Abstract
Genetic structure and diversity of natural populations of prickly lettuce (Lactuca serriola) were studied using AFLP markers and then compared with the diversity of the RGC2 disease resistance gene cluster. Screening of 696 accessions from 41 populations using 319 AFLP markers showed that eastern Turkish and Armenian populations were the most diverse populations and might be located in the origin and center of diversity of L. serriola. Screening 709 accessions using the microsatellite MSATE6 that is located in the coding region of most RGC2 homologs detected 366 different haplotypes. Again, the eastern Turkish and Armenian populations had the highest diversities at the RGC2 cluster. The diversities at the RGC2 cluster in different populations were significantly correlated with their genomewide diversities. There was significant variation of copy number of RGC2 homologs in different populations, ranging from 12 to 22 copies per genome. The nucleotide diversities of two conserved lineages (type II) of RGC2 genes (K and L) were not correlated with diversities calculated using the MSATE6 or AFLP data. We hypothesize that the high genomewide diversity and diversity of the RGC2 cluster in eastern Turkish and Armenian populations resulted from high abiotic and biotic stresses in the regions of origin of L. serriola.
MAXIMAL use of wild genetic resources requires a detailed understanding of the genetic structure, diversity, and divergence of the wild progenitor species. Understanding the nature, organization, geographical structure, and differentiation of a wild species is critical for its biological conservation and is also an important aspect of evolutionary genetics (Nevo 1998). Little is known about these aspects of prickly lettuce (Lactuca serriola; 2x = 2n = 18), the progenitor of cultivated lettuce, L. sativa (Lindqvist 1960; Kesseli et al. 1991; Zohary 1991; Hill et al. 1996).
The wild and cultivated lettuces are sexually compatible and lettuce geneticists and breeders have made extensive use of this wild species, mainly for disease resistance, including resistance against downy mildew caused by Bremia lactucae (Crute 1992; Lebeda et al. 2002; Beharav et al. 2006). At least eight functional resistance genes (R-genes) with different specificities against downy mildew have been mapped to the RGC2 cluster, the major cluster of R-genes in lettuce (Farrara et al. 1987; Kesseli et al. 1994). A functional R-gene, Dm3, conferring resistance against downy mildew, was cloned from the RGC2 cluster (Meyers et al. 1998; Shen et al. 2002). Several other Dm specificities as well as resistance to root aphids have since been shown to be conferred by members of the RGC2 family using RNAi (Wroblewski et al. 2007). The RGC2 cluster varies considerably in copy number, ranging from ∼10 to >30 copies in a haplotype (Kuang et al. 2004). The RGC2 cluster in natural populations of L. serriola is very diverse, as shown by the numerous haplotypes identified by a microsatellite marker located in intron 3 of many RGC2 homologs (Sicard et al. 1999).
Fragments from >300 RGC2 homologs from multiple lettuce cultivars and wild Lactuca species were sequenced in previous studies (Kuang et al. 2004, 2006). Sequence analysis indicated that RGC2 homologs exhibit heterogeneous rates of evolution and could be classified into two types: type I and type II R-genes. Type I R-genes are extensive chimeras generated by frequent sequence exchanges among paralogs, while type II R-genes evolve independently and are highly conserved in different genotypes or closely related species (Kuang et al. 2004). The sequence exchanges between type I R-gene homologs generate a large number of distinct R-gene candidates in a plant population or species (Kuang et al. 2006). However, the frequent sequence exchanges between type I R-gene homologs make it difficult to study their evolutionary and population genetics.
Previous studies of evolutionary and population genetics of other R-genes were done mainly on single-copy R-genes that had no sequence exchange with paralogs (Caicedo et al. 1999; Mauricio et al. 2003; Caicedo and Schaal 2004; Bakker et al. 2006; Rose et al. 2007). These single-copy R-genes have evolutionary patterns similar to those of type II R-genes. The single-copy R-gene, if present, is highly conserved and maintains an obvious allelic relationship in different genotypes. Some single-copy R-genes in Arabidopsis were shown to be under balancing selection (Stahl et al. 1999; Tian et al. 2002; Mauricio et al. 2003). Balancing selection was even detected on R-genes with null alleles (Stahl et al. 1999; Tian et al. 2002). Null alleles are common for single-copy R-genes and type II R-genes (Stahl et al. 1999; Tian et al. 2002; Kuang et al. 2004; Shen et al. 2006). Old null alleles were hypothesized to be maintained in a species due to a fitness cost of the corresponding functional alleles (Tian et al. 2003). Although type II R-genes in R-gene clusters appear to have evolved similarly to single-copy R-genes, a comprehensive study of their population genetics is lacking.
The alleles of resistance gene RPP13, which is a single-copy R-gene in Arabidopsis, show much higher nucleotide diversity (π = 0.04 for the whole gene and π = 0.10 for the leucine-rich repeat (LRR)-encoding region) than those for other single-copy R-genes (Rose et al. 2004; Bakker et al. 2006). The RPP13 homologs are extensive chimeras, similar to type I R-genes. High nucleotide diversity and chimeric structure were also discovered for the alleles of the L resistance gene in flax (Ellis et al. 1999). It remains unknown how the high diversity of these single-copy R-genes was generated and maintained in natural populations.
We assessed the genetic structure of natural populations of L. serriola using AFLPs as genetic markers. The population genetics of the RGC2 resistance gene cluster was studied in detail using a microsatellite located in the coding region of most RGC2 genes. Nucleotide diversity of two lineages of type II genes at the RGC2 locus was also studied. The diversity of the RGC2 cluster in different populations was compared with genomewide diversity. Linkage disequilibrium and evolution of the RGC2 cluster were investigated. We found extensive AFLP polymorphism within and between populations climaxing in the populations from eastern Turkey and Armenia, suggesting that this region is the center of origin of the wild lettuce L. serriola.
MATERIALS AND METHODS
Plant materials:
Seeds from 993 individuals (accessions) of L. serriola populations were collected during the summers of 1993–1998 from 49 populations from 11 countries, with the majority included in previous studies (supplemental Table 1 at http://www.genetics.org/supplemental/; Sicard et al. 1999; Kuang et al. 2006). Each collection was made from a single individual and subsequently treated as an independent accession. The individuals collected from within a population were growing at least 1 m apart. Twenty-five accessions were collected from five different patches 500–3000 m apart in Wageningen, The Netherlands, and are considered one population (Wageningen) in this study. Five to 10 seedlings from each accession were pooled to extract DNA representing the progenitor's genotype. DNA was extracted following the method of Sicard et al. (1999). Each individual is considered homozygous because L. serriola is predominantly self-pollinated (Prince and Carter 1977).
AFLP analysis:
AFLP was performed following the procedures described by Vos et al. (1995). Genomic DNA was digested with restriction enzymes EcoRI and MseI. Adaptors were ligated to the digested fragments, and the ligated fragments were preamplified using the primer combination E + 0 and M + 0 (Table 1). Selective amplifications were performed with two primer combinations: E + ACA/M + CAC and E + ACT/M + CGA (Table 1). The primers derived from EcoRI adaptors were labeled with 1 μCi [γ33P]ATP. PCR products were resolved electrophoretically in a 5% polyacrylamide gel and the AFLP images were scored manually. Fragments that were amplified by the same primer combination and had identical mobility are considered to be the same allele at a locus; accessions with no amplification at the locus are considered to have the alternate allele (Rouppe van der Voort et al. 1997).
TABLE 1.
Oligonucleotide primer sequences
| Name | Sequence 5′–3′ |
|---|---|
| AdaptorE | CTCGTAGACTGCGTACC … CTGACGCATGGTTAA-5′ |
| AdaptorM | GACGATGAGTCCTGAG … TACTCAGGACTCAT-5′ |
| E + 0 | CTCGTAGACTGCGTACCAATTC |
| E + ACA | CTCGTAGACTGCGTACCAATTCACA |
| E + ACT | CTCGTAGACTGCGTACCAATTCACT |
| M + 0 | GACGATGAGTCCTGAGTAA |
| M + CAC | GATGAGTCCTGAGTAACAC |
| M + CGA | GATGAGTCCTGAGTAACGA |
| 5E6 | AATGAAAGTGATWGTGAAG |
| 3E6 | TCWTCCCCAAGAAGAA |
| K-for | CAAGTGCTGAATATATACAGGTG |
| K-rev | AAGGCGAGCAAGTGTTACAGTT |
| Ex4b | GCATTGTCAAGTGTGATTCCAT |
| L-rev | TTGCCAATAGATTCTTCTTCC |
Microsatellite MSATE6:
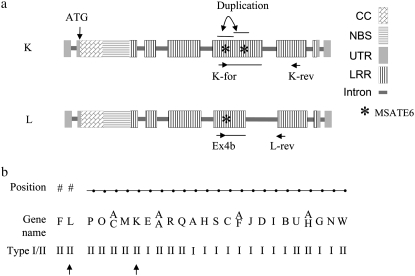
The compound microsatellite MSATE6 comprises three trinucleotide repeat motifs, (ACA)x, (TCA)y, and (TCT)z, and is located in exon 5 of most RGC2 genes (Figure 1). Primers 5E6 and 3E6 were designed to conserved sites flanking the trinucleotide repeats and were expected to amplify PCR products from the majority (>80%) of known RGC2 genes (Table 1). PCR was performed in a 20-μl reaction volume containing 2 mm MgCl2, 0.25 mm dNTPs, 50 mm KCl, 10 mm Tris, 1 unit Taq, 50 ng genomic DNA, 0.5 μm of primer 3E6, and 0.05 μm primer 5E6 that was labeled with 1 μCi [γ33P]ATP. PCR products were resolved electrophoretically in a 5% polyacrylamide gel.
Figure 1.—
Positions of PCR primers and MSATE6 in RGC2 K and RGC2 L. (a) The horizontal lines after K-for and Ex4b indicate the region sequenced and analyzed in this study. (b) Positions of genes TDK and TDL in the RGC2 cluster in cv. Diana are indicated by arrows. Beads in a string indicated the determined physical positions. Genes TDF and TDL were located outside the string. “TD” was omitted for all gene names. Modified from Kuang et al. (2004).
Due to the three trinucleotide repeats, various insertion/deletions between the two primers, and amplification from multiple paralogs, the MSATE6 profile of each accession displayed complex informative patterns containing multiple fragments (Figure 2). Most amplified MSATE6 fragments were 122–221 bp in length. Each MSATE6 fragment may represent alleles of the same gene or belong to different paralogs. Each fragment with a different size in the sequencing gel is called an “array” in this study and named “E6-” followed by its length, for example, array E6-122. A few large arrays (>500 bp), which were amplified from the RGC2 genes with unequal crossover in exon 5, were also observed for most genotypes (Figure 1; Kuang et al. 2004). These large arrays were excluded from further analysis since the genes from which they were amplified also amplified arrays of ∼130 bp in length.
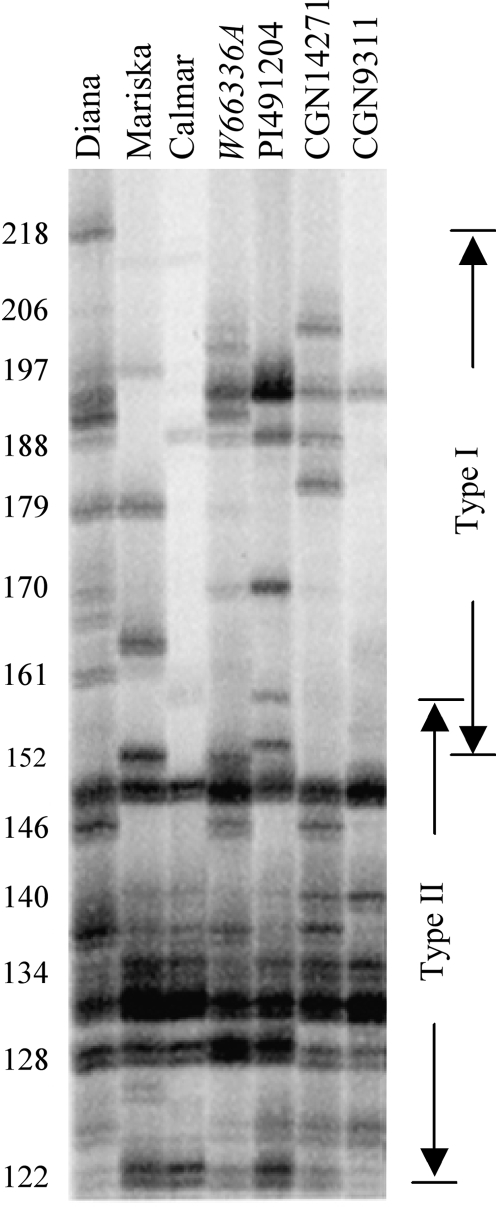
Figure 2.—
MSATE6 profiles of seven Lactuca genotypes. MSATE6 fragments in type I RGC2 genes are 152 bp or larger, while MSATE6 fragments in type II genes are 158 bp or shorter. Numbers on the left show the sizes (bp) of fragments. Modified from Figure 2 of Kuang et al. (2004).
AFLP and microsatellite marker data analysis:
The AFLP marker data were used to estimate Nei's diversity index and genetic distance using the software Popgene (Nei 1973, 1978; Yeh and Boyle 1997). Genetic diversity in the total population (HT) was decomposed into genetic diversity within and between populations (HC and DCT). The relative degree of gene differentiation among populations was measured as GST = DCT/HT (Nei 1973). Nei's genetic distance was calculated using Popgene (Nei 1978; Yeh and Boyle 1997). Correlation between genetic distance and geographic distance was tested using the Mantel test (Simier et al. 1998).
Sequencing alleles of TDK and TDL:
RGC2 homologs TDK and TDL from the lettuce (L. sativa) cultivar Diana belong to two different lineages of type II RGC2 genes in Lactuca (Kuang et al. 2004). TDL is located at one edge of the cluster and is separated by at least four RGC2 genes from TDK (Figure 1; Kuang et al. 2004). It is estimated that TDL and TDK in cultivar Diana are at least 200 kb apart. To study the evolution and population genetics of these two genes, fragments from the LRR-encoding region were amplified from 124 and 83 accessions of L. serriola using primer combinations K-for/K-rev and Ex4b/L-rev, respectively (Table 1; Figure 1). The PCR products were treated with ExoSAP-IT (USB, Cleveland) and then sequenced directly. The alleles of these two genes are called RGC2 K alleles and RGC2 L alleles, respectively, in this article.
Sequences of RGC2 K and L alleles, respectively, were aligned using ClustalX (Thompson et al. 1997). Nucleotide diversity (π), recombination (Hudson and Kaplan 1985), Tajima's D test (Tajima 1989), Fu and Li's D test (Fu and Li 1993), Fu's F test (Fu 1997), and linkage disequilibrium were calculated using DnaSP (Rozas and Rozas 1999).
RESULTS
Genomewide diversity shown by AFLP markers:
Screening of 696 L. serriola accessions from 41 populations using AFLP primer combinations E + ACA/M + CAC and E + ACT/M + CGA identified 168 and 151 loci (markers), respectively. Of the 319 AFLP markers, 294 (92.2%) were polymorphic. Using the AFLP data, the genetic diversity of the whole species population was estimated to be HT = 0.184. The majority of the genetic diversity was attributed to the diversity between populations (DCD = 0.117), with a genetic differentiation of GST = 0.636.
The most diverse populations shown by AFLP markers were from Armenia and eastern Turkey, while European and Israeli populations were the least polymorphic (Table 2). Six of the 10 Turkish populations (Istanbul, Bolu, Kayseri, Kayseri–Sivas, Sivas, and Ankara), three of the five Armenian populations (Chor, Abovian, and Artanish), and two of the four Californian populations (Yolo landfill and Road 29) had a genetic diversity of He > 0.1. The most polymorphic population was Abovian (Armenia) with He = 0.132. By contrast, all European and Israeli populations had genetic diversity with He < 0.1. The highest diversity present in eastern Turkish and Armenian populations suggests that Turkey/Armenia or surrounding regions might be the center of diversity and origin of L. serriola.
TABLE 2.
Genetic diversity of natural populations of wild lettuce
| AFLP
|
MSAT E6
|
RGC2 K
|
RGC2 L
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | Coll. year | N | He | N | No. Hap. | Hap div. | Array no. | He | N | π | N | π |
| Turkey | ||||||||||||
| Goreme | 98 | 19 | 0.081 | 18 | 11 | 2.08 | 13.1 | 0.103 | ||||
| Istanbul | 98 | 20 | 0.109 | 7 | 6 | 1.75 | 11.7 | 0.223 | ||||
| Bolu | 98 | 20 | 0.103 | 16 | 14 | 2.60 | 12.8 | 0.255 | 7 | 0.008 | 6 | 0.010 |
| Ankara | 98 | 10 | 0.113 | 8 | 7 | 1.91 | 12.4 | 0.284 | ||||
| Yakakent | 98 | 19 | 0.052 | 18 | 5 | 1.33 | 11.7 | 0.131 | ||||
| Bergama | 98 | 21 | 0.048 | 16 | 7 | 1.45 | 12.2 | 0.252 | ||||
| Kayseri | 98 | 20 | 0.120 | 19 | 14 | 2.48 | 14.4 | 0.263 | 9 | 0.011 | 6 | 0.003 |
| Kayseri–Sivas | 98 | 18 | 0.102 | 20 | 9 | 1.86 | 11.4 | 0.113 | ||||
| Sivas | 98 | 20 | 0.101 | 8 | 3 | 0.87 | 11.1 | 0.144 | ||||
| Afyon | 98 | 20 | 0.070 | 19 | 7 | 1.38 | 11.4 | 0.132 | ||||
| Armenia | ||||||||||||
| Chor | 98 | 20 | 0.104 | 20 | 16 | 2.74 | 12.9 | 0.162 | 11 | 0.008 | 15 | 0.001 |
| Abovian | 98 | 20 | 0.132 | 17 | 13 | 2.43 | 14.5 | 0.219 | ||||
| Areni | 98 | 19 | 0.080 | 17 | 15 | 2.96 | 16.2 | 0.199 | ||||
| Sevan | 98 | 16 | 0.098 | 12 | 3 | 0.76 | 11.5 | 0.081 | ||||
| Artanish | 98 | 18 | 0.125 | 6 | 6 | 1.75 | 12.8 | 0.172 | ||||
| Israel | ||||||||||||
| N. Nazareth | 94 | 11 | 0.052 | 19 | 6 | 1.49 | 11.4 | 0.154 | ||||
| S. Nazareth | 94 | 31 | 0.034 | |||||||||
| Haifa Freud | 94 | 43 | 0.045 | 50 | 8 | 1.55 | 11.9 | 0.158 | 16 | 0.004 | 10 | 0.000 |
| Shefayim | 94 | 12 | 0.000 | 11 | 1 | 0.00 | 10.0 | 0 | ||||
| Be'er Sheva | 94 | 15 | 0.056 | 36 | 22 | 3.04 | 13.5 | 0.129 | 22 | 0.000 | ||
| Tel Hanan | 95 | 22 | 0.043 | 16 | 13 | 2.31 | 13.8 | 0.111 | ||||
| Netanya | 95 | 6 | 0.089 | 13 | 8 | 1.97 | 14.0 | 0.099 | ||||
| Haifa Oren Road | 95 | 22 | 0.035 | 22 | 15 | 2.60 | 14.9 | 0.174 | ||||
| Europe | ||||||||||||
| Salamanca | 94 | 9 | 0.041 | 8 | 3 | 0.74 | 10.9 | 0.006 | ||||
| Alcanices | 94 | 6 | 0.028 | 6 | 6 | 1.79 | 12.7 | 0.173 | ||||
| Pyrenees | 97 | 12 | 0.041 | 6 | 1 | 0 | 12.2 | 0 | ||||
| Aix les Bains | 97 | 17 | 0.049 | 6 | 4 | 1.39 | 11.5 | 0.149 | ||||
| Mont Blanc | 97 | 14 | 0.027 | |||||||||
| Nancy 1 | 97 | 7 | 0.084 | 10 | 5 | 1.56 | 13.8 | 0.197 | ||||
| Nancy 2 | 97 | 9 | 0.072 | |||||||||
| Pavia | 97 | 16 | 0.095 | |||||||||
| Luzern | 97 | 19 | 0.019 | 17 | 4 | 1.26 | 13.9 | 0.082 | ||||
| Landau | 96 | 16 | 0.006 | 16 | 2 | 0.23 | 12.9 | 0.003 | ||||
| Koblenz | 96 | 14 | 0.057 | 14 | 6 | 1.23 | 14.9 | 0.156 | ||||
| Rostock | 96 | 7 | 0.030 | 8 | 2 | 0.38 | 12.3 | 0.064 | ||||
| Antwerp | 97 | 8 | 0.003 | 13 | 1 | 0 | 13.0 | 0 | ||||
| Wageningen | 98 | 25 | 0.010 | |||||||||
| California | ||||||||||||
| Yolo landfill | 95 | 32 | 0.112 | 19 | 17 | 2.77 | 11.8 | 0.204 | 23 | 0.001 | 15 | 0.008 |
| Putah Creek | 95 | 13 | 0.087 | 25 | 16 | 2.62 | 12.8 | 0.223 | ||||
| Road 29 | 95 | 15 | 0.117 | 55 | 24 | 2.56 | 11.1 | 0.169 | ||||
| Road 32 | 95 | 15 | 0.091 | 30 | 13 | 1.79 | 14.8 | 0.079 | ||||
| Salinas | 95 | 26 | 14 | 2.32 | 14.7 | 0.217 | 13 | 0.011 | 14 | 0.006 | ||
| Gilroy | 95 | 55 | 37 | 3.43 | 15.0 | 0.258 | ||||||
| Russel | 95 | 7 | 0.098 | |||||||||
N, number of accessions studied, only populations with six or more accessions were included; He, Nei's diversity index; He for MSATE6, calculated considering the RGC2 cluster as 34 loci; No. Hap, number of different MSATE6 haplotypes; Array no., average number of MSATE6 arrays; Hap Div, Shannon's index for haplotype diversity; π, nucleotide diversity.
The least polymorphic populations were Shefayim (Israel), Wageningen (The Netherlands), Landau (Germany), Antwerp (Belgium), all with He < 0.01. The 24 accessions from the Shefayim population (Israel) were monomorphic, suggesting that this population was derived from a single genotype. The Wageningen population also had very low genetic diversity: 23 of the 25 accessions analyzed had the same AFLP profiles. These 23 accessions were located throughout five different patches collected 500–3000 m apart. Thus, the high frequency (23/25) of this AFLP-defined genotype suggests its prevalence in Wageningen rather than sampling errors. The other two accessions from the Wageningen population had different AFLP profiles, giving a total of three genotypes in the 25 accessions from Wageningen. Interestingly, the three genotypes from the Wageningen population were very diverse, with a total of 36 polymorphic AFLP markers among them. Such a genetic structure is consistent with a low outcrossing rate in L. serriola (Prince and Carter 1977). The other two populations with low diversity were Antwerp and Landau, containing only 3 and 12 polymorphic AFLP markers among the 8 and 16 accessions studied, respectively.
MSATE6 polymorphism:
MSATE6 arrays derived from type II genes have higher frequencies than those from type I genes:
The lengths of MSATE6 arrays encoded by type I RGC2 genes differ from those encoded by type II genes. In the seven Lactuca genotypes that were investigated extensively in a previous study (Kuang et al. 2004), all MSATE6 fragments >158 bp were encoded by type I RGC2 genes (Figure 2). In contrast, all MSATE6 fragments <152 bp were encoded by type II RGC2 genes.
We screened a total of 709 accessions from 45 natural populations of L. serriola using MSATE6, yielding 34 arrays ranging from 119 to 221 bp in length. The arrays form ladders composed of 3-bp increments except that an array of 212 bp was not observed in any of the 709 accessions screened. Arrays <152 bp, which were most likely encoded by type II genes, were found in higher frequencies than arrays >155 bp, which were most likely encoded by type I genes. The average of frequency for arrays <152 bp is 0.735, in contrast to only 0.219 for arrays >155 bp. Eight of the 10 arrays <152 bp had frequency >0.5, while only 1 (E6-191) of the 21 arrays >155 bp had frequency >0.5. This is consistent with the previous conclusion that each type I R-gene tends to have low frequencies, while each type II R-gene tends to have high frequencies in natural populations.
The number of arrays in MSATE6 profiles differs significantly among populations:
The number of MSATE6 arrays amplified from an accession varied from 8 to 22, with an average of 13.0 ± 2.4. The majority (81%) of the 709 accessions screened had 10–15 MSATE6 arrays. However, 16 accessions had only 8 arrays, in contrast to 9 accessions with no less than 20 arrays. The average number of arrays is significantly different among populations [F = 7.89, d.f. = (42, 683)]. The average number of arrays for individuals in a population varied from 10.0 ± 0 (Shefayim, Israel) to 16.2 ± 2.99 (Areni, Armenia). Excluding the Shefayim population, which is monomorphic in both AFLP and MSATE6 markers, the Salamanca population (Spain) had the lowest average number of MSATE6 arrays (10.9 ± 0.35). The variation of array number is attributed mainly to variations in the number of arrays >155 bp, i.e., those likely encoded by type I RGC2 genes. The Areni population (Armenia) has an average of 7.2 arrays >155 bp, in comparison to only 3.0 in the Salamanca population.
The relationship between the number of MSATE6 arrays and the total number of RGC2 genes in seven genotypes had been determined previously (Kuang et al. 2004). The best regression between the two variables was found to be y = 4.64 × 1.10x, where y is the total number of RGC2 genes in a genome and x is the number of MSATE6 arrays in that genome. Using this formula, the average number of RGC2 homologs in a genome of an L. serriola accession in the study reported here is estimated to be 16.3 ± 5.9 and the number of RGC2 genes in a L. serriola population is estimated to vary from 12 to 22.
Genetic diversity calculated using MSATE6 data:
To simplify the analysis, each MSATE6 array was considered to be a dominant marker with two alleles: the present allele and the absent allele. Since a total of 34 arrays were detected, the RGC2 cluster was considered a region with 34 tightly linked loci. Genetic diversity of the RGC2 cluster in different populations was calculated using the MSATE6 data. Nei's diversity index for all accessions included in this study was 0.278. Approximately half of the diversity was attributed to diversity between populations, with genetic differentiation of GST = 0.491. The diversity index of a population varied from 0 to 0.284 (Table 2). Again, eastern Turkish populations had high diversity at the RGC2 locus: the Ankara population had the largest genetic diversity (0.284), and the populations of Kayseri, Bergama, and Bolu were among the five populations with a diversity index >0.25. European populations were the least polymorphic, with an average diversity index of 0.088. The populations of Antwerp (Belgium), Mount Blanc (France), and the Pyrenees (France) all had monomorphic MSATE6 profiles.
There are a large number of RGC2 haplotypes in natural populations:
The diversity of the RCG2 locus in a population was also estimated considering each distinct RGC2 cluster as a haplotype. RGC2 haplotypes were considered to be different as long as they varied in one array in the MSATE6 profile. We identified 366 different haplotypes in the 709 accessions analyzed, with an average of one haplotype for every two accessions. Of the 366 haplotypes, 274 haplotypes were observed only once (singletons) and the other 92 were present in >1 accession. Accessions with identical MSATE6 profiles were mainly (96.4%) from the same population and only 13 MSATE6 haplotypes occurred in two or three populations, which were from California or Israel, respectively. The most common haplotype, present in Israeli populations, had a frequency of 3.7%. The four most common haplotypes were present in either Israeli populations or Californian populations, probably because a large number of accessions were sampled from these populations (Table 2). The number of different haplotypes in each population is listed in Table 2. The Gilroy population (California) had 37 different RGC2 haplotypes, while the populations of Shefayim (Israel), the Pyrenees (France), and Antwerp (Belgium) each had only 1 haplotype. Comparisons between the numbers of haplotypes in different populations should be treated with caution because sample sizes varied considerably for these populations. To reduce the effects of sampling size, Shannon's diversity index was calculated for each population (Table 2). There was a significant correlation between the diversities when considering the RGC2 cluster as a single haplotype and the diversities when considering the RGC2 cluster as multiple loci (r = 0.741, P < 0.01). However, the former was not as informative as the latter since different MSATE6 profiles may vary by as few as 1 to as many as 10 arrays (Figure 3). Therefore, the latter scenario (RGC2 cluster as multiple loci) was considered to be more informative and, hereafter, the diversity of the RGC2 cluster was calculated considering the RGC2 cluster as multiple loci.
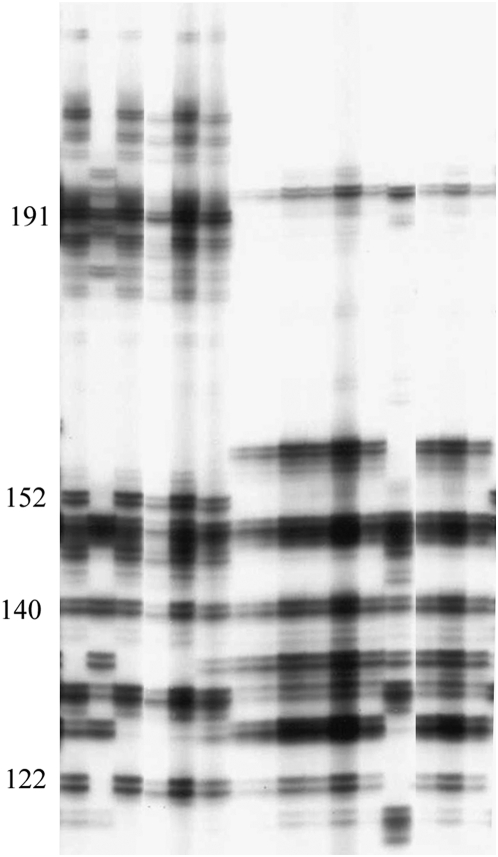
Figure 3.—
Two groups of accessions from population Bergama (Turkey) showing very distinctive MSAT E6 profiles. Numbers on the left show the sizes (bp) of fragments.
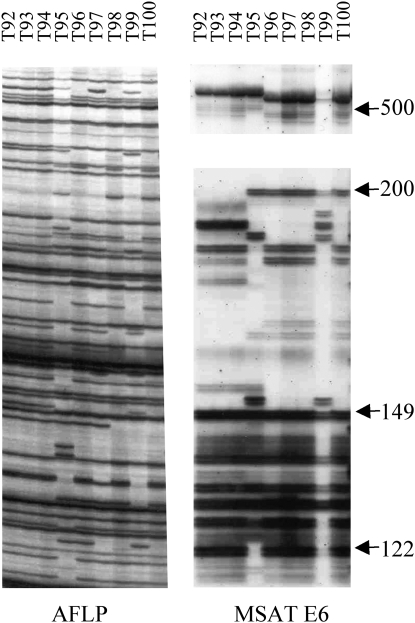
As expected, accessions with identical AFLP profiles always had the same MSATE6 profiles (Figure 4). Some accessions with different AFLP profiles had identical MSATE6 profiles as well; for example, accessions T96, T97, and T98 had different AFLP profiles but had identical MSATE6 profiles (Figure 4). Similarly, accessions T128 and T133 from population Bolu (Turkey) had different AFLP profiles but had the same MSATE6 profiles (data not shown). Sequencing K and L alleles in accessions T128 and T133 showed that they have identical K and L sequences (see below), suggesting that the RGC2 cluster in accessions T128 and T133 are identical. Potentially identical RGC2 clusters (i.e., identical MSATE6 profiles) occurred only in different genotypes from the same population or from geographically close populations, suggesting that the RGC2 clusters were identical by descent and there was gene flow within and locally between different populations.
Figure 4.—
Different genotypes may have identical RGC2 haplotype. Accession T96, T97, and T98 from population Afyon (Turkey) have different AFLP profiles but identical MSATE6 profiles.
Evolution and population genetics of RGC2 K and L:
Nucleotide diversity of RGC2 K:
The RGC2 K alleles/orthologs belong to one of the most distinct lineages of type II RGC2 genes and were investigated in a panel of 40 accessions from different populations in a previous study (Kuang et al. 2004). Here, we extended the study to examine its population structure. Screening 337 accessions using the K-specific primer combination K-for/K-rev showed that RGC2 K had a frequency of 77% in natural populations. To calculate its nucleotide diversity, fragments (1058 bp) of the RGC2 K were PCR amplified and sequenced from 88 accessions. The 88 RGC2 K sequences were combined with the 37 RGC2 K sequences cloned previously from L. serriola, yielding a total of 125 RGC2 K sequences. Twenty-six distinct nucleotide sequences were discovered. The most frequent K allele is present in 41 of the 124 accessions of nine populations; the second most frequent K allele was present in 36 accessions of three Israeli populations; eight K alleles were present in 2 to 7 accessions; and the other 16 alleles were singletons.
There are 62 single nucleotide polymorphisms (SNPs) among the 125 RGC2 K fragments; twenty-two of them are singletons. The total nucleotide diversity for the 125 K alleles was π = 0.010. Nucleotide diversities were calculated for seven populations in which RGC2 K fragments were sequenced from at least six accessions (Table 2). The nucleotide diversities in the seven populations varied from π = 0.00 (Be'er Sheva, Israel) to π = 0.011 (Kayseri, Turkey). All 22 K alleles sequenced from Be'er Sheva (Israel) were identical. The Yolo population (California) had the second lowest nucleotide diversity, with π = 0.001. A total of 23 K alleles were cloned from the Yolo population; 22 of them had identical sequences; the other fragment (Y2301) had 11 SNPs. Among the seven populations studied, the populations of Salinas (California) and Kayseri (Turkey) had the highest nucleotide diversity (π = 0.011).
Nucleotide diversity of the gene L:
We also studied nucleotide diversity of a second lineage of type II RGC2 homologs, the alleles of RGC2 L. Screening 93 accessions using an L-specific primer combination showed that RGC2 L had a frequency of 83% in natural populations. Fragments (565-577 bp) of RGC2 L were PCR amplified and sequenced from 55 accessions, which were combined with the 28 RGC2 L sequences obtained previously (Kuang et al. 2004). Fourteen distinct RGC2 L nucleotide sequences were discovered and ten of them were singletons. The other four sequences were present in 3, 16, 17, and 37 accessions, respectively. There were a total of 22 polymorphic sites (including both SNPs and indels) among the 83 L alleles; seven of them were singletons.
The nucleotide diversities for all 83 RGC2 L fragments was π = 0.010, similar to that for the RGC2 K. Nucleotide diversities were calculated for six natural populations with the RGC2 L gene sequenced from six or more accessions. Nucleotide diversity of RGC2 L varied from π = 0.000 to 0.010 in the six populations (Table 2). The Bolu population (Turkey) had the highest nucleotide diversity for RGC2 L, with π = 0.010. The Haifa Freud Road population (Israel) had the lowest nucleotide diversity (π = 0), and all ten RGC2 L sequences obtained were identical. Nucleotide diversity in the Chor population (Armenia) was near zero. Fifteen RGC2 L sequences were obtained from the Chor population and the only polymorphism among them was a synonymous mutation in accession A6, changing one trinucleotide repeat (TCA) to (TCT) in MSATE6.
Purifying selection acting on RGC2 K and L:
The 125 RGC2 K sequences had 62 SNPs and 23 polymorphic amino acid sites. Its nonsynonymous mutation rate is Ka = 0.0067 and synonymous mutation rate is Ks = 0.0227, with a Ka:Ks = 0.3. Fu and Li's D, Fu's F, and Tajima's D tests all indicated that RGC2 K was under purifying selection, though the parameters are not statistically significant (data not shown). Purifying selection also occurred at the putative solvent exposed residues. The RGC2 K sequences have 48 codons (144 nt encoding solvent exposed residues that had previously been shown to be under significant diversifying selection in comparisons between RGC2 homologs (Kuang et al. 2004). Only eight SNPs were found in the 144 bp sequences of all 125 RGC2 K fragments and only two of them resulted in an amino acid change. The ratio of nonsynonymous to synonymous mutation rates at the codons encoding putative solvent exposed residues is Ka:Ks = 0.067. The Ka:Ks ratio, Fu and Li's D, Fu's F, and Tajima's D tests all suggested that RGC2 K has been under purifying selection. Analysis of the 83 RGC2 L sequences provided similar results indicating that the RGC2 L gene has also been under purifying selection.
Linkage disequilibrium (LD) in the RGC2 cluster:
To investigate linkage disequilibrium in the RGC2 cluster, LD between polymorphic sites within RGC2 K and L genes was measured. Accessions with identical AFLP and MSATE6 profiles were excluded from the LD analysis because they were believed to be the same genotype. A total of 106 nonredundant RGC2 K sequences were available for analysis. The 23 most polymorphic sites in RGC2 K sequences were used for LD analysis. Each nucleotide at these polymorphic sites had frequency >10% in the 106 RGC2 K sequences. Of the 253 possible pairwise combinations, 159 pairs showed significant LD (Bonferroni procedure). Similar results were found for the RGC2 L alleles. LD was investigated between the ten most polymorphic SNPs among 85 RGC2 L sequences. Forty-two of all 45 possible pairwise combinations in the RGC2 L sequences showed significant LD. Using the four-gamete test, twelve and three intragenic recombination events were discovered between RGC2 K and RGC2 L alleles, respectively (Hudson and Kaplan 1985). Therefore, recombination between alleles had not significantly decayed LD between SNPs within the 565 bp of RGC2 K or the 1058 bp of RGC2 L analyzed.
LD between RGC2 K and RGC2 L was also investigated. Both RGC2 K and L sequences were available from 66 accessions. Intergenic LD was investigated using the 23 most informative sites in the RGC2 K sequences and the 10 most informative sites in the RGC2 L sequences of the 66 accessions. None of the 230 possible pairs showed significant LD, suggesting that the RGC2 cluster has been considerably shuffled through recombination. Frequent recombination within the RGC2 locus is consistent with a large number of RGC2 haplotypes observed in natural populations.
Comparison of genomewide diversity and RGC2 cluster diversity:
Correlation between diversities calculated by different methods:
Correlation between diversities calculated using different methods was analyzed. The Shefayim population (Israel) was derived from a single genotype and was excluded from these correlation studies. Significant correlation was detected between genomewide diversity shown by AFLP marker and diversity at the RGC2 cluster calculated from the MSATE6 data (r = 0.67, d.f. = 36, P < 0.001).
The nucleotide diversity of RGC2 K and RGC2 L was not significantly correlated with the RGC2 cluster diversity (calculated by using MSATE6 data) or genomewide diversity (calculated using AFLP data) in different populations. Furthermore, the nucleotide diversity of the K alleles is not significantly correlated with that of the L alleles. For example, the Chor population (Armenia) had high diversities at the RGC2 cluster and genomewide diversity, but its nucleotide diversity for the L alleles was very low (Table 2). The 20 accessions from the Chor population have 20 different AFLP profiles and 16 different MSATE6 profiles; the nucleotide diversity of the K alleles in Chor was moderately high (π = 0.008); however, nucleotide diversity of the L alleles in the Chor population was near zero (see above). By contrast, the Yolo population (California) had high nucleotide diversity of the L allele (π = 0.008) but low nucleotide diversity of the K alleles (π = 0.001). In summary, the nucleotide diversity of one or a few type II R-genes is not indicative of the diversity of the R-gene cluster in which it resides.
Population distance:
Unbiased genetic distance (Nei 1978) among 41 populations was calculated using the AFLP markers. The largest genetic distance was observed between Tel Hanan (Israel) and Alcanices (Spain) (D = 0.270), and the smallest was between Tel Hanan and Haifa Freud Road (D = 0.009), two populations separated by about 15 km on the western and eastern slopes of Mount Carmel, Israel. The Israeli group was least related to other groups, even though it was geographically close to Turkey. The four Davis, California, populations, recent colonizers, were most related to the Turkish populations, and vice versa. Our data suggest that the California populations might be derived from the Turkish or surrounding populations.
A Mantel test was performed to investigate the correlation between genetic distance (calculated from AFLP data) and geographic distance among populations. The correlation was significant when all populations except the California populations were analyzed (P < 0.00001). The Mantel Test was also performed for populations in Europe, Israel, Turkey, and Armenia, independently, and the only significant correlation was found between Israeli populations (P < 0.05). This may suggest that local ecological stresses may prevail and represent the operating selective forces on the local population's genetic structure. Alternatively, complex colonization patterns as well as isolation by geographical barriers may break simple correlations between genetic and geographical distances.
Genetic distance between populations calculated using MSATE6 data varied from 0.0332 (between the Haifa Oren St and Be'er Sheva populations from Israel) to 0.4964 (between the Road 32 population from California and the Landau population from Germany). Populations with very low diversity (Antwerp, Afyon, Landau, Rostock, and Pyrenees) had very large genetic distances from other populations. The Californian populations were most closely related with the Turkish populations, consistent with the AFLP data. The genetic distances calculated using MSATE6 data had a medium correlation coefficient with those calculated using the AFLP data (r = 0.38).
DISCUSSION
The center of genetic diversity and origin of L. serriola:
We assessed the level of genetic diversity in natural populations of L. serriola using AFLP markers. Populations from eastern Turkey and Armenia were more polymorphic than those from Europe and Israel. Our data suggested that eastern Turkey and Armenia (or surrounding regions) might be the center of diversity of L. serriola and possibly its center of origin. The Iran–Turkey region is the distribution center of L. serriola and several other closely related species such as L. aculeata, L. scarioloides, and L. georgica (Zohary 1991; Lebeda et al. 2004). Further sampling of the genetic diversity of L. serriola from other eastern Mediterranean areas, particularly the Iran–Turkey region, would provide better definition of the geographical limits of its center of diversity and origin.
Multiple regression results showed that longitude was significantly correlated with genetic distance (data not shown). This correlation may reflect the origin and dispersion of this species. L. serriola might have spread from its center of origin (including eastern Turkey and Armenia) first to the Mediterranean basin and then to central and western Europe after the glaciers' retreat in the Upper-Pleistocene Holocene periods. Consequently, the peripheral populations in central and western Europe had only part of the diversity of the original populations. Furthermore, recent data showed that diversity of race-specific resistance in L. serriola populations against B. lactucae dramatically decreased from central to western Europe (the Czech Republic to The Netherlands and UK) (Lebeda and Petrželová 2004b).
The center of origin of a species should have most of the alleles that are present in its peripheral populations, and the peripheral populations may have only part of the diversity present in the center of origin. Eastern Turkish populations had a large number of alleles of AFLP markers that were absent in other groups and the majority of alleles found in other populations. Similar numbers of accessions from Turkey/Armenia and Israel were included in this study; 110 alleles were discovered in Turkey but not in Israel, and only 32 alleles were present in Israel but not found in Turkey. A similar pattern was observed for the MSATE6 marker. All 34 arrays of MSATE6 were present in Turkish/Armenian populations, but only 27 different arrays were found in Israeli populations, and 31 in Armenian, European, or Californian populations.
Methodology of studies on diversity of R-gene cluster:
The paucity of studies on R-gene clusters is in part due to the formidable complexity of such studies. The diversity of an R-gene cluster includes variation in overall copy number, presence/absence of type II R-genes, SNPs in type II and type I R-genes, and the chimeric, non-orthologous structures of type I R-genes. Such variety makes the study on diversity of R-gene clusters difficult. If an R-gene cluster contains a few type II genes but no type I genes, its diversity in a population or species can be analyzed more readily. PCR primers specific to each lineage of type II gene can be designed so that individual allelic sequences can be amplified and sequenced directly from each accession. However, for a complex R-gene locus such as the RGC2 cluster in lettuce, it is impractical to sequence all type II genes in the cluster. Sampling a few lineages of type II genes also may not adequately address the problem. We showed in this study that there was no correlation between diversity in two lineages of type II genes. Therefore, diversity of one lineage of type II genes may not represent either the diversity of other lineages of type II genes or the diversity of the R-gene cluster as a whole.
The presence of chimeric type I R-genes in a cluster makes studies on its diversity further complicated. Type I R-genes, if present, can be a large component of the diversity of an R-gene cluster. However, it is nearly impossible to measure the diversity of type I R-genes in a population or a species. First, it is difficult to obtain the sequences of type I R-genes because unlike type II R-genes, type I R-genes can not be reliably PCR amplified using specific primers. Second, the frequent sequence exchanges between type I R-genes abolish allelic relationships between genes in different genotypes and violate the assumptions of most population genetics and evolution tests.
An alternative for measuring the diversity of an R-gene cluster is to use haplotype data. The R-gene cluster can be fingerprinted using locus-specific markers that resolve multiple family members. RFLP markers can be used if there are no close homologs elsewhere in the genome. In this study, we used a highly informative microsatellite, MSATE6, to investigate the diversity of the RGC2 cluster. Such an informative marker is unavailable for most R-gene families. However, the use of haplotypes to estimate population diversities has several limitations. First, different haplotypes contribute equally to the estimation of diversity regardless of how much they differ. Second, different populations may not share any common haplotypes due to the complexity of the R-gene cluster and presence of the fast-evolving type I R-genes. Consequently, the haplotype analysis may provide little information on population structure such as relative diversity in different populations, population distance, differentiation, etc.
Evolution of R-gene cluster:
Previous data indicated that there were numerous type I R-genes in natural populations and each genotype had a different set of them (Kuang et al. 2004). The data from this study were consistent with this hypothesis since a large number of distinct haplotypes of RGC2 clusters was detected in natural populations. Accessions with identical RGC2 haplotypes usually had identical AFLP profiles as well, suggesting that they were derived from the same ancestral genotypes. Only rarely did different AFLP genotypes have the same RGC2 haplotype; these may have been the consequence of recent outcrossing.
Outcrossing, which even occurs in selfing species such as Arabidopsis, wild barley (Hordeum spontaneum) or wild emmer (Triticum dicoccoides) will bring two R-gene haplotypes into one genome and provides the opportunity for the generation of novel haplotypes through recombination and the decay of LD. The lack of LD between RGC2 K and RGC2 L suggested both some level of outcrossing for L. serriola and recombination within the RGC2 cluster. Heterozygosity at an R-gene locus might lead to heterosis and be selected for in natural populations. Rare heterozygosity has previously been observed at the RGC2 locus in L. serriola (Kuang et al. 2004) and resistance gene locus (loci) against Phytophora infestans in Solanum demissum (Black and Gallegly 1957). It will be interesting to compare the relative frequency of heterozygosity at R-gene loci with that at non-R-gene loci in natural populations of selfing species.
Novel R-gene haplotypes can be also generated without outcrossing. First, unequal crossovers between paralogs within a gene cluster expand or contract an R-gene cluster and generate chimeras. Second, gene conversions between paralogs, though not changing the copy number of R-genes in a cluster, generate novel chimeric sequences and therefore a new R-gene haplotypes (Kuang et al. 2004).
Microsatellite in RGC2 locus:
The role of the compound MSATE6, which was used as a marker in this study, on function (i.e., resistance specificity) and evolution of RGC2 genes remains unclear. MSATE6 is located in the coding region of an extended LRR repeat (Kuang et al. 2004) and comprises three trinucleotide repeat motifs that maintain open reading frames. Some RGC2 homologs (such as the K alleles) have no tri-nucleotide repeats in the MSATE6, while others may have as many as 30 repeats. It remains unknown if changes of repeat numbers in MSATE6 affect gene function. Substantial data indicate that microsatellites located in coding regions play important roles in generating genetic variation underlying adaptive evolution (reviewed by Li et al. 2004; Kashi and King 2006). Interestingly, the type I RGC2 genes tend to have more repeats in MSATE6 and greater variation of repeat numbers than type II genes (Figure 2). Presumably, the repeat number in MSATE6 in type I genes, which are under diversifying selection, is subjected to stronger selective pressures than in type II genes.
Genomewide diversity and diversity at R-gene locus:
Genomewide comparison between R-gene loci and non-R-gene loci in Arabidopsis indicated that most R-genes have exceptional polymorphism and often show frequency-dependent selection (Borevitz et al. 2007; Clark et al. 2007). We showed that diversity at the RGC2 locus in different populations is significantly correlated with genomewide AFLP diversity. Genomewide diversity of a wild species is determined by many factors, including its relative location to its origin center and its macrogeographic and microgeographic environmental stresses (Nevo 2001). Recombination frequencies and mutation rates tend to increase under abiotic stresses (Molinier et al. 2006) and, consequently, increase the genomewide diversity of a species in a natural population (reviewed by Nevo 2001). The effects of abiotic stress on the evolution of disease resistance genes remain unknown, but could certainly affect the levels of biotic stresses.
The diversity of the RGC2 cluster might have been influenced by biotic (pathogenic) stresses. The RGC2 cluster contains at least eight functional genes with different specificities against downy mildew caused by B. lactucae (Farrara et al. 1987; Kesseli et al. 1994). B. lactucae is present at least in temperate regions with L. serriola (Lebeda et al. 2001; Lebeda and Petrželova 2004a; Petrzelova and Lebeda 2004), and it was prevalent in the collection sites in eastern Turkey (data not shown). A diverse RGC2 cluster is expected in a L. serriola population if a diverse population of B. lactucae is also present; however, comprehensive surveys of pathogens infecting natural populations of L. serriola are lacking. Furthermore, R-gene diversity reflects host-pathogen co-evolution over evolutionary time and pathogens collected at a single timepoint may have only limited relevance to long-term R-gene evolution. The effects of the pathogen on R-gene evolution are not just limited to selection for genes encoding the corresponding resistance, but also include the changes in frequency of events generating diversity in R-gene homologs (Lucht et al. 2002; Boyko et al. 2007). The high diversity of the RGC2 cluster and genomewide diversity (shown by AFLP analysis) in eastern Turkish and Armenian populations might be the result of high levels of abiotic and biotic stresses in the region of origin of L. serriola, where the RGC2 diversity and the genomewide AFLP diversity evolved together during the long-term evolution of L. serriola.
Acknowledgments
We thank Youchun Li, Avigdor Beiles, Alex Beharav, Alan Templeton, and Alex Lebeda for critical reviewing of this manuscript. This research was supported by U.S.-Israel BARD program grant no. US-2547-95 to R.W.M. and E.N. and by National Science Foundation Plant Genome Program grant #0211923 to R.W.M.
References
- Bakker, E. G., C. Toomajian, M. Kreitman and J. Bergelson, 2006. A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell 18 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beharav, A., D. Lewinsohn, A. Lebeda and E. Nevo, 2006. New wild Lactuca genetic resources with resistance against Bremia lactucae. Genet. Resour. Crop Evol. 53 467–474. [Google Scholar]
- Black, W., and M. E. Gallegly, 1957. Screening of Solanum species for resistance to physiologic races of Phytophthora infestans. Amer. Potato J. 34 273–281. [Google Scholar]
- Borevitz, J. O., S. P. Hazen, T. P. Michael, G. P. Morris, I. R. Baxter et al., 2007. Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104 12057–12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko, A., P. Kathiria, F. J. Zemp, Y. Yao, I. Pogribny et al., 2007. Transgenerational changes in the genome stability and methylation in pathogen-infected plants (virus-induced plant genome instability). Nucleic Acids Res. 35 1714–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo, A. L., and B. A. Schaal, 2004. Heterogeneous evolutionary processes affect R gene diversity in natural populations of Solanum pimpinellifolium. Proc. Natl. Acad. Sci. USA 101 17444–17449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo, A. L., B. A. Schaal and B. N. Kunkel, 1999. Diversity and molecular evolution of the RPS2 resistance gene in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, R. M., G. Schweikert, C. Toomajian, S. Ossowski, G. Zeller et al., 2007. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317 338–342. [DOI] [PubMed] [Google Scholar]
- Crute, I. R., 1992. The role of resistance breeding in the integrated control of downy mildew (Bremia lactucae) in protected lettuce. Euphytica 63 95–102. [Google Scholar]
- Ellis, J. G., G. J. Lawrence, J. E. Luck and P. N. Dodds, 1999. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrara, B., T. W. Ilott and R. W. Michelmore, 1987. Genetic analysis of factors for resistance to downy mildew (Bremia lactucae) in species of lettuce (Lactuca sativa and L. serriola). Plant Pathol. 36 499–514. [Google Scholar]
- Fu, Y. X., 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. X., and W. H. Li, 1993. Statistical tests of neutrality of mutations. Genetics 133 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, M., H. Witsenboer, M. Zabeau, P. Vos, R. Kesseli et al., 1996. PCR-based fingerprinting using AFLPs as a tool for studying genetic relationships in Lactuca spp. Theor. Appl. Genet. 93 1202–1210. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashi, Y., and D. G. King, 2006. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 22 253–259. [DOI] [PubMed] [Google Scholar]
- Kesseli, R., O. Ochoa and R. Michelmore, 1991. Variation at RFLP loci. in Lactuca spp. and origin of cultivated lettuce Lactuca sativa. Genome 34 430–436. [Google Scholar]
- Kesseli, R. V., I. Paran and R. W. Michelmore, 1994. Analysis of a detailed genetic linkage map of Lactuca sativa (Lettuce) constructed from RFLP and RAPD markers. Genetics 136 1435–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang, H., S.-S. Woo, B. C. Meyers, E. Nevo and R. W. Michelmore, 2004. Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell 16 2870–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang, H., O. E. Ochoa, E. Nevo and R. W. Michelmore, 2006. The disease resistance gene Dm3 is infrequent in natural populations of Lactuca serriola due to deletions and frequent gene conversions at the RGC2 locus. Plant J. 47 38–48. [DOI] [PubMed] [Google Scholar]
- Lebeda, A., and I. Petrželova, 2004. a Variation and distribution of virulence phenotypes of Bremia lactucae in natural populations of Lactuca serriola. Plant Pathol. 53 316–324. [Google Scholar]
- Lebeda, A., and I. Petrželová, 2004. b Occurrence of race-specific resistance to Bremia lactucae in Lactuca serriola germplasm originating from four European countries, pp. 113–116 in Genetic Variation for Plant Breeding, edited by J. Vollmann, H. Grausgruber and P. Ruckenbauer. BOKU-University of Natural Resources and Applied Life Sciences, Vienna.
- Lebeda, A., I. Dolezalova, E. Kristkova and B. Mieslerova, 2001. Biodiversity and ecogeography of wild Lactuca spp. in some European countries. Genet. Resour. Crop Evol. 48 153–164. [Google Scholar]
- Lebeda, A., D. A. C. Pink and D. Astley, 2002. Aspects of the interactions between wild Lactuca spp. and related genera and lettuce downy mildew (Bremia lactucae), pp. 85–117 in Advances in Downy Mildew Research, edited by P. T. N. Spencer-Phillips, U. Gisi and A. Lebeda. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Lebeda, A., I. Doležalová, V. Feráková and D. Astley, 2004. Geographical distribution of wild Lactuca species (Asteraceae, Lactuceae). Botanical Review 70 328–356. [Google Scholar]
- Li, Y.-C., A. B. Korol, T. Fahima and E. Nevo, 2004. Microsatellites within genes: Structure, function, and evolution. Mol. Biol. Evol. 21 991–1007. [DOI] [PubMed] [Google Scholar]
- Lindqvist, K., 1960. On the origin of cultivated lettuce. Hereditas 46 319–350. [Google Scholar]
- Lucht, J. M., B. Mauch-Mani, H.-Y. Steiner, J.-P. Metraux, J. Ryals et al., 2002. Pathogen stress increases somatic recombination frequency in Arabidopsis. Nature Genet. 30 311–314. [DOI] [PubMed] [Google Scholar]
- Mauricio, R., E. A. Stahl, T. Korves, D. Tian, M. Kreitman et al., 2003. Natural selection for polymorphism in the disease resistance gene RPS2 of Arabidopsis thaliana. Genetics 163 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B. C., D. B. Chin, K. A. Shen, S. Sivaramakrishnan, D. O. Lavelle et al., 1998. The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10 1817–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier, J., G. Ries, C. Zipfel and B. Hohn, 2006. Transgenerational memory of stress in plants. Nature 442 1046–1049. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1973. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 70 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo, E., 1998. Genetic diversity in wild cereals: Regional and local studies and their bearing on conservation ex-situ and in-situ. Genet. Resour. Crop Evol. 45 355–370. [Google Scholar]
- Nevo, E., 2001. Evolution of genome-phenome diversity under environmental stress. Proc. Natl. Acad. Sci. USA 98 6233–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrželova, I., and A. Lebeda, 2004. Occurrence of Bremia lactucae in natural populations of Lactuca serriola. J. Phytopathol. 152 391–398. [Google Scholar]
- Prince, S. D., and R. N. Carter, 1977. Prickly lettuce (Lactuca serriola L.) in Britain. Watsonia 11 331–338. [Google Scholar]
- Rose, L. E., P. D. Bittner-Eddy, C. H. Langley, E. B. Holub, R. W. Michelmore et al., 2004. The maintenance of extreme amino acid diversity at the disease resistance gene, RPP13, in Arabidopsis thaliana. Genetics 166 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, L. E., R. W. Michelmore and C. H. Langley, 2007. Natural variation in the Pto disease resistance gene within species of wild tomato (Lycopersicon) II. Population genetics of Pto. Genetics 175 1307–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouppe van der Voort, J. N., P. van Zandvoort, H. J. van Eck, R. T. Folkertsma, R. C. Hutten et al., 1997. Use of allele specificity of comigrating AFLP markers to align genetic maps from different potato genotypes. Mol. Gen. Genet. 255 438–447. [DOI] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15 174–175. [DOI] [PubMed] [Google Scholar]
- Shen, J., H. Araki, L. Chen, J.-Q. Chen and D. Tian, 2006. Unique evolutionary mechanism in R-genes under the presence/absence polymorphism in Arabidopsis thaliana. Genetics 172 1243–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, K. A., D. B. Chin, R. Arroyo-Garcia, O. E. Ochoa, D. O. Lavelle et al., 2002. Dm3 is one member of a large constitutively expressed family of nucleotide binding site-leucine-rich repeat encoding genes. Mol. Plant-Microbe Interact. 15 251–261. [DOI] [PubMed] [Google Scholar]
- Sicard, D., S. S. Woo, R. Arroyo-Garcia, O. Ochoa, D. Nguyen et al., 1999. Molecular diversity at the major cluster of disease resistance genes in cultivated and wild Lactuca spp. Theor. Appl. Genet. 99 405–418. [DOI] [PubMed] [Google Scholar]
- Simier, M., J. Thioulouse and J.-M. Olivier, 1998. ADE-4 software: A tool for multivariate analysis and graphical display. Oceanis 24 393–416. [Google Scholar]
- Stahl, E. A., G. Dwyer, R. Mauricio, M. Kreitman and J. Bergelson, 1999. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400 667–671. [DOI] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D., H. Araki, E. Stahl, J. Bergelson and M. Kreitman, 2002. Signature of balancing selection in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 11525–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D., M. B. Traw, J. Q. Chen, M. Kreitman and J. Bergelson, 2003. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423 74–77. [DOI] [PubMed] [Google Scholar]
- Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee et al., 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski, T., U. Piskurewicz, A. Tomczak, O. Ochoa and R.W. Michelmore, 2007. Silencing of the major family of NBS-LRR-encoding genes in lettuce results in the loss of multiple resistance specificities. Plant J. 51 803–818. [DOI] [PubMed] [Google Scholar]
- Yeh, F. C., and T. J. B. Boyle, 1997. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot. 129 157. [Google Scholar]
- Zohary, D., 1991. The wild genetic-resources of cultivated lettuce (Lactuca sativa L). Euphytica 53 31–35. [Google Scholar]