Abstract
Previous quantitative trait locus (QTL) analysis of an intercross involving the inbred mouse strains NZB/BlNJ and SM/J revealed QTL for a variety of complex traits. Many QTL have large intervals containing hundreds of genes, and methods are needed to rapidly sort through these genes for probable candidates. We chose nine QTL: the three most significant for high-density lipoprotein (HDL) cholesterol, gallstone formation, and obesity. We searched for candidate genes using three different approaches: mRNA microarray gene expression technology to assess >45,000 transcripts, publicly available SNPs to locate genes that are not identical by descent and that contain nonsynonymous coding differences, and a mass-spectrometry-based proteomics technology to interrogate nearly 1000 proteins for differential expression in the liver of the two parental inbred strains. This systematic approach reduced the number of candidate genes within each QTL from hundreds to a manageable list. Each of the three approaches selected candidates that the other two approaches missed. For example, candidate genes such as Apoa2 and Acads had differential protein levels although the mRNA levels were similar. We conclude that all three approaches are important and that focusing on a single approach such as mRNA expression may fail to identify a QTL gene.
THE use of animal models and quantitative trait locus (QTL) mapping has greatly aided the identification of genes that affect complex traits (Korstanje and Paigen 2002). However, identifying the causal gene within a QTL remains a nontrivial matter. Whole-transcriptome profiling has advanced the gene discovery process by identifying causal genes on the basis of differential mRNA levels (Aitman et al. 1999; Johannesson et al. 2005). Nonetheless, a QTL may be caused by differences that are not identifiable at the mRNA level, such as protein concentration (Doolittle et al. 1990) or protein function caused by changes in the amino acid sequence (Theuns et al. 2000).
In this study we combine mRNA expression with proteomics and SNP analysis to show that using a global mRNA approach alone is insufficient for identifying QTL candidate genes. Proteomic technologies are still in their infancy relative to mRNA gene expression technologies; nonetheless the ability to interrogate the proteome is improving (Pilch and Mann 2006), and, as we report here, valuable data can be gathered and applied to the complex disease gene discovery process. In addition, mouse strains have been extensively genotyped; indeed, many of the SNPs between strains are known and are publicly available. SNP data can be used to focus on genes in genomic regions that are not identical by descent (IBD) (Peters et al. 2007) and to identify amino acid changes in the protein-coding region. This allows the selection of candidates that may affect the QTL through functional changes rather than quantitative differences in mRNA and protein levels.
Typically, large-scale protein interrogation studies involve either two-dimensional (2D) gel electrophoresis followed by mass spectrometry (MS) or large-scale immunoblotting (BD Biosciences, San Jose, CA). However, these technologies have limitations. The proteins that can be interrogated on 2D gels are limited in size and nature and require considerable effort to identify. Immunoblotting is limited by the availability and cost of antibodies. For the identification of larger-scale complex proteomes, a technology for proteome analysis by MS has emerged (Aebersold and Mann 2003). This method can be extended by coupling liquid chromatography with matrix-assisted laser-induced ionization–tandem time of flight (MALDI–TOF/TOF) MS. Relative and absolute quantitation of proteins by MS is possible with either labeled (Ross et al. 2004; Zhen et al. 2004) or label-free (Ono et al. 2006) technologies.
The maturation of both microarray and proteomic technologies has led to a small number of whole-transcriptome–proteome comparisons (Gygi et al. 1999; Valenzuela et al. 2003; McRedmond et al. 2004; Ribeiro et al. 2004; Ruse et al. 2004; Mijalski et al. 2005; Pastorelli et al. 2006). However, limited quantitative analysis of protein and transcript abundance could be performed as the starting point of these proteomics studies was 2D electrophoresis followed by single protein-by-protein identification using MS. In this study, to complement the microarray data, we employed a quantitative proteomics approach using isobaric tags for relative and absolute quantitation (iTRAQ reagents) in combination with liquid chromatography–MALDI–TOF/TOF technology for extensive protein analysis and quantitation of two inbred mouse strains. We were able to directly identify and simultaneously quantify a total of 943 liver proteins in a gel-free MS analysis. To our knowledge this is the first attempt to correlate differentially regulated proteins with QTL to identify candidate genes for complex quantitative phenotypes.
A systematic search for candidate genes must also include a search for nonsynonymous coding changes. This is possible for 16 inbred mouse strains with the combination of the public sequencing of C57BL/6, the sequencing of A, DBA/2 and 129 by Celera (Mural et al. 2002), and the resequencing of 15 strains by Perlegen (http://mouse.perlegen.com/mouse/). Combining these databases will identify a large proportion of the coding differences. The mouse strains “Small” (SM)/J (SM) and “New Zealand Black” (NZB)/B1NJ (NZB) are currently not on the list of such strains; however, the SNP data can be used to examine the haplotypes, and coding differences between SM and NZB can be inferred since mouse inbred strains share common ancestry (Wade et al. 2002). The SNP data identified many candidate genes that had neither mRNA nor protein abundance differences, yet differed at SNPs in the coding region and hence might affect protein function.
The mouse strains SM (Chai 1956) and NZB (Bielschowsky and Goodall 1970) differ in a number of metabolic phenotypes (http://www.jax.org/phenome) (Bogue et al. 2007). These were used in a large intercross to yield QTL for a number of complex traits including lipid levels (Korstanje et al. 2004), body composition and size (Stylianou et al. 2006), and gallstone susceptibility (Lyons et al. 2005). In this study we evaluate genes within nine QTL for these traits, using all three approaches: mRNA levels, protein levels, and amino acid-changing SNPs in the coding region. The number of candidate genes in each QTL was reduced from hundreds down to a list ranging from 10 to 88 genes depending on the QTL. Some genes were identified by more than one method, but each approach yielded unique genes, showing that it is the combination of methods that provides the best list of candidate genes for further testing.
MATERIALS AND METHODS
Mice, crosses, and tissue collection:
SM and NZB inbred mice were obtained from The Jackson Laboratory, Bar Harbor, Maine. Further details relating to the NZB × SM F2 cross and husbandry conditions have been published (Stylianou et al. 2006). All the QTL identified between SM and NZB are present in both sexes except for two QTL that are female specific (supplemental Table S1); thus only females were interrogated on the microarray and proteomic platforms.
Liver samples from the large lobe of the liver of three female mice for each strain, NZB and SM, were dissected under conditions suitable for RNA. Following a 4-hr morning fast, samples were collected and flash frozen in liquid nitrogen. Two liver samples from each mouse were taken and stored separately to be used for gene and protein analysis. Microarray experiments were performed using all three samples of each strain, but proteomic analysis was performed on two samples of each strain. Additional mice of each sex from both strains were dissected under the same conditions for Western blot analysis. All experiments were performed on 8-week-old mice maintained on a standard chow diet and procedures were approved by The Jackson Laboratory's Animal Care and Use Committee.
SNP and haplotype analysis:
The 138,000 SNPs (∼1 SNP/24 kb) from the Broad Institute database were updated to NCBI Build 36 and used to compare the NZB and SM strains over the nine QTL regions as defined by the 95% confidence interval (C.I.). Where the strains shared at least three common adjacent SNPs, we inferred that the region might be in a haplotype block and be identical by descent. The SNP feature of the Mouse Phenome database was used to search these same QTL regions for all genes carrying a coding-region difference that resulted in an amino acid change. These genes were then compared to the haplotype data, and those genes in regions identical between NZB and SM were eliminated from the list. Examination of the candidate genes for association to their respective traits in published literature was performed as previously described (Stylianou et al. 2006).
Microarrays:
RNA was extracted from frozen samples using TRIzol as previously described (Stylianou et al. 2005), and the quality of the RNA was assessed using a 2100 Bioanalyzer instrument and RNA 6000 Nano LabChip assay (Agilent Technologies, Palo Alto, CA). Following reverse transcription with an oligo(dT)-T7 primer (Affymetrix, Santa Clara, CA), double-stranded cDNA was synthesized with the Superscript double-stranded cDNA synthesis custom kit (Invitrogen, San Diego). In an in vitro transcription (IVT) reaction with T7 RNA polymerase, the cDNA was linearly amplified and labeled with biotinylated nucleotides (Enzo Diagnostics, Farmingdale, NY). Fifteen micrograms of biotin-labeled and fragmented cRNA was then hybridized onto Mouse Genome 430 2.0 GeneChip arrays (Affymetrix) for 16 hr at 45°. Posthybridization staining and washing were performed according to manufacturer's protocols using the Fluidics Station 450 instrument (Affymetrix). Finally, the arrays were scanned with a GeneChip Scanner 3000 laser confocal slide scanner. The data have been deposited in the Gene Expression Omnibus database (series reference GSE4765).
Array images were quantified using GeneChip Operating Software (GCOS) v1.2. Probe level data were imported into the R software environment and expression values were summarized using the Robust MultiChip Average (RMA) function (Irizarry et al. 2003a) in the R/affy package in Bioconductor (version 1.3.25; R. A. Irizarry, L. Gautier, and B. M. Bolstad). Using the R/maanova package (Wu et al. 2003), an analysis of variance (ANOVA) model was applied to the data. The model
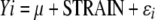 |
(1) |
was used to fit the log-transformed gene expression measures Yi, where μ is the mean for each array, STRAIN is the strain effect, and ɛi captures random error. Strain differences were tested using Fs, a modified F-statistic incorporating shrinkage estimates of variance components (Cui et al. 2005). The proportion of false positives generated was estimated using a false discovery rate (FDR) adjustment described previously (Storey and Tibshirani 2003). Differences between strains were determined at the level FDR < 0.01, which corresponded to permutation P-values <2.6 × 10−3.
TaqMan assays:
mRNA differences of a number of genes that were identified by microarrays were confirmed by TaqMan quantitative real-time PCR assays. Briefly, total RNA (1 μg) was reverse transcribed using the High-Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) according to manufacturer's protocol. A portion of the cDNA was then used in a PCR reaction containing TaqMan Universal PCR Master Mix (Applied Biosystems), which includes AmpliTaq Gold DNA Polymerase, AmpErase UNG, dNTPs with dUTP, Passive Reference 1, and other buffer components. The gene-specific primers and probe sets were obtained from the Applied Biosystems Assay on Demand service and used according to manufacturer's protocols. Real-time PCR was performed in an ABI PRISM 7900HT sequence detection system (Applied Biosystems) with the standard protocol of 95° for 10 min to activate the DNA polymerase followed by 40 cycles of amplification. The threshold cycle (Ct) was determined using the sequence detection system software (SDS2.2) and relative fold change ratios were calculated.
Mass spectrometry:
Two samples for each strain were prepared in parallel. Liver lobes were diced with a sterile scalpel and placed into extraction buffer containing 500 mm triethyl ammonium bicarbonate, 6 m GuHCl, 0.1% Triton X-100, pH 8.5. DNA and tissue were sheared by using a 3-cc syringe fitted with a 20-gauge needle and then sonicated for 20 min. Protein was separated from the buffer by the addition of 6× vol of ice-cold acetone. Samples were then incubated at −20° for 2 hr and then briefly spun before removing the acetone. The protein was solubilized in iTRAQ reagent dissolution buffer with SDS. Protein determination was done using bicinchoninic acid (BCA) (Pierce, Rockford, IL). A total of 100 μg of each sample were labeled with iTRAQ reagent. Reduction and cysteine blocking were done according to standard protocol. Trypsin digestion was done in two steps: half of the recommended amount of trypsin was added to each sample and incubated for 2 hr at 37° and then the second half was added and left overnight at 37°.
Labeling:
The iTRAQ reagents were added to each of the samples as follows: SM no. 1 with 114 (S1), SM no. 2 with 115 (S2), NZB no. 1 with 116 (N1), and NZB no. 2 with 117 (N2). Labeling was performed according to the standard protocol. After labeling the samples were pooled and dried on a Speed-Vac.
HPLC:
iTRAQ reagent-labeled samples were diluted in cation exchange loading buffer to reduce the salt concentration. The pH was adjusted to 3.0 with 1 N phosphoric acid. The sample was loaded onto a poly LC polysulfoethyl A 4.5 × 100-mm, 5-μm, 200-Å column using the Agilent 1100. UV at Å214 was measured. Samples were loaded at 0.5 ml/min and the gradient was run at 1.0 ml/min. The gradient conditions were 0% buffer B (10 mm sodium phosphate, 25% acetonitrile, 500 mm KCl, pH 3.0) to 10% buffer B for 2 min, 20% buffer B to 45% for 2 min, and 45% buffer B to 100% for 5 min. One-milliliter fractions were collected and dried to completeness on Speed-Vac. Fractions were then subjected to reverse phase chromatography, combined with MALDI-matrix, and spotted on a MALDI target plate. MS and MS/MS spectra were acquired using a 4800 Proteomics Analyzer (ABI) MALDI–TOF/TOF mass spectrometer.
Protein mass spectroscopy experiments were performed in duplicate and converted to ratios between samples (i.e., SM2:SM1, NZB1:SM1, NZB2:SM1). Protein measures (analogous to the transcript measures above) were derived from these ratios by first arbitrarily setting the SM1 value equal to one, calculating the corresponding values from the ratio relationships, log transforming each value (SM1, SM2, NZB1, NZB2), and recentering the data by subtracting the column means. The statistical tests for differences in protein levels between strains were performed in the same manner as for transcriptional differences except that the significance level (determined by permutation) was set at P < 0.1. No significant differences would have been found using an FDR as for mRNA due to the limitations of the method (only two biological replicates).
Western analysis for liver proteins:
Western analysis was performed to confirm differentially abundant candidate proteins identified from the MS analysis. Frozen livers were crushed on a stainless steel block while on dry ice and transferred to a glass homogenizer with a Teflon pestle containing lysis buffer (50 mm Tris pH 8.0, 150 mm NaCl, 1 mm EDTA, 1% igepal CA-630 detergent, 1 mm sodium fluoride, 1 mm sodium vanadate, and 1× Sigma protease inhibitor cocktail P2714). Following centrifugation at 16,000 × g for 15 min, the protein concentrations of the supernatants were evaluated using a BCA assay (Pierce). Electrophoresis of 10 μg of each lysate was performed using 4–12% Bis–Tris SDS–PAGE gels with MOPS buffer (Invitrogen). Proteins were transferred to 0.45 μm polyvinylidene fluoride membranes (Invitrogen), using Invitrogen's transfer buffer containing 10% methanol for 1 hr at 30 V and 25°. Membranes were blocked for 1 hr in Tris-buffered saline Tween-20 with 10% skim milk.
Acyl-coenzyme A dehydrogenase, short chain:
Rabbit anti-acyl-coenzyme A dehydrogenase, short chain (ACADS) serum (kindly provided by G. Vockley's lab from the Children's Hospital of Pittsburgh) was used at the dilution of 1:1000 with 5% skim milk for the primary immunostain. Goat anti-rabbit IgG horseradish peroxidase (HRP) (0.4 mg/ml) was used at 1:50,000 for the secondary immunostain (Pierce).
NADH dehydrogenase (ubiquinone) 1 α-subcomplex 9:
Mouse monoclonal against purified mitochondrial complex I of cow (200 μg/ml) (Santa Cruz Biotechnology, Santa Cruz, CA) was used at the dilution of 1:500 with 5% skim milk for the primary immunostain. Goat anti-mouse IgG HRP (10 μg/ml) was used at 1:1000 for the secondary stain (Pierce).
Fumarate hydratase:
Goat polyclonal IgG (Santa Cruz Biotechnology) was used at the dilution of 1:200 with 5% skim milk for the primary immunostain. Donkey anti-goat IgG H + L HRP (1 mg/ml) was used at 1:5000 for the secondary stain (Novus Biologicals, Littleton, CO).
Hydroxysteroid 11-β dehydrogenase 1:
Rabbit polyclonal antibody (Abcam, Cambridge, MA) to human sequence was used at 1:300 for the primary immunostain. Goat anti-rabbit IgG (H + L) HRP (0.4 mg/ml) was used at 1:50,000 dilution for the secondary stain. For a loading control, we used rabbit anti-β-tubulin conjugated with HRP at a 1:200 dilution of 200 mg/ml (Santa Cruz Biotechnology).
For each blot, 3 samples each from a different mouse were run for both strains (SM and NZB) and both sexes, resulting in a total of 12 samples with an N = 3 within-sex–strain comparison and an N = 6 within-strain comparison. The immunoblots were immersed in Pierce's SuperSignal West Dura HRP substrate and imaged using a FujiLas CCD camera. Quantitation was performed using ImageJ (Abramoff et al. 2004).
RESULTS
QTL analysis:
Previous analysis of the SM × NZB intercross population identified 38 main-effect QTL for the phenotypes of high-density lipoprotein (HDL) cholesterol (Korstanje et al. 2004), gallstone susceptibility (Lyons et al. 2005), body weight, adiposity, and obesity (Stylianou et al. 2006), including 2 HDL QTL affected by sex (supplemental Table S1). In this report we focused on the three most significant QTL for each of these traits: Hdlq20, -1, and -24 on chromosomes (Chrs) 1, 5, and 6, respectively for HDL; Lith17, -19, and -21 on Chrs 5, 8, and 10, respectively, for gallstones; and Obwq3, -4, and -5 on Chrs 6, 17, and 19, respectively, for obesity. We reanalyzed the QTL data from the original publications with updated marker positions based on mouse genome Build 36 (Table 1). To obtain the 95% confidence intervals, we used posterior probability distribution as described previously (Sen and Churchill 2001).
TABLE 1.
Selected QTL from the SM × NZB F2 cross for three complex traits
| Trait | QTL | Chr | LOD | Nearest marker | High allele | Mode of inheritance | Mb peak (C.I. start–end) |
|---|---|---|---|---|---|---|---|
| HDL | Hdlq20 | 1 | 10.6 | D1Mit291 | NZB | Dom | 174 (168–184) |
| Hdlq1 | 5 | 10.6 | D5Mit161 | NZB | Add | 122 (116–132) | |
| Hdlq24 | 6 | 6.4 | D6Mit259 | NZB | Add | 128 (115–144) | |
| Gallstone presence | Lith17 | 5 | 7.1 | D5Mit24 | SM | Add | 102 (88–122) |
| Lith19 | 8 | 5.6 | D8Mit155 | SM | Rec | 6 (0–16) | |
| Lith21 | 10 | 5.7 | D10Mit214 | NZB | Rec | 48 (18–58) | |
| QTL for obesity and body weight | Obwq4 | 17 | 6.8 | D17Mit20 | NZB | Add | 54 (34–62) |
| Obwq5 | 19 | 9.3 | D19Mit71 | NZB | Rec | 56 (48–60) | |
| Obwq3 | 6 | 5.8 | D6Mit105 | SM | Rec | 100 (88–122) |
LOD scores are given with sex as an additive covariate except for the body weight and obesity phenotypes. Mb indicates millions of base pairs from the centromere (Build 36). Dom, dominant; Add, additive; Rec, recessive; C.I., 95% confidence interval.
Use of SNP data to reduce the QTL interval and identify genes with coding changes:
QTL regions were reduced by eliminating regions within the 95% C.I. that appeared to be IBD between the two parental strains; these regions are unlikely to contain the causal polymorphisms unless the mutation arose in the last 100 years since the strains were inbred. It is estimated that 97% of mutations are ancestral (Wade et al. 2002). Thus, 3% of mutations, and by inference 3% of QTL, may be due to a recent mutation unique to a single strain. If a mutation is recent, then the QTL is unlikely to be found except in a cross with that particular strain; thus, if the same QTL is found in more than one intercross, the mutation is more likely to be ancestral. For all QTL in this report except one (Lith19 on Chr 8), the QTL have been reported in other crosses, thus increasing greatly the probability that the causal mutation is ancestral and shared among strains. We inferred the IBD regions by directly examining the common polymorphisms between the parental strains, using 130,000 SNPs for SM and NZB from the Broad Institute (http://www.broad.mit.edu/mouse/snp_xls.html). These data were used to exclude large regions within each QTL and reduced the QTL genomic space by between 40 and 72%.
To identify genes that differed within the coding region of the gene, we utilized all the Perlegen SNP data in combination with all SNPs for SM and NZB available through the Mouse Phenome database (http://www.jax.org/phenome) (Bogue et al. 2007). We inferred coding differences between SM and NZB for the nine QTL (Table 2). This resulted in a list of genes that may be causal for each QTL due to coding and hence functional differences (supplemental Tables S2–S10).
TABLE 2.
Inferring codon changing SNPs between SM and NZB by examining SNPs between highly sequenced strains
| Highly sequenced strains, e.g., C57BL/6, DBA/2, etc.
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chr 6 gene | SNP | i | ii | iii | iv | v | vi | SM | NZB | SM and NZB are |
| Syn2 | UTR | A | A | A | G | G | G | G | G | IBD |
| Syn2 | Cs | T | T | T | C | C | C | |||
| Syn2 | Cs | G | G | G | A | A | A | A | G | Non-IBD |
| C | C | C | T | T | T | |||||
| Pparg | UTR | A | A | A | G | G | G | |||
| Pparg | Cn | T | T | T | C | C | C | c | t | Inferred Cn between SM and NZB |
| Pparg | Cs | G | G | G | A | A | A | |||
| Pparg | Cs | C | C | C | T | T | T | |||
| Pparg | Cs | A | A | A | G | G | G | G | A | Non-IBD |
| T | T | T | C | C | C | |||||
| G | G | G | A | A | A | |||||
| Tsen2 | UTR | C | C | C | T | T | T | |||
| Tsen2 | Cs | C | C | C | G | G | G | C | G | Non-IBD |
| Tsen2 | Cs | T | T | T | C | C | C | |||
| Tsen2 | Cs | G | G | G | A | A | A | |||
| Tsen2 | Cs | C | C | C | T | T | T | T | T | IBD |
Italic nucleotides indicate an alternative haplotype present in common inbred strains that can be used to infer the origin of polymorphisms from less well characterized strains. Although SM and NZB are not as well sequenced as other strains, missing SNPs between SM and NZB can be inferred by examining the haplotypes of the other strains. For the Cn in Pparg (underlined), SM is inferred to be “C” and NZB is inferred to be “T.” Cs, coding synonymous; Cn, coding nonsynonymous; IBD, identical by descent.
Transcriptome analysis:
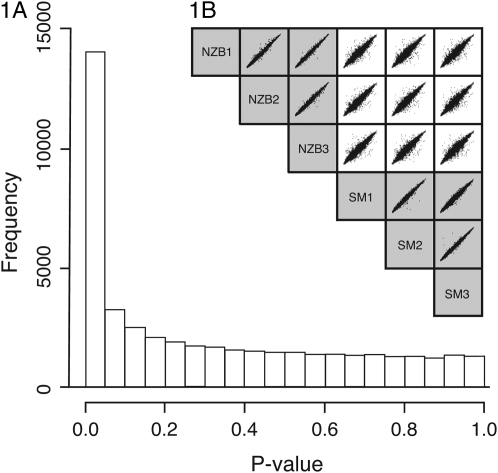
Liver mRNA profiles were compared between three SM and three NZB female mice, using the Affymetrix Mouse Genome 430 2.0 oligonucleotide GeneChip arrays. Statistical diagnostic analysis indicates that the array data generated are highly replicable and hence robust. Genes that are not differentially expressed between the two strains have a characteristically even distribution of nonsignificant P-values (Figure 1A). Scatter plots of mRNA gene expression measures from the six arrays derived from robust multichip analysis (RMA) (Irizarry et al. 2003b) show that within-strain comparisons are tightly distributed and between-strain distributions show differential expression (Figure 1B). To validate the microarray results, we performed real-time PCR using TaqMan assays on 10 differentially expressed genes that had conservative absolute fold changes of <4.0 and FDRs <1 × 10−4. TaqMan assays were consistent with microarray data for all 10 genes; that is, all genes were differentially expressed in the expected direction and the differences were significant after correcting for multiple testing (supplemental Table S11). Using the stringent FDR of <1 × 10−4, we identified 831 differentially expressed (DE) probe sets.
Figure 1.—
Gene expression (mRNA) microarray diagnostics. (A) P-value distribution histogram. F-tests were performed in R/MAANOVA and the number of differentially expressed probes with a false discovery rate (FDR) of q < 0.01 = 6761 and with an FDR q < 0.0001 = 831. (B) Scatter plots of RMA measures. The shaded areas indicate within-strain comparisons while the open areas show between-strain comparisons of the six female liver samples arrayed.
Candidates based on differences in mRNA abundance:
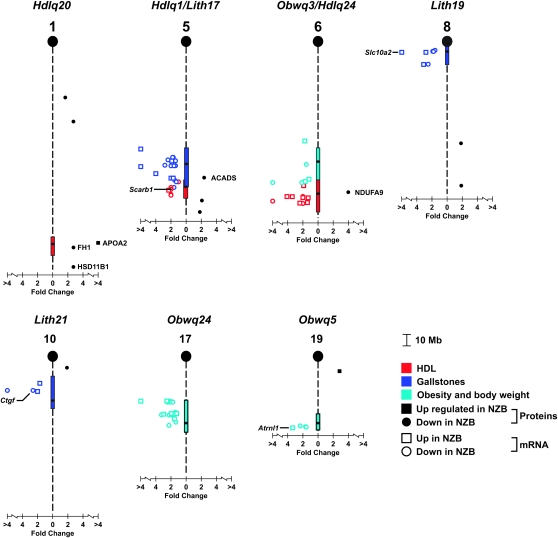
A number of differentially expressed genes were identified from the microarray analysis (Figure 2, supplemental Tables S2–S10 and S12). Several of these are of particular interest because there is evidence that they are involved with the target traits (Table 3). The cholesterol scavenger receptor class B member 1 (Scarb1) is a known receptor for HDL (Acton et al. 1996; Trigatti et al. 2003) and therefore an obvious candidate for Hdlq1 on Chr 5; adiponectin receptor 2 (Adipor2) is a functional candidate for Hdlq24 and Obwq3 on Chr 6. For gallstone QTL, the connective tissue growth factor (Ctgf), shown to have a 40-fold mRNA differential expression in the gallbladders of human cholecystolithiasis patients compared to controls (Koninger et al. 2005), is a candidate for Lith21 on Chr 10; also Slc10a2 on Chr 8 is essential for efficient intestinal absorption of bile acids and therefore may be responsible for the Lith19 QTL (Dawson et al. 2003). Finally, for the obesity QTL on Chr 19, attractin-like 1 (Atrnl1) has a 3.4-fold upregulation in NZB relative to SM. Attractin is part of the agouti pathway that leads to obesity due to aberrant ectopic protein expression (He et al. 2001), and Atrnl1 may similarly be involved in this pathway.
Figure 2.—
Summary of the nine metabolic QTL with locations of differentially regulated genes and proteins. Ninety-five percent confidence intervals and peaks of QTL are given by the vertical bars. All significantly differentially expressed proteins are presented (P < 0.1); however, for clarity only differentially expressed genes within the QTL intervals are presented.
TABLE 3.
Candidate genes for each QTL based on our data and published evidence—the genes to test first
| Evidence
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Trait | QTL | Gene | Chr | Mb | mRNAa | Proteinb | Sequencec | Literatured |
| HDL | Hdlq20 | Apoa2 | 1 | 173.1 | 4.1 | X | Lilja et al. (2002) | |
| Fh1 | 177.5 | −2.8 | X | |||||
| Hsd11b1 | 194.9 | −3.8 | Morton et al. (2001) | |||||
| Hdlq1 | Acads | 5 | 115.3 | −2.6 | X | |||
| Scarb1 | 125.6 | −2.1 | Acton et al. (1996) | |||||
| Hdlq24 | Pparg | 6 | 115.4 | X | Chinetti et al. (2001) | |||
| Rassf4 | 116.6 | −8.2 | ||||||
| Adipor2 | 119.3 | 1.9 | X | Yamauchi et al. (2003) | ||||
| Ndufa9 | 126.8 | −2.6 | X | |||||
| Gallstone presence | Lith17 | Scarb2 | 5 | 93.6 | X | Acton et al. (1996) | ||
| Acads | 115.3 | −2.6 | X | |||||
| Lith19 | Slc10a2 | 8 | 5.1 | 5.2 | Shih et al. (2001) | |||
| Lith21 | Ctgf | 10 | 24.3 | −2.6 | X | Koninger et al. (2005) | ||
| Obesity and body weight | Obwq3 | Pparg | 6 | 115.4 | X | Cock et al. (2004) | ||
| Rassf4 | 116.6 | −8.2 | ||||||
| Adipor2 | 119.3 | 1.9 | X | Yamauchi et al. (2003) | ||||
| Obwq4 | Pgc | 17 | 47.2 | X | Puigserver et al. (1998) | |||
| Obwq5 | Atrnl1 | 19 | 57.7 | 3.4 | Gunn et al. (1999) | |||
Genes listed are in QTL regions that are not identical by descent between SM and NZB (Hsd11b1 maps close to Hldq20). Protein and mRNA fold changes are given where a positive value indicates upregulation and a negative one indicates downregulation in NZB relative to SM.
Fold changes for mRNA are significant (Fs < 1 × 10−4).
Proteins are significantly different as determined by Western analysis (Figure 3), except for APOA2, which is suggestive (P < 0.1) on the basis of MS/MS data.
Genes that contain known or extrapolated codon-changing SNPs leading to potential functional differences are indicated by “X.”
Gene names from supplemental Table S2 were systematically interrogated against the PubMed database using relevant keywords (Stylianou et al. 2006).
Mass-spectrometric proteomic analysis:
We examined liver protein expression differences between the strains using mass spectrometry with iTRAQ reagent chemistry labeling technology (Ross et al. 2004). Samples were used from two of the three biological replicates used for the microarray study. iTRAQ labels are essentially chemical compounds with two parts. The first part, retained by the peptide after scattering, is available at four different masses of 114, 115, 116, or 117 Da. The second part of the label is not retained; this part of the molecule is designed to balance the mass of the four labels. Thus all four labels are isobaric, so that observed differences are not due to the differential mass of the labels. Up to four samples can be labeled so that within a sample every peptide is equally tagged with an additional mass of 114, the peptides of the second sample can be tagged with a mass of 115, and so on. We identified 943 proteins with 95% confidence: ∼7% were ribosomal and 43% represented novel or uncharacterized gene products.
These data are presented as exploratory; the statistical power is not optimal since only a two-by-two comparison is possible on this proteomics platform. Consequently we are using a statistical threshold of P < 0.1, derived from permutation analysis, which we are using to indicate peptides that might be significantly differentially expressed under more reproducible experimental conditions such as Western analysis. Rather than reduce the statistical threshold for this technology and eliminate possible candidates, the higher threshold exposes us to type I errors, which are more acceptable to us than type II errors, that is, to eliminate candidate genes that are responsible for the QTL. For example, apolipoprotein A-II precursor (APOA2), which has a P-value of 0.077 in our analysis, has been shown previously to be differentially abundant with no mRNA difference (Doolittle et al. 1990) and is responsible for the HDL QTL linked to it (Wang et al. 2004).
We identified 45 proteins as differentially regulated between the two strains (P < 0.1) (supplemental Table S13); 33 of these 45 could be linked to corresponding genes. The remaining 12 peptides included 8 peptides that could not be mapped and 4 problem peptides that did not map to a unique location because they were in highly conserved protein families.
Candidates based on differences in protein abundance:
Only a few of the 33 differentially expressed proteins mapped to a QTL (Figure 2). For the HDL QTL, APOA2 (P = 0.077) and fumarate hydratase (FH1) (P = 0.073) are encoded by genes that fall within the Hdlq20 QTL interval on Chr 1. Additionally, hydroxysteroid 11-β dehydrogenase 1 (HSD11B1) maps close to this QTL and knockout mice for this gene have increased HDL cholesterol levels (Morton et al. 2001).
The gene that encodes ACADS (previously known as SCAD) maps within Hdlq1 on Chr 5 (Table 1), and NADH dehydrogenase (ubiquinone) 1 α-subcomplex 9 (NDUFA9) is encoded by a gene within the Hdlq24 QTL on Chr 6. The gallstone QTL Lith17 on Chr 5 overlaps with the HDL QTL Hdlq1, and thus ACADS is also a candidate for this gallstone QTL.
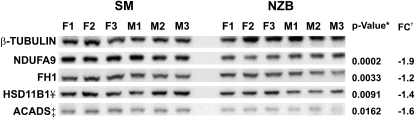
To validate some of the candidates identified from the iTRAQ ABI 4800 Proteomics Analyzer, antibodies suitable for Western analysis were obtained for several proteins. These include FH1 and HSD11B1 on Chr 1, ACADS on Chr 5, and NDUFA9 on Chr 6. Western analysis confirmed the initial results from MS (Figure 3). Significant differences were observed for all four proteins after normalizing to β-tubulin: P = 0.0033 (NDUFA9), P = 0.0002 (FH1), P = 0.0091 (HSD11B1), and P = 0.016 (ACADS).
Figure 3.—
Western blot analysis of ACADS, FH1, NDUFA9, HSD11B1, and control β-tubulin. F and M indicate female and male, and each sex or strain is represented by three biological replicates. As there was no significant difference between sexes for any of the proteins, significance (P-value*) was determined by a t-test of the ratios of the target proteins to β-tubulin between strains (N = 6). †, FC indicates fold change relative to NZB. ¥, an additional NZB male-specific HSD11B1 isoform is apparent at ∼55–60 kDa (supplemental Figure S1); however, females do not have this isoform and consequently NZB females are still significantly decreased compared to SM. ‡, ACADS was probed to a second blot with an identical setup and normalized to β-tubulin probed to the second blot.
DISCUSSION
Often QTL are large, containing hundreds of genes, so different tools and approaches are needed to reduce the number of probable candidate genes that require further testing. Combining three approaches, we generated smaller candidate gene lists for each QTL, and the overall summary for candidate genes identified is given in Table 4. This was achieved by searching for differential amounts of either mRNA or protein abundance and by mining publicly available SNPs to exclude regions of the QTL that are inferred to be identical by descent and to identify genes with protein-coding differences that may indicate functional differences. Each of these three approaches has its limitations and strengths as discussed below.
TABLE 4.
Summary of candidate genes identified by the various methods used
| Method | Significance level | Condition | No. of genes/ proteins | Total no. of genes/ proteinsa |
|---|---|---|---|---|
| Microarray | FDR = 0.05 | Over all chromosomes | 11,223 | 24,530b |
| Microarray | FDR = 0.0001 | Over all chromosomes | 831 | 24,530b |
| Microarray | FDR = 0.0001 | In QTL | 87 | 3,763c |
| Proteomics | P < 0.1 | Over all chromosomes | 45 | 943 |
| Proteomics | P < 0.1 | In QTL region | 5 | Unknownd |
| SNP analysis | NA | In QTL and non-IBD region | 336 | 2,752 |
Gene here also refers to an Affymetrix probe for the microarrays and a peptide linked to a protein entry in the Swiss-Prot or TrEMBL databases for the proteomics.
There are 45,037 noncontrol probe sets on the arrays; however, 20,507 are not expressed in any of the six samples, indicating that genes for those probes are not expressed in adult female mouse liver.
The total number of probes within the total space of the nine QTL including IBD regions between SM and NZB.
Many protein hits could not be mapped to genomic locations so the genes located only in QTL regions cannot be determined.
If these three approaches have not reduced the possible candidate genes to a manageable list, which we consider ≤10 genes, then other approaches are available, some of which have been discussed previously (Dipetrillo et al. 2005; Flint et al. 2005). For example, comparative genomics is a useful approach both for narrowing the QTL if a homologous QTL has been found in other species and for examining the genes found in a human genomewide association study in the mouse. Another approach is to examine the expression databases to determine if the candidate gene is expressed in the relevant tissues. Following this bioinformatics work or global expression studies, we test the small list of candidates by sequencing and by expression analysis. Our goal is to obtain at least three independent lines of evidence for a candidate gene.
mRNA expression data:
Today microarray technology yields reliable, consistent, and reproducible data from probe sets that are well annotated. As with the platform used here (Affymetrix GeneChip Mouse Genome 430 2.0), most technologies report mRNA expression for many thousands of genes, approaching the entire known transcriptome. It is likely, therefore, that if a differentially expressed gene in the liver is causal for any of the nine QTL, we should be able to detect it. Indeed, when we examine the differentially expressed probes in the QTL intervals located in haplotype regions that are not identical by descent between our two strains, we find several genes that are obvious candidates. These include Scarb1 for Hdlq1 on Chr 5, Ctgf for Lith21 on Chr 10, Slc10a2 for Lith19 on Chr 8, and Atrnl1 for Obwq5 on Chr 19 (Figure 3 and Table 3).
Although searching for differential gene expression using microarrays is probably the most advanced technology of the three methods, it has several limitations. Suitable probes for a gene may not be on the microarray and splice variants are difficult to detect. The differential expression could be caused by differential binding of the mRNA due to SNPs in the probe. This can be guarded against by searching the databases for SNPs in the probes and by confirming any expression difference with new probes and RT–PCR. Trans-regulatory elements outside of the QTL may cause differential expression of a gene in the QTL region; similarly, gene interaction or epistasis could cause the parental strains to have similar gene expression even though differential expression was causal for the QTL in an inbred cross. Both errors can be guarded against by measuring gene expression in tissues from selected F2 progeny (comparing mice homozygous for one parent over the QTL region to mice homozygous for the other parent over the same region).
Protein analysis:
The primary aim of microarrays is to identify differentially expressed genes that are representing differentially abundant proteins. Large-scale proteomic techniques have only recently become feasible; however, they are technically and financially restrictive relative to mRNA arrays. Nonetheless, as proteomic technologies improve they may replace microarray gene expression since it will be possible to directly interrogate the presence of differentially abundant proteins. Despite the ability to directly identify 943 proteins, making direct comparisons between mRNA regulation and protein regulation remains difficult, principally because MS/MS proteomics and the accompanying bioinformatics are still at early stages of development. Areas of improvement that will significantly advance the field should include standardized protocols on tissue processing. In this study, we chose a single tissue-processing condition (pH 8.5) that would yield the widest variety of protein peptides from liver; this isolation may miss more basic and more acidic proteins. A more thorough investigation of the liver proteome would include a range of tissue-processing conditions.
Another desirable improvement is peptide annotation, specifically linked annotation between protein/peptides and gene databases. Although we identified 943 proteins, a significant proportion of these (43%) could not be assigned to a gene; thus direct comparisons with Affymetrix probe identifications could not be made. Furthermore, a significant proportion of the 943 proteins, despite a protein identifier, had little or no annotation in public databases and indeed BLAST searches of the peptides to genome databases (Ensembl_mouse) derived no hits at all, even after decoding the peptide into DNA sequence. Finally, standardized analytical methods of MS/MS data are currently lacking and an initiative similar to that seen for mRNA gene analysis is required.
Despite the need for continued technological advancements in the proteomic field, the technology is developed enough to identify changes in protein abundance between samples. ACADS, a protein encoded by a gene in both the HDL QTL Hdlq1 and the gallstone QTL Lith17 on Chr 5, showed significantly increased protein abundance in SM adult female liver relative to the NZB strain (a 2.6- and a 1.6-fold change from MS and Western analysis, respectively), but no difference in mRNA expression (Table 2). The difference in protein abundance was confirmed by Western blot analysis. Similar results were obtained for FH1, HSD11B1, and NDUFA9 (Table 3, Figures 2 and 3). Finally, our MS data indicate a 4-fold protein increase of APOA2 in NZB relative to SM protein abundance with no mRNA expression difference. A difference in the quantity of APOA2 protein but not in mRNA was reported previously between two different strains (Doolittle et al. 1990). We have previously shown that the gene Apoa2 is responsible for the HDL QTL Hdlq5 (Wang et al. 2004) in mice and associated with HDL levels in human studies (Lilja et al. 2002).
The proteomics analysis reveals two important aspects of this study. First, differentially abundant proteins can be identified and may underlie QTL, and second, as others have shown in yeast (Gygi et al. 1999), these differentially expressed proteins are not necessarily accompanied by differences at the mRNA level. Of the 33 named differentially expressed proteins identified with corresponding Affymetrix probe sets, 12 had differentially expressed mRNA in the same direction with an FDR of q < 0.05 (39%) (supplemental Table S13). We expect that differentially abundant proteins whose mRNA levels are similar will have changes at the gene (e.g., 5′- and 3′-UTRs) or the protein level (e.g., phosphorylation) that will affect protein synthesis and degradation. APOA2 is known to have such an amino acid change (Wang et al. 2004).
SNPs and haplotype data:
We used public SNP information to first eliminate those regions that were in the 95% confidence interval of the QTL but were identical by descent between NZB and SM. We then examined the remaining genes by identifying codon-changing SNPs. Once genes with nonsynonymous SNPs were identified between well-characterized strains, they were compared to the haplotypes of SM and NZB, which allowed us to infer if the same nonsynonymous polymorphism might be present between SM and NZB as illustrated in Table 2. This generated a list of genes that may have nonsynonymous changes between the two strains, which may affect the function of the proteins, thus causing the QTL. However, the number of genes identified using this method for some QTL was still prohibitively large for testing (e.g., 88 genes in the Obwq4 QTL). In addition, it has recently been shown that SNPs causing synonymous codon changes, for which the alternate codon is rare, can lead to a difference in protein efficiency or function (Kimchi-Sarfaty et al. 2007).
The method of using SNPs does have several drawbacks that will become less important in the future as SNP databases improve (Reuveni et al. 2007). Using the SNP database to eliminate regions that are identical by descent could lead to error by incorrectly eliminating regions; this type of error will be reduced as the density of the SNPs improves. The SNP database, even for those strains sequenced by Celera and Perlegen, is incomplete because both groups used rigorous criteria to guard against false positives. On the basis of the Perlegen data, ∼50% of the SNPs will be identified if the minor allele is shared by three strains (58% if the minor allele frequency is shared by seven strains) (Yang and Churchill 2007) or a somewhat better percentage if Celera data are included or only classic inbreds (not wild-derived strains) are considered.
Summary:
In spite of the limitations for each method, we were reasonably effective in reducing the number of candidate genes in each QTL region. Some genes had multiple lines of evidence such as Acads for Hdlq1 on Chr 5 and Adipor2 for Obwq3 on Chr 6 (Table 3 and supplemental Table S2) but multiple lines of evidence do not prove that it is the causal gene. In one case we were able to identify a known gene for a QTL (Apoa2); and each method revealed likely candidates that the other methods did not. We conclude that QTL candidate genes should be assessed for changes in coding and both protein and message levels.
Acknowledgments
We give special thanks to Stephen Grubb, Allison Cox, and Sikander Hayat for assistance with bioinformatics; to Douglas Hinerfeld and the Gene Expression and Protein Chemistry Services at The Jackson Laboratory; to G. Vockley's Lab from the Children's Hospital of Pittsburgh for the ACADS antibody; to Applied Biosystems for the kind donation of the TaqMan assays; and to Sarah Langley for assistance in proofreading. This work was supported by The American Heart Association grant 0525816T (to I.M.S.) and by National Institutes of Health grants HL77796 and HL66611 (to B.P.) and GM070683 (to G.C.).
References
- Abramoff, M. D., P. J. Magelhaes and S. J. Ram, 2004. Image processing with ImageJ. Biophotonics Int. 11 36–42. [Google Scholar]
- Acton, S., A. Rigotti, K. T. Landschulz, S. Xu, H. H. Hobbs et al., 1996. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 271 518–520. [DOI] [PubMed] [Google Scholar]
- Aebersold, R., and M. Mann, 2003. Mass spectrometry-based proteomics. Nature 422 198–207. [DOI] [PubMed] [Google Scholar]
- Aitman, T. J., A. M. Glazier, C. A. Wallace, L. D. Cooper, P. J. Norsworthy et al., 1999. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat. Genet. 21 76–83. [DOI] [PubMed] [Google Scholar]
- Bielschowsky, M., and C. M. Goodall, 1970. Origin of inbred NZ mouse strains. Cancer Res. 30 834–836. [PubMed] [Google Scholar]
- Bogue, M. A., S. C. Grubb, T. P. Maddatu and C. J. Bult, 2007. Mouse Phenome Database (MPD). Nucleic Acids Res. 35 D643–D649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, C. K., 1956. Analysis of quantitative inheritance of body size in mice. I. Hybridization and material influence. Genetics 14 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinetti, G., S. Lestavel, V. Bocher, A. T. Remaley, B. Neve et al., 2001. PPAR-alpha and PPAR-gamma activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. Nat. Med. 7 53–58. [DOI] [PubMed] [Google Scholar]
- Cock, T. A., S. M. Houten and J. Auwerx, 2004. Peroxisome proliferator-activated receptor-gamma: too much of a good thing causes harm. EMBO Rep. 5 142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, X., J. T. Hwang, J. Qiu, N. J. Blades and G. A. Churchill, 2005. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics 6 59–75. [DOI] [PubMed] [Google Scholar]
- Dawson, P. A., J. Haywood, A. L. Craddock, M. Wilson, M. Tietjen et al., 2003. Targeted deletion of the ileal bile acid transporter eliminates enterohepatic cycling of bile acids in mice. J. Biol. Chem. 278 33920–33927. [DOI] [PubMed] [Google Scholar]
- DiPetrillo, K., X. Wang, I. M. Stylianou and B. Paigen, 2005. Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet. 21 683–692. [DOI] [PubMed] [Google Scholar]
- Doolittle, M. H., R. C. LeBoeuf, C. H. Warden, L. M. Bee and A. J. Lusis, 1990. A polymorphism affecting apolipoprotein A-II translational efficiency determines high density lipoprotein size and composition. J. Biol. Chem. 265 16380–16388. [PubMed] [Google Scholar]
- Flint, J., W. Valdar, S. Shifman and R. Mott, 2005. Strategies for mapping and cloning quantitative trait genes in rodents. Nat. Rev. Genet. 6 271–286. [DOI] [PubMed] [Google Scholar]
- Gunn, T. M., K. A. Miller, L. He, R. W. Hyman, R. W. Davis et al., 1999. The mouse mahogany locus encodes a transmembrane form of human attractin. Nature 398 152–156. [DOI] [PubMed] [Google Scholar]
- Gygi, S. P., Y. Rochon, B. R. Franza and R. Aebersold, 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19 1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, L., T. M. Gunn, D. M. Bouley, X. Y. Lu, S. J. Watson et al., 2001. A biochemical function for attractin in agouti-induced pigmentation and obesity. Nat. Genet. 27 40–47. [DOI] [PubMed] [Google Scholar]
- Irizarry, R. A., B. M. Bolstad, F. Collin, L. M. Cope, B. Hobbs et al., 2003. a Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis et al., 2003. b Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264. [DOI] [PubMed] [Google Scholar]
- Johannesson, M., L. M. Olsson, A. K. Lindqvist, S. Moller, D. Koczan et al., 2005. Gene expression profiling of arthritis using a QTL chip reveals a complex gene regulation of the Cia5 region in mice. Genes Immun. 6 575–583. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty, C., J. M. Oh, I. W. Kim, Z. E. Sauna, A. M. Calcagno et al., 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315 525–528. [DOI] [PubMed] [Google Scholar]
- Koninger, J., F. F. di Mola, P. Di Sebastiano, A. Gardini, D. R. Brigstock et al., 2005. Transforming growth factor-beta pathway is activated in cholecystolithiasis. Langenbecks Arch. Surg. 390 21–28. [DOI] [PubMed] [Google Scholar]
- Korstanje, R., and B. Paigen, 2002. From QTL to gene: the harvest begins. Nat. Genet. 31 235–236. [DOI] [PubMed] [Google Scholar]
- Korstanje, R., R. Li, T. Howard, P. Kelmenson, J. Marshall et al., 2004. Influence of sex and diet on quantitative trait loci for HDL cholesterol levels in an SM/J by NZB/BlNJ intercross population. J. Lipid Res. 45 881–888. [DOI] [PubMed] [Google Scholar]
- Lilja, H. E., A. Soro, K. Ylitalo, I. Nuotio, J. S. Viikari et al., 2002. A candidate gene study in low HDL-cholesterol families provides evidence for the involvement of the APOA2 gene and the APOA1C3A4 gene cluster. Atherosclerosis 164 103–111. [DOI] [PubMed] [Google Scholar]
- Lyons, M. A., R. Korstanje, R. Li, S. M. Sheehan, K. A. Walsh et al., 2005. Single and interacting QTLs for cholesterol gallstones revealed in an intercross between mouse strains NZB and SM. Mamm. Genome 16 152–163. [DOI] [PubMed] [Google Scholar]
- McRedmond, J. P., S. D. Park, D. F. Reilly, J. A. Coppinger, P. B. Maguire et al., 2004. Integration of proteomics and genomics in platelets: a profile of platelet proteins and platelet-specific genes. Mol. Cell Proteomics 3 133–144. [DOI] [PubMed] [Google Scholar]
- Mijalski, T., A. Harder, T. Halder, M. Kersten, M. Horsch et al., 2005. Identification of coexpressed gene clusters in a comparative analysis of transcriptome and proteome in mouse tissues. Proc. Natl. Acad. Sci. USA 102 8621–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton, N. M., M. C. Holmes, C. Fievet, B. Staels, A. Tailleux et al., 2001. Improved lipid and lipoprotein profile, hepatic insulin sensitivity, and glucose tolerance in 11beta-hydroxysteroid dehydrogenase type 1 null mice. J. Biol. Chem. 276 41293–41300. [DOI] [PubMed] [Google Scholar]
- Mural, R. J., M. D. Adams, E. W. Myers, H. O. Smith, G. L. Miklos et al., 2002. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science 296 1661–1671. [DOI] [PubMed] [Google Scholar]
- Ono, M., M. Shitashige, K. Honda, T. Isobe, H. Kuwabara et al., 2006. Label-free quantitative proteomics using large peptide data sets generated by nanoflow liquid chromatography and mass spectrometry. Mol. Cell Proteomics 5 1338–1347. [DOI] [PubMed] [Google Scholar]
- Pastorelli, R., D. Carpi, R. Campagna, L. Airoldi, R. Pohjanvirta et al., 2006. Differential expression profiling of the hepatic proteome in a rat model of dioxin resistance: correlation with genomic and transcriptomic analyses. Mol. Cell Proteomics 5 882–894. [DOI] [PubMed] [Google Scholar]
- Peters, L. L., R. F. Robledo, C. J. Bult, G. A. Churchill, B. J. Paigen et al., 2007. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat. Rev. Genet. 8 58–69. [DOI] [PubMed] [Google Scholar]
- Pilch, B., and M. Mann, 2006. Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 7 R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver, P., Z. Wu, C. W. Park, R. Graves, M. Wright et al., 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92 829–839. [DOI] [PubMed] [Google Scholar]
- Reuveni, E., V. E. Ramensky and C. Gross, 2007. Mouse SNP Miner: an annotated database of mouse functional single nucleotide polymorphisms. BMC Genomics 8 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro, J. M., R. Charlab, V. M. Pham, M. Garfield and J. G. Valenzuela, 2004. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochem. Mol. Biol. 34 543–563. [DOI] [PubMed] [Google Scholar]
- Ross, P. L., Y. N. Huang, J. N. Marchese, B. Williamson, K. Parker et al., 2004. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics 3 1154–1169. [DOI] [PubMed] [Google Scholar]
- Ruse, C. I., F. L. Tan, M. Kinter and M. Bond, 2004. Integrated analysis of the human cardiac transcriptome, proteome and phosphoproteome. Proteomics 4 1505–1516. [DOI] [PubMed] [Google Scholar]
- Sen, S., and G. A. Churchill, 2001. A statistical framework for quantitative trait mapping. Genetics 159 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, D. Q., M. Bussen, E. Sehayek, M. Ananthanarayanan, B. L. Shneider et al., 2001. Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat. Genet. 27 375–382. [DOI] [PubMed] [Google Scholar]
- Storey, J. D., and R. Tibshirani, 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylianou, I. M., M. Clinton, P. D. Keightley, C. Pritchard, Z. Tymowska-Lalanne et al., 2005. Microarray gene expression analysis of the Fob3b obesity QTL identifies positional candidate gene Sqle and perturbed cholesterol and glycolysis pathways. Physiol. Genomics 20 224–232. [DOI] [PubMed] [Google Scholar]
- Stylianou, I. M., R. Korstanje, R. Li, S. Sheehan, B. Paigen et al., 2006. Quantitative trait locus analysis for obesity reveals multiple networks of interacting loci. Mamm. Genome 17 22–36. [DOI] [PubMed] [Google Scholar]
- Theuns, J., J. Del-Favero, B. Dermaut, C. M. van Duijn, H. Backhovens et al., 2000. Genetic variability in the regulatory region of presenilin 1 associated with risk for Alzheimer's disease and variable expression. Hum. Mol. Genet. 9 325–331. [DOI] [PubMed] [Google Scholar]
- Trigatti, B. L., M. Krieger and A. Rigotti, 2003. Influence of the HDL receptor SR-BI on lipoprotein metabolism and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 23 1732–1738. [DOI] [PubMed] [Google Scholar]
- Valenzuela, J. G., I. M. Francischetti, V. M. Pham, M. K. Garfield and J. M. Ribeiro, 2003. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem. Mol. Biol. 33 717–732. [DOI] [PubMed] [Google Scholar]
- Wade, C. M., E. J. Kulbokas, 3rd, A. W. Kirby, M. C. Zody, J. C. Mullikin et al., 2002. The mosaic structure of variation in the laboratory mouse genome. Nature 420 574–578. [DOI] [PubMed] [Google Scholar]
- Wang, X., R. Korstanje, D. Higgins and B. Paigen, 2004. Haplotype analysis in multiple crosses to identify a QTL gene. Genome Res. 14 1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., K. Kerr and G. Churchill, 2003. MAANOVA: A Software Package for the Analysis of Spotted cDNA Microarray Experiments in the Analysis of Gene Expression Data: An Overview of Methods and Software, edited by G. Parmgiani, E. S. Garret, R. A. Irizarry and S. L. Zeger, pp. 313–431. Springer, New York.
- Yamauchi, T., J. Kamon, Y. Ito, A. Tsuchida, T. Yokomizo et al., 2003. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 423 762–769. [DOI] [PubMed] [Google Scholar]
- Yang, H., and G. Churchill, 2007. Estimating p-values in small microarray experiments. Bioinformatics 23 38–43. [DOI] [PubMed] [Google Scholar]
- Zhen, Y., N. Xu, B. Richardson, R. Becklin, J. R. Savage et al., 2004. Development of an LC-MALDI method for the analysis of protein complexes. J. Am. Soc. Mass Spectrom. 15 803–822. [DOI] [PubMed] [Google Scholar]