.
The inconsistent detection of additional proteins besides SOD1 in DRPs and the apparent low abundance of vimentin creates difficulty in determining the relevance of these data to ALS pathology (see supplemental Table S1 in supplemental information for a complete list of these proteins). Together, the data do suggest that the most abundant species in detergent-resistant aggregates that are associated with ALS is the SOD1 protein and that other proteins are present in very low amounts that are difficult to detect with mass spectrometry.
An SOD1 “elution ruler” is shown at the bottom of the chromatogram overlay in Fig. 2A and illustrates the fractions that were shown, with trypsinization and MS/MS, to contain SOD1 proteins. The elution ruler shows that SOD1 was only detected to be present in ESI-LC-MS fractions that contain full-length, unmodified SOD1 (e.g. fractions 39–41). This implies that no other SOD1 species such as fragmented SOD1 (which would probably elute earlier than full-length SOD1) or ubiquitinated SOD1 (which would probably elute later than full-length SOD1) are present in DRPs in amounts detectable with mass spectrometry. It should be noted that the lower detection limit of the LC MB/MS system is ∼10–20 fmol of protein on column. Thus Western blotting may detect materials that escape detection by this method.
Isolation of Aggregated and Soluble SOD1 from Spinal Cord with SE/IA-LC—The coupled SE/IA-LC method yields information about which proteins are complexed with SOD1 in the spinal cord extracts, whereas the detergent extraction method isolates molecular species that are similarly detergent-resistant and sedimentable but not necessarily aggregated with one another.
The Sephadex G-75 size exclusion media separates globular proteins with molecular masses that range from 3 to 70 kDa. Preparations from hWT or ALS mutant mice consistently yielded two groups of SOD1 immunoreactivity with different molecular masses according to anti-hSOD1 Western blot analysis of SE fractions (Fig. 4; example from analysis of a G37R mouse). The first group of SOD1 eluted from the SE column in the void volume (typically in fractions 22–24) thereby indicating a molecular mass greater than the upper resolving limit (70 kDa) of the medium (Fig. 4). The second group of SOD1 immunoreactivity was detected in fractions that corresponded to the resolving region of the column (typically fractions 30–37) and represents the majority of SOD1 isolated from the spinal cord, as judged by the intensity of the anti-hSOD1 immunostaining. It should be remembered that the supernatant loaded onto the G-75 column is from spinal cord homogenate that was centrifuged at 800 × g as opposed to the 100,000 × g used in the detergent extraction; therefore the supernatant loaded onto the SE column is expected (and has been previously shown) to contain both soluble oligomeric SOD1 as well as dimeric SOD1 (12).
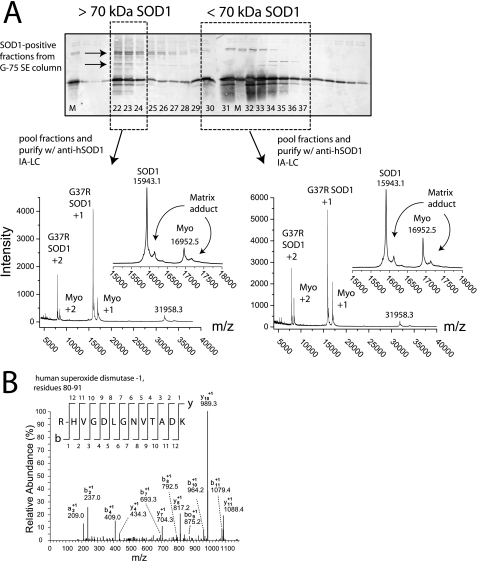
FIGURE 4.
MALDI-TOF and ESI-MS/MS mass spectra of soluble and aggregated G37R SOD1 purified from symptomatic mouse spinal cord with size exclusion and anti-SOD1 immunoaffinity liquid chromatography (SE/IA-LC). A, SDS-PAGE and Western blot analysis of eluted SE fractions reveal SOD1 species with different molecular mass ranges. SOD1 in fractions 22–24 (dashed box on left) eluted in void volume indicating a molecular mass > 70 kDa; SOD1 species that eluted in fractions 31–37 (dashed box on right) have a molecular mass between 7 and 70 kDa (i.e. the resolving range of the G-75 media). Fraction numbers are listed at the bottom of the Western blot (see “Experimental Procedures” for chromatographic parameters), and “M” denotes the hWT SOD1 marker. Arrows on Western blot indicate SOD1 aggregate species that are heat- and SDS-stable. Right spectra: MALDI-TOF mass spectrum of SOD1 species that eluted in resolving region of G-75 column (i.e. soluble SOD1) and were pooled and further purified with anti-SOD1 immunoaffinity chromatography. The theoretical average mass of G37R SOD1 is 15945.8 Da. The 16952 m/z signal corresponds to myoglobin (co-spotted with protein samples as an internal calibrant). The 16153 m/z signal arises from a gas-phase SOD1-matrix adduct (matrix: sinapic acid; molecular mass 224.22 Da). Left spectra: MALDI-TOF spectrum of SOD1 species that eluted in the void volume of a G-75 column (i.e. aggregated SOD1) and were pooled and further purified with anti-SOD1 immunoaffinity chromatography. Insets for both spectra show expanded spectrum from 15,000 to 18,000 m/z. B, ESI-MS/MS spectra of tryptic fragment of G37R SOD1 purified from symptomatic mouse spinal cord with SE/IA-LC. The spectrum shown here was generated from the void volume (>70 kDa) fractions of the G-75 column. A ladder scheme numerates the C and N terminus daughter ions (e.g. y and b daughter ions, respectively) for the tryptic SOD1 peptide (residues 80–91). Daughter ions assigned “bo”or“yo” refer to ions that have undergone the loss of H2O.
As can be seen by the Western blot in Fig. 4, most high molecular mass SOD1 immunoreactive species eluting in the void volume of the G-75 column were dissociated during the SDS boiling process so that the majority of SOD1 migrates at the same molecular mass as the 15.8-kDa SOD1 marker. However, the same void volume fractions also contain higher molecular mass SOD1 immunoreactive species that migrated through the polyacrylamide gel more slowly than the 15.8-kDa SOD1 marker, suggesting that these species remained aggregated even after boiling in SDS and DTT or β-mercaptoethanol. These SDS/DTT-resistant bands were not as abundant in the later, resolving fractions that contain the majority of SOD1 immunoreactivity. Therefore, these high molecular mass bands are probably not artifacts of the SDS/DTT boiling process. These detergent/reductant-resistant, high molecular mass SOD1 species are also present in hWT samples, but in lower amounts, as interpolated from Western blots (data not shown). A recent examination of data from nearly two dozen literature reports has revealed that these thermally stable and reductant/detergent stable species commonly appear with the onset of ALS symptoms in mice and increase in abundance as symptoms progress (1).
Mass Spectral Profile of SOD1 Isolated from Spinal Cord with SE/IA Liquid Chromatography—The SOD1-immunoreactive fractions that eluted from the G-75 column in the void volume (i.e. >70 kDa) and resolving regions (i.e. <70 kDa) were pooled into two separate groups, and each was further purified with anti-SOD1 immunoaffinity chromatography (Fig. 4). The SE/IA-LC-purified SOD1 was then analyzed with MALDI-TOF mass spectrometry instead of ESI-MS due to the low abundance of SOD1 in the purified samples (MALDI-TOF MS is generally more sensitive than ESI-MS by ∼1 order of magnitude). It was found that the SOD1 signal intensity in void volume samples increased when heated to ∼75–80 °C in the presence of 100 mm DTT prior to MS analysis.
The MALDI-TOF mass spectra of SE/IA-LC-purified G37R SOD1 are shown in Fig. 4. As can be seen in both sets of spectra, the predominant signal corresponds to full-length, unmodified G37R SOD1 (molecular mass 15,943.72 Da; the 16952 m/z signal corresponds to the myoglobin calibrant). These spectra show the singly charged MH+ ion for G37R SOD1 at 15943 m/z, as well as the doubly charged ion at 7968 m/z. There is also a small peak at 31,958 m/z that represents dimeric SOD1; this dimer is likely to be a gas-phase artifact of the ionization process and has been observed with numerous non-pathogenic SOD1 proteins from a variety of organisms including, bovine, yeast, and tomato SOD1 (data not shown). Collection of MALDI-TOF MS spectra was carried out in the 1,000–100,000 m/z range, and no discernable ion signals were detected above 40,000 m/z. The 16,153 m/z signal present in spectra from both high and low molecular mass fractions is likely a gas-phase SOD1-matrix adduct (molecular mass of sinapinic acid: 224.22 Da), and a similar adduct was observed to form with the myoglobin calibrant (Fig. 4).
MS/MS Identification of Proteins Co-purifying with SOD1 in SE/IA Liquid Chromatography—Similar to the MS/MS protein identification results of DRPs, there were no proteins consistently detected to co-purify with SOD1 by SE/IA-LC. Fig. 4B shows a typical MS/MS spectrum that was generated from the trypsinization of SE-IA-LC-purified samples. This particular MS/MS spectrum corresponds to a tryptic peptide of G37R SOD1 from the G37R mouse. See supplemental Table S2 in supplemental information for a list of the proteins that were inconsistently identified with MS/MS to elute from the anti-hSOD1 IA column after void volume elution from the G-75 SE column. (See supplemental Fig. S3 in the supplemental information for the MS/MS spectra of peptides from multiple proteins listed in supplemental Table S2.)
DISCUSSION
The detergent-resistant, high molecular mass SOD1-containing species that we isolated have been previously shown to not form in non-transgenic (nTg) mice or mice expressing human wild-type (hWT) SOD1. In addition, the detergent-resistant aggregates are not detectable in ALS mice before the onset of symptoms; the appearance of SOD1 aggregates correlates with the onset of ALS symptoms (9, 12). These DRPs are likely to contain SOD1 aggregates with a broad range of size (e.g. across the nanometer to micrometer scales) with the common property being a high molecular mass that is maintained despite the exposure to high concentrations of detergents. A detergent extraction and sedimentation at 100,000 × g will not isolate small oligomers (e.g. dimers, trimers, or dodecamers). The SE/IA-LC method, however, isolates any soluble oligomer that: 1) has a molecular mass >70 kDa and 2) exhibits a solvent-exposed SOD1 epitope. Together, the SE/IA-LC and detergent extraction methods are likely to isolate the broader aggregated pool of SOD1 from extracellular, cytosolic, or intraorganellar loci, as opposed to isolating a single “type” of SOD1 aggregate, for example a micrometer scale inclusion body or a nanometer scale oligomer.
The SOD1 Protein That Aggregates in ALS-affected Mouse Spinal Cord Is a Full-length Polypeptide—In several neurodegenerative diseases that are linked to protein aggregation, the pathogenic species that undergoes aggregation is not a full-length polypeptide, but a post-translationally generated peptide fragment. For example, the pathogenic forms of the proteins gelsolin (53), which is linked to Finnish-type familial amyloidosis, and amyloid precursor protein (54) (APP), which is linked to Alzheimer's disease, are truncated forms of the full-length (as expressed) protein. Anti-hSOD1 Western blot analysis has shown that truncated forms of SOD1 are present in detergent-stable aggregates from ALS mice (11); however, the predominant SOD1 species appears to be the full-length polypeptide, according to SDS-PAGE (9–11, 32). We find no evidence, from mass spectrometry, for the enrichment of truncated forms of SOD1 within the aggregated pools of SOD1, and SOD1 was only detected with MS/MS to be present in LC-MS fractions that corresponded to the full-length SOD1 protein (Fig. 2A). This result suggests that the majority of SOD1 polypeptides undergo aggregation without prior proteolysis (besides Met-1 removal), and that any truncated forms are present below the limit of detection by ESI-MS.
Aggregated and Soluble SOD1 Proteins in ALS-affected Spinal Cord Tissue Are Unmodified—Native state stability is one factor that contributes to the propensity for a folded protein to aggregate, and the extreme stability of SOD1 is largely dependent on copper and zinc coordination (1). Considering that many ALS variants are not destabilized relative to hWT SOD1 and can coordinate metals properly, it has been hypothesized that ALS variants might undergo chemical modifications that somehow destabilize the native state, such as oxidative modifications to copper-binding histidine residues that decrease the copper binding affinity (28, 29). In vitro studies have shown that, compared with hWT SOD1, ALS variants undergo histidine oxidation at a faster rate in the presence of hydrogen peroxide and bicarbonate, leading to SOD1 inactivation and the presence of oxidative modifications that are +16 and +32 Da in mass. However, the analysis of soluble or aggregated SOD1 with ESI-MS, ESI-MS/MS, and MALDI-TOF-MS presented here clearly demonstrates that ALS variant SOD1 proteins are largely unmodified in ALS-affected spinal cord, apart from the low abundant mass adducts that are commonly observed to form on hWT or mutant SOD1 during purification, handling, and storage in an aerobic environment or during electrospray ionization. Because our samples were treated with high concentrations of DTT, any reversible modifications such as single oxidation of Cys to the sulfenic acid (+16 Da), for example, may not be observed in our analysis.
Additional post-translational modifications to SOD1 that might also destabilize the SOD1 native state and promote aggregation have also been implicated in promoting the aggregation and pathogenesis of SOD1 (4). Immunohistochemical studies have suggested that proteins with advanced glycation end-products such as carboxymethylysine are co-localized with SOD1 in inclusion bodies and that the aggregated SOD1 polypeptide is carboxymethylated or glycated (5) (the carboxymethylation of lysine results in a +58-Da adduct). Again, the mass spectrometric data presented in this work suggests that, if aggregated or soluble SOD1 is glycated or modified with advanced glycation end-products, in ALS mouse spinal cord it is at a level below the detection limits of mass spectrometry.
One very common pathological signature of SOD1-linked familial ALS is the appearance of low molecular mass (<100 kDa) SOD1-containing species that have electrophoretic mobilities equivalent with dimeric or trimeric SOD1, even under completely denaturing SDS-PAGE (1). A recent report has suggested that these SOD1-containing oligomers are held together with covalent bonds and contain covalently modified SOD1 proteins (55). It should be made clear, however, that protein complexes that are stable under the denaturing conditions of SDS-PAGE are not necessarily covalently linked (1). For example, it has been recently suggested that the aggregate species responsible for memory loss in Alzheimer disease is a dodecameric oligomer of the Aβ peptide (56); this species runs as a dodecamer during fully denaturing SDS-PAGE. The dodecamer dissociates, however, in hexafluoroisopropanol (56).
Biochemical Implications for the Lack of Detected Ubiquitin in SOD1 Aggregates—Recent work has demonstrated that ALS variants of SOD1 could promote pathogenesis by inhibiting proteasomal activity, thereby leading to increased concentrations of misfolded or aggregated SOD1, including SOD1 polypeptides that have been tagged with ubiquitin for degradation by the proteasome (57–59). Likewise, proteasomal inhibition has been hypothesized to explain the apparent accumulation of ubiquitinated SOD1 in spinal inclusion bodies (as measured by immunostaining analyses).
Our results show that ubiquitinated forms of SOD1 are not a predominant species in the detergent-resistant aggregates from homogenates of ALS mouse spinal cord tissue (Figs. 3 and 4), being below the limit of detection by mass spectrometry. For example, MS and MS/MS analysis showed that none of the SOD1 proteins isolated by both the detergent extraction and SE/IA-LC methods were found to bear the 8-kDa modification associated with monoubiquitination (i.e. SOD1-Ub1) or the 8n-kDa modifications (i.e. SOD1-Ubn) that would be associated with oligoubiquitination (Figs. 3 and 4). Ubiquitination of SOD1 would have resulted in a 24-kDa or 24 + 8n-kDa species depending upon the number of ubiquitin chains, n. Moreover, no ubiquitin peptides were detected during the protein identification experiments involving trypsinization and LC-ESI-MS/MS of SOD1 proteins isolated with the detergent extraction or SE/IA-LC methods (see supplemental Tables S1 and S2).
The absence of detected ubiquitin in either the DRPs or in SE/IA-LC-isolated samples is not completely surprising in light of previous immunostaining of aggregates from mouse and human spinal tissue. Although anti-ubiquitin immunostaining has shown that ubiquitin is sometimes co-localized with SOD1 in aggregate structures and that the aggregated SOD1 is ubiquitinated (6, 7, 11, 32, 57, 60), many of these studies suggest that ubiquitinated SOD1 is not the predominant form of aggregated SOD1. The inability to detect ubiquitinated SOD1 does not suggest that a perturbation in the ubiquitin proteasome pathway is not involved in the pathogenesis of familial ALS. However, the inability to detect ubiquitinated SOD1 may suggest that ALS variants targeted for proteasomal degradation via ubiquitination do not form aggregate species that can be readily isolated and detected with mass spectrometry. It should be pointed out that trypsinization of proteins or peptides that bear ubiquitin modifications reduce the mono- or poly-Ub protein modification to a +114-Da modification (i.e. a Gly-Gly tag). This 114.0-Da Lys modification was never detected in our LC-ESI-MS/MS proteomic experiments and analyses (which were conducted numerous times on dozens of spinal cords from the three ALS variants G37R, G93A, and H46R/H48Q). One group analyzing SOD1 aggregates from ALS mouse spinal cord that are resistant to Triton-X have reported that some tryptic SOD1 peptides do in fact bear the 114.0-Da lysine modification that is associated with ubiquitination (32). This report, however, did not present mass spectra of the intact, non-trypsinized SOD1 polypeptide showing the 8- or 16-kDa ubiquitin modification to SOD1 (which makes it difficult to judge the relative level of ubiquitinated SOD1 that might have been present in the DRPs).
High Molecular Mass Complexes of Mutant SOD1 Do Not Consistently Contain Other Cellular Proteins—SOD 1 was the only protein that was consistently detected to be present in detergent-resistant aggregate samples and SE/IA purified aggregates from ALS spinal cords. Perhaps most surprising was the absence of molecular chaperones. For example, previous immunohistochemical analyses of G93A and G85R mouse spinal cord tissue have suggested that the heat shock protein Hsc70 is present in SOD1 inclusion bodies (33). In G93A spinal cord cultures, the heat shock protein Hsp70 was detected, via Western blot analysis, to be present with SOD1 in detergent-insoluble fractions (61). However, Hsp70 was also reported to be absent in inclusion bodies from SOD1 transgenic mice, and the presence of Hsp70 in inclusion bodies from ALS patients is reportedly rare (36). The lack of a consistent detection of heat shock proteins, or other molecular chaperones, in DRPs from all ALS mice (Fig. 2; see also supplemental Tables S1 and S2), does not necessarily imply that molecular chaperones do not mediate the formation of SOD1 inclusion bodies, or that molecular chaperones are not somehow involved in SOD1 aggregation. Our findings do demonstrate that molecular chaperones are not irreversibly or stoichiometrically associated with mutant SOD1 in DRPs associated with ALS.
Trace amounts of chaperones that are below the detection limit of ESI-MS could still be relevant to aggregate formation via transient or sub-stoichiometric associations. For example, the molecular chaperone BiP is incorporated into aggregate forms of mutant transthyretin, and stable BiP-transthyretin oligomers have been isolated wherein the BiP:transthyretin ratio can vary from 1:1 for small oligomers (e.g. 100 kDa) up to 1:15 for larger oligomers (e.g. 1000 kDa) (62). Such sequestered chaperones are thought to mediate the formation of large aggregate structures or inclusion bodies and might also facilitate the later disassembly of the inclusion (62).
Autonomous SOD1 Aggregation in ALS Mouse Spinal Cords?—In conclusion, mass spectrometric data has demonstrated that soluble and aggregated forms of ALS variant SOD1 from ALS spinal cord are predominantly full-length and are not ubiquitinated or damaged by oxidative modifications. Clearly, the majority of ALS-variant SOD1 polypeptides that assemble into detergent-stable aggregate structures do so without being irreversibly oxidatively damaged and without co-aggregating with other proteins in a stoichiometric, detergent-stable manner. This is consistent with the idea that the ability to incorporate into a propagating aggregate is an intrinsic property of the SOD1 polypeptide, as in the case of amyloid formation, for example. We cannot, however, rule out the possibility that post-translational modifications or binding interactions with other proteins play a role in initiation of the aggregation process.
Supplementary Material
Acknowledgments
We thank Dr. Se Hui Sohn and Prof. Kym F. Faull for technical advice and Elizabeth Shaw for editing and proofreading the manuscript.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” references, Figs. S1–S3, and Tables S1 and S2.
Footnotes
The abbreviations used are: SOD1, superoxide dismutase 1; ALS, amyotrophic lateral sclerosis; SE, size exclusion; LC, liquid chromatography; IA, immunoaffinity; ESI, electrospray ionization; MS, mass spectrometry; MS/MS, tandem MS; MALDI-TOF, matrix-assisted laser desorption time-of-flight; DRP, detergent-resistant pellet; DTT, dithiothreitol; HPLC, high-performance liquid chromatography.
References
- 1.Shaw, B. F., and Valentine, J. S. (2007) Trends Biochem. Sci. 32 78–85 [DOI] [PubMed] [Google Scholar]
- 2.Wang, J., Xu, G., Gonzales, V., Coonfield, M., Fromholt, D., Copeland, N. G., Jenkins, N. A., and Borchelt, D. R. (2002) Neurobiol. Dis. 10 128–138 [DOI] [PubMed] [Google Scholar]
- 3.Shibata, N., Hirano, A., Kobayashi, M., Siddique, T., Deng, H. X., Hung, W. Y., Kato, T., and Asayama, K. (1996) J. Neuropathol. Exp. Neurol. 55 481–490 [DOI] [PubMed] [Google Scholar]
- 4.Kato, S., Nakashima, K., Horiuchi, S., Nagai, R., Cleveland, D. W., Liu, J., Hirano, A., Takikawa, M., Kato, M., Nakano, I., Sakoda, S., Asayama, K., and Ohama, E. (2001) Neuropathology 21 67–81 [DOI] [PubMed] [Google Scholar]
- 5.Kato, S., Horiuchi, S., Liu, J., Cleveland, D. W., Shibata, N., Nakashima, K., Nagai, R., Hirano, A., Takikawa, M., Kato, M., Nakano, I., and Ohama, E. (2000) Acta Neuropathol. (Berl.) 100 490–505 [DOI] [PubMed] [Google Scholar]
- 6.Bruijn, L. I., Becher, M. W., Lee, M. K., Anderson, K. L., Jenkins, N. A., Copeland, N. G., Sisodia, S. S., Rothstein, J. D., Borchelt, D. R., Price, D. L., and Cleveland, D. W. (1997) Neuron 18 327–338 [DOI] [PubMed] [Google Scholar]
- 7.Kato, S., Hayashi, H., Nakashima, K., Nanba, E., Kato, M., Hirano, A., Nakano, I., Asayama, K., and Ohama, E. (1997) Am. J. Pathol. 151 611–620 [PMC free article] [PubMed] [Google Scholar]
- 8.Kato, S., Shimoda, M., Watanabe, Y., Nakashima, K., Takahashi, K., and Ohama, E. (1996) J. Neuropathol. Exp. Neurol. 55 1089–1101 [PubMed] [Google Scholar]
- 9.Johnston, J. A., Dalton, M. J., Gurney, M. E., and Kopito, R. R. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 12571–12576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonsson, P. A., Ernhill, K., Andersen, P. M., Bergemalm, D., Brannstrom, T., Gredal, O., Nilsson, P., and Marklund, S. L. (2004) Brain 127 73–88 [DOI] [PubMed] [Google Scholar]
- 11.Wang, J., Slunt, H., Gonzales, V., Fromholt, D., Coonfield, M., Copeland, N. G., Jenkins, N. A., and Borchelt, D. R. (2003) Hum. Mol. Genet. 12 2753–2764 [DOI] [PubMed] [Google Scholar]
- 12.Wang, J., Xu, G., and Borchelt, D. R. (2002) Neurobiol. Dis. 9 139–148 [DOI] [PubMed] [Google Scholar]
- 13.Matsumoto, G., Stojanovic, A., Holmberg, C. I., Kim, S., and Morimoto, R. I. (2005) J. Cell Biol. 171 75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez, J. A., Shaw, B. F., Durazo, A., Sohn, S. H., Doucette, P. A., Nersissian, A. M., Faull, K. F., Eggers, D. K., Tiwari, A., Hayward, L. J., and Valentine, J. S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 10516–10521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaw, B. F., Durazo, A., Nersissian, A. M., Whitelegge, J. P., Faull, K. F., and Valentine, J. S. (2006) J. Biol. Chem. 281 18167–18176 [DOI] [PubMed] [Google Scholar]
- 16.Elam, J. S., Taylor, A. B., Strange, R., Antonyuk, S., Doucette, P. A., Rodriguez, J. A., Hasnain, S. S., Hayward, L. J., Valentine, J. S., Yeates, T. O., and Hart, P. J. (2003) Nat. Struct. Biol. 10 461–467 [DOI] [PubMed] [Google Scholar]
- 17.Hough, M. A., Grossmann, J. G., Antonyuk, S. V., Strange, R. W., Doucette, P. A., Rodriguez, J. A., Whitson, L. J., Hart, P. J., Hayward, L. J., Valentine, J. S., and Hasnain, S. S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 5976–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiDonato, M., Craig, L., Huff, M. E., Thayer, M. M., Cardoso, R. M., Kassmann, C. J., Lo, T. P., Bruns, C. K., Powers, E. T., Kelly, J. W., Getzoff, E. D., and Tainer, J. A. (2003) J. Mol. Biol. 332 601–615 [DOI] [PubMed] [Google Scholar]
- 19.Stokin, G. B., Lillo, C., Falzone, T. L., Brusch, R. G., Rockenstein, E., Mount, S. L., Raman, R., Davies, P., Masliah, E., Williams, D. S., and Goldstein, L. S. (2005) Science 307 1282–1288 [DOI] [PubMed] [Google Scholar]
- 20.Vermeiren, C., Hemptinne, I., Vanhoutte, N., Tilleux, S., Maloteaux, J. M., and Hermans, E. (2006) J. Neurochem. 96 719–731 [DOI] [PubMed] [Google Scholar]
- 21.Tortarolo, M., Grignaschi, G., Calvaresi, N., Zennaro, E., Spaltro, G., Colovic, M., Fracasso, C., Guiso, G., Elger, B., Schneider, H., Seilheimer, B., Caccia, S., and Bendotti, C. (2006) J. Neurosci. Res. 83 134–146 [DOI] [PubMed] [Google Scholar]
- 22.Urushitani, M., Kurisu, J., Tsukita, K., and Takahashi, R. (2002) J. Neurochem. 83 1030–1042 [DOI] [PubMed] [Google Scholar]
- 23.Hart, P. J. (2006) Curr. Opin. Chem. Biol. 10 131–138 [DOI] [PubMed] [Google Scholar]
- 24.Okado-Matsumoto, A., and Fridovich, I. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 9010–9014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasinelli, P., Belford, M. E., Lennon, N., Bacskai, B. J., Hyman, B. T., Trotti, D., and Brown, R. H., Jr. (2004) Neuron 43 19–30 [DOI] [PubMed] [Google Scholar]
- 26.Bruijn, L. I., Houseweart, M. K., Kato, S., Anderson, K. L., Anderson, S. D., Ohama, E., Reaume, A. G., Scott, R. W., and Cleveland, D. W. (1998) Science 281 1851–1854 [DOI] [PubMed] [Google Scholar]
- 27.Maatkamp, A., Vlug, A., Haasdijk, E., Troost, D., French, P. J., and Jaarsma, D. (2004) Eur. J. Neurosci. 20 14–28 [DOI] [PubMed] [Google Scholar]
- 28.Rakhit, R., Cunningham, P., Furtos-Matei, A., Dahan, S., Qi, X. F., Crow, J. P., Cashman, N. R., Kondejewski, L. H., and Chakrabartty, A. (2002) J. Biol. Chem. 277 47551–47556 [DOI] [PubMed] [Google Scholar]
- 29.Valentine, J. S. (2002) Free Radic. Biol. Med. 33 1314–1320 [DOI] [PubMed] [Google Scholar]
- 30.Valentine, J. S., Doucette, P. A., and Zittin Potter, S. (2005) Annu. Rev. Biochem. 74 563–593 [DOI] [PubMed] [Google Scholar]
- 31.Elam, J. S., Malek, K., Rodriguez, J. A., Doucette, P. A., Taylor, A. B., Hayward, L. J., Cabelli, D. E., Valentine, J. S., and Hart, P. J. (2003) J. Biol. Chem. 278 21032–21039 [DOI] [PubMed] [Google Scholar]
- 32.Basso, M., Massignan, T., Samengo, G., Cheroni, C., De Biasi, S., Salmona, M., Bendotti, C., and Bonetto, V. (2006) J. Biol. Chem. 281 33325–33335 [DOI] [PubMed] [Google Scholar]
- 33.Watanabe, M., Dykes-Hoberg, M., Culotta, V. C., Price, D. L., Wong, P. C., and Rothstein, J. D. (2001) Neurobiol. Dis. 8 933–941 [DOI] [PubMed] [Google Scholar]
- 34.Niwa, J., Ishigaki, S., Hishikawa, N., Yamamoto, M., Doyu, M., Murata, S., Tanaka, K., Taniguchi, N., and Sobue, G. (2002) J. Biol. Chem. 277 36793–36798 [DOI] [PubMed] [Google Scholar]
- 35.Miyazaki, K., Fujita, T., Ozaki, T., Kato, C., Kurose, Y., Sakamoto, M., Kato, S., Goto, T., Itoyama, Y., Aoki, M., and Nakagawara, A. (2004) J. Biol. Chem. 279 11327–11335 [DOI] [PubMed] [Google Scholar]
- 36.Kabashi, E., and Durham, H. D. (2006) Biochim. Biophys. Acta 1762 1038–1050 [DOI] [PubMed] [Google Scholar]
- 37.Banci, L., Bertini, I., Durazo, A., Girotto, S., Gralla, E. B., Martinelli, M., Valentine, J. S., Vieru, M., and Whitelegge, J. P. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 11263–11267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, J., Xu, G., Li, H., Gonzales, V., Fromholt, D., Karch, C., Copeland, N. G., Jenkins, N. A., and Borchelt, D. R. (2005) Hum. Mol. Genet. 14 2335–2347 [DOI] [PubMed] [Google Scholar]
- 39.Plakoutsi, G., Bemporad, F., Calamai, M., Taddei, N., Dobson, C. M., and Chiti, F. (2005) J. Mol. Biol. 351 910–922 [DOI] [PubMed] [Google Scholar]
- 40.Pieri, L., Bucciantini, M., Nosi, D., Formigli, L., Savistchenko, J., Melki, R., and Stefani, M. (2006) J. Biol. Chem. 281 15337–15344 [DOI] [PubMed] [Google Scholar]
- 41.Wessel, D., and Flugge, U. I. (1984) Anal. Biochem. 138 141–143 [DOI] [PubMed] [Google Scholar]
- 42.Whitelegge, J. P., Katz, J. E., Pihakari, K. A., Hale, R., Aguilera, R., Gomez, S. M., Faull, K. F., Vavilin, D., and Vermaas, W. (2004) Phytochemistry 65 1507–1515 [DOI] [PubMed] [Google Scholar]
- 43.Whitelegge, J. P., Zhang, H., Aguilera, R., Taylor, R. M., and Cramer, W. A. (2002) Mol. Cell Proteomics 1 816–827 [DOI] [PubMed] [Google Scholar]
- 44.Whitelegge, J. P., Gundersen, C. B., and Faull, K. F. (1998) Protein Sci. 7 1423–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez, J. A., Valentine, J. S., Eggers, D. K., Roe, J. A., Tiwari, A., Brown, R. H., Jr., and Hayward, L. J. (2002) J. Biol. Chem. 277 15932–15937 [DOI] [PubMed] [Google Scholar]
- 46.Whitelegge, J. P., Penn, B., To, T., Johnson, J., Waring, A., Sherman, M., Stevens, R. L., Fluharty, C. B., Faull, K. F., and Fluharty, A. L. (2000) Protein Sci. 9 1618–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurahashi, T., Miyazaki, A., Suwan, S., and Isobe, M. (2001) J. Am. Chem. Soc. 123 9268–9278 [DOI] [PubMed] [Google Scholar]
- 48.Wang, J., Xu, G., Slunt, H. H., Gonzales, V., Coonfield, M., Fromholt, D., Copeland, N. G., Jenkins, N. A., and Borchelt, D. R. (2005) Neurobiol. Dis. 20 943–952 [DOI] [PubMed] [Google Scholar]
- 49.de Beus, M. D., Chung, J., and Colon, W. (2004) Protein Sci. 13 1347–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tiwari, A., and Hayward, L. J. (2003) J. Biol. Chem. 278 5984–5992 [DOI] [PubMed] [Google Scholar]
- 51.Zhang, H., Joseph, J., Crow, J., and Kalyanaraman, B. (2004) Free Radic. Biol. Med. 37 2018–2026 [DOI] [PubMed] [Google Scholar]
- 52.Taylor, D. M., Gibbs, B. F., Kabashi, E., Minotti, S., Durham, H. D., and Agar, J. N. (2007) J. Biol. Chem. 282 16329–16335 [DOI] [PubMed] [Google Scholar]
- 53.Kazmirski, S. L., Isaacson, R. L., An, C., Buckle, A., Johnson, C. M., Daggett, V., and Fersht, A. R. (2002) Nat. Struct. Biol. 9 112–116 [DOI] [PubMed] [Google Scholar]
- 54.Chiti, F., and Dobson, C. M. (2006) Annu. Rev. Biochem. 75 333–366 [DOI] [PubMed] [Google Scholar]
- 55.Gruzman, A., Wood, W. L., Alpert, E., Prasad, M. D., Miller, R. G., Rothstein, J. D., Bowser, R., Hamilton, R., Wood, T. D., Cleveland, D. W., Lingappa, V. R., and Liu, J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 12524–12529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lesne, S., Koh, M. T., Kotilinek, L., Kayed, R., Glabe, C. G., Yang, A., Gallagher, M., and Ashe, K. H. (2006) Nature 440 352–357 [DOI] [PubMed] [Google Scholar]
- 57.Cheroni, C., Peviani, M., Cascio, P., Debiasi, S., Monti, C., and Bendotti, C. (2005) Neurobiol. Dis. 18 509–522 [DOI] [PubMed] [Google Scholar]
- 58.Choi, J. S., Cho, S., Park, S. G., Park, B. C., and Lee do, H. (2004) Biochem. Biophys. Res. Commun. 321 574–583 [DOI] [PubMed] [Google Scholar]
- 59.Urushitani, M., Kurisu, J., Tateno, M., Hatakeyama, S., Nakayama, K., Kato, S., and Takahashi, R. (2004) J. Neurochem. 90 231–244 [DOI] [PubMed] [Google Scholar]
- 60.Stieber, A., Gonatas, J. O., and Gonatas, N. K. (2000) J. Neurol. Sci. 173 53–62 [DOI] [PubMed] [Google Scholar]
- 61.Shinder, G. A., Lacourse, M. C., Minotti, S., and Durham, H. D. (2001) J. Biol. Chem. 276 12791–12796 [DOI] [PubMed] [Google Scholar]
- 62.Sorgjerd, K., Ghafouri, B., Jonsson, B. H., Kelly, J. W., Blond, S. Y., and Hammarstrom, P. (2006) J. Mol. Biol. 356 469–482 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.