Abstract
The importance of adult stem cells in the development of neoplastic diseases is becoming increasingly well appreciated. We hypothesized that sarcomas of soft tissue could be categorized by their developmental/differentiation status from stem cell to mature tissue, similar to the hematological malignancies. We conducted gene expression analyses during in vitro differentiation of human mesenchymal stem cells into adipose tissue, as a representative mature connective tissue, and identified genes whose expression changed significantly during adipogenesis. Gene clustering and distance correlation analysis allowed the assignment of a unique time point during adipogenesis that strongly correlates to each of the four major liposarcoma subtypes. Using a novel gene expression strategy, in which liposarcomas are compared to their corresponding adipocytic maturing cells, we identified a group of genes overexpressed in liposarcomas that indicate the stage of differentiation arrest, ie, sharing a similar expression profile to adipocytic cells at a corresponding stage of differentiation, and a distinct set of genes overexpressed in liposarcomas that are not found in the corresponding stage of differentiation. We propose that the latter set is enriched for candidate transformation-associated genes. Our results indicate that a degree of developmental maturity can be quantitatively assigned to solid tumors, supporting the notion that transformation of a solid tumor stem cell can occur at distinct stages of maturation.
Tumorigenesis and tumor progression are the result of sequential genetic aberrancies that ultimately reveal the characteristic phenotype of the cancer cell, including an altered differentiation status, invasiveness, and metastatic potential.1 Established biomarker expression patterns, characteristic of defined developmental time points in hematopoietic lineages, have been associated with distinct leukemia and lymphoma subtypes.2 Functional evidence supporting a developmental model in hematological malignancies is also available. For example, when the AML1-ETO fusion gene, found in 10 to 15% of acute myeloid leukemia patients, is expressed conditionally in hematopoietic stem cells of transgenic mice, they develop a myeloproliferative disorder.3 However, when the same fusion gene is expressed in partially committed myeloid progenitor cells, there appears to be no effect on normal hematopoiesis.4 These data support the notion that specific stages in development are more permissive to critical alterations in any given tumor type.
In the present study we sought to test the above-mentioned hypothesis in a solid tumor, ie, whether subtypes of solid tumors corresponding to the same lineage derive from the same developing cell transformed at different stages of development. We focused on sarcomas and mesenchymal development because: we and others have previously performed gene expression analysis on a representative set of soft-tissue sarcomas, and have shown that hierarchical gene expression does cluster pathologically similar subtypes of sarcomas within the same lineage5,6,7; and established in vitro techniques exist for the maturation of mesenchymal stem cells into mature connective tissue.8 We chose liposarcoma as a model because the four major subtypes,9 although not classified by their degree of maturation, nevertheless exhibit a wide spectrum of adipogenesis, as measured both morphologically and biochemically.10,11
Because the cancer phenotype likely reflects the changes in the expression patterns of myriad genes, identifying the critical events among many differentially regulated ones is difficult. Using a unique comparative gene expression analysis, in which liposarcomas were compared to both corresponding adipocytic maturing cells in an in vitro system of adipogenesis as well as normal fat, two distinct groups of genes could be identified: genes overexpressed in liposarcomas that indicate the stage of differentiation arrest (sharing a similar profile to corresponding adipocytic differentiation cells); and a distinct set of genes overexpressed in liposarcomas that are not found in the corresponding stage of differentiation. The former set, we believe, is analogous to the cell surface antigens CD4 and CD8 used to identify stages of lymphoid differentiation and the corresponding lymphoid neoplasms. The latter set, we propose, would be enriched for candidate tumor genes. Taken together, these data suggest that assigning an objective degree of maturation to distinct liposarcomas is feasible, and that it could provide significant insight into sarcomagenesis, and by extension for other solid tumors.
Materials and Methods
Human Mesenchymal Stem Cells (hMSCs)—Adipogenesis, Retroviral Transduction, Cell Cycle Analysis
hMSCs were kindly provided by Darwin Prockop of Tulane University (New Orleans, LA) and cultured under conditions promoting adipogenesis by continuous culturing in adipocytic medium containing 20% fetal bovine serum (Atlanta Biologicals, Lawrence, GA) in Dulbecco’s modified Eagle’s medium (Life Technologies, Inc., Gaithersburg, MD) and 0.5 μmol/L dexamethasone, 0.5 μmol/L isobutylmethylxanthine, and 50 μmol/L indomethacin for 21 days. Oil-Red-O (Sigma, St. Louis, MO) staining was performed via fixing cells in 60% isopropanol for 1 minute, before adding 60% Oil-Red-O stock [0.5% w/v Oil-Red-O (Sigma-Aldrich, St. Louis, MO) in isopropanol] diluted in dH2O for 30 minutes staining for lipid content. After washing, the cells were counterstained with hematoxylin.
Protein Isolation and Immunoblot Analysis
Immunoblot assays were performed on 25 μg of total protein extracted from differentiating hMSCs along the adipocytic lineage at the indicated time points by published procedures.12 Blots were probed with antibodies directed against human: perlipin A (rabbit anti-perilipin A polyclonal antibody, unconjugated, clone ab3527; Abcam, Cambridge, MA), lipoprotein lipase (mouse anti-lipoprotein lipase monoclonal antibody, unconjugated, clone LPL.A4; Abcam), adiponectin (rabbit anti-Acrp30 polyclonal antibody, unconjugated, Abcam), CD44 (mouse anti-CD44 monoclonal antibody, unconjugated, clone MEM-263), CD54 (mouse anti-CD54 monoclonal antibody, unconjugated, clone 28; BD Biosciences Pharmingen, San Diego, CA), and hepatocyte growth factor [mouse anti-hepatocyte growth factor (HGF) monoclonal antibody, unconjugated, clone 24612.111; Abcam]. Secondary anti-mouse or anti-rabbit (Amersham, Buckinghamshire, UK) antibodies were used to visualize proteins using an enhanced chemiluminescence detection system (Amersham). Ideal concentrations for each antibody were empirically controlled using either mesenchymal stem cells or normal fat as the appropriate control. Working concentrations range from 1:500 to 1:2000 dilutions of recommended stock solutions.
Immunohistochemistry
Sections were deparaffinized, treated with 1% H2O2, immersed in boiling 10 mmol/L citrate buffer for 15 minutes, and incubated in 10% normal horse serum for 30 minutes at room temperature. Anti-perlipin A, lipoprotein lipase, adiponectin, CD44, CD54, and HGF (as above) were used at concentrations ranging from 1:50 to 1:500 as empirically determined. Samples were then incubated with biotinylated anti-mouse IgG at 1:500 dilution (Vector Laboratories, Burlingame, CA) followed by avidin-biotin peroxidase complexes (1:25; Vector Laboratories) for 30 minutes. Diaminobenzidine was used as the chromogen, and hematoxylin as the nuclear counter stain.
RNA Isolation and Gene Expression Profiling
Twenty-five human liposarcomas (corresponding to five samples of each of the five major histological subtypes: well differentiated, dedifferentiated, myxoid/round cell, and pleomorphic) and five samples of human normal fat were subjected to gene expression profiling on U133a oligonucleotide arrays (Affymetrix, Inc., Santa Clara, CA). Tumor tissue collection, analysis, and primary gene expression data as previously described.13 Total cellular RNA was isolated at 0, 3, 7, 10, 14, and 21 days of hMSC-adipocytic differentiation from two separate time courses. Same day samples from different time courses were used as replicates. Total cellular RNA from hMSCs maintained in nondifferentiating medium for an identical period of time were similarly isolated. Total cellular RNA was hybridized on U133a arrays by the Memorial Sloan Kettering Cancer Genomic Core Facility. Unless otherwise indicated gene expression analysis was performed using GeneSpring software (Genespring, Santa Clara, CA). Progression away from stemness was measured using the Find Similar Sample feature in GeneSpring software. In this feature, the gene expression profile of the MSC was set as the baseline pattern, and overall gene expression pattern of each of the differentiating time points was compared against that pattern. A similarity score was assigned to each sample based on the degree to which its overall gene expression pattern resembles the baseline hMSC pattern.
Identification of Adipocytic Differentiation-Specific Genes
A two-phase procedure was used to identify adipogenesis-specific genes. First the genes that were differentially expressed in any of the time points were identified using one-way analysis of variance with time as the parameter. The systems analysis method (SAM)14 was applied using the implementation from the Siggens package of BioConductor (http://www.bioconductor.org). To account for multiple testing the false discovery rate15 was used with a cutoff of 0.05. Second, all data were normalized against the zero time point and a principal component analysis was done to find the dominant changes throughout the time course. We found that the first principal component was a vector that varied monotonically with time and accounted for 78% of the variance (see Supplemental Figure S1 at http://ajp.amjpathol.org). This component was used as a template to further filter the list of genes to find those with the highest correlation.
Distance Correlation Analysis
To place the four liposarcoma types along the differentiation time course we used the genes that were identified in the previous analysis and computed the distance between the liposarcoma types and the adipocytic-maturation samples. The distance was defined using the Pearson correlation function. Given a correlation coefficient of r the distance was defined as d = (1 − r)/2. An average distance was then computed using all samples of a given type to each of the time points. To calibrate this distance scale and approximate the significance of a given distance we randomly permuted the data 1000 times and computed the distances of this random data to the time points. A distance of 0.372 occurred 1% of the time (P value 0.01), a distance of 0.331 occurred 0.1% of the time, and 0.298 occurred 0.01% of the time. We set the significance threshold at 1%, any further distance was considered just a random correlation.
Identification of Liposarcoma-Specific Tumor Genes Accounting for Differentiation Status
To find genes that were specific to the tumorigenesis process and not in the maturation process a list of genes differentially expressed between normal fat tissue and each liposarcoma subtype was generated with stringent cutoff in the false discovery rate of (q <0.1). From each respective list, genes that were differentially expressed between each liposarcoma’s corresponding day of differentiation (as defined by distance mapping analysis) and day 21 (fully mature) adipocytes were removed. Venn diagrams were generated via Gene List Venn Diagram (http://mcbc.usm.edu/genevenn/genevenn.htm). Pathway analysis and gene annotation was performed using the National Institutes of Health DAVID (http://david.abcc.ncifcrf.gov/).
Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) and Primers
SuperScript One-Step RT-PCR with Platinum Taq (Life Technologies, Inc.) was used on 2 μl of total cellular RNA and cDNA synthesis was performed at 55°C for 30 minutes, and PCR amplification consisted of initial denaturing at 94°C, followed by 40 cycles of 15 seconds at 94°C, 30 seconds at the adequate annealing temperature according to each primer couple, and 40 seconds at 72°C. Annealing temperatures ranged from 55°C to 65°C. Primers were designed via Primer3 (Whitehead Institute for Biomedical Research, Cambridge, MA) unless otherwise noted. IL-6: 5′-TACCCCCAGGAGAAGATTCC-3′ and 5′-TTTTCTGCCAGTGCCTCTTT-3′; RAI-3: 5′-TGCTCACAAAGCAACGAAAC-3′ and 5′-TGGTTCTGCAGCTGAAAATG-3′; SEPP1: 5′-CGAGATATGCCAGCAAGTGA-3′ and 5′-GGTGATTGCAGACCCTGTTT-3′; LPL: 5′-GGGCATGTTGACATTTACCC-3′ and 5′-AGCCCTTTCTCAAAGGCTTC-3′;. MAD2 exon 1: 5′-GTGGAAGCGCGTGCTTTTGTTTG-3′ and 5-GGCCTGCGCGAGAACTTACAGAAG-3′ 13; GADPH: 5′-CCCCTTCATTGACCTCAACT-3′ and 5′-CGACCGTAACGGGAGTTGCT-3′.
Results
Human Mesenchymal Stem Cells (hMSCs) and Adipogenesis
hMSCs were cultured into mature adipocytes using standard techniques.8 Total cellular RNA was isolated from hMSCs before (day 0) and at days 3, 7, 10, 14, and 21 days after continual growth in the presence of adipogenesis differentiation medium. Gene expression profiling of total cellular RNA was performed on Affymetrix U133a in conjunction with the Memorial Sloan Kettering Cancer Genomic Core Facility. Terminal differentiation was confirmed by cessation of proliferation (Figure 1A) and acquisition of the mature phenotype, determined via percentage of cells showing fat accumulation as stained with Oil-Red-O (Figure 1B). To confirm progress of maturation at the earlier time points, each time point was compared to the immature state using GeneSpring in which gene expression profile of the hMSC was set as the baseline pattern, and overall gene expression pattern of each of the differentiating time points was compared against the hMSC pattern. A similarity score was assigned to each sample based on the degree to which its overall gene expression pattern resembles the hMSC. A general progression away from stemness was observed the longer cells were maintained in adipocytic medium, confirming maturation at points before those amenable to expression of terminal maturation markers (Figure 1C).
Figure 1.
A: hMSCs were plated at 2 × 104 and cultured in the presence of AM for 21 days as described in the Materials and Methods. At the indicated time points 100 μl of cells in culture were removed and counted via a Coulter counter. B: Separate T25 flasks of hMSCs undergoing adipogenesis were stained with Oil-Red-O at the indicated time points. Bottom panel shows day 0 hMSCs whereas top panel shows hMSCs after 21 days in AM. C: Similarity function (Find Similar Sample) of the GeneSpring package was used to compare each differentiation time point to the hMSCs (day 0). Two replicates were used for each analysis and the average shown.
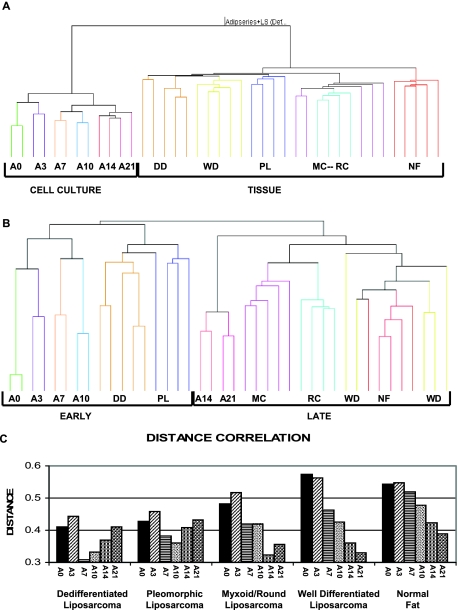
After confirming the maturation of hMSCs into adipocytes, we determined whether the four major liposarcoma subtypes, including dedifferentiated, pleomorphic, myxoid/round cell, and well differentiated lesions, corresponded to different time points of maturation. Not surprisingly, unbiased hierarchical clustering using the full 22.3-K gene base set showed no correlation between liposarcoma samples and the adipocytic-maturation time points (Figure 2A).
Figure 2.
Hierarchical clustering of hMSCs differentiating along the adipocytic lineage in relation to human liposarcomas using the full U133a gene set (A) and using the 69 adipocytic-maturation-specific genes (B). Each hMSC differentiating time point is represented by two replicates and compared to five samples of each of five liposarcoma subtypes as well as to five specimens of normal human fat, visually presented using GeneSpring. C: Distance mapping analysis comparing each liposarcoma subtype and normal fat as a control to adipogenesis time points using the 69 adipocytic-maturation-specific genes as the gene set.
Identification of Adipogenesis-Specific Genes
We hypothesized that the lack of correlation between liposarcoma samples and the maturing mesenchymal stem cells was attributable to the fact that human tumors and normal tissue counterparts have more in common than tumors and cells cultured in vitro. We were able to define a group of adipocytic-specific maturation genes by using a combination of an analysis of variance and a principal component analysis. Using analysis of variance, we first identified genes that showed statistically significant differential expression between the time points specified. However, analysis of variance does not take into account the fact that each point was temporally related. To more accurately define genes that were specifically associated with the differentiation time course, all data were normalized against the zero time point and a principal component analysis was performed to identify the dominant changes throughout the time course. Because the first principal component turned out to be a vector with monotonically changing expression throughout time, and 78% of the total variance was explained by this component, it was used as a template to filter for highly differentiation-specific genes by computing the correlation of each gene found using analysis of variance with the first principal component (also described in Materials and Methods, and schematically represented in Supplemental Figure S1 at http://ajp.amjpathol.org). Only 69 genes that had an absolute value of at least 0.90 of the correlation coefficient were included in subsequent analysis (Table 1). Four genes: LPL, SEPP1, RAI3, and IL6 were chosen as a validation set via RT-PCR (see Supplemental Figure S2 at http://ajp.amjpathol.org). Repeating unsupervised hierarchical clustering analysis using these adipocytic maturation genes revealed that dedifferentiated and pleomorphic liposarcomas associated with early maturation time points; whereas myxoid/round cell and well differentiated associated with late time points (Figure 2B). Normal fat tissue was included in the analysis, as an internal control, and associated with the later adipogenesis time point. Twenty percent of the 69 genes identified in this analysis as specific to adipogenesis overlap significantly with the 67 genes identified in another reported study using an identical adipocytic differentiation protocol but a completely different statistical approach.16
Table 1.
Gene Symbols and Titles
| Gene symbol | Gene title |
|---|---|
| A2M | α-2-Macroglobulin |
| ACDC | Adipocyte, C1Q and collagen domain containing |
| ADH1B | Alcohol dehydrogenase IB (class I), β polypeptide |
| AGC1 | Aggrecan 1 |
| AOC3 | Amine oxidase, copper containing 3 |
| APOD | Apolipoprotein D |
| APOE | Apolipoprotein E |
| ASPN | Asporin (LRR class 1) |
| BEX1 | Brain expressed, X-linked 1 |
| CCL2 | Chemokine (C-C motif) ligand 2 |
| CDH13 | Cadherin 13, H-cadherin (heart) |
| CEBPA | CCAAT/enhancer-binding protein (C/EBP), α |
| CES1 | Carboxylesterase 1 |
| CHRDL1 | Continued from bA814C6.1.1 in Em:AL591489 match |
| CIDE-3 | Cell death activator CIDE-3 |
| CMKOR1 | Chemokine orphan receptor 1 |
| COL11A1 | Collagen, type XI, α1 |
| COL13A1 | Collagen, type XIII, α1 |
| COL21A1 | Collagen, type XXI, α1 |
| COMP | Cartilage oligomeric matrix protein |
| CORIN | Corin |
| CRIP1 | Cysteine-rich protein 2 |
| DF | D component of complement (adipsin) |
| DPT | Dermatopontin |
| FABP4 | Fatty acid-binding protein 4, adipocyte |
| FBLN1 | Fibulin 1 |
| FBXO9 | Fatty acid synthase |
| FRZB | Frizzled-related protein |
| FY | Duffy blood group |
| GDF15 | Growth differentiation factor 15 |
| GPR21 | RAB GTPase activating protein 1 |
| HP | Haptoglobin |
| IBSP | Integrin-binding sialoprotein |
| IER3 | Immediate early response 3 |
| IGF2 | Insulin-like growth factor 2 (somatomedin A) |
| IGSF4B | Immunoglobulin superfamily, member 4B |
| IL6 | Interleukin 6 (interferon, beta 2) |
| KRT18 | Keratin 18 |
| KRTHA4 | Keratin, hair, acidic, 4 |
| LBP | Lipopolysaccharide-binding protein |
| LEP | Leptin (obesity homolog, mouse) |
| LOC283445 | Acetylcoenzyme A carboxylase βa |
| LPL | Lipoprotein lipase |
| MAFF | Human DNA sequence from clone CTA-447C4 |
| MMP13 | Matrix metalloproteinase 13 (collagenase 3) |
| NLGN4X | Neuroligin 4 |
| OLFML2A | Hypothetical protein LOC169611 |
| OLFML2B | Homo sapiens cDNA clone IMAGE:2506318 |
| OLR1 | Oxidized low density lipoprotein (lectin-like) receptor 1 |
| PCK1 | Phosphoenolpyruvate carboxykinase 1 (soluble) |
| PDE1A | Phosphodiesterase 1A, calmodulin-dependent |
| PLAC8 | Placenta-specific 8 |
| PLIN | Perilipin |
| PRELP | Proline arginine-rich end leucine-rich repeat protein |
| PRKAR2B | Protein kinase, cAMP-dependent, regulatory, type II, β |
| PTGDS | Prostaglandin D2 synthase 21 kDa (brain) |
| PTHR1 | Parathyroid hormone receptor 1 |
| RABGAP1 | RAB GTPase-activating protein 1 |
| RAI3 | Retinoic acid-induced 3 |
| RARRES2 | Retinoic acid receptor responder (tazarotene induced) 2 |
| RBP4 | Retinol binding protein 4, plasma |
| S100A4 | S100 calcium-binding protein A4 |
| SEPP1 | Selenoprotein P, plasma, 1 |
| SERPINF1 | Serine (or cysteine) proteinase inhibitor, clade F |
| SLC7A5 | Solute carrier family 7 |
| SORBS1 | Sorbin and SH3 domain containing 1 |
| TNA | Tetranectin (plasminogen-binding protein) |
| TNFAIP6 | Tumor necrosis factor, α-induced protein 6 |
| VEGF | Vascular endothelial growth factor |
Distance Correlation Analysis
To further characterize the relationship between the four types of liposarcomas and mesenchymal stem cell maturation status, we performed a distance correlation analysis. This approach allows assessment of the degree of similarity between each liposarcoma and each maturation time point using the previously identified adipocytic-maturation gene set. In this analysis, the smaller the numerical distance, the greater the correlation, with statistical significance defined at a value of less than 0.37 at which the chance of random data having that small a distance was determined by permutation analysis to be 1%. Examining the distance correlation curves for each liposarcoma subtype demonstrates that: 1) dedifferentiated liposarcomas correlates best with day 7 developing adipocytes; 2) pleomorphic liposarcoma correlates with day 10 developing adipocytes; 3) myxoid/round cell liposarcoma correlates with day 14 developing adipocytes; and 4) well differentiated liposarcoma correlates with day 21 developing adipocytes (Figure 2C). In all liposarcoma correlations to differentiation, values were found to be statistically significant (ie, less than 0.37). As a control, the degree of normal fat association to the in vitro adipogenesis samples clearly demonstrates that normal fat correlates poorly with hMSCs at day 0—before growth in adipocytic medium (A0; Figure 3C). Furthermore, the correlation increases (ie, distance shortens) because normal fat is compared to more maturing adipocytic cells. Statistical significance for normal fat was not reached (ie, the distance is not less than 0.37), implying that in vitro terminally mature adipocytic cells under these conditions may not be as mature as normal fat tissue, although there is a trend for normal fat to associate with more mature adipocytic cells.
Figure 3.
A: Immunoblot analysis of the indicated proteins at the indicated times of adipogenesis. B: Immunohistochemical analysis of a sarcoma tissue microarray examining for expression of the indicated proteins on the indicated representative liposarcoma subtypes. Percentages represent number of liposarcoma subtypes staining positive compared to number present on the tissue microarray.
Confirmation of Differentiation in Liposarcoma Subtypes
Having linked adipocytic differentiation to liposarcoma subtypes, we next sought to test the hypothesis that the four liposarcoma subtypes should express: a relative gain of adipocytic markers and a relative loss of MSC markers as reflective of their maturation status. To examine this possibility, we first performed immunoblot analysis examining the expression patterns of three known adipocytic markers (lipoprotein lipase, adiponectin, and perlipin A) and three known MSC-markers (CD44, CD54, and HGF) in protein extracts obtained from adipocytic maturing cells to confirm their expected behavior in our model system. As shown in Figure 3A, the levels of lipoprotein lipase, adiponectin, and perlipin A increased as cells mature into adipocytes; whereas the levels of the MSC markers CD44, CD54, and HGF decreased as cells become more mature. Next, we performed immunohistochemistry analysis of both the adipocytic and MSC markers on a sarcoma tissue microarray, containing 12 dedifferentiated liposarcomas, 3 pleomorphic liposarcomas, 8 myxoid/round cell liposarcomas, and 10 well differentiated liposarcomas. Figure 3B indicates the typical appearance of a representative liposarcoma of each subtype stained with each antibody. The percentage of liposarcomas staining positive per subtype per antibody is indicated in the corner of each representative caption. These data confirm our earlier results: dedifferentiated and pleomorphic liposarcomas, being the more immature liposarcomas, stain strongest for MSC markers and not at all for adipocytic markers; and myxoid/round cell liposarcomas and well differentiated liposarcomas, being the more mature adipocytic sarcomas, stain strongly for adipocytic markers but not for MSC markers, with more of the well differentiated liposarcomas staining positive than pleomorphic samples for adipocytic markers.
Maturation-Based Differential Gene Expression Analysis
Having assigned a degree of maturation to each liposarcoma subtype, we next sought to perform a unique gene expression analysis that would compare a tumor not to its normal mature counterpart, but to its normal maturing counterpart. We hypothesized that a comparison between tumor and normal counterpart tissue essentially contrasts a transformed, immature, proliferating tissue to a nontransformed, mature, often nonproliferating tissue. The end analysis would reveal genes indicative of the maturation discrepancies between the two tissues, and thus masking genes that are relevant for oncogenic transformation. To overcome this issue, and to generate a list of genes minimizing those that reflect maturation differences, we performed the following three stage analyses (schematically represented on Figure 4A).
Figure 4.
A: Schematic representation of the differential gene expression approach used to eliminate maturation-related genes from potential tumor genes. See text for details. B: Schematic representation of gene lists. Each Venn diagram is a composite of genes either overexpressed (left) or underexpressed (right) in the tumor versus normal fat (red circles) or corresponding differentiating cells versus differentiated cells (yellow circle). The overlapping genes are indicated in orange. C: Percentage of overlapping differentiation genes in the subsets of genes differentially expressed as a function of genes overexpressed (blue), under (red), and total (green) in tumors versus normal fat in tumor versus DD (dedifferentiated), Pl (pleomorphic), M (myxoid), R (round cell) liposarcoma.
In the first stage we identified a group of genes that are significantly (false discovery rate, <0.1) differentially expressed between: 1) day 7 developing adipocytes (ie, the transformation point of dedifferentiated liposarcoma) and day 21 terminally mature adipocytes (day 7 versus day 21); 2) day 10 developing adipocytes (ie, the transformation point of pleomorphic liposarcoma) and day 21 terminally mature adipocytes (day 10 versus day 21); and 3) day 14 developing adipocytes (ie, the transformation point of myxoid/round cell liposarcoma) and day 21 terminally mature adipocytes (day 14 versus day 21) (see Supplemental Table S1; sheets 1 to 3 at http://ajp.amjpathol.org). Replicates were included as discussed in the Materials and Methods.
In the second stage, we identified group of genes that are differentially regulated between: 1) dedifferentiated liposarcoma and normal fat; 2) pleomorphic liposarcoma and normal fat; 3) myxoid liposarcoma and normal fat; and 4) round cell liposarcoma and normal fat, using five liposarcomas per subtype and five normal fat controls (see Supplemental Table S1; sheets 1 to 4; at http://ajp.amjpathol.org) as described by Singer and colleagues.13
In the third stage the gene lists representing the differentially expressed genes between each liposarcoma subtype and normal fat were overlapped with its corresponding gene list representing the degree of differentiation of the liposarcoma to normal fat. Thus: 1) the gene list representing dedifferentiated liposarcoma compared to normal fat was overlapped with the gene list representing day 7 developing adipocytes (ie, the putative transformation point of dedifferentiated liposarcoma) compared to day 21 terminally mature adipocytes (day 7 versus day 21); 2) the gene list representing pleomorphic liposarcoma compared to normal fat was overlapped with the gene list representing day 10 developing adipocytes (ie, the putative transformation point of pleomorphic liposarcoma) compared to day 21 terminally mature adipocytes (day 10 versus day 21); 3) the gene list representing myxoid and round cell liposarcomas as compared to normal fat were separately overlapped with the gene list representing day 14 developing adipocytes (ie, the transformation point of myxoid/round cell liposarcoma) as compared to day 21 terminally mature adipocytes (day 14 versus day 21). These overlaps are schematically represented in Figure 4B. The corresponding gene lists are provided in Supplemental Tables S2 to S5 at http://ajp.amjpathol.org (representing genes overexpressed in tumors versus normal fat, and in corresponding undifferentiated cells versus differentiated cells, columns 1 to 3) and Supplemental Tables S2 to S5 at http://ajp.amjpathol.org (representing genes underexpressed in tumors versus normal fat, and in corresponding undifferentiated cells versus differentiated cells, columns 4 to 6). Note that it was not possible to perform a similar analysis on the well differentiated liposarcoma subtype because it associated strongly with the same point as normal fat (day 21 hMSCs). Repeated attempts to differentiate hMSCs for earlier or later times in an attempt to distinguish normal fat from well differentiated liposarcoma via a distance mapping analysis proved unsuccessful as later day (ie, D24, D27) hMSCs differentiated along the adipocytic lineage yielded identical gene expression patterns to day 21 differentiating cells (data not shown).
Gene Expression Analysis: Dedifferentiated Liposarcoma Versus Normal Fat Accounting for Differentiation
A set of 2026 unique genes was identified to be overexpressed in dedifferentiated liposarcoma versus normal fat (false discovery rate, 0.1) in this analysis. Similarly, 799 unique genes were identified to be overexpressed in hMSCs differentiating along the adipocytic lineage at day 7 versus terminally differentiated adipocytes (day 21). In both gene sets, genes were subdivided into two groups: genes that are overexpressed in either tumor versus normal fat or in day 7 versus day 21; and B genes that are underexpressed in either tumor versus normal fat or in day 7 versus day 21. Because the corresponding differentiation time point of dedifferentiated liposarcoma is day 7 differentiating hMSCs (Figure 2C), we overlapped the two overexpressing gene sets and the two underexpressing gene sets (Figure 2B, first panel; Supplemental Table S2 at http://ajp.amjpathol.org). With this method we found that 277 genes (see Supplemental Table S2, column 3, at http://ajp.amjpathol.org) are shared between a differential analysis that compares overexpressed genes in tumor cells versus normal cells and a differential analysis that compares differentiating cells (that correspond to the differentiation state of the tumor) to differentiated cells. Similarly, we found that 226 genes (see Supplemental Table S2, column 6 at http://ajp.amjpathol.org) are shared between a differential analysis that identifies underexpressed genes in tumor cells versus normal cells and a differential analysis that compares differentiating cells (that correspond to the differentiation state of the tumor) to differentiated cells. These overlapping subsets of genes, comprised within the larger set of genes obtained in comparing tumor cells to normal cells, were termed markers of differentiation (represented by the orange overlap in the Venn diagrams, Figure 4B). Note that in the analysis comparing dedifferentiated liposarcoma to normal fat, these markers of differentiation contribute 13.6% of the overexpressed and 19.7% of the underexpressed genes, making a total of 33.5% of all differentially obtained genes (Figure 4C).
In addition to numerically limiting the list of potential transformation-related genes, this analysis further eliminates pursuing markers of differentiation—genes that otherwise might be assumed to be involved in tumorigenesis. Examples of the latter for the dedifferentiated liposarcoma to normal fat would include: topoisomerase (DNA) IIα (TOPIIa) seemingly overexpressed more than 100 times in dedifferentiated liposarcomas as compared to normal fat, but equally overexpressed to the same extent when immature adipocytes are compared to mature adipocytes (see Supplemental Tables S1 and S6 at http://ajp.amjpathol.org). This analysis would suggest that TOPIIa expression is clearly not related to tumorigenesis, but rather to adipogenesis. This finding potentially explains why liposarcomas are not particularly sensitive to etoposide, a clinically used topoisomerase II inhibitor.17 Alternatively, the overexpression of TOPIIa may simply make liposarcoma cells less sensitive to etoposide compared to tumor cells that lack such overexpression.
Additionally, we have analyzed the pathways represented by genes overexpressed (see Supplemental Table S7, sheet 1, at http://ajp.amjpathol.org) and underexpressed (see Supplemental Table S8, sheet 2, at http://ajp.amjpathol.org) in dedifferentiated liposarcomas versus normal fat after excluding markers of differentiation genes. Even after factoring out the effects of differentiation and the potential contribution of the immature differentiating cell’s cycle in comparison to the normal mature cell, cell cycle genes, and purine metabolism still account for the most up-regulated pathways in these cancer cells (see Supplemental Table S9, sheet 1, at http://ajp. amjpathol.org). Not surprisingly, several fat-associated pathways seem to be lost in dedifferentiated liposarcoma as compared to normal cells after accounting for markers of differentiation. An additional pathway that has not previously been reported to be involved in adipogenesis and whose loss may contribute to sarcomagenesis, namely MAPK signaling, is the most highly represented (see Supplemental Table S10, sheet 1 at http://ajp. amjpathol.org).
Gene Expression Analysis: Pleomorphic Liposarcoma Versus Normal Fat Accounting for Differentiation
Using a similar approach as described in detail for dedifferentiated liposarcoma above, we examined pleomorphic liposarcoma in relation to its differentiation time point at day 10 (Figure 2C). We identified 146 genes of 1688 (8.6%) overexpressed, and 151 genes of 845 (17.9%) underexpressed, which (as above) were termed markers of differentiation (see Supplemental Table S2 at http://ajp.amjpathol.org). Note that in the analysis comparing pleomorphic liposarcoma to normal fat, these markers of differentiation contribute a total of 26.5% of all differentially obtained genes (Figure 4C). As above, identified markers of differentiation genes that otherwise might be assumed to be involved in tumorigenesis, and thus erroneously pursued as such, include: KRAS (a well characterized oncogene)18) expressed 1.8 times in pleomorphic liposarcomas as compared to normal fat (see Supplemental Table S1 at http://ajp.amjpathol.org), but also 1.7 times in day 10 differentiating hMSCs along the adipocytic lineage as compared to differentiated adipocytes (see Supplemental Table S6 at http://ajp.amjpathol.org). Pathway analysis represented by genes overexpressed (see Supplemental Table S7, sheet 2, at http://ajp.amjpathol.org) and underexpressed (see Supplemental Table S8, sheet 2, at http://ajp.amjpathol.org) in pleomorphic liposarcomas versus normal fat after excluding markers of differentiation genes identified similar pathways as those observed in the dedifferentiated liposarcoma analysis, namely, the up-regulation of cell cycle and purine metabolism pathways (see Supplemental Table S9, sheet 2, at http://ajp.amjpathol.org), and the down-regulation of the MAPK signaling pathway (see Supplemental Table S10, sheet 2, at http://ajp.amjpathol.org).
Gene Expression Analysis: Myxoid/Round Cell Liposarcoma Versus Normal Fat Accounting for Differentiation
Despite the observation that the tightest association of any liposarcoma to its stage of differentiation was for the myxoid/round cell liposarcoma subtype with day 14 hMSCs differentiating along the adipocytic lineage (Figure 3C); the contribution of markers of differentiation to the differentially expressed genes between the myxoid/round cell liposarcomas as compared to normal fat was minimum (Figure 4B; panels three and four). For the myxoid liposarcoma analysis, we identified 7 genes of 2841 (0.24%) overexpressed and 7 genes of 1205 (0.58%) underexpressed that were termed myxoid liposarcoma markers of differentiation (see Supplemental Table S3 at http://ajp.amjpathol.org). Note that in this analysis comparing myxoid liposarcoma to normal fat, these markers of differentiation contribute a total of 0.82% of all differentially obtained genes (Figure 4C). Similar analysis for round cell liposarcoma yielded almost identical results (see Supplemental Table S5 at http://ajp.amjpathol.org and Figure 4C). The lack of significant overlap between the differentiation set of genes and the gene set of differentially expressed genes between myxoid/round cell liposarcomas and normal fat is attributable to the small number of genes (ie, 31) obtained in comparing day 14 hMSCs differentiating along the adipocytic lineage to terminally differentiated adipocytes at day 21. This result, taken independently, would suggest that day 14 differentiating hMSCs are fairly mature and that little further adipocytic differentiation occurs at later time points. This also confirms our observations that differentiating hMSCs for longer than 21 days does not identify cells with different gene expression patterns to those observed at day 21. Finally, although only 31 genes were differentially expressed between day 14- and day 21-differentiating cells; approximately half (14 for myxoid and 12 for round cell) overlapped with the respective differentially regulated gene sets.
Dedifferentiated and Pleomorphic Liposarcoma
Our data suggested that ∼30% of genes identified to be differentially regulated between the early differentiation stage liposarcoma subtypes (ie, dedifferentiated and pleomorphic liposarcoma), were as described above markers of differentiation. Accordingly we hypothesized that the remaining set of genes after discounting for markers of differentiation would be enriched for pathways involved in tumorigenesis as described above. In a further attempt to identify genes and pathways that might be broadly representative of pathways involved in tumorigenesis of early differentiation stage liposarcomas, we overlapped the respective differentially expressed gene sets (see Supplemental Tables S11 and S12 at http://ajp.amjpathol.org) and subjected the combined gene set to pathway analysis as described above. The combined analysis re-enforced the results of each independent analysis. Significant up-regulated pathways (see Supplemental Table S7, sheet 3, at http://ajp.amjpathol.org) included cell cycle and purine metabolism; whereas down-regulated pathways (see Supplemental Table S8, sheet 3 at http://ajp.amjpathol.org) included MAPK signaling and insulin signaling.
Discussion
Assigning a Degree of Developmental Maturation to Liposarcoma Subtypes
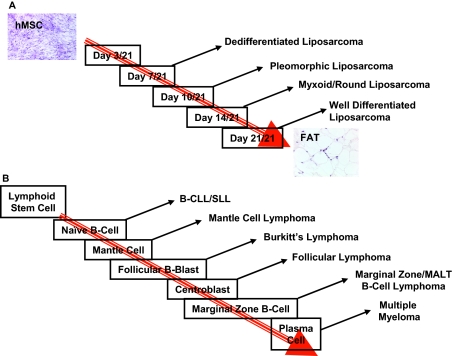
Histopathological classification of solid tumors currently relies heavily on judging a tumor’s degree of differentiation (ie, its approximation of the normal counterpart) versus anaplasia (ie, its difference from the normal counterpart). No comment is made as to the point during normal cellular development at which tumorigenesis occurs. Similarly, models of carcinogenesis, eg, colon adenocarcinoma, which describe tumorigenesis as a step-wise process from polyp, to adenoma, to carcinoma, similarly do not provide insight into the state of maturity of the progenitor cells. We describe in this article a means of assigning a degree of developmental maturation to solid tumors (schematically represented in Figure 5) conceptually similar to the map of lymphocyte maturation and its association with various forms of lymphoma/leukemia. Although hematological malignancies have historically been characterized by their gene expression profiles (ie, antigen patterns) reflecting their maturation status of their precursor cells,2 this has not been considered for solid tumors, owing to the lack of understanding of the genes activated or inactivated during the process of maturation of the normal precursor.
Figure 5.
A: Schematic representation of the correlation of adipogenesis to liposarcoma transformation. B: Schematic representation of the similarity of hematopoietic differentiation to neoplastic formation modified from DeVita, VT, Hellman S, Rosenberg SA: Cancer: Principles and Practice of Oncology, Ed 7, Philadelphia, Lippincott Williams and Wilkins, 2005.
Our results suggest that dedifferentiated liposarcomas, pleomorphic liposarcomas, myxoid/round cell, and well differentiated liposarcomas correspond to transformation of a mesenchymal stem cell at increasingly mature steps in adipogenesis, in agreement with other studies using comparisons of biochemical measurements11,19 of lipid production and fat content among the four types of liposarcomas and a recent differentiation stage-specific model of synovial sarcoma20. Although facilitating diagnosis, stratifying the four types of liposarcomas relative to each other based solely on adipogenesis using biochemical measurements does not easily allow for identification of tumor-related genes. Conversely, characterization of liposarcomas by an assigned transformation time point has several immediately recognizable advantages. First, by correlating each tumor subtype to its developing normal counterpart and using differential gene expression analysis, two distinct gene sets can be identified: genes overexpressed in liposarcomas that mark the stage of differentiation arrest (sharing a similar profile to corresponding adipocytic differentiation cells); and a distinct set of genes overexpressed in liposarcomas that are not found in the corresponding stage of differentiation. The former gene set we propose are analogous to the cell surface antigens CD4 and CD8 used to identify stages of lymphoid differentiation and the corresponding lymphoid neoplasms. The latter set we propose would be enriched for candidate tumor genes. As in vitro techniques improve for the identification and differentiation of epithelial tissue, similar approaches for the characterization of carcinomas could be feasible.
Analysis of Genes that Mark the Degree of Differentiation Arrest Versus Sarcomagenesis
Our analysis identifies two groups of genes: genes overexpressed in liposarcomas that mark the stage of differentiation arrest (sharing a similar profile to corresponding adipocytic differentiation cells); and a distinct set of genes overexpressed in liposarcomas that are not found in the corresponding stage of differentiation. We further propose that this is an important distinction because this approach would allow the elimination of genes that otherwise might be falsely assumed to be involved in tumorigenesis and thus erroneously pursued with therapeutic intent, such as TOPIIa for dedifferentiated liposarcoma and KRAS for pleomorphic liposarcoma (see corresponding Results section).
On the other hand, this type of analysis may also be useful for identifying markers of differentiation that may have diagnostic utility. For example, CD24 shows a biphasic pattern during adipocytic differentiation (data not shown): absent in undifferentiated hMSCs, present at increased levels in early differentiating cells (mainly days 3 and 7) and then again absent in hMSCs further differentiated along the adipocytic lineage. Thus one would predict that if dedifferentiated liposarcomas originated in early differentiating adipocytes, CD24 might be a potential cell surface antigen that may be useful in distinguishing dedifferentiated liposarcomas from pleomorphic liposarcomas as well as other more differentiated subtypes. Indeed CD24 is overexpressed in dedifferentiated liposarcomas as compared to normal fat (see Supplemental Table S2 at http://ajp.amjpathol.org) but not in pleomorphic liposarcomas, or myxoid/round cell liposarcomas (see Supplemental Tables S3, S4, and S5 at http://ajp. amjpathol.org). To date, only one case of clear cell sarcoma has been reported to stain positive for CD24.21 A thorough search characterizing these and other potential markers is in progress.
Even after factoring out differentiation-dependent genes for the early stage differentiation tumors as well as genes that are related to the natural proliferation state of early differentiating cells, a majority of differentially expressed genes are related to the cell cycle and purine metabolism. More interesting in our opinion were the pathways down-regulated in the dedifferentiated and pleomorphic liposarcomas: MAPK and insulin signaling. Although it has been reported that proliferation-activated receptor-γ ligands22 can function as differentiation therapy, it is tempting to speculate that they may work through the insulin signaling pathway given their known functionality as insulin sensitizers.23 Regardless, a study of troglitazone in a phase II study of liposarcoma patients rendered no objective responses,24 indicating blockade of this single pathway is insufficient to mount a measurable change in a group of patients’ tumors.
Model of Solid Tumorigenesis
In this article we propose that tumors can form via one of two mechanisms. The first mechanism involves an initial genetic change occurring in differentiating MSCs (along a given mesenchymal lineage) resulting in hyperplastic/dysplastic MSCs that are limited in their differentiation potential and thus become arrested at a stage of mesenchymal differentiation that morphologically appears as well differentiated amid normal tissue. On further accumulation of genetic changes, the differentiation potential of the same hyperplastic/dysplastic MSCs that gave rise to the well differentiated tumor is further significantly impaired, now giving rise to a dedifferentiated tumor amid the previously formed well differentiated tumor. The case of well differentiated and dedifferentiated liposarcomas are a good example of this model. In this scenario, MSCs accumulate genetic damage manifested as harbor ring and giant marker chromosomes composed of chromosome 12q12-15,25 which harbor several oncogenes such as MDM2, these cells are still capable of initially differentiating to form well differentiated liposarcomas. However, as these MSCs gain further genetic damage, they further lose their differentiation ability and now undergo transformation at a much earlier point of differentiation giving rise to dedifferentiated tumors.
The second mechanism supposes that the initial genetic damage occurring in MSCs results in a primary inhibition of differentiation at a specific point of differentiation. These cells would arrest and transform at a specified stage of differentiation giving rise to tumors that morphologically resemble corresponding differentiating cells. The accumulation of secondary genetic events could further lead to different morphological appearances (resulting in different grade and prognosis) of tumors that arose from the same transformation time point. This model is best exemplified by myxoid/round cell liposarcoma. The presence of the TLS-CHOP fusion protein, characteristic of both myxoid and round cell liposarcoma, has been suggested to interfere with adipogenic differentiation.26 Our model would predict that the initial transformation would likely give rise to myxoid liposarcomas, which with further genetic damage, lead to formation of round cell liposarcoma.
These two mechanisms explain how a developmental model might give rise to different types of solid tumors along a given lineage, and also how morphologically different but genetically similar tumors may originate from different differentiation stages (ie, well differentiated and dedifferentiated); and from the same differentiation stage (ie, myxoid/round cell liposarcomas) depending on whether inhibition is a primary or secondary event. Although this model supports our findings, further work including both in vitro and animal models are needed to fully validate it.
In an attempt to provide initial validation for our model, we used several association studies comparing MSCs differentiating along the adipogenesis lineage and liposarcoma subtypes. This approach has the potential to do more than just associate nonadipocytic cells with other nonadipocytic cells. First, as described in the text we identified a set of genes characteristic of the adipocytic differentiation program. This is not a set of genes that are merely absent early and present late in adipocytic differentiation. These are genes that represent both the loss of the MSC gene expression pattern and the gain of the adipocytic gene expression pattern. Further, our data of course does not exclude the possibility that cells can dedifferentiate—but if they do our data would suggest that in doing so they do not simply lose the mature phenotype and become undifferentiated tumors or some aberrant cell without a mature phenotype. Instead, more mature tumors may actively acquire a greater degree of a stem cell phenotype, and in so doing reverse the normal differentiation program. This, in our opinion, is significantly different from concluding that dedifferentiation is simply the passive loss of a mature phenotype.
Furthermore, as we have highlighted above in the text and in Figure 2C (distance correlation data), a statistically significant association between the in vitro differentiating time points and the liposarcoma subtypes. We have performed similar analysis for other sarcoma subtypes using our previously published gene expression data sets5 and have found no similar associations. The only other statistically significant association was observed between malignant fibrous histiocytoma and the undifferentiated hMSCs27 but not any other early differentiation time point. Finally, our analysis is most informative for the more poorly differentiated liposarcomas, rather than the well differentiated ones, something not predicted if the approach used was simply a commentary on the loss of the mature phenotype and not an active gain of a mesenchymal stem cell phenotype.
Extension to Other Tumor Subtypes
The greater appreciation of stem cell biology could be used to extend these results not only to other mesenchymal lineages, but to other forms of cancer. Such a classification may allow us to design even more rationale approaches to anti-cancer therapy. Finally, application of similar techniques should help us winnow through the chaff of maturation-specific genes to find those that are truly associated with the initiation and progression of cancer.
Acknowledgments
We thank Darwin Prockop and the Tulane University Gene Center for providing us with MSCs as well as detailed instructions regarding their propagation and differentiation; Maria E. Dudas of the Cordon-Cardo laboratory for assisting with immunohistochemistry and cell culture; and Agnes Viale, Ph.D., of the Memorial Sloan Kettering Cancer Genomics Core facility for assistance with U133A Affymetrix array processing.
Footnotes
Address reprint requests to Carlos Cordon-Cardo, M.D., Ph.D., Vice-Chair, Department of Pathology, Associate Director, Herbert Irving Comprehensive Cancer Center, Columbia University, 1130 St. Nicholas Ave., Room 309, New York, NY 10032. E-mail: cc2791@columbia.edu; or Robert G. Maki, M.D., Ph.D., Co-Director, Adult Sarcoma Program, Department of Medicine, Memorial Sloan Kettering Cancer Center, 1275 York Ave., New York, NY. E-mail: makir@mskcc.org.
Supported by the American Cancer Society (to I.M.), the Clinical Scholars/Charles Revson Foundation (to I.M.); the National Cancer Institute (program project CA47179 to R.G.M and C.C.C.), Spin4survival.org (to R.G.M), and The Shuman Family Fund for GIST Research (to R.G.M), and philanthropic funds.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Sieber OM, Tomlinson SR, Tomlinson IP. Tissue, cell and stage specificity of (epi)mutations in cancers. Nat Rev Cancer. 2005;5:649–655. doi: 10.1038/nrc1674. [DOI] [PubMed] [Google Scholar]
- Orfao A, Schmitz G, Brando B, Ruiz-Arguelles A, Basso G, Braylan R, Rothe G, Lacombe F, Lanza F, Papa S, Lucio P, San Miguel JF. Clinically useful information provided by the flow cytometric immunophenotyping of hematological malignancies: current status and future directions. Clin Chem. 1999;45:1708–1717. [PubMed] [Google Scholar]
- Fenske TS, Pengue G, Mathews V, Hanson PT, Hamm SE, Riaz N, Graubert TA. Stem cell expression of the AML1/ETO fusion protein induces a myeloproliferative disorder in mice. Proc Natl Acad Sci USA. 2004;101:15184–15189. doi: 10.1073/pnas.0400751101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Zhou L, Miyamoto T, Iwasaki H, Harakawa N, Hetherington CJ, Burel SA, Lagasse E, Weissman IL, Akashi K, Zhang DE. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc Natl Acad Sci USA. 2001;98:10398–10403. doi: 10.1073/pnas.171321298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal NH, Pavlidis P, Antonescu CR, Maki RG, Noble WS, DeSantis D, Woodruff JM, Lewis JJ, Brennan MF, Houghton AN, Cordon-Cardo C. Classification and subtype prediction of adult soft tissue sarcoma by functional genomics. Am J Pathol. 2003;163:691–700. doi: 10.1016/S0002-9440(10)63696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TO, West RB, Linn SC, Alter O, Knowling MA, O’Connell JX, Zhu S, Fero M, Sherlock G, Pollack JR, Brown PO, Botstein D, van de Rijn M. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet. 2002;359:1301–1307. doi: 10.1016/S0140-6736(02)08270-3. [DOI] [PubMed] [Google Scholar]
- Baird K, Davis S, Antonescu CR, Harper UL, Walker RL, Chen Y, Glatfelter AA, Duray PH, Meltzer PS. Gene expression profiling of human sarcomas: insights into sarcoma biology. Cancer Res. 2005;65:9226–9235. doi: 10.1158/0008-5472.CAN-05-1699. [DOI] [PubMed] [Google Scholar]
- Colter DC, Class R, DiGirolamo CM, Prockop DJ. Rapid expansion of recycling stem cells in cultures of plastic-adherent cells from human bone marrow. Proc Natl Acad Sci USA. 2000;97:3213–3218. doi: 10.1073/pnas.070034097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg AA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: liposarcoma. Cancer Genet Cytogenet. 2004;155:1–24. doi: 10.1016/j.cancergencyto.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Millis K, Weybright P, Campbell N, Fletcher JA, Fletcher CD, Cory DG, Singer S. Classification of human liposarcoma and lipoma using ex vivo proton NMR spectroscopy. Magn Reson Med. 1999;41:257–267. doi: 10.1002/(sici)1522-2594(199902)41:2<257::aid-mrm8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Singer S, Millis K, Souza K, Fletcher C. Correlation of lipid content and composition with liposarcoma histology and grade. Ann Surg Oncol. 1997;4:557–563. doi: 10.1007/BF02305536. [DOI] [PubMed] [Google Scholar]
- Matsushime H, Quelle DE, Shurtleff SA, Shibuya M, Sherr CJ, Kato JY. D-type cyclin-dependent kinase activity in mammalian cells. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer S, Socci ND, Ambrosini G, Sambol E, Decarolis P, Wu Y, O’Connor R, Maki R, Viale A, Sander C, Schwartz GK, Antonescu CR. Gene expression profiling of liposarcoma identifies distinct biological types/subtypes and potential therapeutic targets in well differentiated and dedifferentiated liposarcoma. Cancer Res. 2007;67:6626–6636. doi: 10.1158/0008-5472.CAN-07-0584. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Sekiya I, Larson BL, Vuoristo JT, Cui JG, Prockop DJ. Adipogenic differentiation of human adult stem cells from bone marrow stroma (MSCs). J Bone Miner Res. 2004;19:256–264. doi: 10.1359/JBMR.0301220. [DOI] [PubMed] [Google Scholar]
- Licht JD, Mazanet R, Loehrer PJ, Gonin R, Antman KH. Phase IV trial of daily oral etoposide in the treatment of advanced soft-tissue sarcoma. Cancer Chemother Pharmacol. 1994;34:79–80. doi: 10.1007/BF00686117. [DOI] [PubMed] [Google Scholar]
- Kranenburg O. The KRAS oncogene: past, present, and future. Biochim Biophys Acta. 2005;1756:81–82. doi: 10.1016/j.bbcan.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Chen JH, Enloe BM, Fletcher CD, Cory DG, Singer S. Biochemical analysis using high-resolution magic angle spinning NMR spectroscopy distinguishes lipoma-like well differentiated liposarcoma from normal fat. J Am Chem Soc. 2001;123:9200–9201. doi: 10.1021/ja016182u. [DOI] [PubMed] [Google Scholar]
- Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 2007;11:375–388. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Mechtersheimer G, Tilgen W, Klar E, Moller P. Clear cell sarcoma of tendons and aponeuroses: case presentation with special reference to immunohistochemical findings. Hum Pathol. 1989;20:914–917. doi: 10.1016/0046-8177(89)90106-8. [DOI] [PubMed] [Google Scholar]
- Demetri GD, Fletcher CD, Mueller E, Sarraf P, Naujoks R, Campbell N, Spiegelman BM, Singer S. Induction of solid tumor differentiation by the peroxisome proliferator-activated receptor-gamma ligand troglitazone in patients with liposarcoma. Proc Natl Acad Sci USA. 1999;96:3951–3956. doi: 10.1073/pnas.96.7.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurnell M, Savage DB, Chatterjee VK, O’Rahilly S. The metabolic syndrome: peroxisome proliferator-activated receptor gamma and its therapeutic modulation. J Clin Endocrinol Metab. 2003;88:2412–2421. doi: 10.1210/jc.2003-030435. [DOI] [PubMed] [Google Scholar]
- Debrock G, Vanhentenrijk V, Sciot R, Debiec-Rychter M, Oyen R, Van Oosterom A. A phase II trial with rosiglitazone in liposarcoma patients. Br J Cancer. 2003;89:1409–1412. doi: 10.1038/sj.bjc.6601306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilotti S, Della Torre G, Lavarino C, Sozzi G, Minoletti F, Vergani B, Azzarelli A, Rilke F, Pierotti MA. Molecular abnormalities in liposarcoma: role of MDM2 and CDK4-containing amplicons at 12q13-22. J Pathol. 1998;185:188–190. doi: 10.1002/(SICI)1096-9896(199806)185:2<188::AID-PATH53>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Adelmant G, Gilbert JD, Freytag SO. Human translocation liposarcoma-CCAAT/enhancer binding protein (C/EBP) homologous protein (TLS-CHOP) oncoprotein prevents adipocyte differentiation by directly interfering with C/EBPbeta function. J Biol Chem. 1998;273:15574–15581. doi: 10.1074/jbc.273.25.15574. [DOI] [PubMed] [Google Scholar]
- Matushansky I, Hernando E, Socci ND, Mills JE, Matos TA, Edgar MA, Singer S, Maki RG, Cordon-Cardo C. Derivation of sarcomas from mesenchymal stem cells via inactivation of the Wnt pathway. J Clin Invest. 2007;117:3248–3257. doi: 10.1172/JCI31377. [DOI] [PMC free article] [PubMed] [Google Scholar]