Abstract
The sticky/citron kinase protein is a conserved regulator of cell-cycle progression from invertebrates to humans. While this kinase is essential for completion of cytokinesis, sticky/citron kinase phenotypes disrupting neurogenesis and cell differentiation suggest additional non-cell-cycle functions. However, it is not known whether these phenotypes are an indirect consequence of sticky mutant cell-cycle defects or whether they define a novel function for this kinase. We have isolated a temperature-sensitive allele of the Drosophila sticky gene and we show that sticky/citron kinase is required for histone H3-K9 methylation, HP1 localization, and heterochromatin-mediated gene silencing. sticky genetically interacts with Argonaute 1 and sticky mutants exhibit context-dependent Su(var) and E(var) activity. These observations indicate that sticky/citron kinase functions to regulate both actin–myosin-mediated cytokinesis and epigenetic gene silencing, possibly linking cell-cycle progression to heterochromatin assembly and inheritance of gene expression states.
IN multicellular organisms, development must be coordinated with cell proliferation and differentiation to achieve proper tissue and organ size and morphology. In some cases, differentiation includes dramatic cell-cycle modifications, for example, programmed changes in DNA ploidy (for reviews see Edgar and Orr-Weaver 2001; Lee and Orr-Weaver 2003). By contrast, meiotic divisions lead to decreased ploidy to produce haploid gametes. In each of these examples, the order of cell-cycle events is altered to achieve developmental and tissue-specific outcomes. Moreover, a spectacular range of dynamic changes in nuclear morphology, chromosome condensation, chromosome pairing, and chromatin structure also accompany these variant cell cycles (for reviews see Edgar and Orr-Weaver 2001; Lee and Orr-Weaver 2003; Matzke and Birchler 2005; Wallace and Orr-Weaver 2005; Ivanovska and Orr-Weaver 2006). How cell division, cell differentiation, and changes in chromatin are coordinated remains poorly understood; however, it is clear that these processes must be linked to ensure proper propagation of epigenetic states and maintenance of cell fates (Maurange et al. 2006; Baker 2007; McClure and Schubiger 2007).
As these are fundamental processes ubiquitous to all metazoan, it is of great interest to uncover the factors that link cell-cycle progression to developmental changes in chromatin. Drosophila development, and oogenesis in particular, has proven to be an excellent model for understanding how developmental cues coordinate differentiation with cell-cycle progression (Spradling 1993; Bosco and Orr-Weaver 2002; Lee and Orr-Weaver 2003). For example, heterochromatin packaging and underreplication as well as specific developmentally regulated histone modifications have been described as occurring during Drosophila oogenesis (Lilly and Spradling 1996; Royzman et al. 2002; Aggarwal and Calvi 2004; Ivanovska et al. 2005; Hartl et al. 2007). Therefore, we used this system to screen for mutants that exhibited developmental defects in addition to cell-cycle and chromatin defects. In this report, we focus on the function of the sticky/citron kinase cell-cycle regulator as a possible candidate that links cell-cycle progression to modifications in chromatin structure.
sticky/citron kinase is a member of the AGC family of kinases that include protein kinase B, protein kinase C, Rho-kinase, myotonic dystrophy protein kinase, and myotonin-related Cdc42-binding kinase (D'Avino et al. 2004; Naim et al. 2004; for review see Zhao and Manser 2005). The only known substrate for this kinase is myosin II, the primary motor protein responsible for cytokinesis. Myosin II is activated by phosphorylation of the regulatory light chain (MLC) at Ser19/Thr18. Phosphorylation at this site allows myosin II to interact with actin, resulting in the assembly of an actomyosin complex forming the contractile ring. Several kinases, including citron kinase, have been shown to phosphorylate MLC at Ser19/Thr18, and citron kinase function is essential for cytokinesis in Drosophila as well as in some mammalian cells (for review see Matsumura 2005).
Although it is clear that citron kinase plays a critical function in cytokinesis in many systems, some reports suggest that this kinase may have other functions, particularly during neurogenesis. In mice deficient for citron kinase, death occurs within a few weeks after birth due to severe ataxia and epilepsy, although some non-neuronal cells develop normally (Di Cunto et al. 2000). Citron kinase is also required for neurogenesis (Di Cunto et al. 2003; LoTurco et al. 2003; Ackman et al. 2007). In a Down syndrome mouse model, citron kinase is responsible for inhibiting neurite extension (Berto et al. 2007). Interestingly, this citron-kinase-mediated neurite inhibition is through a direct interaction with tetratricopeptide repeat protein TTC3, a Drosophila ortholog of dTPR2, which suppresses polyglutamine toxicity in a fly Huntington's disease model (Kazemi-Esfarjani and Benzer 2000; Berto et al. 2007). Further supporting a neuro-specific function is the observation that a cleaved form of citron kinase protein, citron-N, directly interacts with the postsynaptic density protein 95 (PSD95) and localizes to synapses (Madaule et al. 2000).
A nuclear and mitotic function for citron kinase also has been described. In mouse-cultured keratinocytes, citron kinase was shown to be important for gene transcriptional regulation and cell differentiation (Grossi et al. 2005). In rat hepatocytes, citron kinase localizes to the nucleus and is important for G2/M progression, suggesting that this protein has a critical nuclear function prior to cytokinesis (Liu et al. 2003). This precytokinesis function is consistent with studies where pharmacological and RNA interference (RNAi) inactivation of myosin II result in mitotic spindle defects in vertebrate and Drosophila cells, although these studies do not directly examine the role of citron kinase in spindle assembly (Somma et al. 2002; Rosenblatt et al. 2004). However, in mouse neuronal explants, live imaging showed that a citron kinase mutation resulted in mitotic defects due to abnormal spindle formation (LoTurco et al. 2003).
Finally, citron kinase has been implicated in retroviral replication: The rubella virus RNA replicase binds directly to citron kinase protein and inactivates its kinase activity (Atreya et al. 2004a,b). By contrast, HIV-1 production is enhanced by citron-kinase-mediated exocytosis (Loomis et al. 2006). Thus, it appears that citron kinase normally inhibits rubella virus RNA replication while promoting HIV-1. How citron kinase affects either RNA virus is not well understood.
The Drosophila sticky (sti) gene is the citron kinase ortholog. Drosophila citron kinase function is essential for viability, and all sti alleles previously studied are homozygous lethal. Mutant phenotypes include neuroblast and spermatocyte cytokinesis defects resulting in multinucleated and polyploid cells (D'Avino et al. 2004; Naim et al. 2004; Shandala et al. 2004). Mutations in sti also result in late telophase defects, including persistent midbodies and abnormal F-actin and anillin structures (Naim et al. 2004). RNAi knockdown of sti in the Drosophila developing eye causes proliferation defects, and in tissue culture cells sti RNAi causes cytokinesis defects, producing multinucleated cells (D'Avino et al. 2004; Echard et al. 2004). Here, we report a novel Drosophila sti allele that is adult viable at low temperatures and lethal at high temperatures. We also show that sti functions in heterochromatin assembly and epigenetic gene silencing, a function previously not known for this gene.
MATERIALS AND METHODS
Drosophila strains:
The sticky gene and sti1 and sti3 mutants were described in Gatti and Goldberg (1991), D'Avino et al. (2004), and Naim et al. (2004). The sti1 and sti3 mutants and all mapping and deficiency stocks were acquired from the Bloomington Stock Center. Mutant lines for the screen were from the Zuker collection and were previously described in Koundakjian et al. (2004). Meiotic recombination mapping was done by using the mapping stock ru, h, th, st, cu, sr, e, and ca. The UAS-Ago1 transgenic line was a gift from Daniela Zarnescu and was previously described (Williams and Rubin 2002; Jin et al. 2004). The Rho1720 and Ago1 mutant lines were obtained from the Bloomington Stock Center; yw; Rho1720/Cyo was previously described (D'Avino et al. 2004). Ago1k08121 (y[1] w[67c23]; P{w[+mC]=lacW}AGO1[k08121]/CyO) and Ago1k00208 (y[1] w[67c23]; P{w[+mC]=lacW}AGO1[k00208]/CyO, P{y[+t7.7] ry[+t7.2]=Car20y}EW1) were previously described (Roch et al. 1998; Spradling et al. 1999; Kataoka et al. 2001). The white+ variegating lines have been previously described as follows: In(1)wm4h was obtained from the Bloomington Stock Center and described in Reuter and Wolff (1981). The DX1-mini-w 7-tandem repeat line was a gift from James Birchler and described in Dorer and Henikoff (1994). The bwD; st variegating line was obtained from Steven Henikoff and was previously described (Slatis 1955; Talbert and Henikoff 2006).
Immunofluorescence and microscopy:
We dissected ovaries in Grace's insect medium (Invitrogen, San Diego). The ovaries were transferred to a 1.5-ml Eppendorf tube and fixed for 5 min in a solution of 4% formaldehyde in buffer B (100 mm KH2PO4/K2HPO4, pH 6.8, 450 mm KCl, 150 mm NaCl, 20 mm MgCl2). Immunostaining was performed as previously described (Royzman et al. 1999; Bosco et al. 2001). The ovaries were incubated for 10 min in a solution of 1 ml PBT (PBS, 0.005% Triton X-100) and 0.5 μl 4′,6-diamidino-2-phenylindole (DAPI; 100 μg/ml in 95% EtOH). This was followed by one 10-min wash in PBT. To stain the cell membranes, mouse antiphosphotyrosine (clone 4G10, Upstate Biotech) was used at 1:75 dilution. Rabbit antihistone H3 dimethyl-lysine-9 (anti-H3-dmK9) was used at 1:150 and obtained from Upstate Biotech. Rabbit anti-heterochromatin protein 1 (HP1) was a gift from Michael Botchan and was used at 1:200 as previously described (Pak et al. 1997).
All samples were mounted in Vectashield (Vector Laboratories, Burlingame, CA). Follicle-cell nuclei, egg chambers, and larval brain mitotic chromosomes were viewed using a Nikon Eclipse E800 microscope. Staged egg chambers were viewed at ×40 and larval brain mitotic chromosomes at ×100. Images were captured using a digital camera (RT Monochrome SPOT Model 2.1.1).
Nuclear diameter measurements:
Approximately 200 DAPI-stained nuclei were imaged as above at ×40 magnification. For each egg chamber, 20–30 nuclei were traced out and selected using the Polygonal Lasso tool, and the diameter of each selected nucleus was determined in pixels using the Image Histogram function of Photoshop 7.0. The average nuclear diameter was calculated for each experimental condition. The values in pixels were converted to microns using the conversion factor of 2.8 pixels/μm. For all of the measurements taken, the standard error was calculated.
Flow cytometry of follicle-cell nuclei:
For DNA content measurements, we dissected 10–20 ovary pairs in Grace's insect medium. Grace's was removed, and 700 μl of filtered ice-cold buffer (200 mm Tris–HCl, ph 7.4, 4 mm MgCl2, 0.1% Triton X) was added to the ovaries. Ovaries were transferred into a 60-mm petri dish with a truncated pipette tip. Ovaries were chopped with a single-edged blade until homogenous. Another 700 μl of buffer was added and ovaries were chopped again. Chopped ovaries were filtered through a small piece of cheesecloth (∼3 cm2). Chopped ovaries were then filtered through 30-μm mesh (Sefar 03-30/18) and placed into flow cytometry tubes (Sarstedt) with 20 μl of DAPI (100 μg/ml) in each tube and left on ice. The petri dish was washed once with 700 μl of buffer and this was filtered through cheese cloth and mesh as above, pooling it in the flow cytometry tube. Samples were kept in the tube on ice with DAPI for 10–60 min before flow cytometry (PARTEC CCA 11-01-3002) and DNA content was analyzed as previously described (Bosco et al. 2007).
Eye-pigment assays and position-effect variegation:
Virgin females from In(1)wm4h and w/w;DX1-mini-w/CyO were crossed to sti3/TM6B, stiZ3-5829/TM6B and yw; Rho1720/Cyo males. The resulting 3- to 4-day-old male progeny that were +/+ (TM6B Tb, Hu, e), sti/+, Rho1720/+, and CyO/+ were separated and processed for eye pigment. In the case of the w/w;DX1-mini-w/CyO cross, only w/Y;DX1-mini-w/+ males were selected. TM6B/+ male progeny from the above crosses were backcrossed to In(1)wm4h females and the resulting male In(1)wm4h; TM6B/+ and In(1)wm4h; +/+ progeny were compared to ensure that TM6B had no Su(var) activity (Figure 6L).
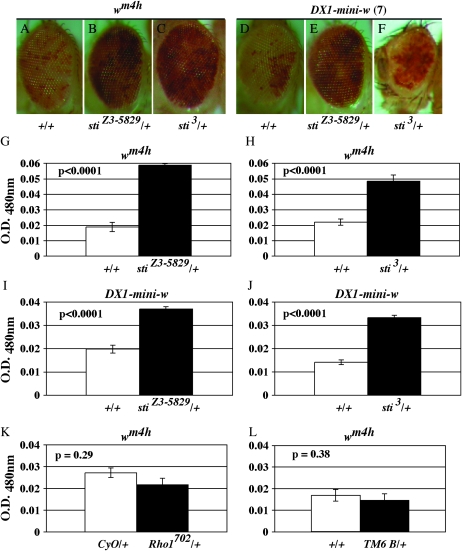
Figure 6.—
Mutations in sticky dominantly suppress position-effect variegation of white. (A) wm4h/Y; +/TM6 B (+/+). (B) wm4h/Y; stiZ3-5829/+. (C) wm4h/Y;sti3/+. (D) w; DX1-mini-w+; +/TM6 B (+/+). (E) w; DX1-mini-w+; stiZ3-5829/+. (F) w; DX1-mini-w+; sti3/+. (G–J) Eye pigment from 10–30 males was extracted and quantified by spectrophotometry at 480 nm (see materials and methods). Open bars are +/+ controls and solid bars are sti/+ heterozygotes as indicated. In each case, highly significant differences were seen as indicated. Controls for (K) wm4h/Y;CyO and wm4h/Y; Rho1702 and (L) wm4h/Y;+/+ and wm4h/Y;TM6B/+ show no significant differences in white gene variegation and expression as indicated. All P-values were calculated by a two-tailed t-test assuming unequal variance.
For white gene eye-pigment analysis, flies were frozen in −80° and ∼10–30 fly heads were used for each assay. Heads were placed in 100 μl/head of methanol and 1 μl/head of 11.6 m HCl. Samples were mixed and allowed to rotate for 1 hr in the dark at room temperature. Samples were neutralized with 8 μl/head of 1.5 m Tris, pH 8.8, mixed briefly, and centrifuged for 3 min in a microcentrifuge at maximum setting. For each sample, 3 μl was analyzed for optical density at 480 nm using a ND-1000 spectrophotometer (NanoDrop). An average of 10 trials and standard error were determined. P-values were calculated by a two-tailed, paired t-test using Microsoft Excel.
For the effect of sticky on brownDominant (bwD) silencing, first bwD/bwD;st/st females were crossed to stiZ3-5829, st, e/TM6B. Second, bwD/bwD;st/st females were also crossed to ru, h, th, st, cu, sr, e, ca males. Eye pigmentation was imaged from bwD/+;stiZ3-5829, st, e/st progeny from the first cross and bwD/+;st/ru, h, th, st, cu, sr, e, ca progeny from the second cross. Finally, bwD/+;stiZ3-5829, st, e/st males were crossed to stiZ3-5829, st, e/TM6B females to obtain bwD/+;stiZ3-5829, st, e/stiZ3-5829, st, e progeny to demonstrate the homozygous effects of stiZ3-5829 on bwD silencing (Figure 7C).
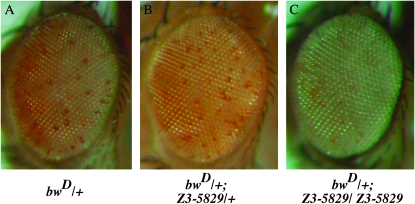
Figure 7.—
The sticky hypomorph mutant enhances silencing at bwD. The hypomorphic stiZ3-5829 mutation had no effect on bwD silencing as a heterozygote (compare A and B); however, 100% (n = 20) of stiZ3-5829 homozygotes strongly enhanced silencing at the bwD locus (C). Flies were reared at 18° to minimize rough-eye phenotype and an individual with little or no eye defect is shown. The relevant genotypes are as follows: (A) bwD/+ ; st/st. (B) bwD/+ ; stiZ3-5829/+, st/st. (C) bwD/+ ; stiZ3-5829/stiZ3-5829, st/st.
Scanning electron microscope imaging:
Adult flies were placed in 70% ethanol and stored up to 1 week. Samples were washed five times with 100% ethanol and dried under vacuum. Dried whole flies were coated with gold and imaged with an Electroscan E3 scanning electron microscope at ×150–200 magnification. Images were either captured on Polaroid film and then scanned or captured directly as digital images.
Temperature-sensitive viability:
To test for temperature-sensitive lethality, stiZ3-5829 e/TM6B Tb, Hu, e females were crossed to sti3/TM6B Tb, Hu, e females and progeny were allowed to develop at either 18° or 25°. All progeny were collected after 32–36 days and 16–18 days for 18° and 25°, respectively.
Western blot analysis:
Levels of sticky protein were determined by Western blot of ovarian extracts from heterozygotes, homozygotes, and trans-heterozygotes raised at room temperature. Ovaries were placed directly into SDS–PAGE loading buffer with 1 mm DTT, homogenized, and incubated at 98° for 5 min. Samples were centrifuged and supernatants were electrophoresed on SDS–PAGE. Approximately 10 ovary-pair equivalents were loaded from 3- to 5-day-old fattened females. The anti-sticky rabbit polyclonal was a gift from David Glover and Western conditions were used as described in D'Avino et al. (2004). Anti-sticky was used at 1:3000 dilution. HRP–anti-rabbit was used as secondary antibody at 1:10,000. This membrane was stripped and reprobed using mouse antilamin (Drosophila lamin Dm0; Developmental Studies Hybridoma Bank ADL84.12) (Stuurman et al. 1995). Antilamin was used at 1:5000 dilution, and secondary HRP–anti-mouse was used at 1:10,000. Secondary antibody signal was visualized by chemiluminescence.
RESULTS
A screen for defects in follicle-cell ploidy:
We performed a genetic screen of female sterile mutants from the Zuker third chromosome collection (Koundakjian et al. 2004) to uncover new genes that function to regulate cell-cycle progression, morphogenesis, and chromatin structure during oogenesis. Ovaries from 1500 homozygous female flies were dissected and stained with DAPI and BrdU (data not shown). Fluorescence microscopy was used to determine whether follicle-cell nuclear morphology and DNA replication patterns were abnormal. We found that seven mutants when homozygous exhibited follicle-cell chromocenter defects and fat nuclei (fnu), which indicated that these may have increased ploidy levels (data not shown). All seven mutants complemented each other in all pairwise combinations with respect to the fnu phenotype. This indicated that they represent seven different complementation groups (data not shown).
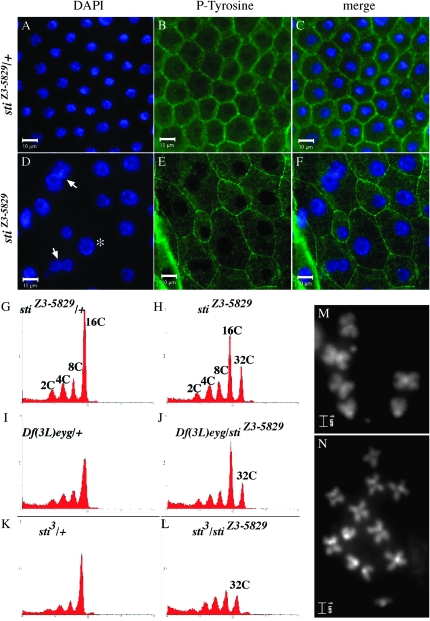
One mutant (Z3-5829) had the most severe nuclear size phenotype, where late stage 12–13 egg chambers accumulated the most extreme fat nuclei as well as multinucleated follicle cells and also had a disorganized follicle-cell layer (Figure 1, A–F). Z3-5829 had increased DNA content to 32c (Figure 1, G–L). Larval brain cell chromosome spreads also exhibited polyploid cells (Figure 1, M and N), thus confirming that Z3-5829 mutants fail to regulate ploidy in polyploid and diploid cells. We therefore focused on the Z3-5829 mutant to characterize it more completely. Further characterization of the other six fnu mutants will be reported in future studies.
Figure 1.—
sticky mutant cells have enlarged nuclei and increased ploidy. Follicle-cell layer of stiZ3-5829/+ (A–C) and stiZ3-5829 homozygotes (D–F) where DAPI-stained nuclei (blue) and cell membranes are stained with antiphosphotyrosine (P-tyrosine, green). (D) sti mutant cells have enlarged nuclei (asterisk) and multinucleated cells (arrows). Follicle-cell size and shape is also disrupted (E). Flow cytometry of DAPI-stained isolated nuclei from wild-type and sticky mutant ovaries: (G) stiZ3-5829/+ follicle-cell nuclei terminate with normal 16c ploidy. (H) stiZ3-5829 homozygous follicle cells have 32c ploidy. (I) Df(3L)eyg/+ follicles have normal ploidy, and (J) stiZ3-5829/Df(3L)eyg have 32c ploidy. (K) sti3/+ follicle-cell nuclei have normal ploidy, and (L) stiZ3-5829/sti3 have 32c ploidy. DAPI-stained larval brain mitotic cells of (M) wild-type stiZ3-5829/+ and (N) stiZ3-5829/Df(3L)ED4483 stained with DAPI. Follicle-cell images taken at ×40; bar, 10 μm. Mitotic chromosome images taken at ×100; bar, 1 μm.
Z3-5829 is a defect in sticky, a novel viable allele of the Drosophila gene encoding citron kinase:
To understand the molecular lesion that caused these hyperpolyploid follicle-cell and fat nuclei phenotypes, we mapped the Z3-5829 mutation. First, we used standard meiotic recombination mapping and followed the large follicle-cell phenotype in addition to the female sterility phenotype for all recombinant progeny. Recombinant progeny showed that the large follicle-cell and ploidy defects mapped to the left arm of chromosome three at ∼36 and ∼11 cM from hairy and ∼7 cM from thread. This placed the Z3-5829 mutation within the 66D-72D1 cytogenetic region delineated by the hairy and thread genes. Deficiency mapping also placed the Z3-5829 mutation in this region (Table 1), and previously described mutants in this region—sticky1 (data not shown) and sticky3 (sti3) as well as the deficiencies Df(3L)eyg, Df(3L)ED4483, and Df(3L)F10—failed to complement the follicle-cell nuclear size and ploidy defects in ovarian follicle cells and larval neuroblasts (Table 1 and Figure 1).
TABLE 1.
Deficiencies used to map Z3-5829
| Deficiency | Region deleted | Complementation of fat nuclei |
|---|---|---|
| Df(3L)eyg | 69A4:69D6 | No |
| Df(3L)ED4483 | 69A4:69D3 | No |
| Df(3L)F10 | 69A2:69D1 | No |
| Df(3L)ED4475 | 68C13:69B4 | Yes |
| Df(3L)ED215 | 69B5:69B4 | Yes |
| Df(3L)Exel6116 | 68F2:69A2 | Yes |
| Df(3L)Exel6117 | 69D1:69E2 | Yes |
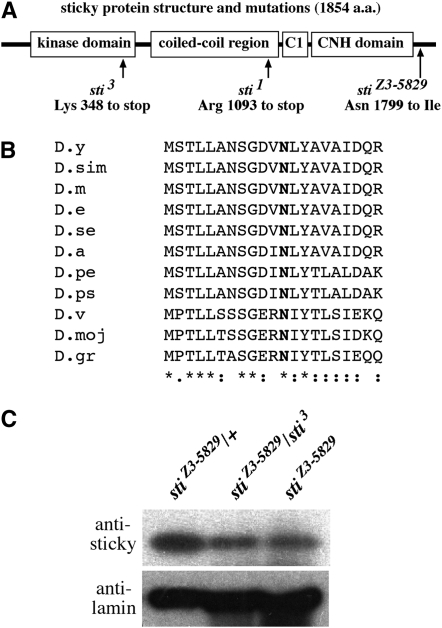
The sti1 and sti3 mutations have been shown to affect the Drosophila gene encoding the citron kinase protein (D'Avino et al. 2004; Naim et al. 2004; Shandala et al. 2004). Sequencing of the entire intron and exon regions of the sticky gene (CG10522) from Z3-5829 homozygotes revealed a single nucleotide mutation that is predicted to cause an asparagine (N)-to-isoleucine (I) change at amino acid position 1799 (N1799I) (Figure 2A). This N1799I mutation occurs at a residue that is conserved in other Drosophila species (Figure 2B). Sequencing of this region from another, unrelated Zuker homozygous mutant (Z3-0019) indicated that this was not a polymorphic site in this genetic background (data not shown). Western blots of ovarian extracts from wild-type and mutant females indicated that sticky protein levels were reduced in the mutants, relative to a nuclear lamin loading control (Figure 2C).
Figure 2.—
stiZ3-5829 mutation in a conserved residue results in reduced sticky protein levels. (A) Gene structure depicting previously described sticky mutations and the position of the stiZ3-5829 N1799I mutation. The sticky gene product encodes a conserved kinase domain, a coiled-coil region, a cysteine-rich (C1) domain, and a citron-N homology (CNH) domain. (B) An alignment of 20 amino acids immediately adjacent to the N1799 site is shown in boldface type. The Drosophila species listed are in the following order: D. yakuba (D.y), D. simulans (D.dim), D. melanogaster (D.m), D. erecta (D.e), D. sechelia (D.se), D. ananasae (D.a), D. persimilis (D.pe), D. pseudoobscura (D.ps), D. virilis (D.v), D. mojavensis (D.moj), and D. grimshawi (D.gr). (C) Western blot of sticky protein levels in heterozygous stiZ3-5829/TM6B control (left lane), stiZ3-5829 homozygotes (middle lane), and stiZ3-5829/stiZ3-5829 (right lane). Nuclear lamin is used as a loading control (bottom). The truncated sticky protein predicted to arise from the heterozygous sti3 extract cannot be detected by this antibody, which was raised against a C-terminal antigen as previously described (D'Avino et al. 2004).
Because we found this molecular change in the sticky gene sequence, and because sti1, sti3, and more than one deficiency in the sticky gene region failed to complement the Z3-5829 mutation, we conclude that the Z3-5829 mutation lies in Drosophila citron kinase encoded by the sticky gene and thus constitutes a novel adult viable allele of this gene. We hereafter will refer to this novel allele as stiZ3-5829. Furthermore, reduced protein levels in this mutant demonstrate that the N1799I mutation alters sticky protein stability, which is likely to be the cause of the hypomorphic phenotypes reported in this study.
The stiZ3-5829 mutant is temperature sensitive:
Interestingly, Drosophila stiZ3-5829 cultured at 18° produced more homozygous adult individuals than those reared at 21° (room temperature) and none were recovered at 25° (data not shown). To determine whether the temperature-sensitive lethality was linked to the sti Z3-5829 mutation, we crossed stiZ3-5829, e/TM6B Hu, e to sti3/TM6B Hu, e and scored the number of three possible progeny classes: (1) stiZ3-5829, e/TM6B Hu, e, (2) sti3/TM6B Hu, e, and (3) stiZ3-5829, e/sti3. At 18°, the stiZ3-5829, e/sti3 class (47% ± 6) appears greater than the expected 33%; however, this was not a statistically significant difference (P = 0.25, n = 338) relative to the sibling controls (Figure 3A). By contrast, at 25° the stiZ3-5829/sti3 class of progeny was significantly less than expected (14% ± 5, P < 0.03, n = 398). When we examined a larger pool of progeny, we observed that this temperature-sensitive decrease in viability was more severe in males than in females (Figure 3B, P = 0.01, n = 4193). This bias against male progeny was not observed in flies grown at 18° (Figure 3B, P = 0.38, n = 3682).
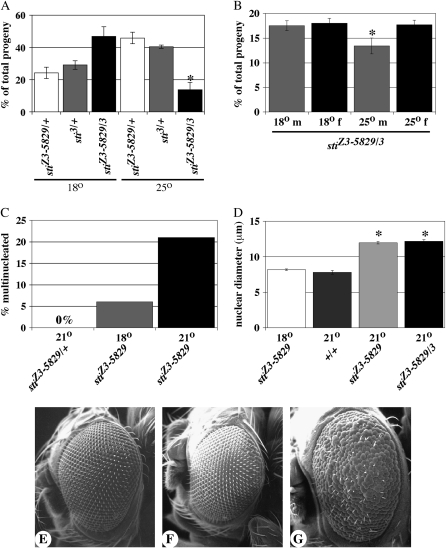
Figure 3.—
The stiZ3-5829 is a temperature-sensitive allele. (A) Graph compares the percentage of total progeny that result from the cross stiZ3-5829/TM6B × sti3/TM6B at 18° and 25°. Open bars, stiZ3-5829/TM6B. Shaded bars, sti3/TM6B. Solid bars, stiZ3-5829/sti3. No significant difference was observed between heterozygous progeny and stiZ3-5829/sti3 (P = 0.25, n = 338) at 18° whereas stiZ3-5829/sti3 progeny were significantly reduced at 25° (asterisk, P < 0.03, n = 398). (B) Both male (m) and female (f) stiZ3-5829/sti3 progeny are equally represented at 18° (P = 0.38, n = 3682) whereas male progeny are significantly reduced at 25° (asterisk, P = 0.01, n = 4193). (C) Multinucleated follicle cells are never observed in stiZ3-5829/+ ovaries (0%, n > 1000 egg chambers) while stiZ3-5829 homozygotes have ∼6% multinucleated cells at 18° (shaded bar, n = 200). stiZ3-5829 homozygotes have ∼22% multinucleated cells at 21° (solid bar, n = 200). (D) Nuclear diameters of sticky mutant follicle cells in stage 12–13 egg chambers at 18° (open bar) and wild type at 21° (+/+, darkly shaded bar) have normal size, ranging from 6 to 11 μm. The nuclear diameters of both stiZ3-5829 homozygotes (lightly shaded bar) and stiZ3-5829/sti3 (stiZ3-5829/3, solid bar) at 21° are significantly larger than stiZ3-5829 homozygotes at the permissive 18° (asterisks, P < 0.001, n = 200 for each genotype). (E and F) Scanning electron microscopy of the eyes of stiZ3-5829/+ (E), sti3/+ (F), and (G) stiZ3-5829/sti3 at 25°. Magnification, ×150.
We also observed the enhancement of three other phenotypes when sticky mutants were grown at higher temperature. First, we examined the frequency of multinucleated follicle cells as well as follicle-cell nuclear size in individuals grown at 18° and 21°. The fraction of multinucleated follicle cells was approximately fourfold higher in flies reared at higher temperature (Figure 3C). In addition, stage 12–13 follicle-cell nuclear diameter in sticky mutants reared at 21° was ∼12 μm ± 0.2, whereas sticky mutants grown at 18° had a normal nuclear diameter of ∼8 μm ± 0.1 (data not shown and Figure 3D). We observed this for both stiZ3-5829 homozygotes and stiZ3-5829/sti3, and for both genotypes this difference was highly significant (P < 0.0001, n = 200).
We observed that the sticky rough-eye phenotype is also temperature sensitive. In stiZ3-5829 homozygotes and stiZ3-5829/sti3, 100% of adults had missing eye bristles and ∼37% had rough eyes when reared at 21°. When grown at 25°, 100% of stiZ3-5829/sti3 adults had missing bristles and rough eyes (Figure 3, E–G, and data not shown). Thus, taken together, these data suggest that the stiZ3-5829 allele of citron kinase is temperature sensitive for viability, follicle-cell division, follicle-cell nuclear size, and rough-eye phenotypes. In addition, the fact that this rough-eye phenotype is similar to an RNAi-induced rough-eye phenotype further supports the idea that the stiZ3-5829 allele is a hypomorph mutant that becomes more penetrant at higher temperatures (D'Avino et al. 2004). Finally, follicle-cell division, follicle-cell nuclear size, and rough-eye phenotypes were never observed in heterozygotes at any temperature, indicating that both the stiZ3-5829 allele and its temperature sensitivity are recessive traits.
Follicle-cell heterochromatin is disrupted in sticky mutants:
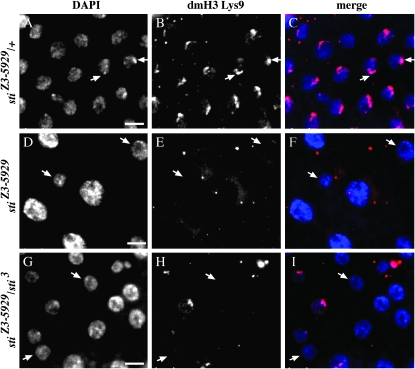
We found it curious that nuclear size increased at higher temperature in the sticky mutant (Figure 3D). Higher temperature did not induce any further increase in ploidy beyond 32C (Figure 1), and thus this increase in nuclear size could not be explained by higher DNA content. Therefore, we speculated that reorganization of chromatin packaging, and not DNA content, was responsible for this dramatic change in nuclear size. As in all Drosophila polyploid tissues, follicle-cell A:T-rich pericentric heterochromatic sequences form an aggregate known as the chromocenter, and this aggregate is where histone H3 lysine-9 (H3 Lys9) methylation and HP1 localization are enriched (Bernstein and Allis 2005; Bosco et al. 2007). First, we probed with anti-H3 dimethylated Lys9 (dmLys9). As expected, the wild-type sibling controls exhibited crescent-like staining in the nucleus that colocalizes with the DAPI-intense chromocenter (Figure 4, B and C, arrows) as previously shown in follicle cells of this developmental stage (Bosco et al. 2007). However, H3 dmLys9 staining was diminished or nearly absent in stiZ3-5829 homozygotes and stiZ3-5829/sti3 trans-heterozygotes (Figure 4, D and E, and F–H). Interestingly, loss of the H3 dmLys9 staining in the sticky mutant was not restricted to the large nuclei within the follicle-cell epithelium as normal-sized nuclei also were affected (Figure 4, arrows). This is important because it suggests that disruption of H3 dmLys9 at the pericentric heterochromatin is not necessarily associated with increased ploidy and nuclear size.
Figure 4.—
Methylation of histone H3-K9 is diminished in sticky follicle cells. Stage 10B/11 follicle cells are shown for (A–C) stiZ3-5829/+, (D–F) stiZ3-5829 homozygotes, and (G–I) stiZ3-5829/sti3 trans-heterozygotes. DAPI-stained nuclei (A, D, and G). Histone H3 dimethyl-Lys9 (B, E, and H). Merged images (C, F, and I) show DAPI in blue and dimethyl-Lys9 in magenta. Arrows in A–C point to chromocenters enriched for H3 dimethyl-Lys9. Arrows in D–F and G–I show both large and normal-sized nuclei with disorganized chromocenters and diminished H3 dimethyl-Lys9 staining. Bars, 10 μm.
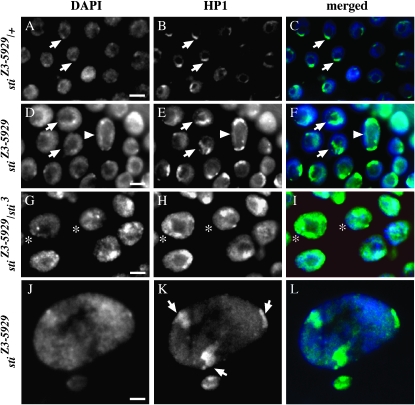
Second, we probed HP1 that is also enriched at the pericentric heterochromatin and requires Lys9 di- and trimethylation on histone H3. In wild-type controls, HP1 was nuclear and clearly enriched at the chromocenter, exhibiting a crescent-shape peri-centric heterochromatin pattern (Figure 5, A–C, arrows). By contrast, stiZ3-5829 homozygotes and stiZ3-5829/sti3 trans-heterozygotes had HP1 mislocalized and diffused throughout the nucleus (Figure 5, D–I). In some cells where the DAPI chromocenter was clearly visible, HP1 was either diffuse throughout the nucleus and/or enriched at nonchromocenter locations (Figure 5, G–I, asterisks). Some stiZ3-5829 homozygous nuclei exhibited multiple regions of HP1 enrichment (Figure 5, D–F, arrowheads), and in extremely large, ∼50-μm, stage 12–13 nuclei, multiple intense regions of HP1 localization were observed (Figure 5, J–L, arrows in K).
Figure 5.—
HP1 is mislocalized in sticky follicle cells. Stage 10B/11 follicle cells are shown for (A–C) stiZ3-5829/+, (D–F and J–L) stiZ3-5829 homozygotes, and (G–I) stiZ3-5829/sti3 trans-heterozygotes. DAPI-stained nuclei (A, D, G, and J). HP1 (B, E, H, and K). Merged image (C, F, I, and L) shows DAPI in blue and HP1 in green. Arrows in A–C point to chromocenters enriched for HP1 in a crescent pattern at the chromocenter. Arrows in D–F show nuclei with disorganized chromocenters and mislocalized HP1 that is not restricted to a crescent shape. Arrowheads in D–F show nucleus with clear double crescent and diffuse HP1 staining. Asterisks in G–I show that in nuclei with or without clearly delineated chromocenters, HP1 still exhibits a diffuse nuclear staining with multiple enrichment sites. In J and K, one normal-size nucleus and one extremely large nucleus are shown. Neither have a clear single chromocenter but multiple sites of HP1 enrichment are observed in K (arrows). Bars, 10 μm.
sticky mutants disrupt position-effect variegation and gene silencing:
The chromatin phenotypes described above in the Drosophila ovary of sticky mutants could be an indirect consequence of cell division defects. For example, the multiple regions of HP1 enrichment (Figure 5) could be due to failure in cytokinesis earlier in follicle-cell development where cells blocked in cytokinesis could have entered endo cycles as mono-nucleated polyploid or multinucleated polyploid cells with multiple chromocenters acting as multiple HP1 nucleation sites. Alternatively, it was possible that citron kinase may function to regulate heterochromatin, independent of its cell-cycle function. We sought to distinguish between these two possibilities by testing whether sticky mutations could function as dominant suppressors of wm4h position-effect variegation (PEV). This was possible because the sticky mutant cytokinesis defect is completely recessive and therefore any dominant modification of variegation could not be associated with cell division defects. In wm4h flies, an X chromosome inversion leads to partial silencing of the white+ gene by pericentric heterochromatin. This heterochromatin silencing is HP1 and Su(var)3-9 (H3-K9 histone methyl transferase) dependent (Grigliatti 1991). We tested two different mutant alleles (stiZ3-5829 and sti3) of the sticky gene, and in both cases observed that one mutant copy of sticky was sufficient to suppress silencing at the wm4h locus (Figure 6, A–C). Extraction and quantitation of eye pigment also confirmed that wm4h gene expression was increased in the sticky heterozygotes (Figure 6, G and H). This Su(var) activity at wm4h was not likely to be due to other mutations in this genetic background since similar tests with heterozygotes for two other Zuker mutants, Z3-0019 and Z3-5163, had no dominant effect on wm4h expression (T. Hartl and G. Bosco, unpublished observation). In addition, a loss-of-function mutation in Rho1, an upstream activator of citron kinase, and the TM6B balancer chromosome had no effect on position-effect variegation in a wm4h background (Figure 6, K and L).
To test whether sticky exhibited Su(var) activity at heterochromatic loci that were not pericentric, we asked whether sticky alleles also dominantly suppressed silencing of tandem arrays of the mini-white+ gene (DX1) (Dorer and Henikoff 1994) present within a euchromatic region of the second chromosome. We found that both stiZ3-5829 and sti3 could dominantly suppress silencing of the DX1-mini-white+ array (Figure 6, D–F, and I and J).
To ensure that the PEV suppression was not an unexpected artifact of sticky/citron kinase regulation of the white gene product, we tested whether sticky also affected silencing at the brown locus (bw). The bwD dominant mutation is an insertion of heterochromatin that disrupts bw gene expression and also results in trans-silencing of the wild-type bw+ allele, and bwD silencing is HP1 dependent (Slatis 1955; Talbert and Henikoff 2006). Therefore, we hypothesized that sticky mutations should disrupt bwD silencing. We found that bwD silencing was unaffected in stiZ3-5829/+ heterozygotes (Figure 7, A and B). However, we were surprised to observe that the stiZ3-5829 homozygotes were strong enhancers of bwD silencing (Figure 7C). This E(var) activity of the stiZ3-5829 homozygote was not likely to be due to other mutations in this genetic background since similar tests with homozygotes for two other Zuker mutants from the same genetic background, Z3-0019 and Z3-5163, exhibit Su(var) activity at bwD (T. Hartl and G. Bosco, unpublished observation).
sticky genetically interacts with Argonaute 1:
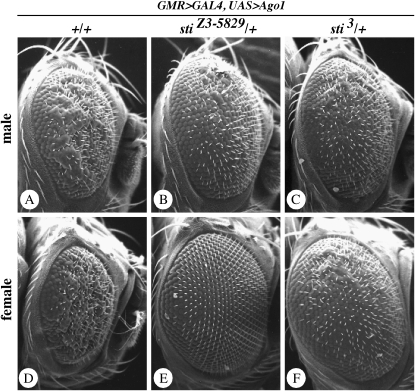
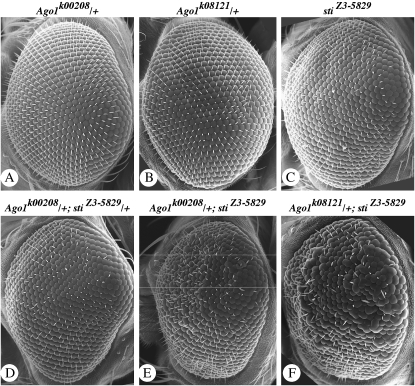
The follicle-cell defects in histone H3 Lys9 methylation and HP1 localization and Su(var) activity for wm4h and DX1-mini-white tandem arrays support a model where sticky kinase functions to regulate transcriptional gene silencing (TGS) through changes in heterochromatin. However, the observation that stiZ3-5829 homozygotes were strong enhancers of bwD silencing suggests that sticky kinase functions to regulate gene silencing in a context-dependent manner, as has been shown for other PEV modifiers, including the RNA-mediated silencing factors piwi and homeless (Pal-Bhadra et al. 2004a). To test this possibility, we examined whether sticky genetically interacts with Argonaute 1 (Ago1) since it also has been shown to regulate multiple RNA-mediated gene-silencing processes, including TGS and heterochromatin formation (for reviews see Nakahara and Carthew 2004; Behm-Ansmant et al. 2006; Moazed et al. 2006; Zofall and Grewal 2006). Gain-of-function mutants of Ago1 cause a rough-eye phenotype in Drosophila, and this is caused by upregulation of the Ago1-mediated functions (Williams and Rubin 2002; Jin et al. 2004). We tested whether sticky hypomorphic and loss-of-function mutations could suppress Ago1 gain-of-function phenotypes. We observed that two different sticky alleles suppressed the rough-eye phenotypes caused by Ago1 overexpression (Figure 8). Strikingly, this was a dominant effect observed in 100% of heterozygotes, and in some cases there was a full suppression of wild-type eye morphology (Figure 8).
Figure 8.—
Mutations in sticky dominantly suppress the Ago1 overexpression eye phenotype. In all cases, GMR>GAL4 is activating expression of UAS>Ago1. (A) Wild-type (+/+) male. (B) stiZ3-5829/+ male. (C) sti3/+ male. (D) Wild-type (+/+) female. (E) stiZ3-5829/+ female. (F) sti3/+ female. Note that sticky mutants suppress both rough- and small-eye phenotypes. We also observed that suppression was consistently stronger in females than in males.
This genetic interaction suggested that Ago1 protein function requires wild-type sticky/citron kinase protein and that sticky mutant phenotypes could be caused, in part, by insufficient Ago1 activity. Therefore, we tested whether Ago1 loss-of-function mutants could enhance sticky rough-eye phenotypes. We found that two different Ago1 loss-of-function mutations dominantly enhanced the stiZ3-5829 rough-eye phenotype (Figure 9). These data provide strong support for a model in which Ago1-mediated processes are sensitive to sticky/citron kinase dosage and dependent on citron kinase wild-type function (Figure 10).
Figure 9.—
Mutations in Ago1 dominantly enhance the sticky rough-eye phenotype. Genotypes are shown above each panel as follows: (A) w; Pw+ Ago1k00208/+. (B) w; Pw+ Ago1k08121/+. (C) yw;stiZ3-5829/stiZ3-5829. (D) w; Pw+ Ago1k00208/+;stiZ3-5829/+. (E) w; Pw+ Ago1k00208/+;stiZ3-5829/stiZ3-5829. (F) w; Pw+ Ago1k08121/+;stiZ3-5829/stiZ3-5829. All flies were grown at 18°. Images were taken at ×150 magnification.
Figure 10.—
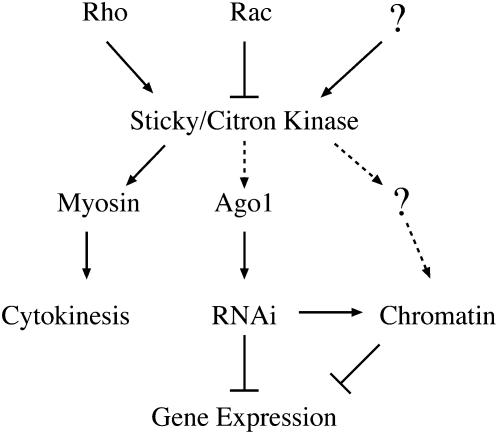
A speculative model for sticky/citron kinase effects on gene expression. As previously shown (D'Avino et al. 2004), Rho is thought to activate while Rac inhibits sticky/citron kinase activity. We speculate that other (question mark) developmental and cellular cues may also impinge upon its activity. Active sticky/citron kinase can phosphorylate myosin to facilitate the completion of cytokinesis. Here we show that Ago1 activity may be dependent on sticky/citron kinase, although this may be indirect (dashed lines). Misexpression of genes in sticky/citron kinase mutants could be due to insufficient Ago1 protein activity, misregulation of RNAi processes, and/or misregulation of other factors (question mark) that modulate chromatin structure. Alterations in gene expression patterns can result from direct (solid arrows) or indirect (dashed arrows) regulation of one or more of these processes by sticky/citron kinase. Although it is not shown in this model, it is possible that cytokinesis can also be regulated by Ago1 and/or other RNAi components (Antar et al. 2005; Deshpande et al. 2005; Meyer et al. 2006). Furthermore, we speculate that sticky/citron kinase may function to link cell-cycle progression to epigenetic maintenance of heterochromatin.
DISCUSSION
sticky/citron kinase is a member of the AGC family of serine/threonine kinases, including protein kinase B, protein kinase C, Rho-kinase, and myotonic dystrophy protein kinase, among others (Zhao and Manser 2005). These kinases can form homodimers where autophosphorylation is important for stimulating kinase activity while N-terminal kinase domains may be inhibited by the C-terminal domain through inter- or intramolecular interactions, and their functions have been implicated in a variety of essential cellular processes (for review see Zhao and Manser 2005 and for a specific example see Bush et al. 2000). However, our lack of understanding of how citron kinase functions in vivo is underscored by the fact that cytoplasmic myosin light chain is the only known substrate for this kinase (for reviews see Matsumura 2005; Zhao and Manser 2005).
The Drosophila sticky/citron kinase gene is essential for viability and sticky mutant alleles reported thus far have precluded analysis of adult functions. The Zuker Drosophila mutants (Koundakjian et al. 2004) have proven to be a valuable collection of adult viable mutations in genes that otherwise would not be recovered in screens for null mutations in essential genes. For example, many cell-cycle genes involved in oogenesis could not be identified if it were not possible to recover adult female hypomorphic mutants in these essential genes (Spradling 1993; Bosco and Orr-Weaver 2002). In a screen of ∼1500 third chromosome mutants, we identified one mutant with dramatic ovarian cell-cycle and chromatin phenotypes. Here, we report the characterization of a new adult viable mutation in the Drosophila sticky (citron kinase) gene. We refer to this new allele as stiZ3-5829. The stiZ3-5829 allele is a temperature-sensitive mutation, and thus this new allele provides us with an excellent genetic tool with which to study the many possible functions of the sticky protein. This kinase almost certainly has other substrates that have yet to be discovered. The complex phenotypes exhibited in mice, rats, humans, and now Drosophila sticky/citron kinase mutant models support the idea that this kinase functions to regulate processes other than cytokinesis.
stiZ3-5829 exhibits temperature-sensitive cytokinesis and nuclear morphology defects:
The stiZ3-5829 homozygous mutant is completely inviable at 25° (data not shown). The stiZ3-5829 homozygotes also show stronger phenotypes at 21°, relative to 18° (Figure 3). Consistent with the stiZ3-5829 allele conferring temperature sensitivity, the stiZ3-5829/sti 3 trans-heterozygotes also exhibits more severe phenotypes at higher temperatures (Figure 3). The rough-eye phenotype (data not shown), viability, cytokinesis, and follicle-cell nuclear size all display temperature sensitivity, where all phenotypes are more severe at higher temperatures. Western analysis confirms that the single-amino-acid substitution of asparagine to isoleucine at position 1799 (N1799I) (Figure 2) results in decreased protein levels, consistent with the hypomorphic behavior of the stiZ3-5829 allele (Figure 2). Although the N1799I mutation is in a small block of amino acids conserved within Drosophila species (Figure 2), it is not clear whether this amino acid substitution makes the sticky protein structurally unstable or if it uncovers an important functional domain critical for sticky protein regulation. If Drosophila sticky kinase forms homodimers and autophosphorylates, as do other AGC kinases, it is then possible that this C-terminal N1799I mutation could be affecting some aspect of auto-regulation. Future studies that further dissect this temperature-sensitive allele will allow an in-depth understanding of this kinase and its in vivo function.
sticky/citron kinase regulates chromatin structure independently of its cytokinesis function:
We show that, in addition to the expected role in cytokinesis, the Drosophila sticky protein functions to regulate nuclear size and chromatin structure during follicle-cell development (Figures 4 and 5). We considered the possibility that misregulation of nuclear morphology and chromatin in the sticky mutants was an indirect consequence of cell division defects. For example, follicle cells with multiple nuclei and/or hyperpolyploid nuclei may have failed to properly establish and organize heterochromatin early in development and subsequently fail to maintain heterochromatin in postmitotic, endoreplicating follicle cells. Since the sticky mutant cytokinesis defect is completely recessive, we sought to ascertain whether sticky mutants had dominant effects on chromatin, using PEV as an assay (Figure 6).
We found that suppression of gene silencing is not an indirect consequence of cell-cycle and cytoskeletal defects in sticky mutants: First, sticky mutants do not have dominant cell-cycle or cytoskeletal defects, whereas in two of the three gene-silencing assays that we tested sticky mutants were dominant suppressors of PEV (Figure 6). Second, a mutation in Rho1 previously shown to disrupt Rho1 activation of sticky/citron kinase does not suppress gene silencing (Figure 6), whereas this Rho1720 mutation does enhance a cytokinesis defect (D'Avino et al. 2004; Shandala et al. 2004). Third, the observation that sticky mutations both suppress and enhance gene silencing depending on the genomic context indicates that citron kinase regulation of heterochromatin is complex and likely due to mislocalization of chromatin factors such as HP1 (Figure 5). Therefore, we favor a model where sticky/citron kinase regulates chromatin independently of its role in actin/myosin regulation during cytokinesis.
How does Drosophila sticky kinase regulate chromatin?
An appealing interpretation of these data is that citron kinase regulates transcription and/or chromatin factors. Its genetic interaction with Ago1 suggests that sticky regulates heterochromatin through one or more RNA-mediated pathways, possibly affecting RNA-induced transcriptional silencing mechanisms (for reviews see Kavi et al. 2005; Talbert and Henikoff 2006). It is intriguing that two other RNA-processing genes, piwi and homeless, also behave as context-dependent suppressors and enhancers of chromatin-mediated gene silencing. Both piwi and homeless mutants enhance transgene-pairing-sensitive silencing whereas mutations in either gene suppress silencing (e.g., DX1-mini-white) of unpaired transgenes (Pal-Bhadra et al. 2004a,b). Silencing at the brown locus involves both heterochromatin and pairing of the bwD and the bw+ alleles (for review see Talbert and Henikoff 2006). Although piwi and homeless mutants have not been tested for their effects on bwD silencing, we speculate that the context-dependent effects of sticky on wm4h, DX1-mini-white and bwD silencing is analogous to the context-dependent Su(var) and E(var) activities of piwi and homeless mutants.
Three previous studies have reported on sticky protein localization, and none of these reports show sticky protein in the nucleus (D'Avino et al. 2004; Naim et al. 2004; Shandala et al. 2004). Although this lack of evidence for nuclear localization does not exclude the possibility, it does raise the question as to how sticky can regulate nuclear chromatin factors. We do not believe that the reported cytoplasmic localization of Drosophila sticky kinase presents a paradox with regards to its novel function in regulating chromatin. First, nuclear envelope breakdown could allow sticky kinase access to chromatin during mitosis, thereby obviating the need for nuclear localization. Second, many examples exist where cytoplasmic localization/function of chromatin proteins and chromatin regulators is the norm. For example, some histone acetyl transferases are strictly cytoplasmic and acetylation of histones facilitates nuclear import required during S-phase (Parthun et al. 1996). Similarly, nuclear import, cytoplasmic retention and/or protein stability may all be affected by phosphorylation of sticky/citron kinase substrates. As these kinase substrates are yet to be identified, it is difficult to say how directly sticky/citron kinase activity affects chromatin factors.
Does citron kinase regulate transcription and chromatin structure through actin/myosin? In mammalian hepatocytes, the citron kinase protein has been localized to the nucleus (Liu et al. 2003). A growing body of evidence also suggests that both myosin and actin proteins may be involved in regulating gene transcription, chromatin remodeling, and nuclear architecture (for reviews see Olave et al. 2002; Grummt 2006). Of particular interest in vertebrates and Drosophila is a nuclear-actin-related protein, Arp6, which physically associates with HP1 and is required for pericentric heterochromatin stability (Kato et al. 2001; Ohfuchi et al. 2006). Recently, nuclear actin has been shown to be directly inhibiting transcription factor activity (Vartiainen et al. 2007). Taken together, these data do suggest that it is possible that nuclearly localized mammalian citron kinase, actin, and myosin could be effectors of gene transcription and chromatin. However, at present there is no evidence to indicate that nuclear actin or myosin is regulated by citron kinase.
sticky/citron kinase activity as a potential link between cell division and maintenance of epigenetic states:
Our observations that sticky/citron kinase modulates heterochromatin structure, affects expression of genes adjacent to or within heterochromatin, and genetically interacts with Ago1 suggest a possible link between cell-cycle progression, RNA-mediated gene silencing, and chromatin remodeling (Figure 10). Although examples of microRNA targeting of cell-cycle genes have been reported (Hatfield et al. 2005; Carleton et al. 2007), it is not clear how cell-cycle progression, aspects of microRNA regulation, and heterochromatin maintenance are coordinated. For example, DNA replication may disrupt epigenetic histone modifications, and how these histone modifications are “inherited” by newly synthesized DNA strands is not known. Recent evidence suggests that disruption of cell-cycle progression, chromatin factors, and epigenetic histone modifications are important for maintenance of Drosophila stem cells and cell fate determination after stem cell division (for reviews see Eissenberg 2006; Maurange et al. 2006; Baker 2007; McClure and Schubiger 2007). Presumably, transfer of epigenetic marks must occur before cell division is completed to propagate an epigenetic state from mother to daughter cell. In addition, mitotic chromosome condensation and decondensation must occur before completion of cytokinesis while also preserving the epigenetic state of heterochromatin. How or if these epigenetic processes are coordinated with the final step of cell division, cytokinesis, is not known. Interestingly, Drosophila Ago1 has been shown to regulate germline stem cell proliferation (Yang et al. 2007). It will be of great interest to determine whether Ago1, other RNA-mediated silencing components, and/or chromatin factors could be direct phosphorylation targets of citron kinase. Furthermore, our observation that sticky mutant males are more susceptible to temperature-sensitive lethality (Figure 3B) also suggests a possible role in dosage compensation. In Drosophila, dosage compensation is yet another RNA- and chromatin-mediated gene activation mechanism (Deng and Meller 2006).
How do we understand mammalian citron kinase function in light of the Drosophila sticky mutant phenotypes? In mammalian models, citron kinase is known to be important for the final step of cell division, cytokinesis, by phosphorylating myosin II (Madaule et al. 1998; Eda et al. 2001; Yamashiro et al. 2003). Some studies have shown citron kinase to be important earlier in the cell cycle as it is required for progression through G2 and mitosis (Liu et al. 2003; LoTurco et al. 2003). Other studies in mice and rats suggest that citron kinase functions during neurogenesis and that a cleaved form of the protein localizes to synapses (Di Cunto et al. 2000, 2003; LoTurco et al. 2003; Zhang and Benson 2006). Consistent with a neurodevelopment function, recent data implicate citron kinase in a mouse Down syndrome model (Berto et al. 2007). Still other studies have reported that citron kinase is important for transcriptional regulation and cell differentiation (Grossi et al. 2005). One possible explanation for many of these citron kinase phenotypes could be that there are many more kinase substrates that are yet to be identified that participate in various processes. We speculate that some of these substrates are chromatin factors. The defects in G2, mitosis, gene transcription, and cell differentiation in citron-kinase-deficient mammalian cells could be explained by a failure to regulate chromatin structure. For example, inefficient packaging of chromatin after replication could lead to G2 arrest/delay, and defects in centric heterochromatin could result in nonfunctional kinetochore and mitotic arrest/delay. Our observation that Drosophila sticky/citron kinase exhibits chromatin defects and epigenetic gene silencing suggests that in Drosophila this is a distinct possibility, but it remains to be determined whether vertebrate citron kinase also regulates chromatin.
Future studies that identify the sticky/citron kinase substrates as well as protein interactors will further elucidate the function of the sticky protein with regards to its role in cytokinesis, chromatin structure, and RNAi proteins. We anticipate that this novel Drosophila stiZ3-5829 mutant and its temperature sensitivity will prove to be an excellent new tool for dissecting the molecular function(s) of the sticky/citron kinase protein and will facilitate identification of its kinase targets as well as sticky genetic interactors in both Drosophila and mammalian systems.
Acknowledgments
We are grateful to Terry Orr-Weaver for fly stocks used in this study and for supporting the initial screen in her lab. We thank Kim Dej, Irena Ivanoska, Laura Lee, and Helena Kashevsky for help in maintaining the Zuker mutant collection. We are grateful to Charles Zuker for sharing the mutant collection. We thank David Glover and Pier Paolo D'Avino for anti-sticky antibodies. We thank David Galbraith for the use of his flow cytometry unit and Joao Torres Leivi-Neto and Barb Carolus of the Arizona Research Lab for assistance with flow cytometry. We thank David Bentley from the Arizona Research Lab for SEM use and preparation of samples. We thank Erin Kelleher, Teri Markow, Tom Hartl, Roy Parker, and Frans Tax for comments on the manuscript and members of the Bosco lab for helpful discussions. S.J.S. was supported in part by the University of Arizona Undergraduate Biology Research Program funded by the Howard Hughes Medical Institute (#71196-521304). This work was supported by a grant to G.B. from the National Institutes of Health (RO1 GM069462).
References
- Ackman, J. B., R. L. Ramos, M. R. Sarkisian and J. J. LoTurco, 2007. Citron kinase is required for postnatal neurogenesis in the hippocampus. Dev. Neurosci. 29 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal, B. D., and B. R. Calvi, 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430 372–376. [DOI] [PubMed] [Google Scholar]
- Antar, L. N., J. B. Dictenberg, M. Plociniak, R. Afroz and G. J. Bassell, 2005. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 4 350–359. [DOI] [PubMed] [Google Scholar]
- Atreya, C. D., S. Kulkarni and K. V. Mohan, 2004. a Rubella virus P90 associates with the cytokinesis regulatory protein citron-K kinase and the viral infection and constitutive expression of P90 protein both induce cell cycle arrest following S phase in cell culture. Arch. Virol. 149 779–789. [DOI] [PubMed] [Google Scholar]
- Atreya, C. D., K. V. Mohan and S. Kulkarni, 2004. b Rubella virus and birth defects: molecular insights into the viral teratogenesis at the cellular level. Birth Defects Res. A Clin. Mol. Teratol. 70 431–437. [DOI] [PubMed] [Google Scholar]
- Baker, N. E., 2007. Patterning signals and proliferation in Drosophila imaginal discs. Curr. Opin. Genet. Dev. 17 287–293. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant, I., J. Rehwinkel and E. Izaurralde, 2006. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb. Symp. Quant. Biol. 71 523–530. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., and C. D. Allis, 2005. RNA meets chromatin. Genes Dev. 19 1635–1655. [DOI] [PubMed] [Google Scholar]
- Berto, G., P. Camera, C. Fusco, S. Imarisio, C. Ambrogio et al., 2007. The Down syndrome critical region protein TTC3 inhibits neuronal differentiation via RhoA and citron kinase. J. Cell Sci. 120 1859–1867. [DOI] [PubMed] [Google Scholar]
- Bosco, G., and T. L. Orr-Weaver, 2002. Regulation of the cell cycle during oogenesis and early embryogenesis in Drosophila, pp. 107–154 in Regulation of Gene Expression at the Beginning of Animal Development, edited by M. DePamphilis. Elsevier, Amsterdam.
- Bosco, G., W. Du and T. L. Orr-Weaver, 2001. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol. 3 289–295. [DOI] [PubMed] [Google Scholar]
- Bosco, G., P. Campbell, J. T. Leiva-Neto and T. A. Markow, 2007. Analysis of Drosophila species genome size and satellite DNA content reveals significant differences among strains as well as between species. Genetics 177 1277–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, E. W., S. M. Helmke, R. A. Birnbaum and M. B. Perryman, 2000. Myotonic dystrophy protein kinase domains mediate localization, oligomerization, novel catalytic activity, and autoinhibition. Biochemistry 39 8480–8490. [DOI] [PubMed] [Google Scholar]
- Carleton, M., M. A. Cleary and P. S. Linsley, 2007. MicroRNAs and cell cycle regulation. Cell Cycle 6 2127–2132. [DOI] [PubMed] [Google Scholar]
- D'Avino, P. P., M. S. Savoian and D. M. Glover, 2004. Mutations in sticky lead to defective organization of the contractile ring during cytokinesis and are enhanced by Rho and suppressed by Rac. J. Cell Biol. 166 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X., and V. H. Meller, 2006. Non-coding RNA in fly dosage compensation. Trends Biochem. Sci. 31 526–532. [DOI] [PubMed] [Google Scholar]
- Deshpande, G., G. Calhoun and P. Schedl, 2005. Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes Dev. 19 1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cunto, F., S. Imarisio, E. Hirsch, V. Broccoli, A. Bulfone et al., 2000. Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron 28 115–127. [DOI] [PubMed] [Google Scholar]
- Di Cunto, F., L. Ferrara, R. Curtetti, S. Imarisio, S. Guazzone et al., 2003. Role of citron kinase in dendritic morphogenesis of cortical neurons. Brain Res. Bull. 60 319–327. [DOI] [PubMed] [Google Scholar]
- Dorer, D. R., and S. Henikoff, 1994. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell 77 993–1002. [DOI] [PubMed] [Google Scholar]
- Echard, A., G. R. Hickson, E. Foley and P. H. O'Farrell, 2004. Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 14 1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eda, M., S. Yonemura, T. Kato, N. Watanabe, T. Ishizaki et al., 2001. Rho-dependent transfer of citron-kinase to the cleavage furrow of dividing cells. J. Cell Sci. 114 3273–3284. [DOI] [PubMed] [Google Scholar]
- Edgar, B. A., and T. L. Orr-Weaver, 2001. Endoreplication cell cycles: more for less. Cell 105 297–306. [DOI] [PubMed] [Google Scholar]
- Eissenberg, J. C., 2006. Divided loyalties: transdetermination and the genetics of tissue regeneration. BioEssays 28 574–577. [DOI] [PubMed] [Google Scholar]
- Gatti, M., and M. L. Goldberg, 1991. Mutations affecting cell division in Drosophila. Methods Cell Biol. 35 543–586. [DOI] [PubMed] [Google Scholar]
- Grigliatti, T., 1991. Position-effect variegation–an assay for nonhistone chromosomal proteins and chromatin assembly and modifying factors. Methods Cell Biol. 35 587–627. [PubMed] [Google Scholar]
- Grossi, M., A. Hiou-Feige, A. Tommasi Di Vignano, E. Calautti, P. Ostano et al., 2005. Negative control of keratinocyte differentiation by Rho/CRIK signaling coupled with up-regulation of KyoT1/2 (FHL1) expression. Proc. Natl. Acad. Sci. USA 102 11313–11318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt, I., 2006. Actin and myosin as transcription factors. Curr. Opin. Genet. Dev. 16 191–196. [DOI] [PubMed] [Google Scholar]
- Hartl, T., C. Boswell, T. L. Orr-Weaver and G. Bosco, 2007. Developmentally regulated histone modifications in Drosophila follicle cells: initiation of gene amplification is associated with histone H3 and H4 hyperacetylation and H1 phosphorylation. Chromosoma 116 197–214. [DOI] [PubMed] [Google Scholar]
- Hatfield, S. D., H. R. Shcherbata, K. A. Fischer, K. Nakahara, R. W. Carthew et al., 2005. Stem cell division is regulated by the microRNA pathway. Nature 435 974–978. [DOI] [PubMed] [Google Scholar]
- Ivanovska, I., and T. L. Orr-Weaver, 2006. Histone modifications and the chromatin scaffold for meiotic chromosome architecture. Cell Cycle 5 2064–2071. [DOI] [PubMed] [Google Scholar]
- Ivanovska, I., T. Khandan, T. Ito and T. L. Orr-Weaver, 2005. A histone code in meiosis: the histone kinase, NHK-1, is required for proper chromosomal architecture in Drosophila oocytes. Genes Dev. 19 2571–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, P., D. C. Zarnescu, S. Ceman, M. Nakamoto, J. Mowrey et al., 2004. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 7 113–117. [DOI] [PubMed] [Google Scholar]
- Kataoka, Y., M. Takeichi and T. Uemura, 2001. Developmental roles and molecular characterization of a Drosophila homologue of Arabidopsis Argonaute1, the founder of a novel gene superfamily. Genes Cells 6 313–325. [DOI] [PubMed] [Google Scholar]
- Kato, M., M. Sasaki, S. Mizuno and M. Harata, 2001. Novel actin-related proteins in vertebrates: similarities of structure and expression pattern to Arp6 localized on Drosophila heterochromatin. Gene 268 133–140. [DOI] [PubMed] [Google Scholar]
- Kavi, H. H., H. R. Fernandez, W. Xie and J. A. Birchler, 2005. RNA silencing in Drosophila. FEBS Lett. 579 5940–5949. [DOI] [PubMed] [Google Scholar]
- Kazemi-Esfarjani, P., and S. Benzer, 2000. Genetic suppression of polyglutamine toxicity in Drosophila. Science 287 1837–1840. [DOI] [PubMed] [Google Scholar]
- Koundakjian, E. J., D. M. Cowan, R. W. Hardy and A. H. Becker, 2004. The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics 167 203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, L. A., and T. L. Orr-Weaver, 2003. Regulation of cell cycles in Drosophila development: intrinsic and extrinsic cues. Annu. Rev. Genet. 37 545–578. [DOI] [PubMed] [Google Scholar]
- Lilly, M. A., and A. C. Spradling, 1996. The Drosophila endocycle is controlled by cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 10 2514–2526. [DOI] [PubMed] [Google Scholar]
- Liu, H., F. Di Cunto, S. Imarisio and L. M. Reid, 2003. Citron kinase is a cell cycle-dependent, nuclear protein required for G2/M transition of hepatocytes. J. Biol. Chem. 278 2541–2548. [DOI] [PubMed] [Google Scholar]
- Loomis, R. J., D. A. Holmes, A. Elms, P. A. Solski, C. J. Der et al., 2006. Citron kinase, a RhoA effector, enhances HIV-1 virion production by modulating exocytosis. Traffic 7 1643–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoTurco, J. J., M. R. Sarkisian, L. Cosker and J. Bai, 2003. Citron kinase is a regulator of mitosis and neurogenic cytokinesis in the neocortical ventricular zone. Cereb. Cortex 13 588–591. [DOI] [PubMed] [Google Scholar]
- Madaule, P., M. Eda, N. Watanabe, K. Fujisawa, T. Matsuoka et al., 1998. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature 394 491–494. [DOI] [PubMed] [Google Scholar]
- Madaule, P., T. Furuyashiki, M. Eda, H. Bito, T. Ishizaki et al., 2000. Citron, a Rho target that affects contractility during cytokinesis. Microsc. Res. Tech. 49 123–126. [DOI] [PubMed] [Google Scholar]
- Matsumura, F., 2005. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 15 371–377. [DOI] [PubMed] [Google Scholar]
- Matzke, M. A., and J. A. Birchler, 2005. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6 24–35. [DOI] [PubMed] [Google Scholar]
- Maurange, C., N. Lee and R. Paro, 2006. Signaling meets chromatin during tissue regeneration in Drosophila. Curr. Opin. Genet. Dev. 16 485–489. [DOI] [PubMed] [Google Scholar]
- McClure, K. D., and G. Schubiger, 2007. Transdetermination: Drosophila imaginal disc cells exhibit stem cell-like potency. Int. J. Biochem. Cell Biol. 39 1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, W. J., S. Schreiber, Y. Guo, T. Volkmann, M. A. Welte et al., 2006. Overlapping functions of argonaute proteins in patterning and morphogenesis of Drosophila embryos. PLoS Genet. 2 e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed, D., M. Buhler, S. M. Buker, S. U. Colmenares, E. L. Gerace et al., 2006. Studies on the mechanism of RNAi-dependent heterochromatin assembly. Cold Spring Harb. Symp. Quant. Biol. 71 461–471. [DOI] [PubMed] [Google Scholar]
- Naim, V., S. Imarisio, F. Di Cunto, M. Gatti and S. Bonaccorsi, 2004. Drosophila citron kinase is required for the final steps of cytokinesis. Mol. Biol. Cell 15 5053–5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara, K., and R. W. Carthew, 2004. Expanding roles for miRNAs and siRNAs in cell regulation. Curr. Opin. Cell Biol. 16 127–133. [DOI] [PubMed] [Google Scholar]
- Ohfuchi, E., M. Kato, M. Sasaki, K. Sugimoto, Y. Oma et al., 2006. Vertebrate Arp6, a novel nuclear actin-related protein, interacts with heterochromatin protein 1. Eur. J. Cell Biol. 85 411–421. [DOI] [PubMed] [Google Scholar]
- Olave, I. A., S. L. Reck-Peterson and G. R. Crabtree, 2002. Nuclear actin and actin-related proteins in chromatin remodeling. Annu. Rev. Biochem. 71 755–781. [DOI] [PubMed] [Google Scholar]
- Pak, D. T. S., M. Pflumm, I. Chesnokov, D. W. Huang, R. Kellum et al., 1997. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91 311–323. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra, M., U. Bhadra and J. A. Birchler, 2004. a Interrelationship of RNA interference and transcriptional gene silencing in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 69 433–438. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra, M., B. A. Leibovitch, S. G. Gandhi, M. Rao, U. Bhadra et al., 2004. b Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303 669–672. [DOI] [PubMed] [Google Scholar]
- Parthun, M. R., J. Widom and D. E. Gottschling, 1996. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell 87 85–94. [DOI] [PubMed] [Google Scholar]
- Reuter, G., and I. Wolff, 1981. Isolation of dominant suppressor mutations for position-effect variegation in Drosophila melanogaster. Mol. Gen. Genet. 182 516–519. [DOI] [PubMed] [Google Scholar]
- Roch, F., F. Serras, F. J. Cifuentes, M. Corominas, B. Alsina et al., 1998. Screening of larval/pupal P-element induced lethals on the second chromosome in Drosophila melanogaster: clonal analysis and morphology of imaginal discs. Mol. Gen. Genet. 257 103–112. [DOI] [PubMed] [Google Scholar]
- Rosenblatt, J., L. P. Cramer, B. Baum and K. M. McGee, 2004. Myosin II-dependent cortical movement is required for centrosome separation and positioning during mitotic spindle assembly. Cell 117 361–372. [DOI] [PubMed] [Google Scholar]
- Royzman, I., R. J. Austin, G. Bosco, S. P. Bell and T. L. Orr-Weaver, 1999. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev. 13 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman, I., A. Hayashi-Hagihara, K. J. Dej, G. Bosco, J. Y. Lee et al., 2002. The E2F cell cycle regulator is required for Drosophila nurse cell DNA replication and apoptosis. Mech. Dev. 119 225–237. [DOI] [PubMed] [Google Scholar]
- Shandala, T., S. L. Gregory, H. E. Dalton, M. Smallhorn and R. Saint, 2004. Citron kinase is an essential effector of the Pbl-activated Rho signalling pathway in Drosophila melanogaster. Development 131 5053–5063. [DOI] [PubMed] [Google Scholar]
- Slatis, H. M., 1955. A reconsideration of the brown-dominant position effect. Genetics 40 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somma, M. P., B. Fasulo, G. Cenci, E. Cundari and M. Gatti, 2002. Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol. Biol. Cell 13 2448–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, A. C., 1993. Developmental genetics of oogenesis, pp. 1–70 in The Development of Drosophila melanogaster, edited by M. Bate and A. Martinez Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Spradling, A. C., D. Stern, A. Beaton, E. J. Rhem, T. Laverty et al., 1999. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman, N., N. Maus and P. A. Fisher, 1995. Interphase phosphorylation of the Drosophila nuclear lamin: site-mapping using a monoclonal antibody. J. Cell Sci. 108(Pt. 9): 3137–3144. [DOI] [PubMed] [Google Scholar]
- Talbert, P. B., and S. Henikoff, 2006. Spreading of silent chromatin: inaction at a distance. Nat. Rev. Genet. 7 793–803. [DOI] [PubMed] [Google Scholar]
- Vartiainen, M. K., S. Guettler, B. Larijani and R. Treisman, 2007. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 316 1749–1752. [DOI] [PubMed] [Google Scholar]
- Wallace, J. A., and T. L. Orr-Weaver, 2005. Replication of heterochromatin: insights into mechanisms of epigenetic inheritance. Chromosoma 114 389–402. [DOI] [PubMed] [Google Scholar]
- Williams, R. W., and G. M. Rubin, 2002. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc. Natl. Acad. Sci. USA 99 6889–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro, S., G. Totsukawa, Y. Yamakita, Y. Sasaki, P. Madaule et al., 2003. Citron kinase, a Rho-dependent kinase, induces di-phosphorylation of regulatory light chain of myosin II. Mol. Biol. Cell 14 1745–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., D. Chen, R. Duan, L. Xia, J. Wang et al., 2007. Argonaute 1 regulates the fate of germline stem cells in Drosophila. Development 134 4265–4272. [DOI] [PubMed] [Google Scholar]
- Zhang, W., and D. L. Benson, 2006. Targeting and clustering citron to synapses. Mol. Cell. Neurosci. 31 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z. S., and E. Manser, 2005. PAK and other Rho-associated kinases∷effectors with surprisingly diverse mechanisms of regulation. Biochem. J. 386 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall, M., and S. I. Grewal, 2006. RNAi-mediated heterochromatin assembly in fission yeast. Cold Spring Harb. Symp. Quant. Biol. 71 487–496. [DOI] [PubMed] [Google Scholar]