Abstract
Aegilops tauschii, the wild relative of wheat, has stronger seed dormancy, a major component of preharvest sprouting resistance (PHSR), than bread wheat. A diploid Ae. tauschii accession (AUS18836) and a tetraploid (Triticum turgidum L. ssp. durum var. Altar84) wheat were used to construct a synthetic wheat (Syn37). The genetic architecture of PHS was investigated in 271 BC1F7 synthetic backcross lines (SBLs) derived from Syn37/2*Janz (resistant/susceptible). The SBLs were evaluated in three environments over 2 years and PHS was assessed by way of three measures: the germination index (GI), which measures grain dormancy, the whole spike assay (SI), which takes into account all spike morphology, and counted visually sprouted seeds out of 200 (VI). Grain color was measured using both Chroma Meter- and NaOH-based approaches. QTL for PHSR and grain color were mapped and their additive and epistatic effects as well as their interactions with environment were estimated by a mixed linear-model approach. Single-locus analysis following composite interval mapping revealed four QTL for GI, two QTL for SI, and four QTL for VI on chromosomes 3DL and 4AL. The locus QPhs.dpiv-3D.1 on chromosome 3DL was tightly linked to the red grain color (RGC) at a distance of 5 cM. The other locus on chromosome 3D, “QPhs.dpiv-3D.2” was independent of RGC locus. Two-locus analysis detected nine QTL with main effects and 18 additive × additive interactions for GI, SI, and VI. Two of the nine main effects QTL and two epistatic QTL showed significant interactions with environments. Both additive and epistatic effects contributed to phenotypic variance in PHSR and the identified markers are potential candidates for marker-assisted selection of favorable alleles at multiple loci. SBLs derived from Ae. tauschii proved to be a promising tool to dissect, introgress, and pyramid different PHSR genes into adapted wheat genetic backgrounds. The enhanced expression of PHS resistance in SBLs enabled us to develop white PHS-resistant wheat germplasm from the red-grained Ae. tauschii accession.
PREHARVEST sprouting (PHS) is the germination of physiologically mature grains in the wheat spike when excessively humid environments persist prior to or during harvest time. PHS susceptibility is a serious problem in many major wheat producing areas around the world including Australia. PHS not only results in yield losses, but also degrades the nutritional and processing quality of the grains, rendering them unsuitable for use in the processing industry. The exposure of grains to wet conditions at ripening triggers a sequence of physiological processes, which among others include the release of hydrolytic enzymes such as α-amylase. Due to the increase in amylase activity, grain carbohydrate reserves will be hydrolyzed, which results in bread wheat quality attributes being affected causing for example sticky crumb and collapsed loaves (Kottearachchi et al. 2006). The financial losses to growers are even greater when grains germinate in the head while attached to the mother plant. In addition to environmental factors such as high moisture and warm temperature, cultural practices such as windrowing, which is a regular practice in North Dakota, also promote the onset of sprout damage in wheat (Gelin et al. 2006).
Due to a consumer preference for white wheats over red wheats in the international market, Australian wheat production like many other countries is targeted for developing white-grained cultivars. Other economic benefits of white-grained wheat are higher flour extraction (McCaig and Depauw 1992) and fewer visible bran specks, which is an important factor to the appearance and acceptability of steam bread and noodle products in the Asian market. The disadvantage is the higher susceptibility of white wheats in general to PHS. At present <2% of the commercial Australian bread wheat cultivars possess PHS resistance. Although red-grained wheats are not always resistant to PHS, red grain color has been recognized as one of the genetic markers for resistance to PHS (Flintham 2000).
Previous studies have implicated other mechanisms in conferring PHSR in wheat. These include vegetative structures of the wheat spike and awns, erectness of spike, openness of florets, tenacity of glumes, and the level of germination inhibitors in the bracts of spikes (Derera and Bhatt 1980; Paterson et al. 1989; Gatford et al. 2002b). Therefore both seed dormancy and PHS are complex traits controlled by many genes or quantitative trait loci (QTL) (Paterson and Sorrells 1990; Mares 1996; Flintham et al. 2002). In wheat, QTL associated with PHS resistance have been identified spanning all 21 chromosomes of the three hexaploid wheat genomes (Anderson et al. 1993; Roy et al. 1999; Zanetti et al. 2000; Kato et al. 2001; Mares and Mrva 2001; Flintham et al. 2002; Groos et al. 2002; Mares et al. 2005; Imtiaz et al. 2006; Kottearachchi et al. 2006; Ogbonnaya et al. 2006), underlying the complexity of PHSR. Four parameters, namely grain dormancy, wetting spikes, falling number (FN), and α-amylase have been used to evaluate and map gene(s)/QTL for PHSR (Zanetti et al. 2000; Kuwal et al. 2005; Mares et al. 2005). In this study we have used grain dormancy and the wetting of spikes (via a rain simulator) to map and characterize gene(s)/QTL associated with PHSR in synthetic backcross-derived lines (SBLs).
We have previously identified a number of Ae. tauschii accessions that displayed a high degree of grain dormancy (Gatford et al. 2002a). In addition to grain dormancy, Ae. tauschii possesses other mechanisms that confer PHSR in wheat, such as the presence of water soluble inhibitors in the bracts surrounding the grain (Gatford et al. 2002b). Modulation of trait expression is also known to occur when traits are transferred from a lower to a higher ploidy species. For example, the expression of cereal cyst nematode resistance genes Cre3 and Cre4 was lower in hexaploid than in diploid wheat species (Eastwood 1995). Once traits have been introgressed into cultivated bread wheat from wild relatives and shown to convey important improvements, it is crucial to detect and characterize trait expression, also with the aim of identifying closely linked markers that will facilitate future introgressions.
Therefore, this study was established to exploit the potential of elevated grain dormancy observed in Ae. tauschii for the improvement of PHSR in cultivated bread wheat. The specific objectives include (i) detect and localize potential Ae. tauschii-derived gene(s)/QTL conferring PHS resistance; (ii) develop PHS-resistant white wheat germplasm derived from SBLs; (iii) characterize QTL for main effects, epistasic effects, and QTL × environment (QTL × E) interactions; and (iv) compare the relative contributions of these genetic components in controlling the expression of PHS resistance in a BC1F7 synthetic backcross-derived wheat population.
MATERIALS AND METHODS
Plant materials:
A mapping population of 271 BC1F7 derived from a cross between “Syn37” and “Janz” was used in this study. Syn37 is synthetic hexaploid wheat (SHW) obtained by crossing a diploid Ae. tauschii accession (AUS18836), having a moderate level of seed dormancy (Gatford et al. 2002a), with a tetraploid (Triticum turgidum L. ssp. durum var. Altar84) wheat. The red-grained Syn37 is resistant to PHS and was used as the female parent, while the recurrent parent Janz, an Australian prime hard white-grained wheat highly susceptible to sprouting, was used as the pollen donor. F1 plants were backcrossed to Janz (as male) to produce 271 BC1F1 plants (Syn37/2*Janz). These plants were grown in the glasshouse and selfed for six generations via single-seed descent (SSD) without selection, producing the BC1F7 population.
Field trials:
A total of 271 BC1F6 and BC1F7 lines with parents were grown in the field from June to December of 2004 and 2005 at the Plant Breeding Centre, Horsham, Victoria, located in the southern Australia wheat belt. The third trial was grown at Wongan Hills, in western Australia, from June to December 2005. Each line was represented by a plot of six rows, 4 m in length, with 0.15 m interrow spacing, in a randomized complete-block design with three replications. The trials were fertilized and maintained free from weeds, insects, and diseases. Trials were sprayed twice with Folicur 430 SC (430 g/liter tebuconazole) at the rate of 29 ml/liter to control foliar diseases such as rusts and were flood irrigated when required.
Evaluation of grain dormancy and PHS:
At physiological maturity (loss of green color in ear and peduncle), 20 heads were taken from each line and air dried at room temperature for 1 week. Ten spikes per line were kept aside for artificial weathering, with the rest handthreshed. Both air-dried spikes and handthreshed seed were placed in storage at −20° to preserve dormancy (Mares 1983). Three measures of preharvest sprouting, further detailed below, were used in the evaluation of the lines—seed dormancy measured as germination index (GI), sprouting index (SI), and visibly sprouted seeds (VI), the latter two following artificial weathering.
Germination tests were carried out 2 months postharvest in a 1.2 ml microtiter polypropylene-sealed tube (Quantum Scientific, Brisbane, Queensland, Australia) filled with 0.577 g of sand (MERCK) and 170 μl of sterile distilled water per tube. Three replicates of 72 (24/replicate) handthreshed seeds were placed crease down on moist sand in microtiter tubes and kept in an incubator with 12 hr of light/darkness at 20° for 14 days. Seeds that displayed pericarp rupture were recorded daily on individual vials during the entire 14-day period. After 14 days of imbibition, ungerminated seeds were induced to germinate with 10 mm gibberellic acid (GA). Seeds that failed to germinate 1 week after treatment with GA were considered nonviable and eliminated from data analysis.
Levels of seed dormancy in the lines were analyzed using a weighted GI (Walker-Simmons and Ried 1988). This index gives maximum weight to grains that germinate rapidly and is calculated from the formula
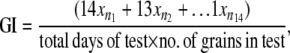 |
where n1, n2, n3, … , n14 are the number of grains that had germinated on day 1, day 2, day 3, … , day 14, respectively. The maximum index is 1.0 if all grains germinate by day 1, while lower indexes are indicative of increasing levels of grain dormancy or reduced germinability.
Spikes from the freezer were subjected to artificial weathering in two replicates for 48 hr using overhead misting of 30-min duration every 6 hr in a rain simulator (Conviron model CMP4030) set to 20° and 98% relative humidity. After 7 days, spikes were removed and a SI (average number of visible sprouting per ear) for each line was determined. Assessment of SI was based on a rating score of 1–6, a modified version of McMaster and Derera (1976), where 1 indicates no visible sprouting and 6 indicates greater than 90% sprouting of single weathered wheat spikes. The heads from each line were then dried for 24 hr to ∼14% moisture and threshed, and the number of visibly sprouted seed (VI) per 200 seeds for each line was recorded.
Estimation of grain color:
For each line (genotype), the grain color was evaluated using both Chroma Meter (Minolta 310) and NaOH methods. The Chroma Meter decomposes color in the L*a*b* color space. In this color space, “L” measures brightness, varying from 0 for black to 100 for white, “a” measures green when negative and red when positive, and “b” measures blue when negative and yellow when positive. The color was measured on a sample of ∼20 g of grains in a 55-mm petri dish. The equipment averages three measures per sample, and for each line, four grain samples were used. For the NaOH method, 30–40 seeds of each line were placed into 100 × 15-mm petri dishes. A 5% NaOH solution was poured over the seeds and they were allowed to soak in the solution overnight. The NaOH solution gives red wheat a dark red color, while white wheat assumes a straw yellow color. On the basis of visual assessments, the BC1F7 population was classified into red, white, and segregating (red:white), when compared to both parental genotypes. Both methods have been shown to be efficient in determining the number of dominant alleles at the R loci of a cultivar (Baker 1981; Wang et al. 1999). Grain color measured through the NaOH method (red:white) was mapped as a qualitative trait while Chroma Meter-generated data was used for quantitative mapping of grain-color components.
Analyses of SSR markers:
Leaf samples were collected from each line in the field, frozen in liquid nitrogen, and stored at −80°. Genomic DNA was extracted from the two parents and 271 BC1F7 lines using the standard phenol/chloroform method as described by Imtiaz et al. (2004). About 433 SSR markers, available in the public domain (http://wheat.pw.usda.gov/GG2/index.shtml), were tested on the parents to determine polymorphism. Unpublished primer sequences are available upon request. Polymorphic SSR markers identified from the parental screening were mapped by genotyping the BC1F7 population comprising 271 lines. For SSR analyses the forward primer from each of the SSR primer pairs was labeled with one of three fluorochrome moieties (FAM, 6-carboxyfluorescein; HEX, hexachloro-6-carboxyfluorescein; or TAMRA, 5-carboxytetramethylrhodamine) (Sigma Genosystems). PCR amplification for SSRs was performed in a final volume of 10 μl containing 5 μl premix D (FailSafe PCR premix; Epicentre Biotechnologies, Madison, WI), 0.5 μm forward primer, 0.5 μm reverse primer, 0.1 μl 2.5 units of FailSafe DNA polymerase, 25–50 ng of template DNA, and dH2O. PCR amplifications were performed on an Eppendorf Mastercycler gradient, using a touchdown cycle consisting of 1 cycle of 94° for 1 min, followed by 18 cycles of 94° for 30 sec, 64° for 30 sec, decreasing 0.5° each cycle, and 72° for 30 sec. An additional 28 cycles followed, consisting of 94° for 30 sec, 55° for 30 sec, and 72° for 30 sec. A final extension of 72° for 5 min was performed before samples were placed at 4°.
To increase the throughput, amplicons generated using FAM, HEX, and TAMRA labels were pooled together. The pooled samples were cleaned with 7.5 m ammonium acetate and precipitated in ethanol before resuspension in 40 μl dH2O. One microliter of cleaned amplicons was mixed with 10 μl of Hi-Di formamide. Triplex PCR products were separated with an ABI 3730 XL 96-channel DNA sequencer (Applied Biosystems, Foster City, CA) and the fragments were sized by means of a ladder labeled with a fourth fluorochrome (ROX, 6-carboxy-X-rhodamine). The allelic size for each SSR and sample were determined in base pairs with GeneMapper 3.5 software (Applied Biosystems).
Statistical analyses:
All data were analyzed by ANOVA, using standard procedures and the residuals were examined for normality. For the combined analysis across environments, the linear model
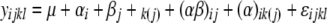 |
was used, where αi, βj, and þk(j) are the main inbred, environment, and replication effects; (αβ)ij is the effect of the inbred × environment interaction; (αþ)ik(j) is the effect of the inbred × replication interaction in the jth environment; and ɛijkl is the error. Two-way analysis of variance was performed for GI, VI, and SI to determine the effect of genotypes, environments, and environment × genotype interaction (G × E) on PHS resistance and to obtain the heritability of PHS. The heritability (H2) estimate (Toojinda et al. 1998) was calculated as
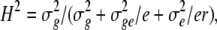 |
where 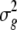 is the variance among BC1F7 lines,
is the variance among BC1F7 lines, 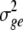 is the genotype × environment variance, r is the number of replicates, and e is the number of environments.
is the genotype × environment variance, r is the number of replicates, and e is the number of environments.
A correlation matrix was derived to study the phenotypic associations among different measures of PHS resistance across three environments. The genetic correlations among GI, VI, and SI were estimated from the equation
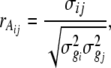 |
where σij is the covariance between traits i and j from an analysis of cross products, and 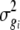 and
and 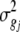 are inbred variances for traits i and j estimated from the ANOVA analyses. The standard errors of genetic correlation were determined following Robertson (1959). Furthermore, chi-square goodness-of-fit tests were used to compare the observed distribution in the BC1F7 population to those predicted by various genetic models for grain color measured with the NaOH method. All statistical analysis was performed using Genstat 5 (Lane et al. 1988).
are inbred variances for traits i and j estimated from the ANOVA analyses. The standard errors of genetic correlation were determined following Robertson (1959). Furthermore, chi-square goodness-of-fit tests were used to compare the observed distribution in the BC1F7 population to those predicted by various genetic models for grain color measured with the NaOH method. All statistical analysis was performed using Genstat 5 (Lane et al. 1988).
QTL analyses:
Linkage maps were constructed using Map Manager QTX version b20 (Manly et al. 2001) and markers were grouped at a log-likelihood (LOD) threshold of 4.00. The genetic distances between markers were estimated using the mapping function of Haldane and Waddington (1931) and the marker order was improved with “ripple” commands. Markers from multilocus primers or those that were different from the reported locus were distinguished with a suffix a, b, c, or d, with the suffix “a” given to the first mapped locus.
Single-locus QTL analyses (SLQA) and composite interval mapping (CIM) were performed using Map Manager QTX version b20 and WinQTL Cartgrapher V2.5 (Basten et al. 1997). The coefficient of determination (R2), which is based on the partial correlation of a putative QTL with the trait adjusted for cofactors in the multilocus model, was estimated to determine the proportion of phenotypic variance explained by a single-marker locus closest to QTL peaks. QTLNetwork 2 and QTLMapper 1.6 (Yang et al. 2005; http://ibi.zju.edu.cn/software/) were employed to determine QTL for additive effects at individual loci, epistatic interactions between two different loci, and interaction between QTL and the environment (QTL × E). The analyses were based on a mixed linear model (MLM) with 1 cM walking speed, 2D genome scan, which refer to map epistatic QTL with or without single-locus effects with 1000 permutations to generate a threshold for the presence of QTL, QTL × E interactions, and a genomewide type I error rate of 5, 1, and 0.1%. The QTL for PHS resistance detected in this study were designated according to the standard nomenclature for QTL designation in wheat. A permutation test (Doerge and Churchill 1996), comprising 1000 permutations, was applied to establish a significance threshold for declaring a QTL. The threshold with LOD ≥ 2.00 was chosen for claiming a putative main or epistatic QTL.
RESULTS
Phenotypic variation for seed dormancy and PHS:
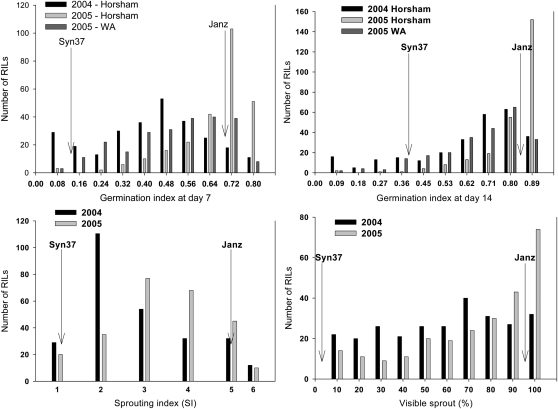
The summary of average values of the parents and the BC1F7 population for the three PHS indexes along with broad sense heritabilities are presented in Table 1. The combined analysis of variance across three environments for GI, SI, and VI showed highly significant (P < 0.001) genotypic differences as well as genotype × environment interaction (G × E). The GI after 7 days (GI-7) for Syn37 was 0.11, 0.12, and 0.17 and after 14 days (GI-14) was 0.29, 0.49, and 0.33 in Horsham in 2004, Horsham in 2005, and Wongan Hills in 2005, respectively, indicative of its PHS resistance. Under the same conditions, Janz exhibited GI-7 values of 0.71, 0.72, 0.73 and GI-14 values of 0.86 in three environments, respectively, proving its high PHS susceptibility. Statistically significant transgressive segregation was evident in the BC1F7 population for GI-14, indicating the presence of BC1F7 lines with phenotypic values outside the range of the parental genotypes (Figure 1). Transgression was not evident for SI and VI at the lower spectrum of the traits; Syn37 had a score of 1 for no visible sprouting (SI) and 1% germinated seed (VI). However, transgression was significant for VI values at the higher end as three BC1F7 lines had 99% VI (Table 1; Figure 1). The heritability estimates were high for GI (0.80–0.92), followed by VI (0.77), and SI (0.62). The lower heritability for SI indicated that environmental factors had more influence on SI compared to VI and GI.
TABLE 1.
Phenotypic data for the parents and BC1F7 population, germination index (GI), whole spike (SI), percentage of visually sprouted seeds (VI), kurtosis, skewness, and 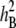 (heritability)
(heritability)
| Parents
|
BC1F7 population
|
||||||
|---|---|---|---|---|---|---|---|
| PHS component | Syn37 mean ± SD | Janz mean ± SD | Min. | Max. | Kurtosis ± SD | Skewness ± SD | 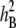 |
| GI-7 | 0.13 ± 0.04 | 0.72 ± 0.04 | 0.00 | 0.70 | 0.49 ± 0.17 | −0.85 ± 0.09 | 0.80 |
| GI-14 | 0.37 ± 0.04 | 0.85 ± 0.04 | 0.03 | 0.86 | 2.80 ± 0.17 | −1.58 ± 0.09 | 0.92 |
| SI | 1.00 ± 0.50 | 6.00 ± 0.50 | 1.00 | 6.00 | −0.64 ± 0.21 | 0.30 ± 0.11 | 0.62 |
| VI | 1.00 ± 0.50 | 92.00 ± 8.00 | 0.50 | 99.00 | −0.77 ± 0.21 | −0.47 ± 0.10 | 0.77 |
Figure 1.—
Frequency distribution for germination index (GI) at days 7 and 14, sprouting index (SI), and visible sprout (VI) derived from BC1F7 population. Level of PHSR in both parents, Syn37 and Janz, are represented by arrows.
Phenotypic correlations between years among GI, VI, and SI are given in Table 2. As expected the correlation was consistently high between GI-7 and GI-14 within each environment (0.95–0.97), while across environments the relationship was moderate, ranging from 0.54 to 0.57 for GI (Table 2), indicating the year/environments effect. A moderately high correlation between 2004 and 2005 SI (0.50) and VI (0.69) was observed. The association of GI with VI was high with correlation coefficient that ranged from 0.54 to 0.67, while GI and SI were the least associated components with a range of 0.30 to 0.51 (Table 2). These highly significant correlations among different indexes of PHS indicated that either the same or closely linked gene(s) are controlling this response. Genetic correlation coefficient (rg) estimated for these indexes showed a similar trend. The expected highest genetic correlation was between GI-7 and GI-14 (rg = 0.81 ± 0.03; standard error). The value of rg between GI and VI was 0.58 ± 0.05 and between GI and SI 0.44 ± 0.05, while that between VI and SI was estimated at 0.77 ± 0.03.
TABLE 2.
Phenotypic correlation coefficient for different PHS components derived from Horsham, Victoria (H), data in 2004 and 2005 and Wongan Hills (WA) in 2005
| PHS component | GI-7-04H | GI-7-05H | GI-7-05WA | GI-14-04H | GI-14-05H | GI-14-05WA | SI-2004 | SI-2005 | VI-2004 | VI-2005 |
|---|---|---|---|---|---|---|---|---|---|---|
| GI-7-04H | 1.00 | |||||||||
| GI-7-05H | 0.54 | 1.00 | ||||||||
| GI-7-05WA | 0.55 | 0.62 | 1.00 | |||||||
| GI-14-04H | 0.95 | 0.54 | 0.50 | 1.00 | ||||||
| GI-14-05H | 0.54 | 0.97 | 0.58 | 0.57 | 1.00 | |||||
| GI-14-05WA | 0.57 | 0.64 | 0.95 | 0.56 | 0.62 | 1.00 | ||||
| SI-2004 | 0.31 | 0.38 | 0.35 | 0.30 | 0.35 | 0.32 | 1.00 | |||
| SI-2005 | 0.36 | 0.51 | 0.51 | 0.34 | 0.48 | 0.51 | 0.50 | 1.00 | ||
| VI-2004 | 0.59 | 0.57 | 0.59 | 0.58 | 0.54 | 0.57 | 0.66 | 0.52 | 1.00 | |
| VI-2005 | 0.59 | 0.67 | 0.67 | 0.55 | 0.65 | 0.67 | 0.45 | 0.71 | 0.69 | 1.00 |
GI-7-04, GI-14-04, germination index at days 7 and 14; SI, sprouting index; and VI, visibly sprouted seeds following artificial weathering for 2004 and 2005. All correlations were significant at P < 0.001.
Grain-color phenotype:
NaOH testing, which enhanced the testa pigmentation, confirmed the visual assessment that both parents, Syn37 and Janz, differed significantly in grain color. The grain L*-values were 56.10 and 52.10, a*-values were 7 and 7.3, and b*-values were 22.9 and 19.2 for Janz and Syn37, respectively. For the BC1F7 population, the grain L*-values varied from 53.8 to 63.7, a*-values between 6.3 and 9.4, and b*-values were in the range of 17.4–26.7. Groos et al. (2002) suggested that the a/L ratio predicts a better relationship between color and PHS. This ratio was 0.12 for Janz and 0.14 for Syn37, while it ranged from 0.10 to 0.17 in the BC1F7 population. Since in this study, grain color was also tested using the NaOH method, we went one step further to estimate the correlation between color and color components and their relation with PHSR. The expected highest correlation was observed between color and L*-values (r = −0.83; P < 0.001), followed by b*-values and a/L ratio (r = −0.78, r = 0.77; P < 0.001), respectively. The lowest association was observed between color and a*-values (r = −0.66; P < 0.001). However, a*-values followed a/L ratio in their relationship with different PHS components. The a/L correlations with GI, VI, and SI were −0.64, −0.48, and −0.37 (P < 0.001), respectively, followed closely by a*-values with −0.58, −0.48, and −0.38 for GI, VI, and SI, consecutively.
As red and white color can be easily distinguished in the presence of NaOH, we have fitted a genetic model to estimate the segregation of grain-color loci. The segregation of white (W) and red (R) (159W:57R) fitted a monogenic ratio of 3:1 (χ2-test probability = 0.85) for the BC1F7 population.
Linkage map:
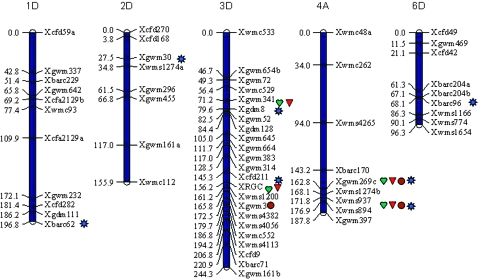
From 400 SSR markers (mostly D-genome specific) tested on Syn37 and Janz, 115 SSRs covering chromosomes 1D–7D were polymorphic between the two parents. These SSRs were supplemented with 30 SSRs randomly selected from A and B genomes of wheat. An additional 33 SSR markers targeting all QTL reportedly linked to PHS on A and B genomes were used in screening for parental polymorphism (Zanetti et al. 2000; Flintham et al. 2002; Tan et al. 2006). The linkage map constructed with the BC1F7 population was composed of 82 loci of which 60 were grouped into five linkage groups (Figure 2), while others either remained unlinked or distantly linked. RGC distinguished as red and white was grouped with SSR markers on chromosome 3D. The total distance covered was 866 cM. Chromosomal identity of these linkage groups was determined on the SSR maps of wheat (Roder et al. 1998; Somers et al. 2004; Song et al. 2005).
Figure 2.—
Linkage maps and distribution of QTL with main effects and epistatic effects for PHS resistance detected by a mixed-model linear approach in the BC1F7 population derived from Syn37 × Janz. Only the linkage maps on which QTL have been involved are presented. SSR names are listed on the right side of the chromosome with the distance (centimorgans) on the left. Red triangle, QTL for seed dormancy (GI); green heart, QTL for VI; brown circle, QTL for whole spike assay (SI); blue sun, QTL involved only in epistatic interactions.
Seed dormancy, PHS, and grain-color QTL:
For GI measured after 7 and 14 days, SLQA detected four genomic regions located on chromosomes 3D and 4A (Figure 2). RGC appeared to be a major contributor to GI in this population, because it accounted for 29 and 43% of the phenotypic variance for GI-7 and GI-14, respectively, in Horsham-2004. The closest marker was wms1200, which explained 25 and 37% variation in GI-7 and GI-14, respectively (Table 3). This QTL although poorly expressed in WA-2005, was stable across three environments in both years and was designated as QPhs.dpivic-3D.1 (Table 3). The second QTL on chromosome 3D designated as QPhs.dpivic-3D.2 accounted for 11% of the phenotypic variance in each of GI-7 and GI-14 over three environments and the nearest marker was gwm341 (Table 3). The QPhs.dpivic-3D.1 explained 15 and 13% variation in VI, Horsham-2004 and Horsham-2005, respectively, while the same genomic region accounted for a maximum of 7% variance in SI for Horsham-2004 and Horsham-2005, consecutively (Table 4). On the contrary, QPhs.dpivic-3D.2 explained only 3% variation in VI, Horsham-2004, while in Horsham-2005 its LOD score threshold was <2. Similarly, SLQA did not detect any association between QPhs.dpivic-3D.2 and SI in either of the years (Table 4). For RGC, the QPhs.dpivic-3D.1 explained 51% variation in a/L ratio with a LOD score of 40 and the wms1200 appeared to be linked with the red-color allele R-D1b at a distance of 5 cM. The QPhs.dpivic-3D.2 did not reveal any significant association with a/L.
TABLE 3.
Putative QTL for grain dormancy measured as the germination index after 7 and 14 days (GI-7 and GI-14) using the composite interval mapping method in a BC1F7 population involving Syn37 and Janz (Syn37/2*Janz), with seed derived from field trials in 2004 (Horsham, Victoria) and 2005 (Horsham and Wongan Hills, western Australia) and the mean across the three environments
| QTL and its location | Closest markers | Mean over three environments
|
Horsham-2004
|
Horsham-2005
|
WA-2005
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LOD | R2(%) | Additive | LOD | R2 (%) | Additive | LOD | R2 (%) | Additive | LOD | R2 (%) | Additive | ||
| GI-7 | |||||||||||||
| QPhs.dpivic-3D.1 | RGC | 14 | 18 | −0.07 | 23 | 29 | −0.12 | 8 | 11 | −0.04 | 3 | 5 | −0.04 |
| QPhs.dpivic-3D.1 | wms1200 | 14 | 18 | −0.07 | 19 | 25 | −0.11 | 7 | 10 | −0.05 | 4 | 5 | −0.04 |
| QPhs.dpivic-3D.2 | gwm341 | 8 | 11 | −0.06 | 6 | 8 | −0.06 | 3 | 4 | −0.03 | 3 | 4 | −0.04 |
| QPhs.dpivic-4A.1 | gwn269c | 13 | 16 | −0.07 | 8 | 8 | −0.07 | 9 | 13 | −0.05 | 15 | 25 | −0.09 |
| QPhs.dpivic-4A.2 | wms894 | 10 | 13 | −0.06 | 5 | 6 | −0.05 | 8 | 13 | −0.04 | 13 | 23 | −0.08 |
| GI-14 | |||||||||||||
| QPhs.dpivic-3D.1 | RGC | 20 | 26 | −0.09 | 36 | 43 | −0.17 | 9 | 13 | −0.04 | 6 | 9 | −0.05 |
| QPhs.dpivic-3D.1 | wms1200 | 19 | 25 | −0.09 | 30 | 37 | −0.16 | 8 | 12 | −0.04 | 5 | 8 | −0.05 |
| QPhs.dpivic-3D.2 | gwm341 | 7 | 11 | −0.06 | 7 | 11 | −0.08 | 4 | 6 | −0.03 | 3 | 4 | −0.03 |
| QPhs.dpivic-4A.1 | gwn269c | 7 | 11 | −0.06 | 6 | 6 | −0.06 | 7 | 9 | −0.04 | 12 | 19 | −0.07 |
| QPhs.dpivic-4A.2 | wms894 | 6 | 8 | −0.04 | 3 | 4 | −0.04 | 7 | 10 | −0.04 | 9 | 15 | −0.06 |
RGC, red grain color; GI, germination index at days 7 and 14.
TABLE 4.
Putative QTL for percentage of visually sprouted seeds (VI) and whole spike (SI), using the composite interval-mapping method in a BC1F7 population involving Syn37 and Janz (Syn37/2*Janz), with seed derived from field trials in 2004 (Horsham, Victoria) and 2005 (Horsham and Wongan Hills, western Australia) and the mean across the three environments
| QTL and its location | Closest markers | Mean over two environments
|
Horsham-2004
|
Horsham-2005
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LOD | R2 (%) | Additive | LOD | R2 (%) | Additive | LOD | R2 (%) | Additive | ||
| VI | ||||||||||
| QPhs.dpivic-3D.1 | RGC | 16 | 17 | −12.98 | 13 | 15 | −13.06 | 10 | 13 | −12.09 |
| QPhs.dpivic-3D.1 | wms1200 | 15 | 15 | −13.08 | 12 | 14 | −13.04 | 10 | 12 | −12.44 |
| QPhs.dpivic-3D.2 | gwm341 | 3 | 3 | −5.40 | 2 | 3 | −5.24 | ND | ND | ND |
| QPhs.dpivic-4A.1 | gwn269c | 17 | 21 | −15.04 | 10 | 14 | −13.25 | 13 | 17 | −14.73 |
| QPhs.dpivic-4A.2 | wms894 | 18 | 24 | −14.03 | 13 | 18 | −13.21 | 12 | 17 | −12.85 |
| SI | ||||||||||
| QPhs.dpivic-3D.1 | gwm3 | 6 | 8 | −0.36 | 4 | 7 | −0.28 | 5 | 7 | −0.43 |
| QPhs.dpivic-3D.1 | wms1200 | 3 | 4 | −0.26 | 2 | 3 | −0.47 | 2 | 3 | −0.30 |
| QPhs.dpivic-4A.1 | gwn269c | 7 | 10 | −0.39 | 4 | 6 | −0.41 | 7 | 11 | −0.52 |
| QPhs.dpivic-4A.2 | wms894 | 9 | 12 | −0.39 | 6 | 9 | −0.45 | 6 | 10 | −0.44 |
VI, visual index; SI, sprouting index; ND, not detected.
SLQA detected two other major regions on chromosome 4A. One was near the known SSR marker gwm269c, which explained 25 and 19% variation in GI-7 and GI-14, respectively, in WA-2005 (Table 3). Unlike QPhs.dpivic-3D.1, the effect of this QTL decreased from GI-7 to GI-14, indicating its better expression in the early part of dormancy breakdown. This QTL named as QPhs.dpivic-4A.1 also accounted for 14 and 17% variation in VI, Horsham-2004 and Horsham-2005, respectively. Similarly its effect on SI was obvious, as it accounted for 6 and 11% trait variance in 2 years (Table 4). The second QTL called QPhs.dpivic-4A.2 explained 23 and 15% variation in GI-7 and GI-14 in WA-2005, respectively, while in Horsham-2004, it accounted for 6 and 4% variation in GI-7 and GI-14, consecutively. The QPhs.dpivic-4A.2 explained 24 and 12% variation in the mean of VI and SI for 2 years, consecutively (Table 4).
Additive effects and additive × E interactions:
In total, 13 QTL for GI-7, GI-14, VI, and SI were detected showing additive main effects (a) and or additive × environment effects (ae; Table 5 and Figure 3a). Of these, only QPhs.dpivic-3D.1 showed significant additive × environment effects for GI-7 and GI-14 across all three environments, while its interactions with environments for VI and SI were not significant. The QPhs.dpivic-3D.2 also showed interaction with Horsham-2004 (ae1) for GI-7 only, while the remaining QTL exhibited only additive effects for GI, VI, and SI. In both cases of significant QTL × E interactions for GI-7 and GI-14, the effects detected in Horsham-2004 were more pronounced than in Horsham-2005 and WA-2005 (Table 5). Syn37 alleles at all QTL decreased GI, VI, and SI values at four genomic regions on chromosomes 3D and 4A, contributing to enhanced expression of resistance to PHS.
TABLE 5.
Estimated additive (a) and additive × environmental interactions (ae) of QTL detected by the mixed linear-model approach for PHS resistance in the BC1F7 population derived from Syn37 and Janz (Syn37/2*Janz)
| QTL name | Flanking interval | LOD | a effect | ae1 | ae2 | ae3 |
|---|---|---|---|---|---|---|
| GI-7 | ||||||
| QPhs.dpivic-3D.1 | Xcfd211–XRGC | 21 | 0.06*** | 0.05*** | −0.02** | −0.04*** |
| QPhs.dpivic-3D.2 | Xgdm8–Xgwm52 | 5 | 0.03*** | 0.02* | NS | NS |
| QPhs.dpivic-4A.1 | Xbarc170–Xgwm269c | 4 | 0.04*** | NS | NS | NS |
| QPhs.dpivic-4A.2 | Xwms937–Xwms894 | 27 | 0.07*** | NS | NS | NS |
| GI-14 | ||||||
| QPhs.dpivic-3D.1 | XRGC–Xwms1200 | 23 | 0.08*** | 0.06*** | −0.03** | −0.04*** |
| QPhs.dpivic-3D.2 | Xgdm8–Xgwm52 | 2 | 0.02*** | NS | NS | NS |
| QPhs.dpivic-4A.1 | Xbarc170–Xgwm269c | 8 | 0.04** | NS | NS | NS |
| QPhs.dpivic-4A.2 | Xwms937–Xwms894 | 12 | 0.07*** | NS | NS | NS |
| VI | ||||||
| QPhs.dpivic-3D.1 | XRGC–Xwms1200 | 12 | 12.79*** | NS | NS | — |
| QPhs.dpivic-4A.1 | Xbarc170–Xgwm269c | 4 | 9.93*** | NS | NS | — |
| QPhs.dpivic-4A.2 | Xwms937–Xwms894 | 24 | 15.85*** | NS | NS | — |
| SI | ||||||
| QPhs.dpivic-3D.1 | Xwms1200–Xgwm3 | 5 | 0.37*** | NS | NS | — |
| QPhs.dpivic-4A.1 | Xbarc170–Xgwm269c | 4 | 0.43*** | NS | NS | — |
a, additive effect; ae1, ae2, and ae3, QTL × environment interaction effects for environments 1 (Horsham, 2004), 2 (Horsham, 2005), and 3 (WA, 2005), respectively. NS, not significant. —, not tested. *P < 0.05; **P < 0.01; ***P < 0.001.
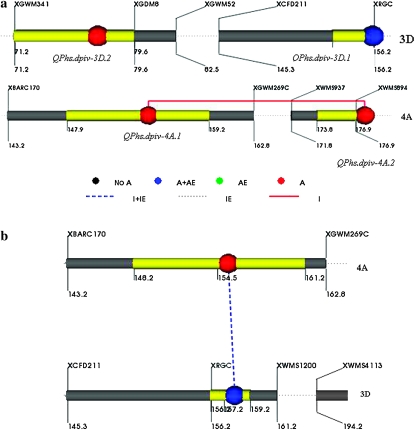
Figure 3.—
The genetic architecture of PHS resistance QTL generated with QTL Network 2.0. (a) Main effect QTL detected on chromosomes 3D and 4A; (b) epistatic interaction between 4A and 3D. A, additive; E, environment; I, interaction.
Epistatic effects and epistasis × E interactions:
In total, seven loci were mapped that were involved in epistatic interaction in 23 digenic combinations (Table 6 and Figures 2 and 3). Four of these seven loci coincide with the QTL showing additive main or additive × environment interaction effects (Tables 5 and 6). Only three loci on chromosomes 1D, 2D, and 6D did not show additive effects, but showed epistatic interactions in combination with one of the QTL with additive effects. From 23 pairwise combinations, only 2 displayed both additive × additive epistatic main effects (aa) and additive × additive epistasis × environment (aae) effects in one or two locations for GI-7 and GI-14 (Table 6; Figure 3, a and b). The remaining loci showed only aa effects for all PHS indexes. The estimated effects of the epistasis were negative at 14 pairs of loci, indicating that recombination of the parental alleles increased PHSR, while at the remaining nine loci, parental allelic combination imparted enhanced PHSR (Table 6).
TABLE 6.
Estimated additive × additive epistatic (aa) and additive × additive epistasis × environment interaction (aae) effects of QTL detected by two-locus analyses using QTLMapper for PHS resistance in the BC1F7 population derived from Syn37 and Janz (Syn37/2*Janz)
| QTLia | Flanking interval | QTLja | Flanking interval | LOD | aiaj effect | aae1 | aae2 | aae3 |
|---|---|---|---|---|---|---|---|---|
| GI-7 | ||||||||
| QPhs.dpivic-1D | Xgdm111–Xbarc62 | QPhs.dpivic-3D.2 | Xgdm8–Xgwm52 | 4 | −0.02* | NS | NS | NS |
| QPhs.dpivic-3D.1 | XRGC–Xwms1200 | QPhs.dpivic-3D.2 | Xgdm8–Xgwm52 | 15 | 0.02** | NS | NS | NS |
| QPhs.dpivic-3D.1 | XRGC–Xwms1200 | QPhs.dpivic-4A.1 | Xbarc170–Xgwm269c | 11 | −0.02* | NS | NS | NS |
| QPhs.dpivic-3D.1 | XRGC–Xwms1200 | QPhs.dpivic-4A.2 | Xwms937–Xwms894 | 32 | −0.03* | NS | 0.03* | NS |
| QPhs.dpivic-4A.2 | Xwms937–Xwms894 | QPhs.dpivic-2D | Xgwm30–Xwms1274a | 23 | 0.02* | NS | NS | NS |
| GI-14 | ||||||||
| QPhs.dpiv-1D | Xgdm111–Xbarc62 | QPhs.dpivic-3D.1 | Xcfd211–XRGC | 19 | −0.02* | NS | NS | NS |
| QPhs.dpivic-3D.1 | XRGC–Xwms1200 | QPhs.dpiv-3D.2 | Xgdm8–Xgwm52 | 14 | 0.02** | NS | NS | NS |
| QPhs.dpivic-3D.1 | XRGC–Xwms1200 | QPhs.dpiv-4A.1 | Xbarc170–Xgwm269c | 21 | −0.03*** | 0.02* | NS | 0.04* |
| QPhs.dpivic-3D.1 | XRGC–Xwms1200 | QPhs.dpiv-4A.2 | Xwms937–Xwms894 | 29 | −0.04*** | NS | NS | NS |
| QPhs.dpiv-6D | Xbarc96–Xwms1166 | QPhs.dpivic-3D.1 | XRGC–Xwms1200 | 18 | 0.02* | NS | NS | NS |
| VI | ||||||||
| QPhs.dpiv-3D.2 | Xgdm8–Xgwm52 | QPhs.dpivic-3D.1 | XRGC–Xwms1200 | 14 | 7.48*** | NS | NS | — |
| QPhs.dpivic-3D.1 | XRGC–Xwms1200 | QPhs.dpiv-2D | Xgwm30–Xwms1274a | 12 | −11.91** | NS | NS | — |
| QPhs.dpivic-3D.1 | XRGC–Xwms1200 | QPhs.dpiv-4A.2 | Xwms894–Xgwm397 | 17 | −5.13* | NS | NS | — |
| QPhs.dpivic-3D.1 | Xwms1200–Xgwm3 | QPhs.dpiv-4A.1 | Xbarc170–Xgwm269c | 13 | −6.58** | NS | NS | — |
| QPhs.dpiv-4A.1 | Xbarc170–Xgwm269c | QPhs.dpiv-4A.2 | Xwms937–Xwms894 | 6 | 5.15*** | NS | NS | — |
| QPhs.dpiv-4A.1 | Xbarc170–Xgwm269c | QPhs.dpiv-2D | Xgwm30–Xwms1274a | 8 | −8.38*** | NS | NS | — |
| QPhs.dpiv-6D | Xbarc96–Xwms1166 | QPhs.dpivic-3D.1 | XRGC–Xwms1200 | 14 | 3.46* | NS | NS | — |
| SI | ||||||||
| QPhs.dpiv-4A.1 | Xbarc170–Xgwm269c | QPhs.dpiv-2D | Xgwm30–Xwms1274a | 7 | −0.39** | NS | NS | — |
| QPhs.dpiv-6D | Xwms1166–Xwms774 | QPhs.dpiv-4A.2 | Xwms937–Xwms894 | 6 | 0.15* | NS | NS | — |
| QPhs.dpiv-1D | Xgdm111–Xbarc62 | QPhs.dpiv-4A.2 | Xwms894–Xgwm397 | 7 | 0.19* | NS | NS | — |
| QPhs.dpivic-3D.1 | XRGC–Xwms1200 | QPhs.dpiv-4A.2 | Xwms937–Xwms894 | 7 | −0.19* | NS | NS | — |
| QPhs.dpivic-3D.1 | Xcfd211–XRGC | QPhs.dpiv-4A.1 | Xbarc170–Xgwm269c | 6 | −0.25* | NS | NS | — |
| QPhs.dpiv-3D.2 | Xgdm8–Xgwm52 | QPhs.dpiv-4A.1 | Xgwm269c–Xwms1274b | 5 | −0.22* | NS | NS | — |
NS, not significant. —, not tested. *P < 0.05; **P < 0.01; ***P < 0.001.
QTLi and QTLj are a pair of QTL. aij is the effect of additive × additive interaction across environments (e1–e3) and positive value means parental type effect is greater than recombinant effect or vice versa.
Pleiotropic effects:
The high coincidence among QTL for GI, VI, and SI indicated that these indexes are controlled by the same genomic regions, which may be due to either pleiotropic or tight linkages. When results of SLQA, CIM, and MLM analyses were combined, three QTL, namely, QPhs.dpivic-3D.1, QPhs.dpivic-4A.1, and QPhs.dpivic-4A.2, were pleiotropic and found to be associated with indexes of PHSR in this study (Tables 3–6). The QTL in the interval RGC–wmc1200 on 3DL and in the genomic regions of gwm269c and wms894 on 4A each showed a positive effect on GI, VI, and SI (Figure 2). The QTL effect is in accordance with high phenotypic correlation values among these indexes (Table 2), which suggested that these QTL were pleiotropic. SLQA detected major loci on chromosomes 3D and 4A only, while MLM detected three minor QTL on chromosomes 1D, 2D, and 6D (Figure 2) in addition to two identified by SLQA.
DISCUSSION
QTL controlling seed dormancy and PHS in wheat:
Resistance to PHS is generally considered to be a complex trait controlled by a number of QTL spread over the A, B, and D genomes of the bread wheat and also to be influenced by the environment. We used three PHS indexes (GI, SI, and VI) jointly in a single study across three environments to characterize QTL for PHS response and then applied the knowledge to select for both red- and white-grained PHS-resistant lines. In all previous studies these indexes were used only individually to map PHS/seed dormancy QTL (Kato et al. 2001; Flintham et al. 2002; Kulwal et al. 2005; Mares et al. 2005; Kottearachchi et al. 2006; Tan et al. 2006). It appeared in this study that GI and VI are the best predictors of resistance to PHS compared to SI, which has subjective aspects when scoring sprouting of susceptible materials after artificial wetting. With SI, sometimes the sprouted grain may be covered in glumes which cannot be seen by the naked eye and the grain may be classified as resistant. The correlation between SI and VI (Table 2) ranged from 0.45 to 0.71, which indicated that SI may not be a good estimator of PHS, especially gauging the level of variability in a range of resistant materials. Zanetti et al. (2000), Shorter et al. (2005), and others also used α-amylase activity and FN to evaluate and map QTL for PHS response. Although both parameters have a high correlation with PHS, this may not represent a casual relationship with PHS, because they measure the damage done to endosperm due to sprouting rather than PHS per se.
This study suggested that true transgressive segregants can only be differentiated statistically at GI-14 (Figure 1). In the present study, seven QTL controlling resistance to PHS were identified on chromosomes 1D, 2D, 3D, 6D, and 4A. The synthetic parent Syn37 contributed resistance on 1D, 2D, 3D, and 4A, while the susceptible cultivar Janz contributed alleles on chromosome 6D, which supports the presence of complementary alleles in the two parents. Furthermore, except for the chromosome 4A regions, none of the other regions on the A and B genomes reportedly linked to PHSR showed any association either in SLQA or MLM analyses in this population.
We also mapped the red grain color (RGC) or R-D1b locus at a distance of 5 cM from the marker wms1200 located on chromosome 3D (Figure 2), which explained the highest level of variation explained by GI (43%), VI (17%), and SI 8% (Tables 3 and 4). Thus Syn37 appears to have a R-D1b allele responsible for red grain color, which is also supported by the observed ratio of white and red grains (159/57) in the BC1F7 population, which fits the expected value for a single independently segregating gene. In previous studies, Groos et al. (2002) and Kulwal et al. (2004) both mapped QTL for PHS in red-grained BC1F7 populations on chromosome 3D, while Mares et al. (2002) used a chromosome-substitution series to postulate the presence of a QTL on chromosome 3D in AUS1408, a white-grained PHS-resistant source. In the current study using a large population of 271 lines, we identified two QTL, QPhs.dpiv-3D.1 and QPhs.dpiv-3D.2, which explained a maximum of 43 and 11% of variation in GI, respectively, which was greater than in previously reported QTL studies. Using graphical genotyping, when PHS indexes for the white-grained lines carrying QPhs.dpiv-3D.2 were compared with those possessing the red grain and QPhs.dpiv-3D.1 loci, the QPhs.dpiv-3D.2 conferred an acceptable level of PHS resistance, which ranged from 0.30 to 0.40 for GI-7, 2 to 3 for SI, and 30 to 45 for VI. Thus, we hypothesize that the markers in the genomic region harboring the putative QTL QPhs.dpiv-3D.2 would be the best candidates to select for white-grained PHS-resistant germplasm, while those markers in the vicinity of QPhs.dpiv-3D.1, such as wms1200 (Figure 2), would be effective for selecting red-grained PHS-resistant germplasm, for cropping regions where white grain color is not an industry preference.
Other color-independent major genomic regions associated with PHS resistance were identified on chromosome 4A, which has been reported in previous studies (Kato et al. 2001; Mares et al. 2005; Kottearachchi et al. 2006). When comparing the level of dormancy between synthetic wheat, Syn37 and its tetraploid (durum variety, Altar84) and diploid (Ae. tauschii) progenitors, Oman et al. (2001) suggested that the lower level of PHS resistance in Syn37 compared to either of its parents was due to the partial expressing of PHS resistance at a higher ploidy level. However, this study clearly demonstrated that Altar84, the A- and B-genome donor in Syn37, carries QTL QPhs.dpivic-4A.1 and QPhs.dpivic-4A.2 for PHS resistance. Although modification in the expression of traits when transferred from the diploid to a higher ploidy level has been reported (Kema et al. 1995; Ma et al. 1995), full expression of PHS resistance was observed in SBLs in the current study (Table 1). Therefore, SBLs proved to be an effective tool to capture the full expression of a trait from its constitutive relatives to enhance germplasm, while characterizing and pyramiding QTL at the same time.
MLM identified two QTL on chromosome 4A (Figure 3, which is in agreement with previously reported results about the presence of two QTL on chromosome 4A; Kato et al. 2001; Mares et al. 2005; Kottearachchi et al. 2006), however, the genomic location of these QTL varies. The discrepancies could be due to the use of different PHS-resistant sources in the different studies, varying methodologies of assessing PHS, or the use of different marker densities.
There is no report we are aware of where seed dormancy QTL have been reported on chromosome 6D, although the only PHS-related QTL on chromosome 6D was reported by Zanetti et al. (2000) for FN and α-amylase activity. The high level of susceptibility to PHS in Janz and at the same time the proven expression of PHS resistance associated with some of its genomic regions in SBLs is in agreement with the expression–suppression phenomenon sometimes observed in wheat, especially in relation to the D-genome, in which gene(s) express in synthetic hexaploids only when the corresponding repressor is absent (Eastwood 1995; Kema et al. 1995; Imtiaz et al. 2003).
The genomic regions linked to two other minor QTL, QPhs.dpivic-1D and QPhs.dpivic-2D, could contribute to the enhancement of PHS resistance. This study reports for the first time the potential of using linked markers (gwm30 and gwm1274a on chromosomes 2D and gdm111–barc62 on chromosome 1D) to select for these regions. Saturation of these regions with more markers would help to further refine these QTL for use in marker-assisted selection.
In addition to SSR markers, candidate genes, TaVP1 (Bailey et al. 1999) and dihydroflavonol reductase (TaDFR-A, TaDFR-B, and TaDFR-D) genes (Himi and Noda 2005) were also targeted in this study (data not presented). We designed primers to cover these candidate regions, but did not find either size or sequence variation between Syn37 and Janz, the parents of our mapping population. Himi and Noda (2005) located the TaDFR-D in a more proximal region than TaVP1 and RGC genes. However, in the current study the only QTL detected within the region was QPhs.dpivic-3D.2 and thus TaVP1 and the QPhs.dpivic-3D.2 are probably collocated. However, further efforts are required to explore this region in more detail.
Environment × QTL interaction:
Phenotypic plasticity, which is the ability of a genotype to change its phenotype in response to changes in the environment, arises in nature due to QTL and environment (Q × E) interactions (Ungerer et al. 2003). In this study, the genetic components of QTL that govern the expression of PHS resistance including additive effects and Q × E interactions, were statistically characterized by two-locus analysis. The only QTL that showed consistent phenotypic plasticity was QPhs.dpivic-3D.1 as its interaction for GI with all three environments was highly significant. However, for VI and SI, this QTL did not show any significant Q × E interaction (Table 5 and Figure 2).
Kottearachchi et al. (2006) compared the GI for red- and white-grained BC1F7 lines grown in the field and under glasshouse conditions. They noted that the difference between the GI of field-grown red- and white-grained wheat was highly significant, while such a large difference was not observed in their glasshouse experiment. They attributed the drastic reduction in the germination of white wheat grown in the glasshouse to the failure of their ability to maintain sufficient dormancy under wet field conditions. It is now evident from this study, that Q × E interaction should be taken into account when exploiting RGC locus for PHSR, although grain color per se does not interact with environment and remains stable across environments (Matus-Cádiz et al. 2003).
Epistasis importance:
The importance of epistatic interactions in trait variation has been reported widely in many crops including for seed dormancy in wheat (Gu et al. 2004; Kulwal et al. 2004; Marwede et al. 2005). Three possible epistatic interactions, as also suggested by Marwede et al. (2005), can be (type I) interactions between two QTL with additive effect, (type II) interactions between a QTL with additive effect and a “background” locus without additive effect, or (type III) interactions between two loci showing epistatic effects only. In this study, 13 of the epistatic interaction were type I and 10 were type II (Table 6), while no type III epistatic interactions were observed. In breeding for PHS resistance such factors should be considered.
In this study, using SBLs derived from Ae. tauschii, both red- and white-grained PHS-resistant bread wheat lines were developed, along with the identification of closely linked molecular markers. These can be used to introgress PHS resistance in breeding. Additionally, the characterization of genetic variation controlling PHS resistance into its genetic components (e.g., additive effects, epistasis, G × E interaction) offers breeders and researchers alike insight into the significance of interacting QTL in conferring acceptable levels of PHS resistance in wheat.
Acknowledgments
The authors thank Erica Steadman, Jayne Wilson, Jacinta Bull, Jacqui Nuttall, and Guoyou Ye for their technical assistance and valuable discussion. This work was supported by Grains Research and Development Corporation, Molecular Plant Breeding Cooperative Research Centre, and the Department of Primary Industries, Victoria, Australia.
References
- Anderson, J. A., M. E. Sorrells and S. D. Tanksley, 1993. RFLP analysis of genomic regions associated with resistance to pre-harvest sprouting in wheat. Crop Sci. 33 453–459. [Google Scholar]
- Bailey, P. C., R. S. McKibbin, J. R. Lenton, M. J. Holdsworth, J. E. Flintham et al., 1999. Genetic map locations for orthologous Vp1 genes in wheat and rice. Theor. Appl. Genet. 98 281–284. [Google Scholar]
- Baker, R. J, 1981. Inheritance of seed coat color in eight spring wheat cultivars. Can. J. Plant. Sci. 61 719–721. [Google Scholar]
- Basten, J. C., B. S. Weir and Z. B. Zeng, 1997. QTL Cartographer: A Reference Manual and Tutorial for QTL Mapping. North Carolina State University, Raleigh, NC.
- Derera, N. F., and G. M. Bhatt, 1980. Germination inhibition of the bracts in relation to pre-harvest sprouting tolerance in wheat. Cereal Res. Commun. 8 199–201. [Google Scholar]
- Doerge, R. W., and G. A. Churchill, 1996. Permutation tests for multiple loci affecting a quantitative character. Genetics 142 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood, R. F., 1995. Genetics of resistance to Heterodera avenae in Triticum tauschii and its transfer to bread wheat (Triticum aestivum). Ph.D. Thesis, Department of Agriculture, University of Melbourne, Parkville, Victoria, Australia.
- Flintham, J. E., 2000. Different genetic components control coat-imposed and embryo-imposed dormancy in wheat. Seed Sci. 10 43–50. [Google Scholar]
- Flintham, J. E., R. Adlam, M. Bassoi, M. Holdsworth and M. Gale, 2002. Mapping genes for resistance to sprouting damage in wheat. Euphytica 126 39–45. [Google Scholar]
- Gatford, K. T., P. Hearnden, F. C. Ogbonnaya, R. F. Eastwood and G. M. Halloran, 2002. a Novel resistance to pre-harvest sprouting in Australian wheat from wild relative Triticum tauschii. Euphytica 126 67–76. [Google Scholar]
- Gatford, K. T., R. F. Eastwood and G. M. Halloran, 2002. b Germination inhibitors in bracts surrounding the grain of Triticum tauschii. Funct. Plant Biol. 29 881–890. [DOI] [PubMed] [Google Scholar]
- Gelin, J. R., E. M. Elias and S. F. Kianian, 2006. Evaluation of two durum wheat (Triticum aestivum L. var. durum) crosses for pre-harvest sprouting resistance. Field Crop Res. 97 188–196. [Google Scholar]
- Groos, C., G. Gay, M. R. Perretant, L. Gervais, M. Bernard et al., 2002. Study of the relationship between pre-harvest sprouting and grain colour by quantitative trait loci analysis in a white × red grain bread-wheat cross. Theor. and Appl. Genet. 104 39–47. [DOI] [PubMed] [Google Scholar]
- Gu, X.-Y., S. F. Kianian and M. E. Foley, 2004. Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa). Genetics 166 1503–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane, J. B. S., and C. H. Waddington, 1931. Inbreeding and linkage. Genetics 16 357–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himi, E., and K. Noda, 2005. Red grain colour gene (R) of wheat is a Myb-type transcription factor. Euphytica 143 239–242. [Google Scholar]
- Imtiaz, M., M. G. Cromey, J. G. Hampton, M. J. Hill, 2003. Inheritance of seedling resistance to stripe rust (Puccinia striiformis f. sp. tritici) in ‘Otane’ and ‘Tiritea’ wheat (Triticum aestivum). N. Z. J. Crop Hort. Sci. 31 15–22. [Google Scholar]
- Imtiaz, M., M. Ahmad, M. G. Cromey, W. B. Griffin and J. G. Hampton, 2004. Detection of molecular markers linked to the durable adult plant stripe rust resistance gene Yr18 in bread wheat (Triticum aestivum L.). Plant Breed. 123 401–404. [Google Scholar]
- Imtiaz, M., J. Bull, J. Wilson, P. Hearnden, J. Oman et al., 2006. Mapping of Aegilops tauschii derived genes controlling seed dormancy and pre-harvest sprouting in wheat. Proceedings of the 13th Australasian Plant Breeding Conference, Christchurch, New Zealand.
- Kato, K., W. Nakamura, T. Tabiki, H. Miura and S. Sawada, 2001. Detection of loci controlling seed dormancy in group 4 chromosomes of wheat and comparative mapping with rice and barley genomes. Theor. Appl. Genet. 102 980–985. [Google Scholar]
- Kema, G. H. J., W. Lange and C. H. V. Silfhout, 1995. Differential suppression of stripe rust resistance in synthetic wheat hexaploid derived from Triticum turgidum subsp. Dicoccoides and Aegilops squarrosa. Phytopathology 85 425–429. [Google Scholar]
- Kottearachchi, N. S., N. Uchino, K. Kato and H. Miura, 2006. Increased grain dormancy in white-grained wheat by introgression of preharvest sprouting tolerance QTLs. Euphytica 152 421–428. [Google Scholar]
- Kulwal, P. L., R. Singh, H. S. Balyan and P. K. Gupta, 2004. Genetic basis of pre-harvest resistance using single-locus and two-locus QTL analysis in bread wheat. Funct. Integr. Genomics 4 94–101. [DOI] [PubMed] [Google Scholar]
- Kulwal, P. L., N. Kumar, A. Gaur, P. Khurana, J. P Khurana et al., 2005. Mapping of a major QTL for pre-harvest sprouting tolerance on chromosome 3A in bread wheat. Theor. Appl. Genet. 111 1052–1059. [DOI] [PubMed] [Google Scholar]
- Lane, P., N. Galway and N. Alvery, 1988. Genstat 5: An Introduction. Oxford University Press, Oxford.
- Ma, H., R. P. Singh and K. A. Mujeeb, 1995. Suppression/expression of resistance to stripe rust in synthetic hexaploid wheat (Triticum aestivum × T. tauschii). Euphytica 83 87–93. [Google Scholar]
- Manly, K. F., R. H. Cudmore, Jr. and J. M. Meer, 2001. Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome 12 930–932. [DOI] [PubMed] [Google Scholar]
- Mares, D. J., 1983. Preservation of dormancy in freshly harvested wheat. Aust. J. Agric. Res. 34 33–38. [Google Scholar]
- Mares, D. J., 1996. Dormancy in white wheat: mechanism and location of genes, pp. 179–184 in Preharvest Sprouting in Cereals, 1995, edited by K. Noda and D. J. Mares. Centre for Academic Societies, Osaka, Japan.
- Mares, D. J., and K. Mrva, 2001. Mapping quantitative trait loci associated with variation in grain dormancy in Australian wheat. Aust. J. Agric. Res. 52 1257–1265. [Google Scholar]
- Mares, D. J., K. Mrva, M-K Tan and P. Sharp, 2002. Dormancy in white-grained wheat: progress towards indentification of genes and molecular markers. Euphytica 126 47–53. [Google Scholar]
- Mares, D., K. Mrva, K. Cheong, K. Williams, B. Watson et al., 2005. A QTL located on chromosome 4A associated with dormancy in wheat- and red-grained wheats of diverse origin. Theor. Appl. Genet. 111 1357–1364. [DOI] [PubMed] [Google Scholar]
- Marwede, V., M. K. Gul, H. C. Becker and W. Ecke, 2005. Mapping of QTL controlling tocopherol content in winter oilseed rape. Plant Breed. 124 20–26. [Google Scholar]
- Matus-Cádiz, M. A., P. Hucl, C. E. Perron and R. T. Tyler, 2003. Genotype × Environment interaction for grain color in hard white spring wheat. Crop Sci. 43 219–226. [Google Scholar]
- McCaig, T. N., and R. M. Depauw, 1992. Breeding for pre-harvest sprouting tolerance in white seed color spring wheat. Crop Sci. 32 19–23. [Google Scholar]
- McMaster, G. J., and N. F. Derera, 1976. Methodology and sample preparation when screening for sprouting damage in cereals. Cereal Res. Commun. 4 251–254. [Google Scholar]
- Ogbonnaya, F. C., M. Imtiaz, P. Hearnden, J. Wilson, R. F. Eastwood et al., 2006. Identification of novel gene for seed dormancy in wheat. Proceedings of the 13th Australasian Plant Breeding Conference, Christchurch, New Zealand.
- Oman, J., F. C. Ogbonnaya, V. Matassa and R. F. Eastwood, 2001. Comparison of dormancy between a synthetic hexaploid bread wheat and its tetraploid and diploid progenitors. Proceedings of the 10th Wheat Breeding Assembly, Mildura, Australia, pp. 205–208.
- Paterson, A. H., and M. E. Sorrells, 1990. Inheritance of grain dormancy in white-kernelled wheat. Crop Sci. 30 25–30. [Google Scholar]
- Paterson, A. H., M. E. Sorrells and R. L. Obendorf, 1989. Methods of evaluation for pre-harvest sprouting resistance in wheat breeding programs. Can. J. Plant Sci. 69 681–689. [Google Scholar]
- Robertson, A., 1959. The sampling variance of the genetic correlation coefficient. Biometrics 15 469–485. [Google Scholar]
- Roder, M. S., V. Korzun, K. Wendehake, J. Plaschke, M.-H. Tixier et al., 1998. A microsatellite map of wheat. Genetics 149 2007–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, J. K., M. Prasad, R. K. Varshney, H. S. Balyan, T. K. Blake et al., 1999. Identification of a microsatellite on chromosomes 6B and a STS on 7D of bread wheat showing an association with pre-harvest sprouting tolerance. Theor. Appl. Genet. 99 336–340. [Google Scholar]
- Shorter, S. C., C. A. Munro and J. Hodgkinson, 2005. Predicting pre-harvest susceptibility in New Zealand wheat cultivars. Euphytica 143 309–312. [Google Scholar]
- Somers, D. J., P. Isaac and K. Edwards, 2004. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 109 1105–1114. [DOI] [PubMed] [Google Scholar]
- Song, Q. J., J. R. Shi, S. Singh, E. W. Fickus, J. M. Costa et al., 2005. Development and mapping of microsatellite (SSR) markers in wheat. Theor. Appl. Genet. 110 550–560. [DOI] [PubMed] [Google Scholar]
- Tan, M. K., P. J. Sharp, M. Q. Lu and N. Howes, 2006. Genetics of grain dormancy in a white wheat. Aust. J. Agric. Res. 57 1157–1165. [Google Scholar]
- Toojinda, T., E. Baird, A. Booth, L. Broers, P. Hayes et al., 1998. Introgression of quantitative trait loci (QTLs) determining stripe rust resistance in barley: an example of marker-assisted line development. Theor. Appl. Genet. 96 123–131. [Google Scholar]
- Ungerer, M. C., S. S. Halldorsdottir, M. D. Purugganan and T. F. C. Mackay, 2003. Genotype–environment interactions at quantitative trait loci affecting inflorescence development in Arabidopsis thaliana. Genetics 165 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker-Simmons, M. K., and J. L. Ried (Editors), 1988. Preharvest Sprouting in Cereals. American Association of Cereal Chemists Press, St. Paul, MN, pp. 21–29.
- Wang, D., F. E. Dowell and R. E. Lacey, 1999. Predicting the number of dominant r alleles in single wheat kernels using visible and near-infrared reflectance spectra. Cereal Chem. 76(1): 6–8. [Google Scholar]
- Yang, J., C. C. Hu, X. Z. Ye and J. Zhu, 2005. QTLNetwork 2.0. Institute of Bioinformatics, Zhejiang University, Hangzhou, China. http://ibi.zju.edu.cn/software/qtlnetwork.
- Zanetti, S., M. Winzeler, M. Keller, B. Keller and M. Messmer, 2000. Genetic analysis of pre-harvest sprouting resistance in a wheat x spelt cross. Crop Sci. 40 1406–1417. [Google Scholar]