Abstract
Insulin genes in mouse and rat compose a two-gene system in which Ins1 was retroposed from the partially processed mRNA of Ins2. When Ins1 originated and how it was retained in genomes still remain interesting problems. In this study, we used genomic approaches to detect insulin gene copy number variation in rodent species and investigated evolutionary forces acting on both Ins1 and Ins2. We characterized the phylogenetic distribution of the new insulin gene (Ins1) by Southern analyses and confirmed by sequencing insulin genes in the rodent genomes. The results demonstrate that Ins1 originated right before the mouse–rat split (∼20 MYA), and both Ins1 and Ins2 are under strong functional constraints in these murine species. Interestingly, by examining a range of nucleotide polymorphisms, we detected positive selection acting on both Ins2 and Ins1 gene regions in the Mus musculus domesticus populations. Furthermore, three amino acid sites were also identified as having evolved under positive selection in two insulin peptides: two are in the signal peptide and one is in the C-peptide. Our data suggest an adaptive divergence in the mouse insulin two-gene system, which may result from the response to environmental change caused by the rise of agricultural civilization, as proposed by the thrifty-genotype hypothesis.
SEVERAL mechanisms have been proposed to be involved in the retention of duplicate genes in genomes (Force et al. 1999; Lynch and Conery 2000; Long et al. 2003; Shiu et al. 2006). Yet, how retrogenes evolve with their parental genes remains an interesting question. Preproinsulins (insulin genes), with critical functions relating to the pathogenesis of diabetes, provide a valuable system to investigate this issue. In contrast to other mammals studied to date, i.e., human and guinea pig (Chan et al. 1984), in which one copy of the insulin gene (Ins) was found, insulin genes in mouse and rat form a two-gene system (Soares et al. 1985; Wentworth et al. 1986). The two-gene system is composed of preproinsulin 2 (Ins2), an ortholog to the insulin genes in the other mammals, and preproinsulin 1 (Ins1), a rodent-specific retrogene. Ins2 and Ins1 are expressed in the pancreas and both encode proinsulin peptides composed of four parts: signal peptide, B chain, C-peptide, and A chain. Ins1 was identified as originating from a reverse-transcribed partially processed mRNA of Ins2 and thus retains only one of the two introns, which is homologous to the first intron of Ins2 (Figure 1) (Soares et al. 1985; Wentworth et al. 1986). Contrary to the origins of most retrogenes, Ins1 carries homologous regulatory regions with Ins2 from aberrant transcription; i.e., the mRNA was transcribed from the upstream region of Ins2 and thus the transcript includes the gene itself and the regulatory regions. In the mouse genome, these two insulin genes are located on different chromosomes, chromosome (ch)7 (Ins1) and ch19 (Ins2) (Wentworth et al. 1986; Davies et al. 1994), while in rat they are on the same chromosome (ch1) but are >100 Mb apart (Soares et al. 1985).
Figure 1.—
Gene structure of Ins2 and Ins1 in the house mouse. Boxes indicate exon regions; solid lines indicate intronic or flanking regions. Ins2 has three exons and two introns, while Ins1 contains only one intron homologous to the first intron of Ins2. Both Ins2 and Ins1 carry 5′- and 3′-untranslated regions (UTR) (hatched boxes) and signal peptide (checkered boxes), B chain (left shaded boxes), C-peptide (solid boxes), and A chain regions (right shaded boxes). The arrows above the genes indicate the EcoRI digestion sites, and the predicted genomic fragment sizes after enzyme digestion are shown. The dotted line flanked by opposing arrows at the bottom illustrates the genomic sequences amplified by two primers (Ins2-952 and Ins2-1997) as a probe for Southern analyses. Primers were designed specific to the house mouse Ins2 sequences.
Recent knockout experiments with nonobese diabetic (NOD) mice revealed that these two insulin genes have different null phenotypes related to the etiology of diabetes (Chentoufi and Polychronakos 2002; Moriyama et al. 2003; Thebault-Baumont et al. 2003; Jaeckel et al. 2004; Nakayama et al. 2005; Babaya et al. 2006). The differing phenotypes between Ins2 and Ins1 knockout NOD mice imply a functional divergence between these two genes. First, without the presence of Ins2 alleles, Ins1-carrying mice (Ins1+/+, Ins2−/− and Ins1+/−, Ins2−/−) were inflicted with insulin deficiency that accelerated the onset of type 1 diabetes, particularly in the male NOD mice. In contrast, no decrease in insulin content was detected in mice carrying Ins2 alleles (Ins1−/−, Ins2+/− or Ins1−/−, Ins2+/+) (Babaya et al. 2006). These observations suggest that the retrogene, Ins1, might exert some negative effects that worsen the diabetic syndrome. Moreover, Ins2 and Ins1 were observed to behave differently under hormone stimulation in rats (Kakita et al. 1982). The nature of the null phenotypes of the insulin two-gene system provides a valuable system for investigating the origin of new genes in association with the common disease, diabetes. However, the evolution of Ins1 remains unknown. Two questions are of immediate interest: (i) When did Ins1 originate and diverge in function from the parental gene, Ins2?, and (ii) Given the seemingly deleterious nature of the Ins1 gene, what selection mechanisms were involved in the origins and evolution?
Despite early sporadic data from insulin genes (Beintema and Campagne 1987), the lack of experimental testing of the actual copy number of insulin genes in rodents has made it difficult to understand the distribution of Ins1 in rodents. To elucidate the above questions, we first conducted a phylogenetic survey of the distribution of Ins1 and Ins2 in the rodent family Muridae by genomic Southern analyses. Muridae, to which mice and rats belong, is a large family with >1300 species and has been divided into ∼12 subfamilies (e.g., Michaux et al. 2001). We examine insulin genes by selecting taxa progressively moving away from mouse and rat, including taxa from subfamilies Murinae, Gerbillinae, Cricetinae, and Arvicolinae. Second, to vary the Ins1 signals detected by the genomic Southern analysis, we sequenced insulin genes in several rodent species by PCR cloning and sequencing. We further investigated the functional constraint on both Ins2 and Ins1 by examining Ka/Ks ratios among species. Finally, we identified selection mechanisms acting on this insulin two-gene system by analyzing distributions of polymorphism in the house mouse populations.
MATERIALS AND METHODS
DNA samples:
Genomic DNA was extracted from a total of nine murid species in this study. All were wild caught in Taiwan, except as noted. Their taxonomic affiliations are as follows. Five species are in the murid subfamily Murinae: Mus musculus (C57BL/6), M. caroli, Rattus losea, Apodemus semotus, and Niviventer coxingi. One species, Meriones unguiculatus (from a pet shop), is in the subfamily Gerbillinae. One species, Mesocricetus auratus (from a pet shop), is in the subfamily Cricetinae. Two species, Eothenomys melanogaster and Microtus kikuchii, are in the subfamily Arvicolinae.
Samples of house mouse natural populations, M. musculus domesticus, were collected from France and Germany (Ihle et al. 2006). Nineteen individuals are used in this study. The final sample sizes for various gene regions shown in Table 2 vary because of the failures of the PCR amplification or sequencing for certain samples due to the likely mutations in the primer regions. However, even the small sample sizes in these gene regions (≥12) are adequate for estimating population genetic parameters, according to the sampling theory of Tajima (1989). In addition, we pooled two populations for analyses because there is no evidence of significant divergence in the two particular insulin loci and the flanking regions [Hst values (Hudson et al. 1992) are 0.06 (not significant) and 0.00 (not significant) for the gene regions of Ins1 and Ins2, respectively, and 0.00 (not significant) and 0.09 (not significant, with the Bonferroni correction of multiple tests) for the flanking regions of Ins1 and Ins2, respectively].
TABLE 2.
Summary statistics of Tajima's D and Fu and Li's D estimations
| Parental gene (Ins2)
|
New gene (Ins1)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | Gene | 5′, 10 kb | 5′, 100 Mb | 3′, 8 kb | 3′, 10 kb | Gene | 5′, 8 kb | 5′, 10 kb | 5′, 12 kb | 3′, 10 kb |
| Tajima's D | −2.1168** | 0.2092 | 0.4482 | 0.0484 | −0.6193 | −1.7289* | −0.2583 | 2.3793** | 0.3347 | −0.0669 |
| Fu and Li's D | −2.9607** | 1.3157* | 1.1864 | 0.3575 | 1.2632 | −1.8817* | −0.4590 | 1.0277 | 0.0219 | −0.0010 |
| l (bp) | 605 | 1379 | 809 | 843 | 1643 | 118 | 1045 | 597 | 1020 | 936 |
| n | 15 | 24 | 20 | 22 | 24 | 14 | 12 | 24 | 14 | 26 |
| S | 20 | 19 | 5 | 5 | 18 | 6 | 2 | 11 | 8 | 10 |
| π | 0.0052 | 0.0041 | 0.0020 | 0.0017 | 0.0024 | 0.0085 | 0.0012 | 0.0091 | 0.0031 | 0.0030 |
| θ | 0.0107 | 0.0039 | 0.0017 | 0.0016 | 0.0029 | 0.0164 | 0.0013 | 0.0054 | 0.0028 | 0.0031 |
l, sequence length; n, sample size; S, number of segregating sites; π, nucleotide diversity; θ, average number of segregating sites. For the gene region, we presented the statistics from the intron region as the gene's representative values, because the intron region is less subject to natural selection than the exon regions. *P < 0.05, **P < 0.01.
Southern-blot analysis and PCR sequencing:
We prepared genomic DNA with a phenol/chloroform extraction of tissues that had been treated with proteinase K and RNase A. Genomic DNA was digested with EcoRI and BamHI, respectively, and the representative images from one of the enzyme digestions in each species are shown in Figure 2B. Digested DNA was separated on a 0.8% agarose gel with 0.5× TBE buffer and transferred to nylon membranes. Probes labeled with [α-32P]dCTP were hybridized to the nylon membranes to confirm copy numbers in different species. The probes were amplified from M. musculus Ins1 using primers Ins2-952 (5′-ACC ACC AGC CCT AAG TGA TCC GCT A-3′) and Ins2-1997 (5′-AAG GTT TTA TTC ATT GCA GAG GGG T-3′) (the probe region is shown in Figure 1). Primers were designed specific to Ins2, which differs from Ins1 by two nucleotides within Ins2-952 and one nucleotide within Ins2-1997.
Figure 2.—
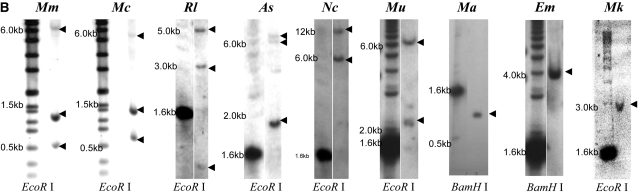
Southern-blot analyses of insulin two-gene systems. (A) Species tree (Michaux et al. 2001; Steppan et al. 2004) of 11 rodent species and human. Shaded branches represent species carrying both Ins2 and Ins1 genes in their genomes. Solid branches represent those with only the Ins2 ortholog gene, Ins or INS, in their genomes. Estimated divergence time of selected species is shown along the x-axis. The scale bar for divergence time is independent of the tree's branch lengths. Southern blot places the origin of Ins1 before the mouse–rat split, ∼20 MYA, but later than the divergence of the Murinae from the Gerbillinae. (B) Southern-blot results from nine species of rodents, including the house mouse as reference. The entire genomic DNA was digested separately by EcoRI and BamHI to confirm insulin gene copy numbers, but only one digestion per species is presented. The selected sizes of DNA ladders are shown on the left of each image; arrowheads on the right indicate positive signals. The darkest signal band in the Mm blot is from Ins1 (1.3 kb) and the two lighter bands are from Ins2 (0.5 and 6.0 kb), as predicted from their genomic sequences (Figure 1). Mm, Mus musculus; Mc, M. caroli; Rl, Rattus losea; As, Apodemus semotus; Nc, Niviventer coxingi; Mu, Meriones unguiculatus; Ma, Mesocricetus auratus; Em, Eothenomys melanogaster; and Mk, Microtus kikuchii.
To obtain sequences of insulin genes, i.e., insulin genes in the rodent species and the mouse populations, we cloned the PCR product followed by sequencing at least three clones. Only identical nucleotides between these clones were selected for the evolutionary analysis. Although the genes are on the autosomes (chromosomes 7 and 19) and may have heterozygote sites, we chose only one allele from each diploid individual.
Evolutionary analysis:
PCR products corresponding to Ins2 and Ins1 were amplified by the Ins2-952 and Ins2-1997 primers from M. caroli, R. losea, A. semotus, and N. coxingi, as well as a single product, Ins, from Mer. unguiculatus and Mi. kikuchii. The Ins2-952 and Ins2-1997 primers were designed from the transcripts in the conserved regions and are able to amplify homologous genes in other rodent species. The PCR products of these insulin genes were then cloned from six rodent species followed by sequencing. For the insulin genes of each species, we sequenced at least three clones to eliminate PCR or sequencing errors. Sequences were analyzed only when they appeared identically in at least two clones. Ins2 and Ins1 genes in the house mouse (M. musculus) and the rat (R. norvegicus) were retrieved from GenBank (accession nos. X04724, X04725, J00748, and J00747). The outgroup, human (Homo sapiens) insulin gene, INS, was also retrieved from GenBank (accession no. X70508).
Coding regions of preproinsulin genes from human and various rodent species and sequence data sets obtained from Ins2 and Ins1 in the house mouse population were aligned by Clustal W version 1.83 (Thompson et al. 1994). To analyze the phylogenetic relationship of the two insulin genes and Ins2 homologous ancestral genes, Ins, in other rodents, we used coding-region sequences to reconstruct a neighbor-joining tree implemented in MEGA3 (Kumar et al. 2004) with 1000 bootstrap repeats. The functional constraints were estimated by Ka/Ks ratios implemented in PAML (Yang 1997). The estimated pairwise Ka/Ks ratios were calculated between the eight rodent species, including six species carrying both Ins2 and Ins1 and two species carrying Ins. Twice the log-likelihood difference between the estimated Ka/Ks ratio and the fixed Ka/Ks ratio (=1) was compared with a χ2-distribution with d.f. = 1 to test whether the estimated Ka/Ks ratio was significantly <1. We eliminated those ratios with extremely small Ks values to reduce stochastic bias.
The spectra of distribution of allele frequencies at segregating sites [i.e., Tajima's D (Tajima 1989) and Fu and Li's D (Fu and Li 1993)] were calculated for indications regarding strength and type of selection implemented by DnaSP 4.0 (Rozas et al. 2003). The significance (P-values) of each of Tajima's D values as well as Fu and Li's D values was estimated by coalescent simulations with 10,000 replicates. To investigate the evolutionary forces acting on Ins1 and Ins2, we examined their gene regions and flanking regions. The four flanking regions for each insulin gene were chosen randomly with an 8-kb to 100-Mb distance from the gene region and the repeated sequences were avoided (Table 2 and Figure 4).
Figure 4.—
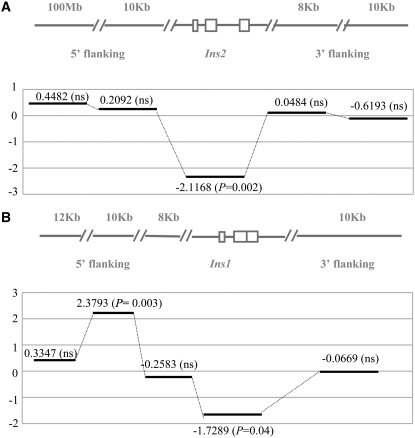
Tajima's D values in Ins2 (A) and Ins1 (B) gene regions and flanking regions. Also shown are flanking regions 8 kb–100 Mb away from these two gene regions. Refer to Table 2 for estimated parameters for Tajima's D. ns, not significant.
To further understand the selective force on residues, we conducted analyses by performing model M3 (three ratios) and model M8 (β and ω), respectively, in PAML to test whether there was an acceleration of evolutionary rates (Yang and Nielsen 2002; Yang 2006). In addition, M3 and M8 were compared with M0 (one ratio) and M7 (β), respectively, by performing log-likelihood-ratio tests. The input phylogenetic tree was based on Figure 3 while running different models.
Figure 3.—
Single origin of Ins1 confirmed by a neighbor-joining tree from insulin genes of eight murid species and human INS as an outgroup. The phylogenetic tree was reconstructed using Kimura two-parameter distances with human INS as the outgroup. The numbers at the branch nodes indicate bootstrap values. The cluster formed by Ins2 and Ins1 implies a single origin of Ins1 in the murine species.
RESULTS AND DISCUSSION
Origin of the duplicate retrogene, Ins1:
The copy numbers of certain insulin-coding genes have been confirmed in certain mammalian species: two copies of insulin genes, Ins2 and Ins1, have been identified in the genomes of house mouse (M. musculus) and rat (R. norvegicus) and a single copy in the genomes of human (H. sapiens, INS) and guinea pig (Cavia porcellus, Ins), which are orthologs of Ins2 (Chan et al. 1984; Soares et al. 1985; Wentworth et al. 1986). We selected eight rodent species to date the origin of Ins1 precisely. We chose eight species from subfamilies Murinae (R. losea, N. coxingi, A. semotus, and M. caroli), Gerbillinae (Mer. unguiculatus), Cricetinae (Mes. auratus), and Arvicolinae (Mi. kikuchii, E. melanogaster). The phylogenetic relationships between the four species with known insulin gene sequences and our eight selected rodent species with unknown copy numbers are illustrated in Figure 2A.
We then carried out Southern blot analyses in the eight rodent species, together with the genomic DNA of house mouse as a positive control. The results revealed that Ins1 exists only in the subfamily Murinae (Figure 2B). As predicted by the distribution of restriction sites (Figure 1), we detected three signals in the house mouse genome (Figure 2B), 0.5 and 6.0 kb from Ins2 and 1.4 kb from Ins1. Three signals were also detected for species that are closely related to the house mouse: M. caroli, R. losea, and A. semotus. Two large bands were detected for N. coxingi. PCR cloning and sequencing revealed that the restriction patterns in these four species were derived from the restriction sites in the two copies of insulin genes, Ins1 and Ins2. One restriction site is missing in Ins2 in N. coxingi, explaining the two signals in this species.
Only one genomic Southern signal was detected in Mes. auratus, E. melanogaster and Mi. kikuchii, which suggests that there is a single copy of the insulin-coding gene in these genomes. However, the copy number in the Mer. unguiculatus genome was unclear because the two signals were detected in the genomic Southern analysis (Figure 2B). We conducted PCR sequencing and observed that only a single copy of the insulin gene, which is the orthologous copy of the Ins2 gene in the house mouse, is present in that genome. One EcoRI restriction site was identified in the Mer. unguiculatus insulin gene, which results in two signal bands in this species. In summary, we conclude that only murine rodents, i.e., species in the subfamily Murinae, possess two copies of the insulin genes.
To further confirm the origin of Ins1, we analyzed the evolutionary relationships of Ins1 and Ins2 using the sequence data from the six Murinae species and Ins in Mer. unguiculatus and Mi. kikuchii generated from the PCR cloning and sequencing experiments. We observe that the gene structures of both Ins2 and Ins1 remain identical in all the Murinae species we analyzed: two introns appear in Ins2 and only one intron in Ins1. With human INS as an outgroup, we constructed a neighbor-joining tree using the protein-coding sequences (330 bp) (Figure 3). As expected, Ins2 and Ins1 in the murine rodents formed a distinct clade (the bootstrap support of the Ins1–Ins2 cluster is >95% when subtracting the sequence of Mi. kikuchii from the data set, data not shown). This indicates that the evolution of a two-gene system in murine species is unique and differs from that in other murid species (i.e., nonmurine rodents) carrying only a single copy of Ins (orthologous to human INS). These results further confirm the single origin of Ins1, which occurred in the most recent common ancestor of the Murinae. By mapping these results onto existing phylogenies, we estimate that the retroposition event took place before the mouse–rat split and after the divergence of the Murinae from the Gerbillinae, ∼20 million years ago (O'Huigin and Li 1992; Michaux et al. 2001). Thus, Ins1 is a relatively young gene and presumably a Murinae-specific retrogene with newly evolved functions in the glucose metabolic pathways.
Functionality of Ins2 and Ins1 in rodents:
To determine the functional constraint on the insulin-coding genes in these rodent species, we used a well-developed comparative analysis of synonymous (Ks) and nonsynonymous substitutions (Ka) (Li 1993; Nekrutenko et al. 2002). In general, a Ka/Ks ratio that is significantly lower than unity is considered to indicate functional constraint. We performed pairwise orthologous comparisons of Ins2 and Ins of eight murid species and of Ins1 in six murine species. Also, we performed Ka/Ks ratio tests for the entire coding regions as well as for the B + A chain and C-peptides of both genes, respectively, because insulin peptides are composed of four subfunctional parts. All comparisons revealed unexpectedly small Ka/Ks ratios (significantly <1) (Table 1). Note that not only the insulin functional peptides, B and A chains, but also the C-peptide of both Ins1 and Ins2 appear to be highly constrained in all species examined. Our data are consistent with the evidence from the previous literature: in addition to the critical role in the protein structure assembly, C-peptides serve important functions in the endocrine systems (reviewed in Steiner 2004). Overall, the above analyses demonstrate the selective constraints in all insulin subfunctional regions, implying the functional importance of the insulin two-gene system in murine species.
TABLE 1.
The Ka/Ks ratios of Ins2 and Ins1 and their subfunctional parts
| M. musculus | M. caroli | A. semotus | R. norvegicus | R. losea | N. coxingi | Mer. unguiculatus | |
|---|---|---|---|---|---|---|---|
| Ins2 | |||||||
| A. semotus | 0.2258 | 0.2037 | |||||
| R. norvegicus | 0.1704 | 0.1618 | 0.1642 | ||||
| R. losea | 0.2588 | 0.2351 | 0.2651 | ||||
| N. coxingi | 0.1807 | 0.1430 | 0.1405 | 0.0955 | 0.1179 | ||
| Mer. unguiculatus | 0.3965 | 0.3022 | 0.2839 | 0.1994 | 0.1994 | 0.2098 | |
| Mi. kikuchii | 0.1397 | 0.1354 | 0.1257 | 0.1291 | 0.1291 | 0.1029 | 0.1346 |
| Ins2 A and B chain | |||||||
| A. semotus | 0.0010 | 0.0010 | |||||
| R. norvegicus | 0.0010 | 0.0010 | 0.0010 | ||||
| R. losea | 0.0281 | 0.0181 | 0.0311 | ||||
| N. coxingi | 0.0010 | 0.0010 | 0.0010 | 0.0010 | 0.0408 | ||
| Mer. unguiculatus | 0.1879 | 0.1197 | 0.1555 | 0.0841 | 0.1099 | 0.1015 | |
| Mi. kikuchii | 0.0251 | 0.0161 | 0.0264 | 0.0224 | 0.0388 | 0.0222 | 0.0545 |
| Ins2 C-peptide | |||||||
| Mer. unguiculatus | 0.3569 | 0.2980 | 0.1787 | 0.1329 | 0.2375 | 0.1671 | |
| Mi. kikuchii | 0.0580 | 0.0651 | 0.1140 | 0.0864 | 0.1822 | 0.1620 | 0.1899 |
| Ins1 | |||||||
| M. caroli | 0.1731 | ||||||
| A. semotus | 0.2601 | 0.2349 | |||||
| R. norvegicus | 0.1515 | 0.1443 | 0.1859 | ||||
| R. losea | 0.1776 | 0.1651 | 0.2645 | ||||
| N. coxingi | 0.1719 | 0.1615 | 0.2132 | 0.1184 | 0.2819 | ||
| Ins1 A and B chain | |||||||
| A. semotus | 0.0444 | ||||||
| R. norvegicus | 0.0010 | 0.0301 | 0.0010 | ||||
| R. losea | 0.0010 | 0.0372 | 0.0010 | ||||
| N. coxingi | 0.0010 | 0.0365 | 0.0010 | 0.0010 | |||
| Ins1 C-peptide | |||||||
| A. semotus | 0.1039 | 0.1255 | |||||
| R. norvegicus | 0.1505 | 0.1736 | 0.1411 | ||||
| R. losea | 0.0968 | 0.1286 | 0.2308 | ||||
| N. coxingi | 0.1349 | 0.1542 | 0.2034 | ||||
Adaptive evolution of the insulin two-gene system:
Our analyses indicate that insulin retrogenes had a single recent origin and that both Ins2 and Ins1 maintain important functions in murine rodents. Although homologous regulatory sequences have been found in the two-gene system of rodents, recent studies propose that possible new functions have evolved in the two insulin-coding genes; i.e., NOD mice with either Ins1 or Ins2 resulted in different phenotypes in the onset of diabetes (Chentoufi and Polychronakos 2002; Moriyama et al. 2003; Thebault-Baumont et al. 2003; Jaeckel et al. 2004; Nakayama et al. 2005; Babaya et al. 2006). It is interesting to know whether the two-gene system is subject to selection for new functions, as was shown for many other types of new genes in various organisms (Long et al. 2003). The conventional whole-gene-based method of Ka/Ks-ratio analysis, which is usually used with a large number of substitutions suggesting strong selection, may lack adequate power to detect the varied selection effects among the differing residues because of the small number of substitutions that have occurred in the short divergence time between Ins1 and Ins2. Therefore, we used two approaches to test evolutionary forces in both genes in mouse populations: (i) molecular population genetics to detect the signature left by any recent selection sweep and (ii) a site-specific test of positive selection using site-specific Ka/Ks ratios.
Genetic variation of DNA sequences in natural populations can be estimated by two different parameters: the number of segregating sites (S) and the average number of nucleotide differences using a pairwise comparison (π). Tajima's D tests were performed by estimating the difference between these two parameters (Tajima 1989). If strong positive selection is acting on a given gene sequence, there will be an excess of rare alleles (e.g., singletons) (Kimura 1983). We thus sequenced the Ins2 and Ins1 introns, which are assumed to be evolving neutrally, from the population of a subspecies of house mice (M. musculus domesticus). Remarkably, the polymorphic spectrum was significantly biased toward rare variants in both genes (Tajima's D = −2.1168, P = 0.0030 and Tajima's D = −2.2454, P = 0.000 for the intron and exon regions of Ins2, respectively, and D = −1.7289, P = 0.040 for the intron region of Ins1. For the exon region of Ins1, although Tajima's D is negative but not significant (Tajima's D = −0.6348, P = 0.300), the bias in the spectrum measured by Fu and Li's method is significant: Fu and Li's D = −1.9301, P = 0.045) (Table 2 and Figure 4). Polymorphic distributions are shown in Figure 5. The data indicate that the insulin two-gene system is subject to positive selection in the mouse populations.
Figure 5.—
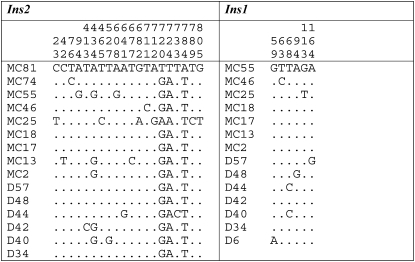
Nucleotide variations of Ins2 and Ins1 gene surrounding regions in wild house mouse populations. Only introns were extracted from genes examined. The numbers for the positions, e.g., 23 and 59, indicate the positions of polymorphic sites. Dots indicate the identical nucleotide as in individuals MC81 and MC55 for Ins2 and Ins1, respectively.
Although the significant D values we observed may result from positive selection acting on these gene regions, alternative interpretations should be also considered, e.g., a recent bottleneck effect or the hitchhiking effect of linkage to adjacent regions subject to positive selection. These alternatives could also create a skewed spectrum of polymorphisms imitating positive selection (Braverman et al. 1995; Nurminsky et al. 1998). We thus investigated sequence variation in four regions in 5′- and 3′-flanking sequences that are 8 kb–100 Mb away from the gene region of Ins2 and Ins1. Tajima's D values are 0.2092 (not significant, NS) and 0.4482 (NS) for the two 5′-upstream regions and are 0.0484 (NS) and −0.6193 (NS) for the two 3′ downstream regions of Ins2 (Figure 4A and Table 2). These different flanking regions show no bias in the frequency spectra, suggesting a different evolutionary history and thus precluding the alternative hypotheses. We also investigated the polymorphisms in the flanking sequences of Ins1. Tajima's D's are 2.3793 (P = 0.004), 0.3347 (NS), and −0.2583 (NS) for the three 5′-upstream regions and −0.0669 (NS) for the 3′-downstream regions (Figure 4B and Table 2). Once again, these four flanking regions clearly do not follow the same evolutionary history as the coding region of Ins1. These results rule out a genomewide bottleneck or hitchhiking effects in Ins2 and Ins1. Furthermore, Fu and Li's D test statistic is consistent with the conclusions drawn by Tajima's D values (Table 2). All of the above evidence reveals that Darwinian positive selection is the predominant selection mechanism contributing to the retention of Ins1 and the evolution of the two-gene insulin system in the mouse populations. Did the positive selection act on the regulatory region or on the protein-coding regions? The spectrum-based population genetic tests do not provide direct discrimination for the two possibilities. However, on the basis of the elevated Tajima's D's in the closest flanking regions, the selection would be more likely to occur in the protein-coding regions. This conjecture is supported by the following substitution analyses of the gene sequences.
To determine whether or not the amino acids evolve nonuniformly in Ins2 and Ins1 peptides, we analyzed the two-gene system in six murine species by using the human insulin gene as an outgroup, including 13 coding sequences (see Figure 3 for their phylogenetic relationships). The statistical results showed that model 3 (M3, three ratios) and model 8 (M8, β and ω) fit the data significantly better than model 0 (M0, one ratio) and model 7 (M7, β) (P < 0.01), respectively. In both M3 and M8, positive selection was detected in three amino acid residues (Table 3): two are located in the signal peptide and the third one in the C-peptide. This reinforces our hypothesis that the coding regions of insulin two-gene systems are subjected to positive selection. Thus, in conjunction with the recent functional analyses in the literature, our data reveal an adaptively evolved insulin two-gene system with diverged functions in the mouse genome. Interestingly, our recent study also demonstrated that positive selection on young retrogene pairs evolves novel functions (Shiao et al. 2007). This suggests that the advantage of retrogenes carrying novel functions may be a universal phenomenon of genomes.
TABLE 3.
Positively selected amino acid residues shared by Ins2 and Ins1 in murid species
| Region | Positive selected amino acid* |
|---|---|
| Signal peptide | 15 (P = 0.891), 20 (P = 0.921) |
| B chain | NA |
| C-peptide | 10 (P = 0.992) |
| A chain | NA |
The statistical significances were estimated by M8 under naive empirical Bayes analysis. *P < 0.01.
Scenario of evolution of the insulin two-gene system:
In conclusion, the retroposed preproinsulin gene, Ins1, was generated in the most recent common ancestor leading to murine species. In general, the gene has been subject to strong selective constraints on all functional parts of the insulin peptides. Interestingly, we detected unexpected significant recent positive selection on both Ins2 and Ins1 in the mouse populations. This, then, raises a particular question of why Ins1, which may be responsible for the development of type 1 diabetes in mice (Moriyama et al. 2003; Babaya et al. 2006), is subject to positive selection in the mouse population. We hypothesize that the evolution of Ins1 in the mouse populations may be explained by an ancestral-susceptibility model (Di Rienzo and Hudson 2005) and that the protective property of Ins2 could result from the risk of being exposed to diabetes due to a defect of Ins1.
On the basis of recent studies, Ins1 may be responsible for the development of type 1 diabetes in mice (Moriyama et al. 2003; Babaya et al. 2006). However, Ins1 not only is fixed in the wild populations but also is subject to positive selection. This seems to be contradictory to the conventional concept that only genes/alleles that provide an advantageous effect would be adaptive in natural populations. To explain this unexpected observation in Ins1 in mice, we hypothesize that the preservation and adaptation of Ins1 may follow an extended form of the thrifty-genotype hypothesis that accounts for the evolution of diabetes-related genes in some human populations (Neel 1962). According to this hypothesis, some alleles that increase the risk to common diseases may likely be ancestral alleles in the populations. The derived alleles protect individuals against common diseases and became advantageous recently (Fullerton et al. 2002; Vander Molen et al. 2005). It was proposed that a shift in environment and lifestyle increases the risk of individuals carrying the ancestral alleles in modern populations. In addition to type 2 diabetes, the susceptibility to certain common diseases, e.g., Alzheimer's disease (Corder et al. 1993; Strittmatter et al. 1993), has been determined to result from carrying ancestral alleles at one genetic locus that, under a shift in lifestyle, confer an unfavorable increased risk of disease. In contrast, the derived alleles confer protective functions and are subject to positive selection in the same populations.
Although Ins2 and Ins1 are two independent genetic loci, we may apply this model to explain the adaptive evolution of these two genes. We propose that, on the basis of the above model derived from the thrifty-genotype hypothesis, the fixation and preservation of the retrogene, Ins1, likely resulted from the advantageous effect under an ancient lifestyle (e.g., an efficient utilization of the intake of energy from the scant food resources in ancient environments). As environments changed, those individuals carrying Ins1 were exposed to an increasing risk of developing type 1 diabetes, because of more abundant foods available when the agricultural civilization arose. However, as a newly evolved retrogene, Ins1 in the existing mouse populations is subject to positive selection for improving its functions. Meanwhile, as an evolutionary response to the recently emerging disadvantageous effect of Ins1, the Ins2 copy might have been positively selected for the protection of individuals from developing diabetes and evolved adaptively in these populations as well.
Acknowledgments
Y.-C. Chan and C.-H. Yu in H.-T. Yu's laboratory offered technical support and discussion; members of M. Long's lab offered valuable discussion. B. Harr sent us mouse DNA samples from her collection. We thank R. Arguello, J. Spofford, T. M. Martin, K. Bullaughey, and B. R. Stein for reading of the manuscript and providing valuable comments. Grant support was provided by the National Science Council (Taiwan) to H.-T.Y. and by the National Science Foundation (USA) and the National Institutes of Health (USA) to M.L. The Goodwill Foundation (Taiwan) granted a fellowship to M.-S.S.
References
- Babaya, N., M. Nakayama, H. Moriyama, R. Gianani, T. Still et al., 2006. A new model of insulin-deficient diabetes: male NOD mice with a single copy of Ins1 and no Ins2. Diabetologia 49 1222–1228. [DOI] [PubMed] [Google Scholar]
- Beintema, J. J., and R. N. Campagne, 1987. Molecular evolution of rodent insulins. Mol. Biol. Evol. 4 10–18. [DOI] [PubMed] [Google Scholar]
- Braverman, J. M., R. R. Hudson, N. L. Kaplan, C. H. Langley and W. Stephan, 1995. The hitchhiking effect on the site frequency spectrum of DNA polymorphisms. Genetics 140 783–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S. J., V. Episkopou, S. Zeitlin, S. K. Karathanasis, A. Mackrell et al., 1984. Guinea pig preproinsulin gene: An evolutionary compromise? Proc. Natl. Acad. Sci. USA 81 5046–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chentoufi, A. A., and C. Polychronakos, 2002. Insulin expression levels in the thymus modulate insulin-specific autoreactive T-cell tolerance: the mechanism by which the IDDM2 locus may predispose to diabetes. Diabetes 15 1383–1390. [DOI] [PubMed] [Google Scholar]
- Corder, E., A. M. Saunders, W. J. Strittmatter, D. E. Schmechel, P. C. Gaskell et al., 1993. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261 828–829.8346434 [Google Scholar]
- Davies, P., C. Poirier, L. Deltour and X. Montagutelli, 1994. Genetic reassignment of the Insulin-1 (Ins1) gene to distal mouse chromosome 19. Genomics 21 665–667. [DOI] [PubMed] [Google Scholar]
- Di Rienzo, A., and R. R. Hudson, 2005. An evolutionary framework for common diseases: the ancestral-susceptibility model. Trends Genet. 21 596–601. [DOI] [PubMed] [Google Scholar]
- Force, A., M. Lynch, F. Pickett, A. Amores, Y. Yan et al., 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., and W. Li, 1993. Statistical tests of neutrality of mutations. Genetics 133 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton, S., A. Bartoszewicz, G. Ybazeta, Y. Horikawa, G. I. Bell et al., 2002. Geographic and haplotype structure of candidate type 2 diabetes susceptibility variants at the calpain-10 locus. Am. J. Hum. Genet. 70 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., D. D. Boos and N. L. Kaplan, 1992. A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 9 138–151. [DOI] [PubMed] [Google Scholar]
- Ihle, Ã. Â., S. Ravaoarimanana, M. Thomas and D. Tautz, 2006. An analysis of signatures of selective sweeps in natural populations of the house mouse. Mol. Biol. Evol. 23 790–797. [DOI] [PubMed] [Google Scholar]
- Jaeckel, E., M. A. Lipes and H. von Boehmer, 2004. Recessive tolerance to preproinsulin 2 reduces but does not abolish type 1 diabetes. Nat. Immunol. 5 1028–1190. [DOI] [PubMed] [Google Scholar]
- Kakita, K., S. Giddings and M. A. Permutt, 1982. Biosynthesis of rat insulins I and II: evidence for differential expression of the two genes. Proc. Natl. Acad. Sci. USA 79 2803–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, UK.
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Li, W. H., 1993. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 36 96–99. [DOI] [PubMed] [Google Scholar]
- Long, M., E. Betran, K. Thornton and W. Wang, 2003. The origin of new genes: glimpses from the young and old. Nat. Rev. Genet. 4 865–875. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and J. S. Conery, 2000. The evolutionary fate and consequences of duplicate genes. Science 290 1151–1155. [DOI] [PubMed] [Google Scholar]
- Michaux, J., A. Reyes and F. Catzeflis, 2001. Evolutionary history of the most speciose mammals: molecular phylogeny of muroid rodents. Mol. Biol. Evol. 18 2017–2031. [DOI] [PubMed] [Google Scholar]
- Moriyama, H., N. Abiru, J. Paronen, K. Sikora, E. Liu et al., 2003. Evidence for a primary islet autoantigen (preproinsulin 1) for insulitis and diabetes in the nonobese diabetic mouse. Proc. Natl. Acad. Sci. USA 100 10376–10381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, M., N. Abiru, H. Moriyama, N. Babaya, E. Liu et al., 2005. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature 435 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel, J., 1962. Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am. J. Hum. Genet. 14 353–362. [PMC free article] [PubMed] [Google Scholar]
- Nekrutenko, A., K. Makova and W. Li, 2002. The K(A)/K(S) ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res. 12 198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurminsky, D. I., M. V. Nurminskaya, D. D. Aguilar and D. L. Hartl, 1998. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature 396 572–575. [DOI] [PubMed] [Google Scholar]
- O'Huigin, C., and W. H. Li, 1992. The molecular clock ticks regularly in muroid rodents and hamsters. J. Mol. Evol. 35 377–384. [DOI] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sánchez-Delbarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19 2496–2497. [DOI] [PubMed] [Google Scholar]
- Shiao, M.-S., P. Khil, R. D. Camerini-Otero, T. Shiroishi, K. Moriwaki et al., 2007. Origins of new male germ-line functions from X-derived autosomal retrogenes in the mouse. Mol. Biol. Evol. 24 2242–2253. [DOI] [PubMed] [Google Scholar]
- Shiu, S. H., J. K. Byrnes, R. Pan, P. Zhang and W. H. Li, 2006. Role of positive selection in the retention of duplicate genes in mammalian genomes. Proc. Natl. Acad. Sci. USA 103 2232–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares, M. B., E. Schon, A. Henderson, S. K. Karathanasis, R. Cate et al., 1985. RNA-mediated gene duplication: the rat preproinsulin I gene is a functional retroposon. Mol. Cell. Biol. 5 2090–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, D. F., 2004. The proinsulin C-peptide—a multirole model. Exp. Diabesity Res. 5 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppan, S. J., M. R. Akhverdyan, E. A. Lyapunova, D. G. Fraser, N. N. Vorontsov et al., 2004. Molecular phylogeny of the marmots (Rodentia: Sciuridae): tests of evolutionary and biogeographic hypotheses. Syst. Biol. 48 715–734. [DOI] [PubMed] [Google Scholar]
- Strittmatter, W., A. M. Saunders, D. Schmechel, M. Pericak-Vance, J. Enghild et al., 1993. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. USA 90 1977–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thebault-Baumont, K., P. Krief, J. P. Briand, P. Halbout, K. Vallon-Geoffroy et al., 2003. Acceleration of type 1 diabetes mellitus in proinsulin 2-deficient NOD mice. J. Clin. Invest. 111 851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Molen, J., L. M. Frisse, S. M. Fullerton, Y. Qian, L. Del Bosque-Plata et al., 2005. Population genetics of CAPN10 and GPR35: implications for the evolution of type 2 diabetes variants. Am. J. Hum. Genet. 76 548–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentworth, B. M., I. M. Schaefer, L. Villa-Komaroff and J. M. Chirgwin, 1986. Characterization of the two nonallelic genes encoding mouse preproinsulin. J. Mol. Evol. 23 305–312. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 2006. On the varied pattern of evolution of two fungal genomes: a critique of Hughes and Friedman. Mol. Biol. Evol. 23 2279–2282. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and R. Nielsen, 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19 908–917. [DOI] [PubMed] [Google Scholar]