Abstract
The conduction mechanism of Escherichia coli AmtB, the structurally and functionally best characterized representative of the ubiquitous Amt/Rh family, has remained controversial in several aspects. The predominant view has been that it facilitates the movement of ammonium in its uncharged form as indicated by the hydrophobic nature of a pore located in the center of each subunit of the homotrimer. Using site-directed mutagenesis and a combination of biochemical and crystallographic methods, we have investigated mechanistic questions concerning the putative periplasmic ammonium ion binding site S1 and the adjacent periplasmic “gate” formed by two highly conserved phenylalanine residues, F107 and F215. Our results challenge models that propose that NH4+ deprotonation takes place at S1 before NH3 conduction through the pore. The presence of S1 confers two critical features on AmtB, both essential for its function: ammonium scavenging efficiency at very low ammonium concentration and selectivity against water and physiologically important cations. We show that AmtB activity absolutely requires F215 but not F107 and that removal or obstruction of the phenylalanine gate produces an open but inactive channel. The phenyl ring of F215 must thus play a very specific role in promoting transfer and deprotonation of substrate from S1 to the central pore. We discuss these results with respect to three distinct mechanisms of conduction that have been considered so far. We conclude that substrate deprotonation is an essential part of the conduction mechanism, but we do not rule out net electrogenic transport.
Ammonium transport is facilitated by the Amt proteins, found in all domains of life from bacteria to man. In animals they are represented by the Rhesus (Rh) proteins, which have been implicated in ammonium homeostasis and possibly in CO2 transport (1, 2). Escherichia coli AmtB is the best studied Amt protein, both functionally and structurally (3). AmtB is a homotrimer, and each subunit contains a narrow pore connecting periplasmic and cytoplasmic vestibules (4, 5). The pore appears sterically blocked on its periplasmic side by a constriction or “gate” formed by the partly stacked phenyl rings of F107 and F215 (4, 5).
Based on structural insights it has been proposed that NH4+ is recruited at a periplasmic binding site (S1 or Am1), becomes deprotonated, and then diffuses as NH3 through the hydrophobic pore (4, 5). The results of an in vitro assay using proteoliposomes reconstituted with AmtB were consistent with ammonia conduction (5), but others could not reproduce them (3), and the issue of whether Amts are NH4+ transporters or NH3 channels has remained controversial. Electrogenic ammonium transport has been reported for some plant Amts in the oocyte system, and it has been proposed that ammonia and a proton might transit the pore separately (for review see ref. 6). More recently, the group of Kustu has provided evidence that AmtB also mediates net transport of the ionic form (7). Thus, in the absence of a reliable in vitro assay, the conduction mechanism remains poorly understood (3). Important mechanistic questions concern (i) the biological role of the external NH4+ binding site in substrate recruitment (8) and in discrimination against water and physiologically important cations such as Na+ or K+ (9–11) and (ii) the nature of the deprotonation process if ammonium does transit the pore as NH3.
A plethora of molecular dynamics (MD) simulations have attempted to shed light on mechanistic aspects of ammonium permeation through Amt proteins. These studies have addressed the strength and specificity of the periplasmic binding site, the nature of ammonium deprotonation and ammonia reprotonation in the vestibule regions, the dynamics and possible function of the highly conserved Phe gate, NH4+ versus NH3 conduction through the channel pore, and channel hydration (12–19). Overall, they agree in predicting strong binding of an ammonium ion at S1, in supporting the view of a highly dynamic Phe gate, and, very importantly, in rejecting the possibility of NH4+ conduction through the pore because of a high energy barrier. They have not explicitly addressed the possibility of separate conduction paths for NH3 and H+. Some of the studies suggest an important role for the Phe gate in deprotonation or in discrimination of substrate versus water. They disagree in other aspects, particularly with respect to the site and mechanism of deprotonation.
In a previous study, we experimentally investigated, through a structure/function analysis, the role of the two highly conserved pore histidines of AmtB (20). We have now used the same approach to investigate the role of other conserved residues that define both the putative periplasmic ammonium binding site and the Phe gate. Our results provide new information on the characteristics of the periplasmic ammonium binding site and suggest an essential role of F215 in substrate conduction.
Results
Mutagenesis and Inhibition at the Periplasmic NH4+ Binding Site.
The putative NH4+ binding site S1 is located at the base of the periplasmic vestibule and is structurally defined by a hydrogen-bonding interaction with the hydroxyl group of S219 and by cation–π interactions with the aromatic rings of F107 and W148 (4, 5). All three residues are highly conserved in Amt proteins whereas only F107 is conserved in Rh proteins. To clarify the biological and mechanistic role of the ammonium binding site in Amt proteins we examined the effects of modifying the three residues shaping S1 and searched for substrates that bind to S1 but are not conducted to separate effects on binding from those on transport.
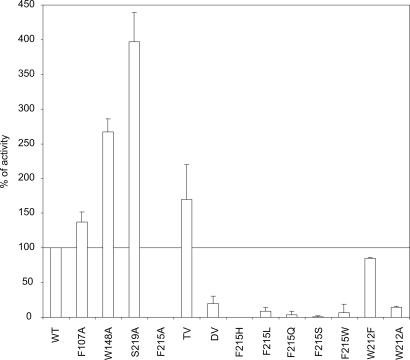
First, the activities of the alanine variants F107A, W148A, and S219A and of the triple variant F107A/W148A/S219A were measured by using our previously established MA transport assay (21). All variants behaved like wild-type AmtB in being correctly targeted to the inner membrane, in running as one major trimeric species with an apparent molecular mass of ≈90 kDa on SDS/PAGE, and in their expression level, as checked by Western blotting [see supporting information (SI) Figs. S1 and S2]. Cells expressing F107A and the triple mutant had MA transport activities comparable to wild type, whereas those expressing W148A and S219A had activities, respectively, 2.5 and four times higher than wild type (Fig. 1).
Fig. 1.
AmtB variant activity in vivo. [14C]Methylammonium accumulation (AmtB activity) was measured in GT1000 (ΔglnK ΔamtB) expressing plasmid-encoded variant AmtB by adding 20 μM (final concentration) [14C]methylammonium at time 0. Activities are percentages of the activity in GT1000 (ΔglnK ΔamtB) expressing wild-type AmtB. The level in GT1000 (ΔglnK ΔamtB) has been subtracted. TV, triple variant (F107A/W148A/S219A); DV, double variant (F107A/F215A).
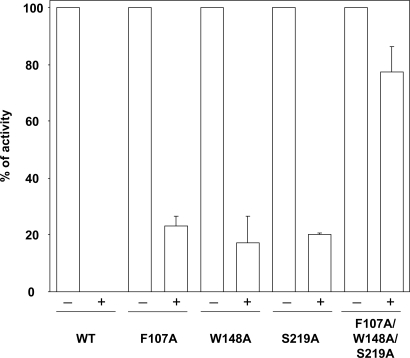
To corroborate, and assess more directly, the loss of binding affinity in these variants we investigated the effect of inhibitors likely to act by competitive binding at S1. Our structural analysis of wild-type AmtB crystallized in the presence of 150 mM imidazole at pH 6.0 (3) showed strong density at S1, compatible with imidazole, most likely in its protonated state (Fig. S3). Indeed imidazole does inhibit MA conduction (Fig. S4), supporting the concept of competitive binding to S1, but 10–100 mM imidazole is needed for significant inhibition. 50 mM Na+ or K+ showed no inhibition. The monovalent cations Cs+ and Tl+ have been shown to bind to the ammonium binding site of glutamine synthetase (22). A concentration of 50 mM Cs+ was required for full inhibition of AmtB activity (data not shown), but Tl+ effected complete inhibition at 0.5 mM (Fig. 2), indicating strong binding to S1. This interpretation was substantiated by the observation that the triple variant (F107A/W148A/S219A) showed only 15–20% inhibition at the same concentration of Tl+ (Fig. 2). By contrast, cells expressing any one of the single variants showed ≈80% inhibition (Fig. 2), indicating that here the binding affinity for Tl+ is not strongly diminished.
Fig. 2.
AmtB activity inhibition by thallium. [14C]Methylammonium accumulation (AmtB activity) was measured by adding 20 μM (final concentration) [14C]methylammonium at time 0 without (−) or with (+) 0.5 mM TI+. For each variant, activities are a percentage of the activity measured without TI+. The level in GT1000 (ΔglnK ΔamtB) has been subtracted.
To confirm binding of Tl+ to S1, wild-type AmtB was crystallized in the presence of 1 mM thallium chloride and diffraction data to 2.3-Å resolution were collected (Table S1). Unfortunately, crystals could be obtained only in the presence of 100 mM ammonium sulfate, which was expected to interfere strongly with Tl+ binding, and, indeed, no significant difference electron density peak was observed. However, an anomalous difference electron density map from a data set collected at the Tl+ absorption edge (λ = 0.97822 Å) had its highest peak (5σ) precisely at the S1 site (Fig. 3). The next highest peaks were all observed at the sulfur atoms of cysteine and methionine residues corroborating the significance of the Tl+ peak. The low signal corresponds to an occupancy of at best 10% indicating efficient competition by the much more abundant ammonium ion. Together, these results validate the presence of a cationic ammonium, or MA, binding site at the periplasmic entrance of AmtB.
Fig. 3.
Binding of thallium at the S1 position. A cut perpendicular to the membrane plane through an AmtB monomer, represented by selected structural elements, is shown together with the surface of the solvent-accessible space delineating the pore. The anomalous difference map (blue mesh, contoured at 3σ) has its highest peak exactly at the binding site S1.
Although the effects on MA transport of binding site modification are not dramatic, they indicate that S219 and W148 play a more significant role than F107 in stabilizing the ion, in accord with computational studies (15). However, F107 might affect substrate conduction in a more complex way (see below). Moreover, the results show that an intact binding site is not necessary for MA transport under the chosen conditions, suggesting that S1 may be biologically significant only at much lower substrate concentrations. In accordance with this hypothesis, Amt proteins in plants and microorganisms are expressed only at very low external ammonium concentrations (23).
Functional and Structural Studies of Phe Gate Variants.
Two partially stacked phenyl rings, F107 and F215 in AmtB, are seen at the periplasmic pore entrance in all Amt structures (4, 5, 24, 25) and appear to severely constrict the substrate pathway (Fig. 3). Their strict conservation in conducting Amt/Rh proteins suggests an important biological or mechanistic role, primary candidates being ammonium deprotonation or control of substrate access to the pore. MD simulations (26) have shown that their rotation, permitting passage of an NH4+ ion, can occur at low energy cost. The constriction is thus a highly dynamic gate, and steric hindrance is unlikely to be rate-limiting for substrate conduction.
As already discussed F107 does not play an essential role in substrate transport, but, in contrast, the activity of F215A is below detection (Fig. 1). To explore the stringency for phenylalanine at this position a series of additional F215 variants (H, L, Q, S, and W) were investigated (Fig. 1 and Figs. S1 and S2). All were inactive indicating a very specific role of this phenyl ring. We also probed another highly conserved residue, W212, located below F215 and opposite the pore residue H168 (Fig. 3). W212A was inactive whereas W212F retained 80% of wild-type activity (Fig. 1 and Figs. S1 and S2). Therefore, an aromatic ring at this position is sufficient for activity, and the hydrogen bond donor function of the tryptophan side chain is not essential.
To verify the essential function of F215 in the conduction of the true substrate, ammonium, we used the ammonium-dependent growth phenotype of a Saccharomyces cerevisiae strain lacking all of its Amt proteins (Mep1–3) (21). Whereas E. coli AmtB is able to restore the normal growth phenotype (Fig. S5), the F215A variant cannot, either on minimal medium agar plates containing 1 mM NH4+ at pH 4.0 or pH 6.0 (Fig. S5A) or in minimal liquid medium containing 3 mM ammonium as sole nitrogen source (Fig. S5B). Thus, AmtB F215A is unable to conduct ammonium or MA.
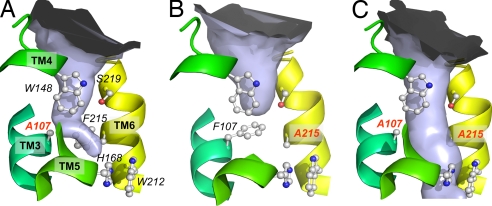
To analyze the structural consequences of Phe gate modification, the F107A, F215A, and F107A/F215A variants were crystallized in the previously reported hexagonal form (4), which permitted their structure determination to a resolution of 2.4 Å or better (Table S1). Structural changes were restricted to the altered residues with very minor changes in the conformation of neighboring ones (Fig. 4 and Fig. S6). Therefore, the inactivity of F215 variants is due to a specific local effect at the substitution site and not to a change in overall structure. Analysis of residual electron density peaks around the gate and in the pore of AmtB for the variant proteins revealed some unexpected features. In the wild type, such peaks have been assigned to an NH4+ ion or to imidazole at S1 (Fig. S6) and as partly occupied ammonia/water sites (S2, S3 and S4) in the pore (4, 5, 20). For F107A and F215A continuous density above 4σ extends from the cytoplasmic vestibule to about residue 215 (Fig. S6 A and B) and is very well explained by the presence of an LDAO detergent molecule. Although a crystallization artifact, the accommodation of a long alkyl chain in the pore is a striking illustration of its hydrophobic nature. In both single variants, the preserved phenyl ring shows some reorientation, which appears to be induced by accommodation of LDAO. In the active F107A, the phenyl ring of F215 is nearly perpendicular to the orientation in the wild type, a conformation that is likely to be close to that of a transiently open gate.
Fig. 4.
Periplasmic pore constriction of AmtB wild type and variants. The solvent-accessible space shown as a light blue surface was calculated by using the program CAVER, with a water-omitted structure. (A) F107A. (B) F215A. (C) F107A/F215A. Selected, highly conserved residues are shown in ball-and-stick representation for the ammonium binding site, phenylalanine gate, and central pore. The substitutions are labeled in red.
The pore of the F107A/F215A variant is not occupied by LDAO but shows a 4σ peak at S1 and similarly high peaks at S3 and S4 (Fig. S6C) indicating partial occupancy by water/ammonia molecules. Well defined water molecules typically show peak heights around 8σ. A large density peak of 10σ was observed near S2 in the space freed by the F215A substitution (Fig. S6C). We have tentatively assigned this to an imidazole molecule, which fits the density very well and whose refined B factors are not much higher than those of the closest protein atoms.
Removal of the Phe gate constriction in AmtB generates a protein with an open pore (Fig. 4C), which in principle could be more permeable than wild-type AmtB to NH4+, NH3, water, and perhaps other small ions. The inactivity of this variant in MA transport (Fig. 1) corroborates the special function of F215 in substrate conductance. If substrate was conducted as NH4+ and its uptake was driven by the membrane potential the inactivity of the open-pore variant is difficult to rationalize because the intrinsic pore characteristics are not markedly changed.
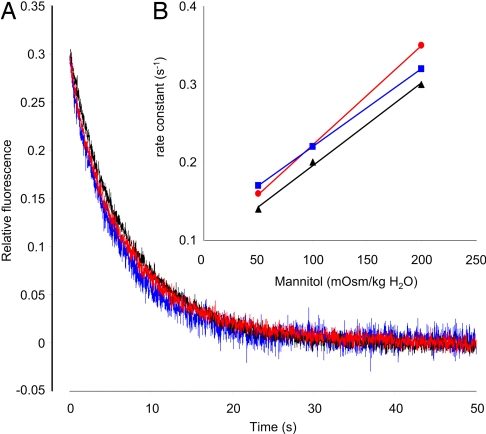
If the AmtB channel conducts NH3, one possible explanation could be that the channel has lost its ability to discriminate against water, which is present in huge excess and which now outcompetes MA. We therefore measured the water permeability in reconstituted proteoliposomes containing purified wild-type or F107A/F215A AmtB for comparison with that of liposomes. Control liposomes and proteoliposomes exhibited similarly low osmotic water permeabilities, P′f, determined by measuring the equilibration rate constant after osmotic shock (Fig. 5A). The rates varied linearly with the magnitude of the osmotic shock (Fig. 5B). Arrhenius activation energies (Ea) for the three systems were also very comparable (Fig. S7) and in accordance with other studies (27). We conclude that water is conducted neither through the pore of wild type nor through the pore of F107A/F215A AmtB at a rate sufficient to increase measurably the permeability of proteoliposomes.
Fig. 5.
Water conductance of liposomes and proteoliposomes. (A) CF self-quenching time course of liposomes (blue trace) and proteoliposomes containing wild-type AmtB (black trace) or the double variant F107A/F215A (red trace) when applying an inwardly directed 50 mOsm/kg H2O mannitol gradient at t = 0. The osmotic water permeabilities P′f, measured as described in the text, were 3.7 × 10−4, 3.3 × 10−4, and 3.3 × 10−4 cm/s for liposomes and proteoliposomes containing wild-type AmtB or the double variant, respectively. (B) Measured rate constants for the time course self-quenching of CF-containing liposomes (blue trace) and proteoliposomes containing wild-type AmtB (black trace) or the double variant F107A/F215A (red trace) as a function of the osmotic shock applied by adding 50, 100, or 200 mOsm/kg H2O mannitol at t = 0. Experiments were performed at 10°C.
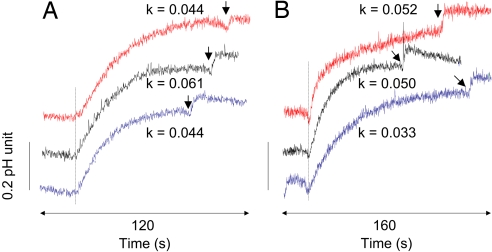
In contrast to uncharged water, ammonia, or methylamine, small ions are very poorly membrane-permeable, and even a small leakage through a protein channel should yield a measurable signal. We therefore assessed whether F107A/F215A proteoliposomes show increased permeabilities for H+ or K+ ions. After an external alkalinization pulse of 1 pH unit the change in internal liposome pH was monitored via a pH-sensitive fluorescent dye. The build-up of a potential counteracting H+ release was prevented by addition of valinomycin, permitting rapid K+ equilibration. FCCP was added at the end of the experiment to verify that pH equilibration was complete (Fig. 6A). The apparent P′H values observed for liposomes, wild-type AmtB, and F107A/F215A proteoliposomes were all low and of comparable magnitude (Fig. 6A). The same assay was then used to measure apparent K+ permeability. In this case, FCCP was added first to make pH equilibration dependent on the rate of K+ exchange and valinomycin was added at the end to verify complete equilibration (Fig. 6B). Again, the measured P′K values for the three systems were low and of comparable magnitude. These data demonstrate that neither H+ nor K+ ions are conducted at measurable rates by wild-type or F107A/F215A AmtB.
Fig. 6.
Proton and potassium conductance of liposomes and proteoliposomes. Shown is pH variation of liposomes (blue trace) and proteoliposomes containing wild-type AmtB (black trace) or the double variant F107A/F215A (red trace) when applying a pH pulse of 1 unit by adding 5 mM KOH. The dashed line indicates addition of 1 μM valinomycin in A and 1 μM FCCP (carbonyl cyanide-p-trifluoromethoxyphenyl-hydrazone) in B. The arrow on each trace indicates the addition of 1 μM FCCP in A and 1 μM valinomycin in B. The alkalanization rate constant (k, s−1) resulting from the charge equilibration due to the equilibration of H+ (A) and K+ movement (B) is noted above each trace. The apparent proton permeabilities P′H, measured as described in the text, were 2.0 × 10−8, 2.8 × 10−8, and 2.0 × 10−8 cm/s for liposomes and proteoliposomes containing wild-type AmtB or the double variant, respectively. The apparent K+ permeabilities P′K were 1.5 × 10−8, 2.3 × 10−8, and 2.5 × 10−8 cm/s. Experiments were performed at 20°C.
Together these results indicate that NH4+ deprotonation is mandatory for conduction and that F215 plays an essential role in this process.
Discussion
Three mechanisms of substrate conduction, which we designate SH+, S/H+, and S, have been considered for Amt proteins. Originally, transport of NH4+ (SH+) driven by the electrochemical gradient was considered most likely. This conclusion, drawn in the absence of detailed studies at the molecular level, was mainly based on the apparent accumulation of the substrate analogue MA (28). Facilitated diffusion of NH3 (S) was proposed when it was found that the apparent accumulation was due to metabolic or acid trapping of MA (29, 30), and the crystal structure of AmtB strongly supported this view, based on the hydrophobic nature of the pore (4, 5). However, at least for some plant Amts, electrogenic transport was convincingly shown in the oocyte system (31). To reconcile this observation with the mainly hydrophobic pore of AmtB and the high sequence identity between plant and bacterial Amts it was proposed that neutral NH3 and the dissociated proton might cross the channel separately (S/H+) (6, 18, 24). Our present studies with AmtB variants of highly conserved residues at the periplasmic NH4+ binding site and of the Phe gate do not directly address the issue of the nature of the conducted species, but they are not compatible with mechanism SH+.
The Periplasmic NH4+ Binding Site.
The functional and structural results with Tl+ demonstrate that an ion does indeed bind to this site, which thus far has not been experimentally shown. The concept that ammonium transporters might conduct NH3 but bind NH4+ at the cell surface goes back to the first characterization of an ammonium transport system in filamentous fungi (32). It was based on the fact that competitive inhibition of MA transport yielded a Ki of ≈2.5 × 10−7 M for NH4+ and 2–3 orders of magnitude lower when calculated for NH3 at the experimental pH. Such a high NH3 affinity was considered hardly possible and would indeed be incompatible with a reasonable transport rate (33). More direct evidence for an NH4+ binding site (S1 or Am1) at the periplasmic pore entry was inferred much later from structural studies of E. coli AmtB (4, 5). NH4+ stabilization and selectivity were indicated by the possibility of tetrahedral coordination in this highly conserved site by hydrogen bonds to the S219:Oγ atom and to a water molecule, and by cation–π interaction with the indole ring of W148 and the phenyl ring of F107. Subsequent computational studies on AmtB (14, 15, 17) confirmed a preference for NH4+ at this site.
Two inhibitors, Tl+ and imidazole, that are not substrates but that compete for binding at S1, were clearly less effective in variant proteins, where S1 is perturbed or destroyed (S219A, W148A, F107A, and a triple variant) consistent with a significant reduction or complete loss of binding at S1. Perturbation or destruction of S1 did not reduce but rather increased the MA transport rate, up to 4-fold for the S219A variant. This result may indicate that dissociation from this binding site into the Phe gate region is rate-limiting at 20 μM MA, a concentration that is likely to saturate the binding site (5). Thus, reducing the binding affinity could lower the rate-limiting barrier and result in an increased transport rate as long as the site occupancy is not correspondingly decreased. Calculations of the potential of mean force for ammonium or for ammonia along the conduction path support such an interpretation (13). The highest energy barrier is where ammonium enters the Phe gate region from an energy minimum at the S1 site up to the equivalence region where deprotonation is considered to take place. However, the increase in MA transport would be expected to be largest for the triple variant, which is not the case. Additional effects thus appear to affect the MA transport rate of the variants to a different extent. An increased transport rate has recently been reported for an W148L variant of EcAmtB, and these authors concluded that transport is electrogenic without discussing a specific mechanism (7).
Independent of the mechanism, the high-affinity NH4+ binding site may serve to increase the scavenging efficiency at very low ammonium concentrations (<1 μM) whereas it must discriminate against other physiologically important cations such as K+ or Na+. MD simulation studies on AmtB have reported that the relative binding free energies favor NH4+ binding versus other ions by several kilocalories per mole (15). The unusual combination of π–cation stabilization and hydrogen bonding to form a tetrahedral coordination strongly favors NH4+, because Na+ and K+ prefer octahedral and higher coordination, respectively. A similar combination of hydrogen bonding and electrostatic interactions has been successfully used to develop synthetic chelators for NH4+ with a selectivity of 1,000 over K+ (34, 35). The NH4+ binding site can also provide strong discrimination against water, which is present in huge excess under the conditions where Amt proteins are needed. If ammonium indeed passes the pore in the uncharged form after deprotonation (mechanism S or S/H+), such discrimination may be important because sufficient selectivity between H2O and NH3 may be difficult to achieve elsewhere along the conduction path.
The Phe Gate Residue F215 Is Essential for Substrate Conductance.
The Phe gate residues F107 and F215 are strictly conserved in conducting Amt/Rh proteins, suggesting an important functional role. F107 forms part of the NH4+ binding site, and under the conditions tested we found it not to be essential for MA conduction. However, F215 is absolutely required. Fundamental to our ability to draw this conclusion is the fact that, with one exception, all variants are expressed at the wild-type level, are correctly targeted to the E. coli inner membrane, and maintain a native trimeric structure, as assessed either indirectly in vivo or, in some cases, directly by structure determination.
We are strongly inclined to rule out mechanism SH+ in AmtB, (i) because of energetic considerations disfavoring ion conduction through a hydrophobic pore as confirmed by all published MD simulations and (ii) because the open-pore variant is inactive in MA conduction and does not increase the permeability to H+ or K+ ions. The open-pore mutant proteoliposomes used for measuring these permeabilities also showed no increase in water permeability. For aquaporin proteoliposomes the measured permeability increase is typically <100-fold compared with control liposomes (27). Considering the nearly diffusion-limited conduction rates of 108 to 109 water molecules per second per aquaporin channel it can be estimated that a rate >106 molecules per second would be required to produce a measurable effect. The AmtB pore is much more hydrophobic than that of aquaporins, which explains, in our opinion, that the water conduction rate of the open-pore variant is several orders of magnitude lower that that of aquaporins. Given NH4+ binding at S1, deprotonation has to occur in the Phe gate region before entering the central part of the pore between the two conserved histidines. Deprotonation either at S1, as previously considered (8), or by S219, as suggested by computational studies (36), appears to be ruled out by our mutagenesis study of S1 residues.
Several computational studies suggest that deprotonation takes place in the Phe gate region, but they differ in detail and in the nature of the suggested base accepting the proton (12, 13, 15–17). In accordance with mechanism S these calculations all discuss deprotonation with the view that the released proton will not transit the pore. Proton conduction via a file of water molecules in a Grotthus-type mechanism has been proposed (18). The observed binding of imidazole in the inactive double variant F107A/F215A within H-bonding distance of the main-chain carbonyl oxygen of A162 and the Nε of H168 suggests that, in a transiently open state of the wild-type Phe gate, NH4+, NH3, or H2O could be stabilized in a similar location. Interestingly, a water molecule is observed in the same location in the structure of the Nitrosomonas europaea Rhesus protein in which the Phe gate constriction is widened compared with Amt structures (37). However, it is now well established that the related Rhesus family proteins are not electrogenic (6).
For mechanism (S/H+), a role of H168 as transient proton acceptor appears attractive and would be compatible with the observation that all H168 mutants with the exception of H168E have been found to be inactive (20). Clearly, it is now urgent to resolve the uncertainty on whether AmtB and Amts in general are electrogenic or not to further progress in our mechanistic understanding. Although it is not obvious to us how a separate path for NH3 and H+ would significantly reduce the electrostatic barrier to charge conduction, such a mechanism would certainly be fascinating for further investigation.
Materials and Methods
Strains, Plasmids, and Culture Conditions.
Strains, plasmids and oligonucleotides are listed in Table S2 and Table S3. All mutants were derivatives of plasmid pJT6E (20) and were constructed as described in ref. 38. Growth of E. coli (39) and S. cerevisiae (21) was carried out as previously described.
Fractionation, AmtB Purification, and Western Blotting.
Fractionation, AmtB purification, and Western blotting were performed as previously described (39).
Protein Quantification.
Expression of each AmtB variant was quantified as previously described (20).
[14C]Methylamine Transport Assays.
These were performed as previously described (21).
Production, Crystallization, and Structure Analysis of Wild-Type and Variant AmtB Proteins.
These were performed essentially as described earlier (20). One-step affinity purification was used, and the eluted AmtB-containing fractions, after concentration, were directly used to set up crystallization trials. To study Tl+ binding to AmtB, TlCl (1 mM final concentration) was added to the crystallization drops. Data collection and final refinement statistics are in Table S1.
Proteoliposome Reconstitution.
Proteoliposomes were prepared by using polar E. coli lipids (Avanti) solubilized in β-octylglucopyranoside (protein/lipid ratio 1/100 wt/wt) in 10 mM Hepes (pH 6.8), 50 mM K2SO4, and either 10 mM 6-carboxyfluorescein (CF) or 0.15 mM pyranine as described previously (3). After detergent trapping with Bio-Beads (SM-2; Bio-Rad), external CF was removed by purifying proteoliposomes on gel-filtration G25 prepacked columns (Amersham Pharmacia). External pyranine was removed by filtration across an anionic exchange column (AG 1-X8; Bio-Rad). Control liposomes were prepared in a similar manner, but protein was omitted. Protein insertion was controlled by electron micrographs of freeze-fractured samples (3), and homogeneity of the size distribution (diameter of 250 nm) was checked by static light scattering. The theoretical protein density in the proteoliposomes, given the protein/lipid ratio, the liposome size, and assuming 100% protein incorporation, is 10 AmtB trimers per proteoliposome, which is in accordance with the density observed by electron microscopy (3).
Liposome Permeability Measurements.
The osmotic behavior of the proteoliposomes and control liposomes was analyzed by following the intensity of the fluorescence quenching as a function of the time in a stopped-flow apparatus (SFM400; Bio-logic): λexc, 485 nm; λem, up to 520 nm. The liposomes were subjected to an inwardly directed mannitol gradient (as indicated in Fig. 5) at 10°C or different temperatures for the Ea determination. Data from 5–10 time courses were averaged and fitted to single exponential function by using the Simplex method of the Biokine software (Bio-logic). The pH variation for K+ and H+ permeability measurement was performed as previously described (40). P′f (cm/s), the osmotic water permeability, was measured according to the equation P′f = k × (V0/S) × (1/Ww) × (1/C0). k (s−1) is the measured rate constant for the fluorescence quenching, V0/S is initial vesicle volume to surface ratio, Ww is the partial molar volume of water (18 cm3), and C0 is the external osmolarity. The apparent permeability P′ for H+ and K+ was measured according to the equation P′ = k × (V0/S). k (s−1) is the measured alkalinization rate constant.
Acknowledgments.
We thank X. D. Li and S. Bernèche for helpful comments on the manuscript, D. Kostrewa for help with crystallographic analysis, and J. P. Cartron and Y. Colin for access to stopped-flow spectrofluorometric facilities. Crystallographic data were collected at the Swiss Light Source, Paul Scherrer Institut, Switzerland, and we gratefully acknowledge the excellent help of T. Tomizaki. A.J., T.F., and M.M. acknowledge support from the Biotechnology and Biological Sciences Research Council (U.K.), and A.J., D.L., and F.K.W. acknowledge support by the Swiss National Science Foundation within the framework of the National Center for Competence in Research Structural Biology program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3C1H, 3C1I, 3C1J, and 3C1G).
This article contains supporting information online at www.pnas.org/cgi/content/full/0711742105/DCSupplemental.
References
- 1.Soupene E, Inwood W, Kustu S. Lack of the Rhesus protein Rh1 impairs growth of the green alga Chlamydomonas reinhardtii at high CO2. Proc Natl Acad Sci USA. 2004;101:7787–7792. doi: 10.1073/pnas.0401809101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang CH, Peng J. Evolutionary conservation and diversification of Rh family genes and proteins. Proc Natl Acad Sci USA. 2005;102:15512–15517. doi: 10.1073/pnas.0507886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Javelle A, et al. Structural and mechanistic aspects of Amt/Rh proteins. J Struct Biol. 2007;158:472–481. doi: 10.1016/j.jsb.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Zheng L, Kostrewa D, Bernèche S, Winkler FK, Li X-D. The mechanism of ammonia transport based on the crystal structure of AmtB of E. coli. Proc Natl Acad Sci USA. 2004;101:17090–17095. doi: 10.1073/pnas.0406475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khademi S, et al. Mechanism of ammonia transport by Amt/MEP/Rh: Structure of AmtB at 1.35 Å. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 6.Ludewig U. Ion transport versus gas conduction: function of AMT/Rh-type proteins. Transfus Clin Biol. 2006;13:111–116. doi: 10.1016/j.tracli.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 7.Fong RN, Kim KS, Yoshihara C, Inwood WB, Kustu S. The W148L substitution in the Escherichia coli ammonium channel AmtB increases flux and indicates that the substrate is an ion. Proc Natl Acad Sci USA. 2007;104:18706–18711. doi: 10.1073/pnas.0709267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winkler FK. Amt/MEP/Rh proteins conduct ammonia. Pflugers Arch. 2006;451:701–707. doi: 10.1007/s00424-005-1511-6. [DOI] [PubMed] [Google Scholar]
- 9.Westhoff CM, Ferreri-Jacobia M, Mak DO, Foskett JK. Identification of the erythrocyte Rh-blood group glycoprotein as a mammalian ammonium transporter. J Biol Chem. 2002;277:12499–12502. doi: 10.1074/jbc.C200060200. [DOI] [PubMed] [Google Scholar]
- 10.Bakouh N, et al. NH3 is involved in the NH4+ transport induced by the functional expression of the human Rh C glycoprotein. J Biol Chem. 2004;279:15975–15983. doi: 10.1074/jbc.M308528200. [DOI] [PubMed] [Google Scholar]
- 11.Marini A-M, Soussi-Boudekou S, Vissers S, Andre B. A family of ammonium transporters in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4282–4293. doi: 10.1128/mcb.17.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bostick DL, Brooks CL., III On the equivalence point for ammonium (de)protonation during its transport through the AmtB channel. Biophys J. 2007;92:L103–L105. doi: 10.1529/biophysj.107.109165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bostick DL, Brooks CL., III Deprotonation by dehydration: The origin of ammonium sensing in the AmtB channel. PLoS Comput Biol. 2007;3:e22. doi: 10.1371/journal.pcbi.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Hu X. Molecular determinants for binding of ammonium ion in the ammonia transporter AmtB-A quantum chemical analysis. J Phys Chem A. 2006;110:1375–1381. doi: 10.1021/jp054261c. [DOI] [PubMed] [Google Scholar]
- 15.Luzhkov VB, Almlof M, Nervall M, Aqvist J. Computational study of the binding affinity and selectivity of the bacterial ammonium transporter AmtB. Biochemistry. 2006;45:10807–10814. doi: 10.1021/bi0610799. [DOI] [PubMed] [Google Scholar]
- 16.Lin Y, Cao Z, Mo Y. Molecular dynamics simulations on the Escherichia coli ammonia channel protein AmtB: Mechanism of ammonia/ammonium transport. J Am Chem Soc. 2006;128:10876–10884. doi: 10.1021/ja0631549. [DOI] [PubMed] [Google Scholar]
- 17.Nygaard TP, Rovira C, Peters GH, Jensen MO. Ammonium recruitment and ammonia transport by E. coli ammonia channel AmtB. Biophys J. 2006;91:4401–4412. doi: 10.1529/biophysj.106.089714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamoureux G, Klein ML, Berneche S. A stable water chain in the hydrophobic pore of the AmtB ammonium transporter. Biophys J. 2007;92:L82–L84. doi: 10.1529/biophysj.106.102756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao Z, Mo Y, Thiel W. Deprotonation mechanism of NH4+ in the Escherichia coli ammonium transporter AmtB: Insight from QM and QM/MM calculations. Angew Chem Int Ed Engl. 2007;46:6811–6815. doi: 10.1002/anie.200701348. [DOI] [PubMed] [Google Scholar]
- 20.Javelle A, et al. An unusual twin-his arrangement in the pore of ammonia channels is essential for substrate conductance. J Biol Chem. 2006;281:39492–39498. doi: 10.1074/jbc.M608325200. [DOI] [PubMed] [Google Scholar]
- 21.Javelle A, Thomas G, Marini AM, Kramer R, Merrick M. In vivo functional characterisation of the E coli ammonium channel AmtB: Evidence for metabolic coupling of AmtB to glutamine synthetase. Biochem J. 2005;390:215–222. doi: 10.1042/BJ20042094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liaw SH, Kuo I, Eisenberg D. Discovery of the ammonium substrate site on glutamine synthetase, a third cation binding site. Protein Sci. 1995;4:2358–2365. doi: 10.1002/pro.5560041114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Wirén N, Merrick M. Regulation and function of ammonium carriers in bacteria, fungi and plants. Trends Curr Genet. 2004;9:95–120. [Google Scholar]
- 24.Andrade SL, Dickmanns A, Ficner R, Einsle O. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc Natl Acad Sci USA. 2005;102:14994–14999. doi: 10.1073/pnas.0506254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conroy MJ, et al. The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc Natl Acad Sci USA. 2007;104:1213–1218. doi: 10.1073/pnas.0610348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H, Xu Y, Zhu W, Chen K, Jiang H. Detailed mechanism for AmtB conducting NH4+/NH3: Molecular dynamics simulations. Biophys J. 2007;92:877–885. doi: 10.1529/biophysj.106.090191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borgnia MJ, Agre P. Reconstitution and functional comparison of purified GlpF and AqpZ, the glycerol and water channels from Escherichia coli. Proc Natl Acad Sci USA. 2001;98:2888–2893. doi: 10.1073/pnas.051628098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleiner D. Bacterial ammonium transport. FEMS Microbiol Rev. 1985;32:87–100. [Google Scholar]
- 29.Soupene E, He L, Yan D, Kustu S. Ammonia acquisition in enteric bacteria: Physiological role of the ammonium/methylammonium transport B (AmtB) protein. Proc Natl Acad Sci USA. 1998;95:7030–7034. doi: 10.1073/pnas.95.12.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soupene E, Lee H, Kustu S. Ammonium/methylammonium transport (Amt) proteins facilitate diffusion of NH3 bidirectionally. Proc Natl Acad Sci USA. 2002;99:3926–3931. doi: 10.1073/pnas.062043799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludewig U, von Wirén N, Frommer WB. Uniport of NH4+ by the root hair plasma membrane ammonium transporter LeAMT1;1. J Biol Chem. 2002;277:13548–13555. doi: 10.1074/jbc.M200739200. [DOI] [PubMed] [Google Scholar]
- 32.Hackette SL, Skye GE, Burton C, Segel IH. Characterization of an ammonium transport system in filamentous fungi with [14C]-methylammonium as the substrate. J Biol Chem. 1970;245:4241–4250. [PubMed] [Google Scholar]
- 33.Booth IR, Edwards MD, Miller S. Bacterial ion channels. Biochemistry. 2003;42:10045–10053. doi: 10.1021/bi034953w. [DOI] [PubMed] [Google Scholar]
- 34.Chin J, et al. A rational approach to selective recognition of NH4+ over K+ Angew Chem Int Ed Engl. 1999;38:2756–2759. [PubMed] [Google Scholar]
- 35.Chin J, et al. Tuning and dissecting electronic and steric effects in ammonium receptors: Nonactin vs artificial receptors. J Am Chem Soc. 2002;124:5374–5379. doi: 10.1021/ja0174175. [DOI] [PubMed] [Google Scholar]
- 36.Ishikita H, Knapp EW. Protonation states of ammonia/ammonium in the hydrophobic pore of ammonia transporter protein AmtB. J Am Chem Soc. 2007;129:1210–1215. doi: 10.1021/ja066208n. [DOI] [PubMed] [Google Scholar]
- 37.Lupo D, et al. The 1.3-Å resolution structure of Nitrosomonas europaea Rh50 and mechanistic implications for NH3 transport by Rhesus family proteins. Proc Natl Acad Sci USA. 2007;104:19303–19308. doi: 10.1073/pnas.0706563104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Javelle A, Severi E, Thornton J, Merrick M. Ammonium sensing in E. coli: The role of the ammonium transporter AmtB and AmtB-GlnK complex formation. J Biol Chem. 2004;279:8530–8538. doi: 10.1074/jbc.M312399200. [DOI] [PubMed] [Google Scholar]
- 40.Paula S, Volkov AG, Van Hoek AN, Haines TH, Deamer DW. Permeation of protons, potassium ions, and small polar molecules through phospholipid bilayers as a function of membrane thickness. Biophys J. 1996;70:339–348. doi: 10.1016/S0006-3495(96)79575-9. [DOI] [PMC free article] [PubMed] [Google Scholar]