Abstract
The nematodes Caenorhabditis elegans and C. briggsae independently evolved self-fertile hermaphroditism from gonochoristic ancestors. C. briggsae has variably divergent orthologs of nearly all genes in the C. elegans sex determination pathway. Their functional characterization has generally relied on reverse genetic approaches, such as RNA interference and cross-species transgene rescue and more recently on deletion mutations. We have taken an unbiased forward mutagenesis approach to isolating zygotic mutations that masculinize all tissues of C. briggsae hermaphrodites. The screens identified loss-of-function mutations in the C. briggsae orthologs of tra-1, tra-2, and tra-3. The somatic and germline phenotypes of these mutations are largely identical to those of their C. elegans homologs, including the poorly understood germline feminization of tra-1(lf) males. This overall conservation of Cb-tra phenotypes is in contrast to the fem genes, with which they directly interact and which are significantly divergent in germline function. In addition, we show that in both C. briggsae and C. elegans large C-terminal truncations of TRA-1 that retain the DNA-binding domain affect sex determination more strongly than somatic gonad development. Beyond these immediate results, this collection of mutations provides an essential foundation for further comparative genetic analysis of the Caenorhabditis sex determination pathway.
UNLIKE many other developmental processes, such as anterior/posterior patterning and appendage specification, sex determination varies greatly between different phyla (e.g., De Rosa et al. 1999; Zarkower 2001; Panganiban and Rubenstein 2002). Both environmental sex determination and genetic sex determination (GSD) exist, the latter being the better characterized of the two since it is used by most genetic model species. However, even within GSD systems, diversity is extreme. For example, in both mammals and Drosophila, females are XX and males are XY, but in mammals the male dominant Y chromosome determines sex through the action of SRY (Sinclair et al. 1990; Koopman et al. 1991; Graves 2002), while in Drosophila the X-to-autosome (X:A) ratio determines sex through the differential splicing of dsx (Schutt and Nothiger 2000). In hymenopteran insects such as the honeybee, males are haploid for the entire genome, with heterozygosity at a highly polymorphic locus determining femaleness (Beye et al. 2003; reviewed by Cook 1993). Most nematodes employ an XX/XO system in which females (or hermaphrodites) are XX and males are XO. Like Drosophila, sex determination in nematodes proceeds through assessment of the X:A ratio (Nigon 1951; Madl and Herman 1979). However, unlike Drosophila, the nematode responds to X dosage through a cell-nonautonomous negative regulatory cascade that probably represents a highly modified version of the hedgehog-signaling pathway (Cline and Meyer 1996; Haag and Pilgrim 2005).
Despite the above variety, the discovery that dsx in Drosophila melanogaster, mab-3 in Caenorhabditis elegans, and DMRTI in humans are all homologs with sex determination functions (Raymond et al. 1998) suggests that the various sex determination systems are derived from a common ancestral system and that this deep homology has become virtually unrecognizable at both the sequence and pathway levels (Zarkower 2001; Matsuda et al. 2002; Miller et al. 2003; Haag and Doty 2005). Because of its organismal importance and rapid change, sex determination presents a fascinating opportunity for studying the evolution of development. However, given the rapidity with which sex determination evolves, comparisons between different phyla yield little or no information about the process of change. In contrast, comparisons between closely related species are ideal for addressing this. C. elegans is one of the best-studied sex determination models, and its congeners offer such comparisons on both the molecular and the developmental genetic level (reviewed by Haag 2005; Haag and Pilgrim 2005). Further, because these relatives include both ancestrally gonochoristic (male/female) and derived androdioecious (male/hermaphrodite) mating systems, Caenorhabditis presents an excellent system in which to study the adaptive evolution of an organismally important reproductive trait.
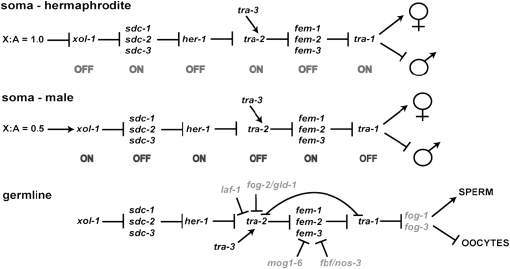
In C. elegans, various autosomal and X-linked loci exert their influence on sex determination by regulating the expression of xol-1. It is transcriptionally repressed in response to an X:A ratio of 1, while a ratio of 0.5 promotes transcription (Meyer 2000). As summarized in Figure 1, low levels of XOL-1 indirectly promote female fate through the transcriptional inhibition of her-1, while high levels of XOL-1 indirectly promote male fates by allowing expression of the secreted protein HER-1 (Hodgkin 1980; reviewed by Trent et al. 1991; Rhind et al. 1995; Meyer 2000). In the absence of HER-1, the membrane protein TRA-2, in conjunction with TRA-3, represses the FEM proteins, which allows the Gli/Ci-related zinc-finger transcription factor TRA-1 to promote female development and repress male development (reviewed by Goodwin and Ellis 2002). In XO males, HER-1 binding to TRA-2 renders the latter incapable of inhibiting the FEM proteins, which can then repress TRA-1 by facilitating its ubiquitin-mediated proteolysis, allowing activation of male fates (Kuwabara 1996a; Goodwin and Ellis 2002; Hamaoka et al. 2004; Starostina et al. 2007).
Figure 1.—
The sex determination pathway of C. elegans. The top and middle represent somatic sex determination in hermaphrodites and males, respectively, while the bottom shows hermaphrodite germline modifications necessary to produce sperm and eggs. Genes required only for germline sex determination are in gray. C. briggsae has orthologs of all pathway components with the exception of fog-2, which has only distant relatives with no clear role in sex determination (Nayak et al. 2005), and fbf-1 and fbf-2, which are most closely related to a clade of three C. briggsae homologs (Lamont et al. 2004; Haag 2005).
Since C. elegans hermaphrodites are essentially females that make sperm prior to oogenesis, the germline must transiently downregulate tra-1 in spite of its XX karyotype. This is dependent upon the post-transcriptional regulation of the tra-2 and fem-3 mRNAs via their 3′-UTR sequences (Figure 1; Doniach 1986; Graham and Kimble 1993; Graham et al. 1993; Zhang et al. 1997; Gallegos et al. 1998; Jan et al. 1999; Kraemer et al. 1999; Clifford et al. 2000). In addition, a TRA-2/TRA-1 protein interaction is necessary for sperm production in hermaphrodites (Lum et al. 2000; Wang and Kimble 2001).
Recent molecular phylogenies of Caenorhabditis (Cho et al. 2004; Kiontke et al. 2004) suggest that C. elegans and C. briggsae evolved self-fertility independently. Functional characterization of C. briggsae sex determination genes has generally supported this convergent evolution scenario. Interspecies transgenesis or RNA interference (RNAi) were used to demonstrate overall functional conservation of Cb-tra-1 (de Bono and Hodgkin 1996) and Cb-tra-2 (Kuwabara 1996b; Jan et al. 1997), Cb-her-1 (Streit et al. 1999), Cb-fog-3 (Chen et al. 2001), Cb-fem-2 (Stothard et al. 2002), and Cb-fem-3 (Haag et al. 2002). However, these methods produce weakly penetrant somatic phenotypes and, in the case of the Cb-fem genes, no germline phenotype was seen at all. In addition, C. briggsae has no fog-2 ortholog, and its well-conserved gld-1 homolog has a major sperm-repressing function not seen in C. elegans gld-1 (Nayak et al. 2005). Taken together, these experiments suggested that some aspects of sex determination differed between C. briggsae and C. elegans, especially in the germline.
Given the incomplete sexual transformation seen in the above studies, we have generated loss-of-function mutations in C. briggsae sex determination genes to obtain more definitive results. Our first study analyzed the fem class of male-promoting genes and demonstrated that Cb-fem-2 and Cb-fem-3 truly are dispensable for XX spermatogenesis, while still performing a conserved, essential role in male somatic development (Hill et al. 2006). In addition, XO Cb-fem mutants are transformed into self-fertile hermaphrodites, and not into self-sterile females as in C. elegans. This intriguing difference in germline fem function implies that C. briggsae hermaphrodite development is regulated downstream of the Cb-fem genes, prompting us to examine the female-promoting tra genes that lie immediately upstream and downstream. In this study, we use the historically successful approach of identifying tra mutations, which transform XX hermaphrodites into pseudomales (Hodgkin and Brenner 1977). These mutations help clarify which aspects of tra function are conserved, inform structure–function relationships, enable both assessment of genetic interactions and modifier screens not possible with RNAi, and set the stage for future comparisons between C. briggsae and its gonochorisitc sister species.
MATERIALS AND METHODS
Nematode husbandry and strains:
General nematode propagation and storage procedures outlined by Brenner (1974) and Wood (1988) were employed to maintain all C. briggsae strains. Incubations were at 15°–25°, and for conditional alleles the temperature is specifically stated. The genetic nomenclature of C. briggsae is currently under revision. For orthologs of C. elegans genes, this article follows the established practice of using the C. elegans name with a “Cb” prefix added to it. For mutations in unknown genes, provisional names that combine a class name matching that of C. elegans, as well as an allele number, are used, but without specifying a gene number [e.g., dpy(nm4), a mutation with a Dumpy phenotype, but of unknown homology]. Although class names are subject to future revision, allele numbers will not change. Chromosomes are named according to the provisional genetic map of phenotypic mutants curated by B. Gupta of McMaster University, which assumes extensive synteny with C. elegans (see http://wormlab.caltech.edu/briggsae/). Assignments of Cb-tra genes and linked markers to specific chromosomes have been confirmed by a DNA polymorphism linkage map (Hillier et al. 2007; see also http://snp.wustl.edu/snp-research/c-briggsae/genetic-map-of-c-briggsae.html).
Mutations described and/or used in this study include:
LGII: Cb-tra-2(nm1, nm9ts, ed23ts, nm29, nm31, nm36ts), dpy(nm4), dpy(sy5148)
LGIII: Cb-tra-1 (nm2, nm10, nm30), dpy(sy1281), let(nm28), dpy(sy5022)
LGIV: Cb-tra-3 (nm65, ed24ts), dpy(sy5027).
F2 mutagenesis screens:
Late L4 C. briggsae AF16 hermaphrodites were placed in M9 salts containing 50 mm ethyl methanesulfonate (for nm alleles) or 1 mm N-nitroso-N-ethylurea (for ed alleles) and incubated on a rocker at room temperature for 4 hr. Following mutagenesis, worms were washed with M9, transferred to NGM plates, and allowed to produce F1 self-progeny (at 25° for temperature-sensitive screens). Either 30 (for alleles nm1 and nm2) or 2 F1 L4 hermaphrodites were transferred to new NGM plates and allowed to self. Each plate was subsequently scored for the presence of F2 Tra animals, and Cb-tra alleles were initially maintained through sib selection. Each was outcrossed with unmutagenized AF16 mates at least four times before stocks were archived and characterization began. Cb-tra-1(nm10) was also maintained as a stock (strain CP10) in the background in which it was originally isolated due to the fortuitous presence of a linked lethal mutation in trans, let(nm28), which serves as a helpful pseudobalancer. The let(nm28) mutation was later used to stabilize the outcrossed Cb-tra-1(nm2) allele in the strain CP38.
Linkage to candidate genes via DNA polymorphisms:
Easily scored polymorphisms in Cb-tra-1, Cb-tra-2, and Cb-tra-3 were identified by resequencing of introns in the HK104 strain, which was known to be genetically distinct (Graustein et al. 2002; Cutter et al. 2006b). For Cb-tra-1, a 37-nt insertion–deletion polymorphism in intron 1 was used. Cb-tra-2 linkage was assessed using a polymorphic BstBI restriction site in intron 2. The Cb-tra-3 polymorphism in intron 1 was scored using an affected SpeI site. Cb-tra hermaphrodites carrying mutant alleles were mated with wild-type HK104 males. Several F1 hermaphrodites from each plate were singled and selfed (at nonpermissive temperature where relevant), and F2 Tra animals were isolated for single-worm PCR (SW–PCR) using one or more of the three PCR conditions described below. Single-worm templates were prepared using the protocol of Barstead et al. (1991). To verify that the F1 were of hybrid origin, one of the PCR assays was performed on either the F1 hermaphrodites or the non-Tra F2 siblings. AF16 and HK104 animals were also used in the SW–PCR assays below as positive controls.
For the Cb-tra-1 indel assay, the PCR reaction mixtures contained the following: 0.25 mm dNTP, 1× Thermo Pol PCR buffer [2 mm Mg2+, New England Biolabs (NEB)], 0.5 μm of each primer (EH15/EH16), and 1 unit of Taq DNA polymerase per 10 μl. The cycling conditions were as follows: 95° denature (2 min) [95° denature (30 sec), 55° annealing (35 sec), 72° extension (45 sec)], with the bracketed subroutine repeated for 32 cycles, followed by a 72° extension (4–7 min) and a 4° hold. The PCR products (183 nt for AF16 vs. 220 nt for HK104) were separated using a 1.8% TBE agarose gel. For the Cb-tra-2 “snip-SNP” assay, the PCR cocktail was prepared as above, but with 2.5 mm Mg2+, EB2/EB3 primers, and 57° annealing temperature. The 250-nt PCR products were restriction digested with BstBI (NEB) and separated using a 1.8% TBE agarose gel. For the Cb-tra-3 HK104/AF16 assay, the PCR reactions were as for Cb-tra-1, but used primers DK48 and DK49 and a 62° annealing temperature. The 550-nt PCR products were restriction digested with SpeI (NEB) and separated using a 0.8% TBE agarose gel. Primer sequences may be found in the supplemental materials at http://www.genetics.org/supplemental/.
Linkage of Cb-tra(ed24ts) to Cb-tra-3 was further assessed using VT847 as the mapping strain. Resequencing of all Cb-tra-3 introns identified one SNP, a G-to-A transition mutation in the 3′-most intron. F2 Tra progeny from ed24ts/+ F1 hybrids sired by VT847 males were used for single-worm PCR with the primers EH33 and EH34. The cycling conditions were as follows: 95° denature (2 min) [95° denature (30 sec), 55° annealing (30 sec), 72° extension (30 sec)], with the bracketed subroutine repeated for 30 cycles, followed by a 72° extension (5 min) and a 4° hold. PCR products (25 μl) were prepared for direct sequencing by addition of a mixture of 0.4 μl phosphatase (Antarctic phosphatase, NEB) and 0.4 μl E. coli exonuclease I (NEB) in 5 μl of 1× phosphatase buffer and incubation for 30 min at 37° and 15 min at 80° (the latter to inactivate the enzymes). One-half of a microliter of each reaction (∼25 ng DNA) was used as template in EH33-primed 10-μl sequencing reactions (ABI Big Dye) on ABI Prism 3100 and 3730 instruments.
Complementation tests:
For likely Cb-tra-2 alleles, either spontaneous ed23ts XO (for nm1) or ed23ts/+ (for nm9ts, nm21, nm29, and nm36ts) males were mated either to homozygous XX mutants at permissive temperature (for the conditional alleles nm9ts and nm36ts) or to heterozygous carriers (for the nonconditional alleles nm1, nm21, and nm29) marked with the tightly linked dpy(nm4) mutation in trans. The presence of abundant wild-type males, few or no Dpy animals, and a large number of Tra worms (clearly distinguished from XO males by their incomplete masculinization) in the progeny of the test cross indicated noncomplementation.
For Cb-tra-3, ed24ts XX hermaphrodites were crossed at permissive temperature with single males that potentially carried one copy of the Cb-tra-3(nm65) deletion mutation. Uncertainty in the male's genotype required that many such crosses be set up. After mating, the males were genotyped by SW–PCR for the presence of the deletion mutation, and progeny of those carrying the mutation were grown at 25° and examined for the Tra phenotype.
Sequencing of candidate genes in mutants:
Cb-tra-2:
For nm1 and nm9ts alleles, all Cb-tra-2 exons were amplified from homozygous mutants, using three to five pooled SW–PCR reactions per fragment and primers DK1-DK25, T2AIR, T2BIR, T2CIL, and T2COR (sequences and locations are given in supplemental materials at http://www.genetics.org/supplemental/). PCR products >500 bp were gel purified before sequencing. Larger PCR fragments were cloned using the original TA cloning kit (Invitrogen, San Diego) and sequenced using the GPS-1 genome priming system (NEB). Mutations were subsequently verified by resequencing of more mutants. For ed23ts, total RNA was extracted from homozygotes grown at 16° and used as template for cDNA synthesis with Superscript II reverse transcriptase (Invitrogen) and 10 pmol of random (adaptor) primer (GIBCO, Grand Island, NY). This cDNA was then used as template for a PCR reaction with primers Cbtra-2RTCF and Cbtra-2RTCR to amplify a 920-bp fragment containing the region corresponding to exons 8–14 of the Cb-tra-2 gene. The PCR product was cloned into pGEM-T and sequenced using Cbtra-2RTCF and Cbtra-2RTCR. The mutation was verified by direct sequencing of genomic DNA of both mutant and wild-type animals.
Cb-tra-1:
PCR reactions using overlapping primer pairs from the set DK26-DK41 (see supplemental materials for sequences and locations) flanking the exons were performed on three to five nm2 animals per fragment with the high-fidelity Optimase polymerase (Transgenomics). For nm10 and nm30, these products were directly sequenced. For nm2, corresponding PCR products from both the Tra and AF16 animals were created and tested for mismatch using the Surveyor mutation detection kit (Transgenomic). The Cb-tra-1 PCR fragment identified as containing the nm2 lesion was then sequenced.
Cb-tra-3:
The deletion in Cb-tra-3(nm65) was localized by sequencing of the diagnostic PCR product used to identify the mutation in the screen with the inner primer set AD49 and AD50.
Isolation of Cb-tra-3(nm65) deletion mutation:
A single screen for deletion mutations in Cb-tra-3 was generally performed as described by Hill et al. (2006). The outer primer set was DK42 and DK45, and the inner set AD49 and AD50. For genotyping potential Cb-tra-3(nm65) individuals via SW–PCR, AD49/50 was used to detect the nm65 deletion allele, while primers within the deleted region, AD57 and AD58, were used to positively identify the wild-type allele (see Figure S1 at http://www.genetics.org/supplemental/ for sequences and approximate locations).
Reverse transcription PCR:
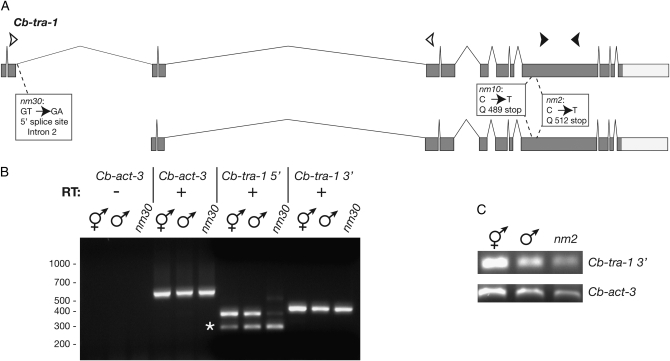
Groups of five similarly aged adult worms were picked into 5-μl drops of nuclease-free water in the lid of a microcentrifuge tube and then transferred to the bottom by a brief centrifugation. This procedure was repeated until 30–50 worms were obtained. Two hundred microliters of TRI reagent (Molecular Research Center) was added to each sample, after which the worms were frozen at −80° for at least 2 hr. After thawing, pelleted worms were lysed with a plastic pestle in a 1.6-ml centrifuge tube. Following the addition of 800 μl of TRI reagent and 4 μl of polyacryl carrier (Molecular Research Center), the RNA was extracted according to the manufacturer's protocol. Each sample was resuspended at a concentration of 1 μl/worm and then used in the AccessQuick reverse transcription–polymerase chain reaction (RT–PCR) system (Promega, Madison, WI) with 5–10 μl of RNA/50-μl reaction. R. Ellis kindly provided the primer sequences used for the Cb-act-3 control. Primers DK52 and DK57 amplify a fragment of both long and short forms of the Cb-tra-1 mRNA (de Bono and Hodgkin 1996), while primers EH29 and EH30 amplify a 5′ cDNA fragment specific to the longer, predominant form that encodes Cb-TRA-1A (Figure 4).
Figure 4.—
Sequence lesions in Cb-tra-1 mutant alleles. (A) Diagram of the two alternative Cb-tra-1 mRNAs (de Bono and Hodgkin 1996) with locations of mutations in the nm2, nm10, and nm30 alleles indicated in boxes. Open arrowheads represent the locations of 5′ oligonucleotide primers (DK52 and DK57) and solid arrowheads the 3′ primers (EH29 and EH30), used for RT–PCR in B. Both long and short mRNAs are trans-spliced to SL1 (de Bono and Hodgkin 1996). (B) Cb-tra-1 transcripts in nm30 homozygotes as assayed by RT–PCR. The longer form mRNA is specifically diminished in nm30 mutants, and a novel, higher-molecular weight species is formed at low levels. Sequence analysis shows that the band marked by an asterisk in the Cb-tra-1 5′ reactions is a nonspecific amplicon from another locus (data not shown). Cb-act-3 (actin control) and Cb-tra-1 reactions all employed the same RNA preparations, but the former used half as much as the latter. (C) Detection of Cb-tra-1 transcripts by RT–PCR with mRNA from nm2 homozygotes, using the 3′ primers.
Cb-tra-1 RNAi:
Mated AF16 hermaphrodites and virgin Cb-tra(nm2)/Cb-let(nm28) hermaphrodites were microinjected in either the rachis or the gut with Cb-tra-1 double-stranded RNA at a concentration of 1 μg/μl, which was generated by in vitro transcription of a T7-tailed PCR product with the Megascript kit (Ambion). The template was amplified with primers EH23 and EH24 and represents the entire 1.7-kb exon 10 of the Cb-tra-1A mRNA (de Bono and Hodgkin 1996). Successfully injected worms were rescued and transferred to successive NGM plates over several 24-hr laying windows. The F1 progeny from each laying window were scored for sexual transformation. As Cb-tra-1(RNAi) produces only partial somatic transformation in XX animals, the observation of a complete male tail was used to identify XX Tra homozygotes produced from virgin Cb-tra(nm2)/Cb-let(nm28) hermaphrodites and XO males produced from inseminated AF16 hermaphrodites.
Cell lineage analysis:
Cell lineages were followed as described (Sulston and Horvitz 1977). Wild-type males and tra-1 pseudomales were identified by their enlarged B-cell nucleus. Some individuals were observed through the L1 stage for wild-type and Cb-tra-1(nm2), but most were followed only through completion of the somatic gonad precursor (SGP) divisions. All mutant strains were backcrossed to AF16 at least two additional times before lineaging.
Construction of various double mutants:
Cb-tra-2; Cb-tra-3:
Cb-tra-2(ed23ts) dpy(sy5148) hermaphrodites were crossed with spontaneous Cb-tra-3(ed24ts) XO males at 16°. Many F2 Dpy worms derived from selfing of F1 hermaphrodites were singled at 16°. F2 hermaphrodites that produced Cby Tra F3 progeny at 16° were rescued and crossed with Cb-tra-3(ed24ts) and Cb-tra-2(ed23ts) males and the cross-progeny were grown at 25° to confirm the presence of both tra alleles in the genotype. Lines that showed 100% Tra XX worms in the cross-progeny in both crosses at 25° were selected and maintained at 16°.
Cb-tra-1; Cb-tra-2:
This double mutant was observed via tra-1-like Tra animals produced in strains segregating the Cb-tra-1(nm2) mutation in the Cb-tra-2(ed23ts) homozygous background. Cb-tra-1(nm2)/let(nm28) hermaphrodites were crossed with Cb-tra-2(ed23ts) dpy(sy5148) spontaneous males. F1 XX worms were singled and F2 lines scored for the presence of Dpy Tra transformed XX worms at 16°, interpreted as Cb-tra-1(nm2); Cb-tra-2(ed23ts) dpy(sy5148). Dpy siblings of these triple homozygotes, whose genotype is Cb-tra-2(ed23ts) dpy(sy5148) and potentially Cb-tra-1(nm2), were then examined at 25° to look for the more complete Tra phenotype of Cb-tra-1(nm2) mutants.
Cb-tra-1; Cb-tra-3:
Cb-tra-1(nm2)/let(nm28) hermaphrodites were crossed with Cb-tra-3(ed24ts) spontaneous males. Hermaphrodite progeny were backcrossed twice with Cb-tra-3(ed24ts) males, and lines that showed both 100% tra(ed24ts)-like imperfect masculinization and ∼25% Cb-tra-1-like XX Tra animals at 25° were analyzed.
Cb-tra-2; Cb-fem-2:
The strategy to isolate the tra-2; fem-2 double mutant was based on the assumed suppression of the Cb-tra-2(ed23ts) phenotype at the restrictive temperature.
Cb-fem-2(nm27) hermaphrodites were crossed with Cb-tra-2(ed23ts) dpy(sy5148)/Cb-tra-2(ed23ts) + males. F1 were singled and F2 Dpy worms selected and allowed to expand at 16°. Ten eggs were transferred from each F2 line to 25°. Adult hermaphrodites from three lines with fertile Dpy hermaphrodites were crossed with tra-2(ed23) males. Cross-progeny with 100% Tra worms at 25° indicated that suppression of the Tra phenotype seen in these lines was due to the presence of the fem allele (tra-2 cby-15; fem-2) and not to a recombinant + cby-15 chromosome. SW–PCR genotyping of the Cb-fem-2 locus (Hill et al. 2006) also confirmed the strain DP369.
Cb-tra-3; Cb-fem-2:
Cb-fem-2(nm27) hermaphrodites were crossed with Cb-tra-3(ed24ts) spontaneous males, and F1 males were backcrossed with Cb-fem-2(nm27) hermaphrodites. Fifty percent of the resulting progeny lack a Cb-tra-3(ed24ts) allele, and another 25% are Cb-fem-2(nm37); Cb-tra-3(ed24ts)/+ and will potentially fail to produce Tra progeny due to suppression by Cb-fem-2(nm27). Thus, with suppression, only 25% of the F2 broods will have any Tra animals, but if Cb-tra-3 is not suppressed by Cb-fem-2, 50% will produce Tra's. Twenty-nine of 36 singled F2 hermaphrodites produced only wild-type worms when selfed, while Tra worms were seen in 7. Cb-fem-2(nm27); Cb-tra-3(ed24ts) double mutants (strain DP372) were subsequently identified by crossing tester hermaphrodites to tra(ed24ts) males and checking for plates with 100% Tra worms in the cross-progeny grown at 25°. As with Cb-tra-2; Cb-fem-2 worms, Cb-tra-3(ed24ts); Cb-fem-2 mutants are fertile hermaphrodites that can be maintained at 25°.
RESULTS
Isolation of Cb-tra mutants in forward screens:
Since spontaneous males are produced by virgin hermaphrodites at a low frequency (∼0.2%), mutations that transform XX hermaphrodites into males are easily identified (Figure 2). F2 screens of ∼17,000 haploid genomes yielded 10 recessive Cb-tra mutants (Table 1). In addition, we found two recessive mutations causing both a dumpy (Dpy) and a Tra phenotype, which may be involved in dosage compensation as well as sex determination (Delong et al. 1993), and two dominant Him (high incidence of males) mutations that may be caused by X-specific translocations. We will not discuss these mutants further here. Of the canonical tra mutants, three complementation groups mapping to three different chromosomes have been identified. Below, we provide evidence indicating that each corresponds to an ortholog of one of the three C. elegans tra genes.
Figure 2.—
Mutations in Cb-tra-2. (A) Wild-type C. briggsae AF16 XX hermaphrodite, with oocytes (o), sperm (s), embryo (e), vulva (v), and gut (g) labeled (anterior to left). (B) Wild-type XO AF16 male. (C) Cb-tra-2(nm1) XX Tra mutant, with fluid-filled vacuole (fv) in the distal gonad and an incomplete male tail (asterisk). (D) Cb-tra-2(ed23ts) XX Tra grown at nonpermissive temperature (25°). (E) Linkage of nm1 to a molecular polymorphism within Cb-tra-2. F3 nm1 homozygotes and their non-Tra siblings were obtained from the self-progeny of heterozygotes resulting from two rounds of outcrossing with the mapping strain HK104. Individuals of each phenotype were then used as template in single-worm PCR reactions. The amplified fragment cuts with the enzyme BstBI in AF16, but not in HK104. (F) Cb-tra-2 sequence lesions in nm1, ed23ts, and nm9ts. Bar in A, 100 μm; magnifications are approximately equal in A–D.
TABLE 1.
Summary of masculinizing C. briggsae mutations
| Allele | Tra phenotype | Linkage group [marker allele] | Candidate linkage | Fails to complement | Sequence lesion |
|---|---|---|---|---|---|
| nm2 | Very strong, male mating behavior; XX homozygotes produce “ooids” late, infertile | III [Cb-cby-1 and let(nm28)] | Cb-tra-1 | ND | Q512stop |
| nm10 | Incomplete, blunt tail, some mating behavior, infertile, dominant male Fog | III [Cb-let(nm28)] | Cb-tra-1 | ND | Q489stop |
| nm30 | Incomplete, XX homozygotes produce oocytes, occasional embryos | ND | Cb-tra-1 | ND | Intron 2 5′ splice site |
| ed23ts | Incomplete, occasional Pvl, complete rescue at 15° | II [Cb-cby-15)] | Cb-tra-2 | nm1 | D587A |
| nm1 | Incomplete | II [Cb-dpy(nm4)] | Cb-tra-2 | ed23ts | R1197stop |
| nm9ts | Incomplete, frequent Pvl, complete rescue at 15° | II [Cb-dpy(nm4)] | ND | ed23ts | P1214L |
| nm21 | Incomplete, dominant at 25° | II [Cb-dpy(nm4)] | ND | ed23ts | ND |
| nm29 | Incomplete | II [Cb-dpy(nm4)] | Cb-tra-2 | ed23ts | ND |
| nm36ts | Incomplete, frequent Pvl, complete rescue at 15° | II | Cb-tra-2 | ed23ts | ND |
| ed24ts | Incomplete, strongly zygotic phenotype, partial rescue at 15° | IV [Cb-cby-7] | Cb-tra-3 | Cb-tra-3 (nm65) | None found |
| nm65 | Incomplete, maternally rescued | IVa | Targeted deletion | ed24ts | Deletes exons 2 and 3 |
Fog, feminized germ cells; ts, temperature sensitive; Pvl, protruding vulva.
Linkage group was inferred from the position of the fingerprint contig containing Cb-tra-3 (cb25.fpc4250) in the preliminary C. briggsae polymorphism map.
Cb-tra-2 alleles:
Six C. briggsae tra mutants were genetically mapped to LGII using either dpy(nm4) or dpy(sy5148). They possess the same phenotypic characteristics as C. elegans tra-2 mutants, such as a one-armed gonad with only sperm, an abnormal male tail with reduced sensory rays, and a total lack of male mating behavior (Hodgkin and Brenner 1977) (Table 1 and Figure 2). On the basis of the synteny with C. elegans, the association of tra-2-like mutants with chromosome 2 suggested that they were mutations in Cb-tra-2. Using a polymorphic restriction site in Cb-tra-2 that differs between the mutagenized AF16 strain and the mapping strain HK104, four alleles (nm1, nm29, ed23ts, and nm36ts) were shown to be tightly linked to Cb-tra-2 (Figure 2E; Table 1). In addition, these alleles and two others (nm9ts and nm21) all form a complementation group, suggesting that they all affect the same gene.
Cb-tra-2 was sequenced in three reference alleles, and a missense or nonsense mutation was found in each (Figure 2 and Table 1). The two conditional alleles have missense changes: ed23ts changes conserved amino acid 587 (Kuwabara et al. 1992; Haag and Kimble 2000), located in the second transmembrane loop of the protein, from aspartic acid to alanine. nm9ts changes the nonconserved amino acid 1214, located in the cytoplasmic FEM-3-binding domain of TRA-2, from a proline to a leucine. In contrast, the lesion in the nonconditional allele nm1 is a premature stop approximately one-third of the way into the FEM-3-binding domain. We conclude that nm1, nm9ts, ed23ts, and the other three alleles in their complementation group are all loss-of-function Cb-tra-2 mutants.
C. elegans tra-2 mutations have no effect on XO males (Hodgkin and Brenner 1977). To determine whether this is also true for Cb-tra-2 males, XO ed23ts homozygotes were produced in quantity by crossing ed23ts/+ males with ed23ts hermaphrodites and rearing the progeny at 25°. These crosses yield the same number of completely male (as opposed to Tra pseudomale) progeny as a wild-type cross (53%, N = 119), and all of those examined with DIC microscopy (N = 34) appeared normal in all respects. In addition, spontaneous XO ed23ts males mate successfully at the restrictive temperature. We conclude that the XO Cb-tra-2 phenotype is indistinguishable from that of wild-type males, consistent with the XO phenotype of Ce-tra-2.
Cb-tra-1 alleles:
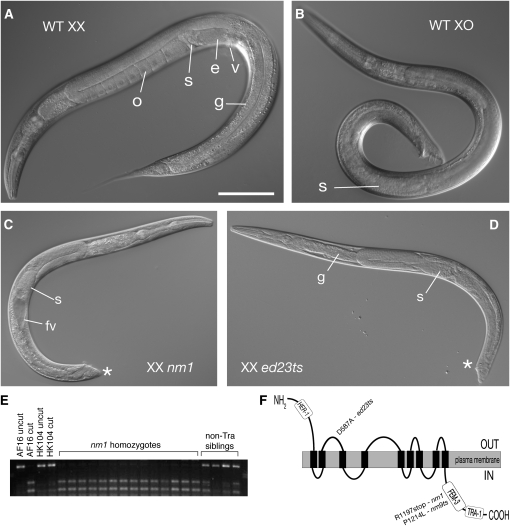
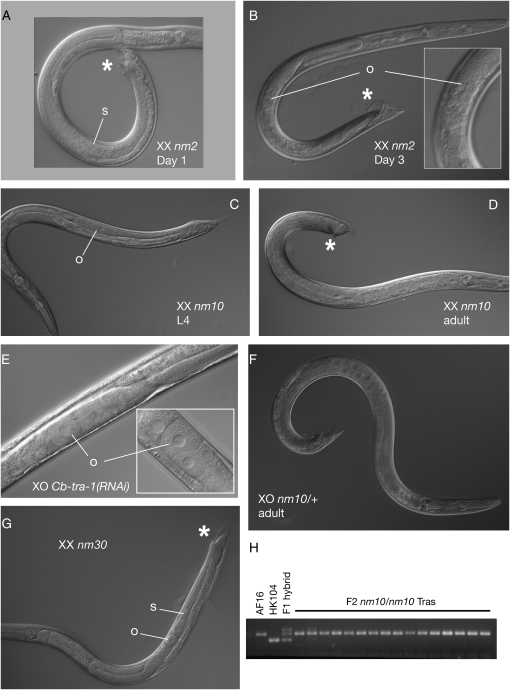
Three Cb-tra alleles mapped to LGIII and possess similar phenotypic characteristics as C. elegans tra-1 mutants, such as one-armed gonads, masculinized tails, and, in one case, vigorous male mating behavior (Hodgkin and Brenner 1977; Hodgkin 1987) (Table 1 and Figure 3). The allele showing the strongest masculinization is nm2, which has a perfect male soma and normal male mating behavior. Young XX nm2 Tra males can sire cross-progeny when mated to hermaphrodites with genetically feminized germlines (i.e., true females), but are far less fertile than wild-type XO males (data not shown). The two other alleles, nm10 and nm30, have imperfect somatic masculinization and little male mating behavior. nm30 homozygotes are also distinguished by the frequent production of both sperm and robust oocytes (i.e., large, morphologically normal, and in a single file; Figure 3) and, in some cases, bear embryos inside a somatically male gonad.
Figure 3.—
Phenotypes of Cb-tra-1 mutations. (A) Cb-tra-1(nm2) XX Tra worm, typical of those on the first day of adulthood, has a complete male tail (asterisk) and produces only sperm (s). (B) By the third day of adulthood, all Cb-tra-1(nm2) XX animals have ceased spermatogenesis and produce poorly formed oocytes (o). (C) L4 larval Cb-tra-1(nm10) XX homozygote, with a developing male somatic gonad and tail, but only oocytes (o) and no sperm evident. (D) Adult nm10 homozygote, showing blunted male tail. Differentiated germ cells, partly out of focus in this image, are imperfect oocytes. (E) Cb-tra-1(RNAi) XO male, with partially feminized germline containing sperm (s) and oocytes (o). (F) XO Cb-tra-1(nm10)/+ male with extreme germline feminization (most have more disorganized oogenesis). (G) Cb-tra-1(nm30) XX homozygote, with both sperm and robust oocytes developing in a male somatic gonad. This young adult animal retains larval cuticle over its partially masculinized tail (asterisk). (H) Linkage of nm2 to Cb-tra-1 via a polymorphic indel. Two small indels distinguish the Cb-tra-1 intron 1 of AF16 and HK104, which are easily resolved by agarose electrophoreses of single-worm PCR reactions with flanking primers.
On the basis of synteny with Ce-tra-1 and placement of its supercontig on a recently published linkage map (Hillier et al. 2007), Cb-tra-1 should be toward the right end of chromosome 3. As expected, tra(nm2) was loosely linked to dpy(sy1281), which is also in that region. To strengthen the link between tra(nm2) and the Cb-tra-1 locus, an assay that exploits a natural DNA polymorphism within the Cb-tra-1 locus of the AF16 and HK104 strains was developed (Figure 3H). Using the assay, no recombinants were observed between nm2, nm10, and nm30 and Cb-tra-1 in 50–100 haploid genomes (Table 1), indicating that all lie ≤2 cM from this candidate locus.
Resequencing of Cb-tra-1 identified single DNA lesions in nm2, nm10, and nm30 homozygotes. Both nm2 and nm10 have glutamine-to-nonsense mutations at codons 512 and 489, respectively. These sites lie in the region encoding conserved region 6 (de Bono and Hodgkin 1996), just C-terminal to the zinc-finger domain of Cb-TRA-1. These premature stop codons are predicted to eliminate nearly 50% of the full-length TRA-1A protein (Figure 4A) and even more of the N-terminally truncated Cb-TRA-1B isoform. Two plausible explanations for the strong Tra phenotype of nm2 and nm10 exist. First, the premature stop codon may create a lack of TRA-1 protein function due to the loss of over one-half of the TRA-1 sequence (including the TRA-2-binding domain). Alternatively, a potentially functional truncated protein may not be made due to the transcript being a substrate for nonsense-mediated decay (NMD), which can greatly reduce the abundance of transcripts bearing a premature stop codon (Pulak and Anderson 1993; Cali et al. 1999). However, Cb-tra-1 transcripts are easily detected by RT–PCR in nm2 homozygotes, although at somewhat reduced levels compared to wild type (Figure 4C). Given the modest reduction, the strong loss-of-function nm2 phenotype is more likely a consequence of Cb-TRA-1 truncation than of NMD. This interpretation is supported by our unsuccessful attempts to enhance the nm2 phenotype with Cb-tra-1 RNAi (data not shown).
Sequencing of nm30 revealed a T-to-C transition mutation in the 5′ splice site of intron 2 (Figure 4A), an intron unique to the longer form of Cb-TRA-1 (Cb-TRA-1A). Since the shorter form (Cb-TRA-1B) may be either transcribed from its own promoter in intron 2 or processed from a full-length pre-mRNA via SL1 trans-splicing (neither requiring a functional 5′ splice site), we reasoned that perhaps only the Cb-TRA-1A mRNA would be affected. Indeed, RT–PCR experiments show that normal splicing of the full-length mRNA is greatly reduced and a novel splice form is produced (probably due to intron retention), yet normal amounts of the shorter Cb-TRA-1B-encoding mRNA are still made (Figure 4B). This suggests that the phenotypic consequences of nm30 are due primarily to the loss of the longer transcript. Interestingly, these effects seem to be restricted largely to the soma. We note that the truncated forms of TRA-1 produced in C. elegans and C. briggsae are truncated at opposite ends and are thus not likely to be homologous (Zarkower and Hodgkin 1992; de Bono and Hodgkin 1996).
The evidence presented thus far is consistent with nm2, nm10, and nm30 representing strong loss-of-function alleles of Cb-tra-1, and for the rest of this article we will treat them as such. Repeated attempts at transformation rescue by microinjections of Cb-tra-1-containing BAC clones (necessary owing to the large size of all three Cb-tra genes) have thus far failed to produce any stable transgenic lines (even though the same cocktail could be used to transform C. elegans). Since plasmid constructs have been successfully transformed into C. briggsae by other workers (Jan et al. 1997; Wang and Chamberlin 2002), the reasons for this are unclear. Standard complementation tests, used to show allelism of C. elegans tra-1 mutants (Hodgkin and Brenner 1977), have also not been possible for two reasons. First, as will be discussed below, Cb-tra-1 (lf)/+ XO males are often partially or completely infertile, which prevents key test crosses. Second, despite their good mating behavior and early sperm production, XX nm2 Tra males are not able to inseminate self-fertile Cb-tra-1/+ hermaphrodites well enough to give definitive results.
Germline feminization in Cb-tra-1 mutants:
Although strong C. elegans tra-1(lf) mutations fully masculinize the XX soma and often produce abundant sperm, both XX and XO mutants often produce oocytes as well (Hodgkin 1987; Schedl et al. 1989). This puzzling result suggests that in the germline, unlike in the soma, tra-1 has a role in promoting or sustaining male fates. To address whether this is a conserved aspect of tra-1 function, we examined Cb-tra-1(nm2) XX Tra males at various ages. We find that, although most individuals produce sperm on their first day of adulthood, this ceases by day 2, and by the third day of adulthood all nm2 XX males are producing various approximations of oocytes (Figure 3).
We have not yet devised a method to examine the phenotype of XO Cb-tra-1 homozygotes. However, we noted during post-mutagenesis outcrossing that Cb-tra-1(nm10)/+ males could mate but not sire progeny, whereas XX heterozygotes could pass the mutation to both self- and cross-progeny (data not shown). This suggested that there is a subtle dominant phenotype of nm10 that is specific to XO males. Indeed, when the XO progeny of nm10/+ hermaphrodites mated with AF16 males were examined with DIC microscopy, one-half (presumably the nm10/+ individuals) showed strong germline feminization, usually none or at best 10–20 sperm being produced (Figure 3). This effect can also be mimicked by a Cb-tra-1(RNAi) treatment, in which the XO progeny of mated AF16 hermaphrodites show robust oogenesis in a normal male soma. These results indicate that partial reduction in tra-1 activity strongly feminizes germ cells in males, but not in hermaphrodites. Surprisingly, similar dominant germline feminization was not seen in Cb-tra-1(nm2)/+ XO males.
Gonad development in Cb-tra-1 XX males:
Another defect often observed in C. elegans tra-1 mutant males (both XX and XO) is in the development of the somatic gonad. Wild-type C. elegans males have a J-shaped gonad, whereas tra-1 mutants often have a small and variably misshapen gonad. This defect is due to a loss of gene function and is most penetrant (up to 50%) in the strong loss-of-function alleles, e1099 and e1835 (Hodgkin 1987; Schedl et al. 1989). Like C. elegans males, C. briggsae males have a J-shaped gonad. C. briggsae tra-1 mutant males also typically had a normal J-shaped gonad. This observation suggested that there might be differences in the function of tra-1 in the two species.
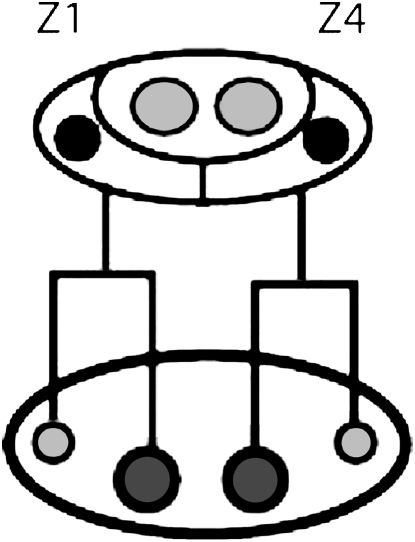
The somatic gonad develops from two SGPs in all described nematodes (Chitwood and Chitwood 1950). In C. elegans, the SGPs divide asymmetrically to generate daughter cells with different fates and sizes (Kimble and Hirsh 1979). The SGPs divide with opposite polarity such that their smaller daughters are at the distal poles of the developing gonad (Figure 5). These distal daughters are the presumptive distal-tip cells, which promote germline proliferation. In C. elegans tra-1 mutants, both SGPs divide with the same polarity to position their distal-tip cells to the posterior (Mathies et al. 2004). In addition, the posterior SGP (Z4) often migrates toward the anterior of the gonad primordium prior to its division. It is not known which, if any, of these early lineage defects correlates with the disorganized adult gonad in C. elegans tra-1 mutants. We determined the early gonadal lineage of C. briggsae wild-type males and Cb-tra-1mutants and scored two features: (1) polarity of the SGP divisions and (2) migration of Z4 prior to its division (Table 2). The wild-type C. briggsae male lineage was similar to that described for C. elegans males: the two SGPs divided with opposite polarity to generate daughter cells of different sizes and the SGPs remained at the poles of the primordium until their division. Next, we examined the three C. briggsae tra-1 mutants. Surprisingly, the two strongest loss-of-function alleles (nm2 and nm10) had early lineage defects: Z1 always divided with reversed polarity and we occasionally observed early migration of Z4 (Table 2). Therefore, C. briggsae tra-1 mutants have early SGP lineage defects, but are generally able to recover from these defects to generate a grossly normal male somatic gonad.
Figure 5.—
The SGP and their first division. Z1 and Z4 (solid nuclei) wrap around the primordial germ cells, Z2 and Z3 (shaded nuclei). Both Z1 and Z4 divide asymmetrically and with reversed polarity, generating mirror-image symmetry that is maintained in hermaphrodites but soon broken in males.
TABLE 2.
Early gonadal lineages in C. briggsae tra-1 mutants
| Reversed polarity
|
|||
|---|---|---|---|
| Genotype | Z1 | Z4 | Z4 migration |
| C. briggsae AF16 | 0/4 | 0/4 | 0/4 |
| Cb-tra-1(nm2) | 8/8 | 0/7 | 0/7 |
| Cb-tra-1(nm10) | 5/5 | 0/5 | 1/5a |
| Cb-tra-1(nm30) | 0/6 | 0/6 | 0/6 |
| Ce-tra-1(e1781) | 5/10 | 2/10 | 1/10 |
Z4 was not positioned at the posterior pole, but was not seen migrating.
C. briggsae tra-1 mutants rarely have adult somatic gonad defects, yet they often have early SGP lineage defects. Therefore, either C. briggsae and C. elegans tra-1 have minor differences in their function during somatic gonad development or the C. briggsae tra-1 alleles retain some activity and are able to recover from the early lineage alterations. To distinguish between these possibilities, we reexamined the existing C. elegans tra-1 mutants. In his initial description of the tra-1 alleles, Hodgkin (1987) classified some alleles as having a “large male somatic gonad.” The molecular lesion for one of these, e1781, is a premature stop codon downstream of the zinc fingers, comparable to the lesion found in Cb-tra-1 nm2 and nm10 alleles. We examined the SGP lineage of C. elegans tra-1(e1781) and found that it was qualitatively similar to C. briggsae tra-1(nm2) in that the SGPs divided with reversed polarity and Z4 seldom migrated before its division. Together these results suggest that premature migration of Z4 is a factor contributing to the disorganization of the adult gonad in C. elegans tra-1(null) mutants. Furthermore, they indicate that C. briggsae tra-1 functions similarly to C. elegans tra-1 during somatic gonadal development.
Cb-tra-3 alleles:
Another mutation, the conditional allele ed24ts, incompletely masculinizes XX animals at restrictive temperature (Figure 6) and mapped to LGIV. The C. briggsae ortholog of tra-3, which also has an incomplete Tra phenotype, is predicted to be on LGIV as well and was thus the obvious candidate gene. Consistent with this, ed24ts was linked to an AF16/HK104 polymorphism in the Cb-tra-3 gene (CBG21580; data not shown). However, while performing the mapping crosses, we noted that only ∼3% of the F2 progeny of AF16/HK104 F1 hybrids were Tra (Table 3), even though 25% were predicted to be Cb-tra(ed24ts) homozygotes. In addition, many intersexual animals were observed among the F2.
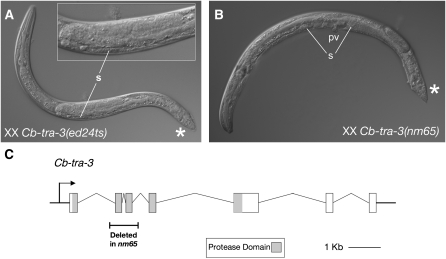
Figure 6.—
Mutations in Cb-tra-3. (A) At 25°, Cb-tra-3(ed24ts) possesses a one-armed gonad that makes only sperm (s) and an abnormal male tail (*) with reduced sensory rays and spicules and often displays a protruding vulva phenotype (not shown). (B) nm65 deletion homozygote, F3 Tra (m-, z-). These mutants are less completely masculinized than are ed24ts homozygotes and often have two-armed gonads and protruding partial vulvae (pv). (C) Intron–exon diagram of the Cb-tra-3 locus, showing the extent of the nm65 deletion.
TABLE 3.
Strain-specific effects on F2 expression of the zygotic Tra phenotype of Cb-tra-3(ed24ts)
| P0 father | Wild-type hermaphrodites | Tra (%) | Intersexual (%) |
|---|---|---|---|
| AF16 | 358 | 81 (18.5)a | 0 (0) |
| HK104 | 469 | 13 (2.7)b | 6 (1.2) |
| VT847 | 339 | 102 (23.1) | 0 (0) |
P0 mothers were AF16-derived Cb-tra-3(ed24ts) homozygotes for both crosses.
Differs significantly from the 25% Mendelian expectation (χ2 = 5.63, 1 d.f., P < 0.025). This reproducible effect is due to slower growth and reduced viability of Tra animals.
Reduced Tra frequency relative to AF16 is highly significant (χ2 = 62.22, 1 d.f., P < 0.001).
As HK104 appears to be part of a clade of temperate strains that is genetically distinct from tropical strains like AF16 (Graustein et al. 2002; Cutter et al. 2006a), we surmised that the unexpected phenotypic ratios above might not be seen when more closely related mapping strains are used. Indeed, when the tropical strain VT847 served as the father of the hybrids, the expected proportion of F2 Tra progeny was produced (Table 3). We therefore resequenced the VT847 Cb-tra-3 locus's introns to identify a mapping tool and found a single SNP. As this SNP could not be scored by restriction digest or amplicon size, we used resequencing of single-worm PCR products to genotype a large number of potential recombinant F2 Tra progeny. In all, 208 Tra progeny and 5 non-Tra control siblings were genotyped, with all of the former being homozygous for the AF16 allele and the latter being either heterozygous or homozygous for the VT847 allele. The ed24ts lesion is therefore no >100/(2 × 208) = 0.240 cM (SE ± 0.240 cM) from Cb-tra-3 (Grandillo and Fulton 2002). Cb-tra-3 is located at ∼2.1 Mbp from the left end of LGIV, part of the left “arm” region of relatively high recombination, where 1 cM corresponds to ∼230 kbp of genomic DNA (Hillier et al. 2007). This indicates that the physical distance between the ed24ts mutation and the Cb-tra-3 SNP is ∼ ≤55 kb (95% C.I. 0–166 kb), consistent with ed24ts affecting Cb-tra-3 function.
Although the above results supported its being an allele of Cb-tra-3, ed24ts differs from even the strongest C. elegans tra-3(lf) mutations in that it was found in an F2 screen and is thus not maternally rescued (Hodgkin and Brenner 1977). We suspected that this maternal rescue would be conserved in conventional Cb-tra-3(lf) mutants, as Cb-tra-3(RNAi) knockdown has no discernible phenotype (data not shown). To obtain a straightforward null allele of Cb-tra-3, we supplemented forward genetic screens with a reverse genetic screen for gene-specific deletions, as was previously employed for Cb-fem-2 and Cb-fem-3 (Hill et al. 2006). This yielded the deletion allele Cb-tra-3(nm65), which deletes all of exons 2 and 3, including roughly one-third of the calpain protease domain (Figure 6). As expected, nm65 causes partial masculinization only in homozygous mutants whose mothers were also homozygotes.
With the likely null nm65 allele in hand, we assessed whether ed24ts might be allelic to Cb-tra-3 through a complementation test. If the ed24ts mutation eliminates maternal rescue, it may differ from a conventional loss-of-function allele in encoding a weakly antimorphic gene product. However, because it is also recessive, we reasoned that a truly null allele in trans should still allow manifestation of the ed24ts zygotic phenotype. Indeed, when ed24ts hermaphrodites were crossed (at permissive temperature) with Cb-tra-3(nm65)/+ males and their progeny grown at restrictive temperature, 50% of the XX offspring (N = 100) were Tra. This result is thus also consistent with ed24ts being an allele of Cb-tra-3, and for the remainder of this article we will treat it as such. However, sequencing of all predicted Cb-tra-3 codons, most of the introns (including sequence flanking all exons), and 2 kb of 5′ upstream DNA failed to identify a mutation.
Phenotypes of various double mutants:
To determine whether the C. briggsae tra genes act in a pathway similar to that of their C. elegans homologs, we examined a number of double-mutant combinations (Table 4). Just as with Cb-tra single mutants, they generally behave similarly to their C. elegans equivalents (Hodgkin 1980). Cb-tra-2; Cb-tra-3 mutants do not show an enhanced masculinization relative to the single mutants. Cb-tra-1(nm2), whose loss-of-function phenotype is very strongly Tra, predominates in combination with the weaker masculinization of Cb-tra-2 and Cb-tra-3. Finally, both Cb-tra-2 and Cb-tra-3 are completely suppressed (to self-fertile hermaphrodites) by Cb-fem-2. A detailed analysis of germline development in Cb-tra-1; Cb-fem-2/3 XX double mutants will be presented elsewhere (R. C. Hill, unpublished data), but they have the strongly masculinized soma predicted from C. elegans research (Hodgkin 1986).
TABLE 4.
Summary of various double mutants
| Genes affected | Genotype assayed | XX phenotypea | XO phenotypea | Stable strain? (no.) |
|---|---|---|---|---|
| Cb-tra-2 and Cb-tra-3 | Cb-tra-2(ed23ts) dpy(sy5148) II; Cb-tra-3(ed24ts) IV | Imperfectly Tra pseudomales | Normal, fertile males | Yes (DP368) |
| Cb-tra-2 and Cb-tra-1 | Cb-tra-2(ed23ts) dpy(sy5148) II; Cb-tra-1(nm2) III | Strongly Tra pseudomales | ND | No |
| Cb-tra-3 and Cb-tra-1 | Cb-tra-3(ed24ts) IV; Cb-tra-1(nm2) III | Strongly Tra pseudomales | ND | No |
| Cb-tra-2 and Cb-fem-2 | Cb-tra-2(ed23ts) dpy(sy5148) II; Cb-fem-2(nm27) III | Hermaphrodite | Hermaphrodite | Yes (DP369) |
| Cb-tra-3 and Cb-fem-2 | Cb-tra-3(ed24ts) IV; Cb-fem-2(nm27) III | Hermaphrodite | Hermaphrodite | Yes (DP372) |
Assayed at 25° when any temperature-sensitive alleles were present. ND, no data.
DISCUSSION
Conserved roles for Cb-tra genes:
The Cb-tra-1, Cb-tra-2, and Cb-tra-3 mutants described in this article are quite similar to their counterparts in C. elegans (Hodgkin and Brenner 1977; Hodgkin 1980, 1986), both in phenotype and in how they interact genetically with each other. This result would be frankly unremarkable were it not for two further observations. First, the sequences of both TRA-1 (de Bono and Hodgkin 1996) and TRA-2 (Kuwabara 1996b; Haag and Kimble 2000) have diverged to a much greater extent than is typical of orthologs from these species (Stein et al. 2003). Molecular sequence divergence is therefore not correlated with functional divergence as assayed by genetic criteria. Second, the C. briggsae fem genes, while functioning similarly to their C. elegans orthologs in male somatic sex determination, are dispensable for hermaphrodite spermatogenesis (Hill et al. 2006). In addition, their loss leads to a male-to-hermaphrodite transformation (the Her phenotype) rather than strict feminization (Fem). As the fem genes lie between tra-2/tra-3 and tra-1 in the epistasis pathway (Figure 1) and the self-fertility of C. briggsae fem mutants implies that hermaphrodite development is controlled downstream of the Cb-fem genes, the similar germline phenotypes of C. elegans and C. briggsae tra-1 strong loss-of-function mutants is puzzling.
As with C. elegans tra-1 mutants, Cb-tra-1(nm2) XX males are essentially perfectly formed, exhibit full male mating behavior, and under certain circumstances can sire progeny before their germlines become feminized. For example, when XX animals are rendered truly female by the germline feminizing mutation nm38, such that no self sperm are present, XX Cb-tra-1(nm2) males sire Cb-tra-1(nm2)/+ progeny (A. V. Doty, unpublished results). This suggests that tra-1's role in the germline is similar in C. elegans and C. briggsae and that the differential requirement for the fem genes in hermaphrodite spermatogenesis is due to other germline factors that regulate tra-1 or its targets. Overall, it appears that the complexities of tra-1 germline function are conserved in Caenorhabditis, and the discovery that tra-1 mutants of the much more distantly related Pristionchus pacificus also show strong somatic masculinization with intersexual germlines (Pires-Dasilva and Sommer 2004) indicates that it is a fundamental feature that remains even after massive sequence divergence. Recent studies have found that males have a small but detectable amount of full-length TRA-1A protein, whereas hermaphrodites have this plus a much more abundant, C-terminally truncated form termed TRA-1100 (Schvarzstein and Spence 2006). Perhaps the full-length form plays a positive role in spermatogenesis, similar to the activating role for unprocessed forms of the Hedgehog pathway transcription factors Gli and Cubitus interruptus, the homologs of TRA-1 in mammals and Drosophila, respectively (Huangfu and Anderson 2006).
One possible exception to the congruence of Cb-tra-1 and C. elegans tra-1 phenotypes is the Cb-tra-1(nm10) allele, which exhibits a recessive somatic masculinization that is weaker than the nm2 allele, yet also produces a strong dominant feminizing effect on XO males. Despite the highly variable phenotypes found in C. elegans tra-1 alleles (Hodgkin 1987; Schedl et al. 1989), no such combination of recessive and dominant phenotypes has been reported. However, the recessive Ce-tra-1(e1781) allele (class A3 in the classification scheme of Hodgkin 1987) has been examined in XO homozygotes, where it also strongly feminizes the germline (Hodgkin 1987). As noted above in the discussion of gonad development, the e1781 amber mutation lies in the same conserved domain as nm10 and nm2 [five residues C-terminal to Cb-tra-1(nm2) in the alignment of de Bono and Hodgkin 1996]. The nm10 mutation is a premature stop 23 codons 5′ to the nm2 mutation, yet their phenotypes are significantly different. Paradoxically, the slightly longer protein appears to retain less Cb-tra-1 function than the shorter protein. Taken together, this indicates that minor differences in the point of truncation can have a large impact on the phenotypic consequences of tra-1 mutations and that this complexity is conserved between species. Perhaps differences in the stability of these mutant gene products are important here. In addition, seemingly minor differences in truncation point may differentially affect a domain required for TRA-1 autoregulation or interaction with other factors, perhaps in tissue-specific ways. For example, nm10 may remove a site of negative regulation that is particularly important in the germline (explaining its dominance), yet be more stable in the soma than the nm2 product. The resolution of these questions will require the development of new tools for characterizing TRA-1's post-translational regulation in multiple species.
Functions of Cb-tra-1 splice variants:
We have shown that the Cb-tra-1(nm30) allele induces a splice-form-specific reduction of Cb-tra-1a mRNA (Figure 4). Interestingly, nm30 homozygotes typically have transformed somas but hermaphrodite germlines (distinct from nm2 in having robust oocytes). This suggests that, of the two splice variants, Cb-TRA-1A is particularly important for somatic development, while Cb-TRA-1B is sufficient for normal hermaphrodite germline development. Consistent with this, the Cb-tra-1b mRNA becomes detectable on Northern blots from L3 onward, increasing in relative abundance as the germ cells proliferate (de Bono and Hodgkin 1996). It is also possible, however, that this is primarily the result of different dosage requirements, where germline development requires only a small amount of tra-1 activity and the soma needs more. Since the two 5′-most exons of the mRNA encoding Cb-TRA-1A are not found in the shorter mRNA, this may enable isoform-specific assays to determine whether they differ in their tissue specificity.
TRA-1 and gonadogenesis:
The consistently normal male gonads in Cb-tra-1(nm2) homozygotes initially led us to suspect that the role for tra-1 in gonadogenesis demonstrated in C. elegans (Mathies et al. 2004) was not conserved in C. briggsae. However, a comparable C. elegans tra-1 allele whose premature stop is also just C-terminal to the zinc-finger DNA-binding domain also has an early lineage defect with little effect on the adult organ. We conclude that, in both species, mutant forms of TRA-1 that are generally devoid of sex-determining activity exist that still allow enough function to enable development of a normal male gonad. This suggests that there may be a domain in the C-terminal half of the protein that is indispensable for sex determination, whereas the N-terminal half containing the zinc fingers is sufficient for performance of most of TRA-1's gonadogenesis role. Consistent with this, a recent study has demonstrated that the C terminus is a site of normal sex-specific processing in C. elegans (Schvarzstein and Spence 2006)
Unusual properties of Cb-tra-3(ed24ts):
The temperature sensitivity and the weakly antimorphic properties of ed24ts led us to expect that a missense mutation had occurred that created a heat-instable protein variant that also acts to antagonize small amounts of wild-type maternal Cb-TRA-3. However, resequencing of the coding sequences in homozygous mutants found no such mutation. Preliminary RT–PCR experiments and Northern hybridizations indicate that ed24ts does produce Cb-tra-3 transcripts at detectable levels (data not shown). There is a precedent for a temperature-dependent phenotype in sex determination genes in putatively null nonsense alleles of C. elegans fem-2 (Hodgkin 1986; Pilgrim et al. 1995), although this is not the case with null alleles of C. briggsae fem-2 (Hill et al. 2006). More recently, an amino acid polymorphism in C. elegans tra-3 has been associated with natural variation in the developmental response of body size to temperature (Kammenga et al. 2007), and the authors note that expression of C. elegans tra-3 mRNA is elevated at 24° relative to 12°. These studies suggest that the temperature-dependent phenotype of Cb-tra-3(ed24ts) may result from an inherent temperature-dependent modulation of, or requirement for, tra-3 activity rather than from the instability of a mutated gene product. Alternatively, temperature-dependent alleles of C. elegans her-1 that alter transcription exist (Perry et al. 1994). Perhaps a variant Cb-tra-3 transcript is produced at a nonpermissive temperature that has reduced function.
Regarding the lower-than-expected number of Tra animals found in mapping crosses involving the HK104, at least three potential explanations exist. One is that there is an HK104 locus that dominantly suppresses the Tra phenotype, such that only Cb-tra-3(ed24ts) animals that are also homozygous for the AF16 allele of the putative modifier locus are affected. In this case, the “missing” Tra animals are alive, but phenotypically hermaphroditic. A second is that the HK104 version of TRA-3 is largely insensitive to the antimorphic properties of the ed24ts product. Finally, there may be a recessive synthetic-lethal locus in AF16 that is linked to Cb-tra-3. In this scenario, the paucity of Tra progeny in the F2 is due to their death from simultaneous homozygosity of this synthetic-lethal mutation. The few viable Tra progeny are thus attributed either to rare recombination between ed24ts and the synthetic-lethal locus or to absence (through independent assortment) of one or more key interacting HK104 loci required for lethality. All three scenarios are consistent with the number of F2 Tra progeny seen. However, they differ in that the first two predict the presence of a large number of cryptic ed24ts homozygotes. Distinguishing between them could in principle be accomplished with a marker tightly linked to Cb-tra-3(ed24ts) or by resequencing Cb-tra-3 alleles in many F2 individuals should the causative mutation be found. In the absence of such data, the presence of intersexual animals leads us to favor the suppression hypothesis, as these may represent incomplete suppression.
Utility of Cb-tra mutations:
The above discussion has focused on the ways in which having analogous mutants in homologous tra genes can directly assist in clarifying the evolution of their roles in sex determination. However, as was historically the case for C. elegans research, the relatively easily obtained tra mutants also set the stage for modifier screens capable of identifying feminizing (fem) mutations that are less easily obtained through direct screening (Hodgkin 1986). Indeed, the self-fertility of Cb-fem mutants (Hill et al. 2006) means that this direct route would have been completely impossible in C. briggsae. We recently reported results of suppressor screens using the Cb-tra-2(ed23ts) and Cb-tra-2(nm9ts) alleles (Hill et al. 2006), which identified 75 suppressors in at least three loci, all of which are self-fertile. In addition, similar suppressors of Cb-tra-3(ed24ts) have also been identified (C. E. de Carvalo and D. Pilgrim, unpublished data), at least some of which fail to suppress Cb-tra-2.
Finally, mutants in the Cb-tra genes enable the placement of other mutations into the sex determination pathway. For example, we and others are currently identifying loci that feminize the C. briggsae hermaphrodite germline (A. V. Doty and E. S. Haag, unpublished results; R. Ellis, personal communication). The existence of strongly masculinized mutants at two different points in the pathway will be invaluable for determining where these genes act. These Cb-tra mutants thus represent a sort of evolutionarily stable reference point with which to make sense of the potentially novel genes that act in the germline.
Acknowledgments
We thank Bhagwati Gupta and Paul Sternberg for sharing valuable unpublished marker mutants, Zigrida Smith and Edward Large for the deletion mutant library, Brittany Trogen for sequencing assistance, and Steve Mount and various members of the Haag and Pilgrim labs for fruitful discussions. This work was supported by a Natural Sciences and Engineering Research Council of Canada grant to D.P., startup funds from the University of Maryland and National Science Foundation grant IBN-0414512 to E.S.H., and a University of Maryland/Howard Hughes Medical Institute Undergraduate Research Fellowship to M.L.
References
- Barstead, R. J., L. Kleiman and R. H. Waterston, 1991. Cloning, sequencing, and mapping of an alpha-actinin gene from the nematode Caenorhabditis elegans. Cell Motil. Cytoskeleton 20 69–78. [DOI] [PubMed] [Google Scholar]
- Beye, M., M. Hasselmann, M. K. Fondrk, R. E. Page and S. W. Omholt, 2003. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell 114 419–429. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali, B. M., S. L. Kuchma, J. Latham and P. Anderson, 1999. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics 151 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P., S. Cho, S. Jin and R. Ellis, 2001. Specification of germ cell fates by FOG-3 has been conserved during nematode evolution. Genetics 158 1513–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood, B., and M. Chitwood, 1950. Introduction to Nematology. University Park Press, Baltimore.
- Cho, S., S. W. Jin, A. Cohen and R. E. Ellis, 2004. A phylogeny of Caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res. 14 1207–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford, R., M. Lee, S. Nayak, M. Ohmachi, F. Giorgini et al., 2000. FOG-2, a novel F-box-containing protein, associates with the GLD-1 RNA-binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development 127 5265–5276. [DOI] [PubMed] [Google Scholar]
- Cline, T., and B. Meyer, 1996. Vive la difference: males vs. females in flies vs. worms. Annu. Rev. Genet. 30 637–702. [DOI] [PubMed] [Google Scholar]
- Cook, J., 1993. Sex determination in the Hymenoptera: a review of models and evidence. Heredity 71 421–435. [Google Scholar]
- Cutter, A. D., M. A. Felix, A. Barriere and D. Charlesworth, 2006. a Patterns of nucleotide polymorphism distinguish temperate and tropical wild isolates of Caenorhabditis briggsae. Genetics 173 2021–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter, A. D., M. A. Felix, A. Barriere and D. Charlesworth, 2006. b Patterns of nucleotide polymorphism distinguish temperate and tropical wild isolates of Caenorhabditis briggsae. Genetics 173 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bono, M., and J. Hodgkin, 1996. Evolution of sex determination in Caenorhabditis: unusually high divergence of tra-1 and its functional consequences. Genetics 144 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong, L., J. D. Plenefisch, R. D. Klein and B. J. Meyer, 1993. Feedback control of sex determination by dosage compensation revealed through Caenorhabditis elegans sdc-3 mutations. Genetics 133 875–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rosa, R., J. K. Grenier, T. Andreeva, C. E. Cook, A. Adoutte et al., 1999. Hox genes in brachiopods and priapulids and protostome evolution. Nature 399 772–776. [DOI] [PubMed] [Google Scholar]
- Doniach, T., 1986. Activity of the sex-determining gene tra-2 is modulated to allow spermatogenesis in the C. elegans hermaphrodite. Genetics 114 53–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos, M., J. Ahringer, S. Crittenden and J. Kimble, 1998. Repression by the 3′UTR of fem-3, a sex-determining gene, relies on a ubiquitous mog-dependent control in Caenorhabditis elegans. EMBO J. 17 6337–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, E., and R. Ellis, 2002. Turning clustering loops: sex determination in Caenorhabditis elegans. Curr. Biol. 12 R111–R120. [DOI] [PubMed] [Google Scholar]
- Graham, P., and J. Kimble, 1993. The mog-1 gene is required for the switch from spermatogenesis to oogenesis in Caenorhabditis elegans. Genetics 133 919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, P. L., T. Schedl and J. Kimble, 1993. More mog genes that influence the switch from spermatogenesis to oogenesis in the hermaphrodite germ line of Caenorhabditis elegans. Dev. Genet. 14 471–484. [DOI] [PubMed] [Google Scholar]
- Grandillo, S., and T. Fulton, 2002. Approaches to gene mapping, pp. 101–136 in Plant Molecular Biology, edited by P. Gilmartin and C. Bowler. Oxford University Press, Oxford.
- Graustein, A., J. M. Gaspar, J. R. Walters and M. F. Palopoli, 2002. Levels of DNA polymorphism vary with mating system in the nematode genus Caenorhabditis. Genetics 161 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves, J. A. M., 2002. Evolution of the testis-determining gene: the rise and fall of Sry, pp. 86–96 in The Genetics and Biology of Sex Determination, edited by D. Chadwick and J. Goode. J. Wiley & Sons, New York. [PubMed]
- Haag, E., 2005. The evolution of nematode sex determination, in WormBook, edited by The C. elegans Research Community. WormBook. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Haag, E., and A. Doty, 2005. Sex determination across evolution: connecting the dots. PLoS Biol. 3 e21–e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag, E., and J. Kimble, 2000. Regulatory elements required for development of Caenorhabditis elegans hermaphrodites are conserved in the tra-2 homologue of C. remanei, a male/female sister species. Genetics 155 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag, E., and D. Pilgrim, 2005. Harnessing Caenorhabditis genomics for evolutionary developmental biology. Curr. Genomics 6 579–588. [Google Scholar]
- Haag, E., S. Wang and J. Kimble, 2002. Rapid coevolution of the nematode sex-determining genes fem-3 and tra-2. Curr. Biol. 12 2035–2041. [DOI] [PubMed] [Google Scholar]
- Hamaoka, B. Y., C. E. Dann, III, B. V. Geisbrecht and D. J. Leahy, 2004. Crystal structure of Caenorhabditis elegans HER-1 and characterization of the interaction between HER-1 and TRA-2A. Proc. Natl. Acad. Sci. USA 101 11673–11678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, R., C. Egydio de Carvalho, J. Salogiannis, B. Schlager, D. Pilgrim et al., 2006. Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev. Cell 10 531–538. [DOI] [PubMed] [Google Scholar]
- Hillier, L. W., R. D. Miller, S. E. Baird, A. Chinwalla, L. A. Fulton et al., 2007. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 5 e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., 1980. More sex-determination mutants of Caenorhabditis elegans. Genetics 96 649–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., 1986. Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics 114 15–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., 1987. A genetic analysis of the sex-determining gene, tra-1, in the nematode Caenorhabditis elegans. Genes Dev. 1 731–745. [DOI] [PubMed] [Google Scholar]
- Hodgkin, J., and S. Brenner, 1977. Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics 86 275–287. [PMC free article] [PubMed] [Google Scholar]
- Huangfu, D., and K. V. Anderson, 2006. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development 133 3–14. [DOI] [PubMed] [Google Scholar]
- Jan, E., J. W. Yoon, D. Walterhouse, P. Iannaccone and E. B. Goodwin, 1997. Conservation of the C. elegans tra-2 3′UTR translational control. EMBO J. 16 6301–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan, E., C. K. Motzny, L. E. Graves and E. B. Goodwin, 1999. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 18 258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammenga, J., A. Doroszuk, J. Riksen, E. Hazendonk, L. Spiridon et al., 2007. The Caenorhabditis elegans wild type defies the temperature-size rule owing to a single nucleotide polymorphism in tra-3. PLoS Genet. 3 e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble, J., and D. Hirsh, 1979. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70 396–417. [DOI] [PubMed] [Google Scholar]
- Kiontke, K., N. P. Gavin, Y. Raynes, C. Roehrig, F. Piano et al., 2004. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl. Acad. Sci. USA 101 9003–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman, P., J. Gubbay, N. Vivian, P. Goodfellow and R. Lovell-Badge, 1991. Male development of chromosomally female mice transgenic for Sry. Nature 351 117–121. [DOI] [PubMed] [Google Scholar]
- Kraemer, B., S. Crittenden, M. Gallegos, G. Moulder, R. Barstead et al., 1999. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol. 9 1009–1018. [DOI] [PubMed] [Google Scholar]
- Kuwabara, P. E., 1996. a A novel regulatory mutation in the C. elegans sex determination gene tra-2 defines a candidate ligand/receptor interaction site. Development 122 2089–2098. [DOI] [PubMed] [Google Scholar]
- Kuwabara, P. E., 1996. b Interspecies comparison reveals evolution of control regions in the nematode sex-determining gene tra-2. Genetics 144 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara, P. E., P. G. Okkema and J. Kimble, 1992. tra-2 encodes a membrane protein and may mediate cell communication in the Caenorhabditis elegans sex determination pathway. Mol. Biol. Cell 3 461–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont, L. B., S. L. Crittenden, D. Bernstein, M. Wickens and J. Kimble, 2004. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev. Cell 7 697–707. [DOI] [PubMed] [Google Scholar]
- Lum, D., P. Kuwabara, D. Zarkower and A. Spence, 2000. Direct protein-protein interaction between the intracellular domain of TRA-2 and the transcription factor TRA-1A modulates feminizing activity in C. elegans. Genes Dev. 14 3153–3165. [PMC free article] [PubMed] [Google Scholar]
- Madl, J. E., and R. K. Herman, 1979. Polyploids and sex determination in Caenorhabditis elegans. Genetics 93 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathies, L. D., M. Schvarstein, K. Morphy, R. Blelloch, A. Spence et al., 2004. TRA-1/GLI controls development of somatic gonadal precursors in C. elegans. Development 131 4333–4343. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., Y. Nagahama, A. Shinomiya, T. Sato, C. Matsuda et al., 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417 559–563. [DOI] [PubMed] [Google Scholar]
- Meyer, B. J., 2000. Sex in the worm counting and compensating X-chromosome dose. Trends Genet. 16 247–253. [DOI] [PubMed] [Google Scholar]
- Miller, S. W., D. C. Hayward, T. A. Bunch, D. J. Miller, E. E. Ball et al., 2003. A DM domain protein from a coral, Acropora millepora, homologous to proteins important for sex determination. Evol. Dev. 5 251–258. [DOI] [PubMed] [Google Scholar]
- Nayak, S., J. Goree and T. Schedl, 2005. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 3 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigon, V., 1951. Polyploide experimentale chez un nematode libre, Rhabditis elegans Maupas. Bull. Biol. Fr. Belg. 85 187–225. [Google Scholar]
- Panganiban, G., and J. L. Rubenstein, 2002. Developmental functions of the Distal-less/Dlx homeobox genes. Development 129 4371–4386. [DOI] [PubMed] [Google Scholar]
- Perry, M. D., C. Trent, B. Robertson, C. Chamblin and W. B. Wood, 1994. Sequenced alleles of the Caenorhabditis elegans sex-determining gene her-1 include a novel class of conditional promoter mutations. Genetics 138 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim, D., A. McGregor, P. Jackle, T. Johnson and D. Hansen, 1995. The C. elegans sex-determining gene fem-2 encodes a putative protein phosphatase. Mol. Biol. Cell 6 1159–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires-daSilva, A., and R. J. Sommer, 2004. Conservation of the global sex determination gene tra-1 in distantly related nematodes. Genes Dev. 18 1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak, R., and P. Anderson, 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7 1885–1897. [DOI] [PubMed] [Google Scholar]
- Raymond, C., C. Shamu, M. Shen, K. Seifert, B. Hirsch et al., 1998. Evidence for evolutionary conservation of sex-determining genes. Nature 391 873–881. [DOI] [PubMed] [Google Scholar]
- Rhind, N. R., L. M. Miller, J. B. Kopczynski and B. J. Meyer, 1995. xol-1 acts as an early switch in the C. elegans male/hermaphrodite decision. Cell 80 71–82. [DOI] [PubMed] [Google Scholar]
- Schedl, T., P. L. Graham, M. K. Barton and J. Kimble, 1989. Analysis of the role of tra-1 in germline sex determination in the nematode Caneorhabditis elegans. Genetics 123 755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt, C., and R. Nothiger, 2000. Structure, function and evolution of sex-determining systems in Dipteran insects. Development 127 667–677. [DOI] [PubMed] [Google Scholar]
- Schvarzstein, M., and A. M. Spence, 2006. The C. elegans sex-determining GLI protein TRA-1A is regulated by sex-specific proteolysis. Dev. Cell 11 733–740. [DOI] [PubMed] [Google Scholar]
- Sinclair, A. H., P. Berta, M. S. Palmer, J. R. Hawkins, B. L. Griffiths et al., 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346 240–244. [DOI] [PubMed] [Google Scholar]
- Starostina, N. G., J. M. Lim, M. Schvarzstein, L. Wells, A. M. Spence et al., 2007. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev. Cell 13 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, L., Z. Bao, D. Blasiar, T. Blumenthal, M. R. Brent et al., 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1 166–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard, P., D. Hansen and D. Pilgrim, 2002. Evolution of the PP2C family in Caenorhabditis: rapid divergence of the sex-determining protein FEM-2. J. Mol. Evol. 54 267–282. [DOI] [PubMed] [Google Scholar]
- Streit, A., W. Li, B. Robertson, J. Schein, I. Kamal et al., 1999. Homologs of the Caenorhabditis elegans masculinizing gene her-1 in C. briggsae and the filarial parasite Brugia malayi. Genetics 152 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston, J. E., and H. R. Horvitz, 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56 110–156. [DOI] [PubMed] [Google Scholar]
- Trent, C., B. Purnell, S. Gavinski, J. Hageman, C. Chamblin et al., 1991. Sex-specific transcriptional regulation of the C. elegans sex-determining gene her-1. Mech. Dev. 34 43–55. [DOI] [PubMed] [Google Scholar]
- Wang, S., and J. Kimble, 2001. The TRA-1 transcription factor binds TRA-2 to regulate sexual fates in Caenorhabditis elegans. EMBO J. 20 1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., and H. Chamberlin, 2002. Multiple regulatory changes contribute to the evolution of the Caenorhabditis lin-48 ovo gene. Genes Dev. 16 2345–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, W. B. (Editor), 1988. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Zarkower, D., 2001. Establishing sexual dimorphism: Conservation amidst diversity? Nat. Rev. Genet. 2 175–185. [DOI] [PubMed] [Google Scholar]
- Zarkower, D., and J. Hodgkin, 1992. Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell 70 237–249. [DOI] [PubMed] [Google Scholar]
- Zhang, B., M. Gallegos, A. Puoti, A. Durkin, S. Fields et al., 1997. A conserved RNA binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390 477–484. [DOI] [PubMed] [Google Scholar]