Abstract
To investigate the role of jasmonate in the defense of plants against fungal pathogens, we have studied a mutant of Arabidopsis, fad3–2 fad7–2 fad8, that cannot accumulate jasmonate. Mutant plants were extremely susceptible to root rot caused by the fungal root pathogen Pythium mastophorum (Drechs.), even though neighboring wild-type plants were largely unaffected by this fungus. Application of exogenous methyl jasmonate substantially protected mutant plants, reducing the incidence of disease to a level close to that of wild-type controls. A similar treatment with methyl jasmonate did not protect the jasmonate-insensitive mutant coi1 from infection, showing that protective action of applied jasmonate against P. mastophorum was mediated by the induction of plant defense mechanisms rather than by a direct antifungal action. Transcripts of three jasmonate-responsive defense genes are induced by Pythium challenge in the wild-type but not in the jasmonate-deficient mutant. Pythium species are ubiquitous in soil and root habitats world-wide, but most (including P. mastophorum) are considered to be minor pathogens. Our results indicate that jasmonate is essential for plant defense against Pythium and, because of the high exposure of plant roots to Pythium inoculum in soil, may well be fundamental to survival of plants in nature. Our results further indicate that the fad3–2 fad7–2 fad8 mutant is an appropriate genetic model for studying the role of this important signaling molecule in pathogen defense.
Keywords: jasmonic acid, root rot, defense signaling, Pythium
Plants defend themselves against fungi and other microbial pathogens by the induction of both localized and systemic responses. Typically, a pathogen interacting with a resistant host plant triggers a localized hypersensitive response, the intensity and spread of which are regulated by complex molecular mechanisms (1, 2). At the same time, long-distance signals initiated at the infection site lead to the induction of specific pathogenesis-related (PR) genes in uninfected parts of the plant—a process termed “systemic acquired resistance” or SAR (3). Signaling molecules such as salicylic acid, methyl jasmonate, ethylene, hydrogen peroxide, and superoxide radicals have been proposed to be involved in the induction and coordination of these plant responses (3, 4). However, salicylic acid has been ascribed a central role in both localized responses and SAR (1–3, 5, 6). The complexities of the relationship between the hypersensitive response, salicylic acid, and SAR are beginning to be appreciated and understood through the characterization of several families of mutants (2–4, 7) and the availability of salicylic acid-deficient plants expressing the bacterial NahG gene encoding the salicylate hydroxylase enzyme (6). In particular, the increased susceptibility of NahG plants to a range of fungal and bacterial pathogens establishes the biological relevance of salicylic acid (6).
It has been known for some time that some fungal elicitors, such as oligogalacturonides and chitosan, also activate wound-response genes (8, 9). This point suggests that there is cross-talk from the pathogen response pathway into the wound-signaling pathway, which uses jasmonic acid as the requisite localized effector of defense responses aimed at chewing insects (10, 11). More recent evidence suggests that jasmonic acid is involved in the induction of genes that act primarily in defense against pathogens rather than insects. For example, fungal elicitors induce transient accumulation of jasmonic acid as well as the synthesis of several classes of phytoalexins in suspension cell cultures of a number of plant species (12). Exogenous application of jasmonates induces the same antimicrobial compounds, apparently by transcriptional activation of genes that encode the biosynthetic enzymes involved (12). The defensin gene PDF1.2 of Arabidopsis, which encodes a protein with demonstrated antifungal activity, is induced strongly by either pathogen challenge or methyl jasmonate but not by salicylic acid (13). The same is true of the thionin encoded by the Arabidopsis Thi2.1 gene (14, 15). Finally, jasmonate and ethylene are synergistic in inducing members of the PR1 and PR5 gene families, which encode pathogenesis-related proteins (16).
Interpreting the significance of these observations is made difficult by the complexities inherent in plant–pathogen interactions. For example, other defensin and thionin genes closely related to PDF1.2 and Thi2.1 are not induced by jasmonate in Arabidopsis (13–15), and thionin genes in barley can be induced by salicylic acid as well as jasmonate (17). Furthermore, the jasmonate-induced genes also might be induced by pathogen attack through parallel pathways that do not involve jasmonate. Only in the case of the Arabidopsis PDF1.2 gene has a dependence on jasmonate been shown through the inability of the fungal pathogen Alternaria brassicola to induce expression of PDF1.2 in the coi1 mutant, which is deficient in jasmonate signaling (13, 18). These caveats notwithstanding, there appears to be a strong case for a second, jasmonate-dependent pathway that mediates some plant responses to pathogens.
Until now, it has not been possible to demonstrate the biological relevance of jasmonate signaling in a host–pathogen interaction. For this reason, the practical importance of jasmonate signaling in pathogen defense has remained unclear. For example, the jasmonate-induced genes might have a relatively minor role in supplementing the hypersensitive response and/or SAR. Here, we use a mutant of Arabidopsis that is deficient in jasmonate synthesis to demonstrate that jasmonate-signaling is essential for protection against the soil-borne pathogenic fungus Pythium mastophorum (Drechs.).
MATERIALS AND METHODS
Plant Materials.
Arabidopsis thaliana plants used were descended from the Columbia wild type in which mutations were produced by treatment with ethyl methane sulfonate. The fad3–2 fad7–2 fad8 triple mutant and the coi1 mutant are described elsewhere (11, 18, 19). Plants were grown routinely in a controlled environment growth chamber in commercial peat-based rooting medium pasteurized with aerated steam at 60°C for 40 min to eliminate pathogens. Arabidopsis plants used for experiments with Pythium under axenic conditions and for induction of defense genes were grown in Petri dishes on sterile 1% agar medium containing Gamborg’s B5 basal mixture (Sigma). Seeds of wild-type and mutant plants were surface-sterilized in a 0.5% sodium hypochlorite solution containing 0.1% Tween 20 (Bio-Rad) and washed four times in sterile distilled water before being planted in parallel rows on the medium. The plates were oriented vertically under continuous illumination (100 μE/m2/s) at 22°C for 20 days to encourage root growth on the agar surface.
Isolation and Identification of Pathogen.
The pathogen, a natural inhabitant of the peat-based medium, initially was isolated from diseased fad3–2 fad7–2 fad8 plants grown in the unpasteurized rooting medium. Roots from wild-type and fad3–2 fad7–2 fad8 plants were excised and washed thoroughly in running de-ionized water for 12 h to remove attached soil particles. Root pieces subsequently were surface-sterilized in 0.5% sodium hypochlorite solution for 3 min and washed four times in sterile distilled water before being plated on water agar plates containing 1.5% Bacto Agar (Difco). Transfers were made from the edges of fungal colonies growing out of the roots onto potato dextrose agar medium. The isolates were identified as Pythium mastophorum (Drechs.) based on morphological characteristics and comparison of rDNA internal transcribed sequences (ITS I and ITS II) from all of the type and neotype cultures of Pythium species described in Van der Plaats-Niterink (20). Cultures of Pythium mastophorum were maintained (sub-cultured every 30 days) on 1/5 strength potato dextrose agar medium (Difco) supplemented with Difco agar at 15 g/liter.
Infection of Arabidopsis with Pythium mastophorum.
Pythium-infested rooting medium was prepared by finely slicing 5-day-old cultures of P. mastophorum grown on 10-cm-diameter plates and mixing the pieces uniformly with 1.8 kg of pasteurized rooting medium (sufficient to fill five pots). Healthy wild-type Columbia and mutant plants were grown separately on pasteurized soil plugs (1 cm diameter × 6 cm long) for 20 days. Five wild-type and five mutant plants were selected randomly and transplanted along with their soil plugs into each pot containing Pythium-infested rooting medium. One milliliter of an aqueous methyl jasmonate solution (13 or 45 μM) containing 10 ppm of Tween 20 was applied to the roots of individual plants each day as a soil drench. The control plants received an equal volume of water containing Tween 20. The treatment began 2 days before the transplantation into the Pythium-infested soil and continued for the duration of the experiment. Root rot was recognized easily in the experimental plants by the severe leaf wilt and collapse of the plants caused by destruction of the roots by Pythium. Disease development was estimated each day by scoring the number of plants with more than one wilted leaf in the rosette.
The dilution of added jasmonate solution in soil was determined by applying 1 ml of a tracer dye dissolved in water to the soil surface in the same way that experimental plants were treated with jasmonate. The dye was observed to spread into a 6-ml volume of soil that contained 3.5 ml of water. Therefore, the effective concentration of jasmonate in the soil for the 45- and 13-μM treatments was calculated as 13 and 4 μM, respectively.
Inoculation of axenically grown plants with P. mastophorum was initiated by placing a slab of water agar colonized by the fungus between the parallel rows of wild-type and fad3–2 fad7–2 fad8 plants grown on Gamborg agar medium in 15-cm Petri dishes. Control plants received a mock inoculation with nutrient agar. The plates then were incubated under continuous illumination for 7 days at 22°C. One set of plates received a daily spray of 1 ml of 45 μM methyl jasmonate solution and 10 ppm Tween 20 in sterile water, and another set was sprayed with a 10-ppm Tween 20 solution only.
Estimation of Root Infection Levels.
The fibrous roots of wild-type and mutant Arabidopsis plants used in the jasmonate protection studies were washed gently in tap water and subsequently cleaned in running deionized water for 12 h. Particles of rooting medium still attached to the roots were removed with a brush. A random sample of 9–14 roots each excised 5–10 mm behind the tip was stained with acid fuchsin lactophernol and mounted on microscopic slides for observation of Pythium within the tissue.
Transcript Levels of Pathogen-Activated Defense Genes in Wild-Type and fad3–2 fad7–2 fad8 Plants.
Wild-type and fad3–2 fad7–2 fad8 mutant Arabidopsis plants were grown aseptically on vertically oriented Gamborg agar plates for 15 days before harvesting control (unchallenged) plants. Surface-exposed roots were challenged with the pathogen by placing on them water agar slabs harboring extensive hyphal growth of P. mastophorum. Total RNA was extracted by the method of Dong and Dunstan (21) from seedlings harvested 24 and 48 h after inoculation. Total RNA (30 μg) from each sample was separated electrophoretically on a denaturing agarose gel, blotted onto nylon membrane, and probed with the inserts from Arabidopsis cDNA clones of the following genes obtained from the expressed sequence collection of the Arabidopsis Biological Resource Center, Ohio State University, Columbus, OH: PDF1.2 encoding an Arabidopsis homolog of plant defensin (13, 15); naringenin chalcone synthase (CHS) (22); and lipoxygenase 2 (LOX2) (23, 24). The LOX2 probe also cross-hybridized with the root-specific LOX1 transcript (data not shown). The visualization and quantification of the transcripts was carried out on a phosphorimager (Bio-Rad GS 525 molecular imaging system).
RESULTS
Mutant Plants Are Highly Susceptible to Pythium Infection.
The fad3–2 fad7–2 fad8 mutant of Arabidopsis is unable to accumulate jasmonate because it is deficient in linolenic acid, the lipid precursor of jasmonate (11). We successfully had grown mutant plants for several years but noted classic symptoms of severe root rot after changing to a new source of commercial rooting medium. Mutant plants wilted and eventually died while wild-type plants in adjacent pots remained healthy and free of disease symptoms. When the rooting medium was pasteurized with moist heat at 60°C for 40 min, the mutant plants remained healthy like their wild-type counterparts, indicating that a soil-borne pathogen was responsible.
Roots from wild-type and mutant plants grown in unpasteurized rooting medium were harvested, surface-sterilized, and placed on water agar. Sixteen hours later, fungal mycelia were observed emerging from sections of fad3–2 fad7–2 fad8 roots but not from the wild type. The fungus, transferred to 1/5 potato dextrose agar, was identified as Pythium mastophorum by using DNA sequence analysis of the internal transcribed sequence regions within the ribosomal RNA gene cluster. Sequence differences within these regions have been used as the basis for species identification within the genus Pythium (25). When mycelia from axenic cultures of P. mastophorum were used to infest pasteurized soil, the jasmonate-deficient mutant plants developed the same disease symptoms whereas wild-type Arabidopsis grown in the same pots remained largely unaffected. P. mastophorum was reisolated from these diseased mutant plants.
Jasmonate Is Necessary for Plant Defense.
The inability to synthesize jasmonate in the fad3–2 fad7–2 fad8 line (11) provided a plausible explanation for the susceptibility of mutant plants to infection by P. mastophorum. However, these initial experiments could not exclude other possibilities. For example, additional mutations inadvertently maintained in the triple-mutant line might be responsible. To investigate such possibilities, and to quantify the consequences of defective pathogen-defense in the mutant, we carried out experiments in which healthy wild-type and mutant plants were transplanted as mixed stands into the same pots containing rooting medium infested with P. mastophorum. In control pots treated with water, disease symptoms quickly developed in mutant plants but not in wild-type plants growing in the same pots. By the 12th day after transfer, >90% of mutant plants were severely diseased whereas only 6% of wild-type plants showed strong symptoms (Fig. 1 A and B). In pots treated with an aqueous solution of 45 μM methyl jasmonate, the proportion of dead or dying mutant plants was reduced to <15% and most plants remained healthy and free of symptoms (Fig. 1 A and C). A lower concentration of methyl jasmonate, 13 μM, also substantially reduced the incidence of disease in fad3–2 fad7–2 fad8 plants. Thus, complementing the jasmonate-deficiency of mutant plants with external jasmonate was sufficient to greatly limit Pythium damage. Tests with tracer dyes and analyses of soil water content indicated that the 1 ml of jasmonate solution applied to the soil surface rapidly dispersed into a volume of soil containing 3.5 ml of water. This result indicates that the concentration of jasmonate in the root zone of treated plants would have been 13 and 4 μM respectively, after application of 45 and 13 μM jasmonate solution to the soil surface.
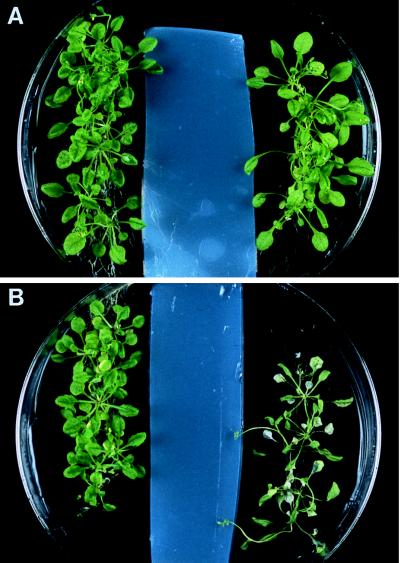
Figure 1.
Disease development and jasmonate-induced resistance in mutant plants inoculated with Pythium mastophorum: (A) Wild-type and fad3–2 fad7–2 fad8 plants were grown on pasteurized soil for 15 days and then transferred to pots containing soil infested with P. mastophorum. Starting 2 days before transfer, the roots of each plant were drenched each day with 1 ml of methyl jasmonate solution or an equal volume of water. The number of disease-free plants from 32 wild-type (•), 28 mutant plants treated with water (○), and 36 mutant plants treated with 45 μM methyl jasmonate (□) were scored daily. Disease development in wild-type plants treated with methyl jasmonate was the same as in the wild-type controls. (B and C) Photographs of plants taken on day 12. (B) Wild-type controls (Left) and mutant plants (Right) treated with water. (C) Wild-type (Left) and mutant plants (Right) treated with 45 μM methyl jasmonate.
The pasteurization procedure we used will effectively eliminate fungal pathogens and Gram-negative bacteria but will allow survival of heat-tolerant microorganisms including actinomycetes and spore-forming bacteria. The presence of these surviving microorganisms, as well as the possibilities for inadvertent contamination of the pasteurized soil during the course of the experiment, somewhat complicates interpretation of our results. For example, jasmonate-mediated defenses in Arabidopsis might be directed primarily at other organisms whose growth in or around roots of the jasmonate-deficient mutant might precondition roots to infection by Pythium. To confirm that Pythium alone was causing the infection, we conducted an experiment using P. mastophorum and plants grown under sterile conditions. Wild-type and mutant Arabidopsis plants were grown from surface-sterilized seeds sown in two rows on Petri dishes of agar medium. When the plants were 20 days old, a strip of water agar containing a pure culture of P. mastophorum was placed in the Petri dish between the two rows. Forty-eight hours later, mycelia growing from the agar strip had made contact with roots of the plants. Mutant plants developed disease symptoms, including chlorosis and collapse of their leaf tissues, within 2 days of this initial contact. By contrast, wild-type plants remained indistinguishable from uninoculated controls even after 7 days of exposure to the pathogen (Fig. 2). Low levels of methyl jasmonate (13 or 45 μM applied as a spray) provided protection to the mutant plants grown on Petri dishes (not shown).
Figure 2.
Extreme susceptibility of fad3–2 fad7–2 fad8 plants to infection by Pythium mastophorum. Wild-type (Left) and fad3–2 fad7–2 fad8 plants (Right) were grown axenically on agar medium for 20 days before being inoculated with P. mastophorum. (A) Mock inoculation with sterile agar slab. (B) Inoculation with P. mastophorum. The plants were photographed 7 days after inoculation.
Taken together, these results indicate that wild-type Arabidopsis is highly resistant to infection by P. mastophorum and that jasmonate-mediated signaling is both necessary and sufficient to provide protection of the fad3–2 fad7–2 fad8 mutant plants against this pathogen.
Jasmonate Has no Direct Effect on Fungal Growth or Infectivity.
We investigated the possibility that methyl jasmonate might directly affect the growth or infectivity of P. mastophorum by two different approaches. In the first experiment, circular plugs were taken from a uniform mycelial culture and transferred to 1/5 potato dextrose agar supplemented with different concentrations of methyl jasmonate. Growth of the fungus on the jasmonate-containing medium was measured each day for 5 days (Table 1). There was no effect of jasmonate on the growth of Pythium up to a concentration of 45 μM; even at 130 μM, the growth rate (1.27 ± 0.12 cm/day) was decreased by <10% compared with the average growth rate (1.41 ± 0.14 cm/day) of the three lower concentrations (Table 1). Thus, mycelial growth of P. mastophorum is unaffected by jasmonate concentrations that are ≈10-fold higher than that around the roots of plants shown in Fig. 1C.
Table 1.
Effect of methyl jasmonate on growth rate of Pythium mastophorum
| Jasmonate concentration, μM | Average growth rate, cm/day |
|---|---|
| 0 | 1.41 (±0.03) |
| 13 | 1.45 (±0.06) |
| 45 | 1.38 (±0.08) |
| 130 | 1.27 (±0.05) |
The data represent the average of five separate colonies at each concentration of jasmonate. The numbers within brackets represent SE.
Although our results indicated that jasmonate did not limit mycelial growth of P. mastophorum, it was still possible that jasmonate could directly affect the infection process itself (26). To test this possibility, we repeated the experiment described in Fig. 1 but included the jasmonate-insensitive coi1 mutant of Arabidopsis (13, 18), along with the wild-type and fad3–2 fad7–2 fad8 plants. In this experiment, the wild-type and fad3–2 fad7–2 fad8 plants exhibited a similar response to both the pathogen and the jasmonate treatment as shown in Fig. 1. However, by the 12th day after Pythium inoculation, 85% of the water-treated and 95% of the jasmonate-treated coi1 plants were severely diseased (Table 2). Rootlets of randomly selected wild-type and mutant plants collected on the 12th day after Pythium inoculation and washed clean of rooting medium were stained and observed under a microscope for direct evidence of root infection by P. mastophorum. Typical ornamented oospores of P. mastophorum normally were found in a 5- to 10-mm apical zone of young rootlets. An average of 87% of root segments from the water-treated and jasmonate-treated wild-type plants had no oospores, and the remaining 13% of the root segments had only a few oospores. All root segments from the water-treated fad3–2 fad7–2 fad8 plants were heavily laden with Pythium oospores. However, 90% of the root segments obtained from jasmonate-treated fad3–2 fad7–2 fad8 plants were devoid of oospores. In contrast, roots of coi1 mutant plants were filled with oospores, whether or not they were treated with jasmonate (Fig. 3). These results clearly demonstrate that externally applied methyl jasmonate was not protecting the fad3–2 fad7–2 fad8 mutant by a direct effect on the infectivity of P. mastophorum but was instead acting to induce plant defenses against the fungus. These results also provide direct confirmation that root infection by P. mastophorum is responsible for the severe wilting in the experimental plants.
Table 2.
Inability of jasmonate to protect the jasmonateinsensitive mutant coi1 from infection by Phythium mastophorum
| Healthy plants, %
|
||
|---|---|---|
| No jasmonate | 45 μM jasmonate | |
| Wt | 90 | 87 |
| fad3-2 fad7-2 fad8 | 0 | 88 |
| coi1 | 15 | 5 |
Wild-type (Wt) and mutant Arabidopsis plants were treated as described in Fig. 1. The percentage of disease-free plants was calculated 12 days after inoculation.
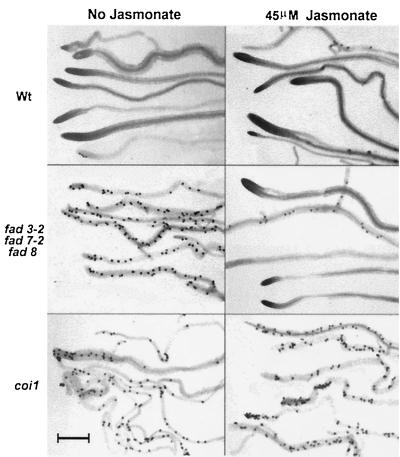
Figure 3.
Oospore formation by Pythium mastophorum in infected roots of Arabidopsis as indicators of susceptibility of the fad3–2 fad7–2 fad8 and coi1 mutants and the protection of fad3–2 fad7–2 fad8 by 45 μM jasmonate. Wild-type and mutant plants were treated as described in Fig. 1. Microphotographs of representative root segments were taken on the 12th day after inoculation with the pathogen as described in Materials and Methods. The dark stained bodies are oospores of P. mastophorum. (Bar = 1 mm.)
Pythium Induces Jasmonate-Responsive Genes in Wild-Type but Not Mutant Plants.
Because our results indicate that jasmonate signaling is essential for effective defense against Pythium infection, the fad3–2 fad7–2 fad8 mutant provides a new means to investigate the genes involved and to determine the levels of expression of these genes required for meaningful protection. As a first step, we measured the transcript levels of three genes induced by jasmonate in Arabidopsis. Genes encoding lipoxygenase are induced 5- to 10-fold by jasmonate (23, 24) in a response that may further increase jasmonate levels and lead to the production of oxygenated lipid derivatives thought to be active in plant defense (27). Chalcone synthase catalyses a key step in the synthesis of many secondary compounds with demonstrated antifungal activity (28). The Arabidopsis PDF1.2 gene encodes a 5-kDa plant defensin protein with demonstrated antifungal activity and is induced at least 10-fold by jasmonate (13). When 20-day-old Arabidopsis seedlings were inoculated with P. mastophorum, wild-type plants showed substantial increases in the transcript levels of all three genes (Fig. 4). By contrast, mutant plants exhibited no increase in any of the transcripts over the low levels constitutively present in control plants. These results demonstrate that the fad3–2 fad7–2 fad8 mutant is indeed defective in fungal induction of known defense genes whose increased expression depends on jasmonate signaling.
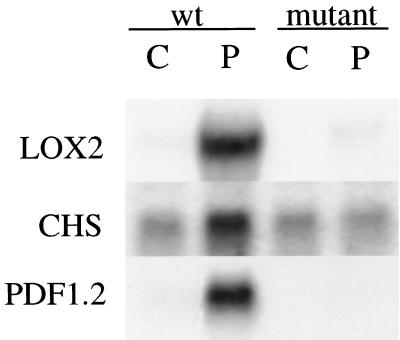
Figure 4.
Pathogen induction of gene expression in wild-type (wt) and fad3–2 fad7–2 fad8 (mutant) Arabidopsis plants. Transcript levels are shown in tissue from uninoculated seedlings (C) and seedlings inoculated (P) with P. mastophorum 48 h earlier (24 h for LOX). Results are shown for genes encoding lipoxygenase 2 (LOX2), a plant defensin gene (PDF1.2), and chalcone synthase gene (CHS).
DISCUSSION
Pythium Species Are Ubiquitous in Soil and Root Habitats Worldwide.
Of the estimated 130 species (20), most are “minor pathogens,” defined by Salt (29) as having the ability to invade the meristematic tips, epidermis, and cortex but not the endodermis and vascular tissues of roots. Pythium mastophorum is among those considered to be minor pathogens. Neither the presence of a Pythium species in the untreated peat-based rooting medium used in our studies nor the ability of the wild-type Arabidopsis seedlings to grow normally in this rooting medium was surprising, but the severe root rot and rapid wilting of the jasmonate-deficient and jasmonate-insensitive mutants in the presence of this minor pathogen was unexpected. However, our experiments conducted under axenic conditions and with pasteurized rooting medium demonstrate the susceptibility of fad3–2 fad7–2 fad8 mutant plants to P. mastophorum and the ability of exogenous jasmonate to substantially protect the mutant plants. The concentration of jasmonate required to protect fad3–2 fad7–2 fad8 plants did not result in any significant reduction in growth or infectivity of P. mastophorum, suggesting that jasmonate acts solely as a signal to activate the required plant defenses. Consistent with this conclusion, we observed that the coi1 mutant of Arabidopsis that is defective in jasmonate perception (18) also is killed by P. mastophorum but cannot be protected by application of exogenous jasmonate. The apparent resistance of wild-type plants to P. mastophorum, as well as the high degree of susceptibility of the mutant plants to this species, points clearly to the critical importance of jasmonate in plant defense against Pythium. Recently, Knoester et al. (30) showed that transgenic tobacco plants rendered ethylene-insensitive through expression of the mutant etr1–1 gene from Arabidopsis are also highly susceptible to Pythium infection. This finding and other results (13, 16) raise the possibility that jasmonate and ethylene may both contribute to a signaling pathway required for broad defense against potential fungal pathogens.
Soils typically contain 102 to 103 oospores and other propagules of Pythium per gram, present mainly in the top 10–20 cm of the soil profile and supported by plant roots and fresh organic matter. At these population densities, virtually every germinating seed and developing root is exposed to infection by one or more Pythium species, which can explain why the root necrosis and other minor root damage caused by Pythium affects virtually 100% of any given population of plants within a field (31). Several investigators over the past half century have proposed or shown that the almost-universal increased growth response of plants to soil fumigation is caused by elimination of the minor but uniform root damage caused by the ubiquitous Pythium species (32). Our results would now suggest that, were it not for the defense system mediated by jasmonate, the minor damage responsible for slight stunting or plant “growth stasis,” as proposed by Wilhelm (33), would become major if not lethal damage to plants. Indeed, considering the uniformity of Pythium infections and the extreme susceptibility of the jasmonate-deficient and -insensitive mutants of Arabidopsis, it would appear that the jasmonate-mediated defense system is fundamental to survival of plants in nature.
It is significant that the jasmonate-mediated response demonstrated in our studies provides defense against a fungus that otherwise would be considered too minor to merit attention as a plant pathogen. The great majority of research on plant defense systems, including research on the salicylic-acid-mediated SAR, has focused on recognized and often host-specific pathogens rather than weak parasites with broad host ranges, which might explain why the jasmonate-mediated defense system has not attracted more attention as a basis for understanding host–pathogen interactions. Inoculation of plants with P. mastophorum led to the induction of several jasmonate-regulated genes in wild-type Arabidopsis but not in the fad3–2 fad7–2 fad8 mutant. It is expected that other genes also will be involved in protection of Arabidopsis from Pythium infection, and our results indicate that the fad3–2 fad7–2 fad8 mutant, as well as other mutants in jasmonate signaling, will provide a means to identify them. In particular, transgenic expression in the mutant background can be used to test candidate defense genes for their ability to measurably reduce damage from pathogen attack. Finally, a search for mutations that suppress the susceptibility of fad3–2 fad7–2 fad8 Arabidopsis to Pythium can be used to identify components of the signaling pathway that act downstream of jasmonate. Because genetic determinants of resistance to soil-borne pathogens such as Pythium are seen rarely in crop plants (34), these approaches may prove valuable in the development of crop plants resistant to Pythium and similar pathogens.
Acknowledgments
We are grateful to Kaye Peterman (Wellesley College, MA) for a LOX1 cDNA, to John Turner (University of East Anglia, U.K.) for seeds of coi1, and to the Arabidopsis Biological Resource Center, Ohio State University. We thank Annick Stinzi for helpful discussion and advice on plant–pathogen interactions. This work was supported by grants from the United States Department of Energy (DE-FG06-92ER20077), the National Science Foundation (IBN-9407902), and by the Agricultural Research Center, Washington State University; Agricultural Research Service, U.S. Department of Agriculture and Agriculture and Agri-Food Canada.
ABBREVIATION
- SAR
systemic acquired resistance
References
- 1.Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar S P. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 2.Dangl J L, Dietrich R A, Richberg M H. Plant Cell. 1996;8:1793–1807. doi: 10.1105/tpc.8.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryals J A, Neuenschwander U H, Willits M G, Molina A, Steiner H-Y, Hunt M D. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammond-Kosack K E, Jones J D G. Plant Cell. 1986;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryals J, Lawton K A, Delaney T P, Friedrich L, Kessmann H, Neuenschwander U, Uknes S, Vernooij B, Weymann K. Proc Natl Acad Sci USA. 1995;92:4202–4205. doi: 10.1073/pnas.92.10.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delaney T, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 7.Jabs T, Dietrich R A, Dangl J L. Science. 1996;273:1853–1856. doi: 10.1126/science.273.5283.1853. [DOI] [PubMed] [Google Scholar]
- 8.Doares S H, Syrovets T, Weiler E W, Ryan C A. Proc Natl Acad Sci USA. 1995;92:4095–4098. doi: 10.1073/pnas.92.10.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farmer E E, Ryan C A. Trends Cell Biol. 1992;2:236–241. doi: 10.1016/0962-8924(92)90311-a. [DOI] [PubMed] [Google Scholar]
- 10.Howe G A, Lightner J, Browse J, Ryan C A. Plant Cell. 1996;8:2067–2077. doi: 10.1105/tpc.8.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McConn M, Creelman R A, Bell E, Mullet J E, Browse J. Proc Natl Acad Sci USA. 1997;94:5473–5477. doi: 10.1073/pnas.94.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blechert S, Brodschelm W, Hölder S, Kammerer L, Kutchan T M, Mueller M J, Xia Z-Q, Zenk M H. Proc Natl Acad Sci USA. 1995;92:4099–4105. doi: 10.1073/pnas.92.10.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penninckx I A M A, Eggermont K, Terras F R G, Thomma B P H J, De Samblanx G W, Buchala A, Métraux J-P, Manners J M, Broekaert W F. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epple P, Apel K, Bohlmann H. Plant Physiol. 1995;109:813–820. doi: 10.1104/pp.109.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epple P, Apel K, Bohlmann H. FEBS Lett. 1997;400:168–172. doi: 10.1016/s0014-5793(96)01378-6. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y, Chang P-F L, Liu D, Narasimhan M L, Raghothama K G, Hasegawa P M, Bressan R A. Plant Cell. 1994;6:1077–1085. doi: 10.1105/tpc.6.8.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kogel K-H, Ortel B, Jarosch B, Atzorn R, Schiffer R, Wasternack C. Eur J Plant Pathol. 1995;101:319–332. [Google Scholar]
- 18.Feys B J F, Benedetti C E, Penfold C N, Turner J G. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McConn M, Browse J. Plant Cell. 1996;8:403–416. doi: 10.1105/tpc.8.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van der Plaats-Niterink A J. Studies in Mycology No. 21. Baarn, The Netherlands: Centraal Bureau voor Schimmel Cultures; 1981. pp. 1–242. [Google Scholar]
- 21.Dong J-Z, Dunstan D I. Plant Cell Reports. 1996;15:516–621. doi: 10.1007/BF00232985. [DOI] [PubMed] [Google Scholar]
- 22.Creelman R A, Tierney M L, Mullet J E. Proc Natl Acad Sci USA. 1992;89:4938–4941. doi: 10.1073/pnas.89.11.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melan M A, Dong X, Endara M E, Davis K R, Ausubel F M, Peterman T K. Plant Physiol. 1993;101:441–450. doi: 10.1104/pp.101.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell E, Mullet J E. Plant Physiol. 1993;103:1133–1137. doi: 10.1104/pp.103.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lévesque C A, Harlton C, Cock W A M d. Phytropathology. 1998;88:213–222. doi: 10.1094/PHYTO.1998.88.3.213. [DOI] [PubMed] [Google Scholar]
- 26.Schweizer P, Gees R, Mösinger E. Plant Physiol. 1993;102:503–511. doi: 10.1104/pp.102.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Croft K P C, Juttner F, Slusarenko A J. Plant Physiol. 1993;101:13–24. doi: 10.1104/pp.101.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahlbrock K, Scheel D. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:347–369. [Google Scholar]
- 29.Salt G A. In: Soil-Borne Plant Pathogens. Schippers B, Gams W, editors. London: Academic; 1979. pp. 289–312. [Google Scholar]
- 30.Knoester M, van Loon L C, van den Heuevel J, Hennig J, Bol J F, Linthorst H J M. Proc Natl Acad Sci USA. 1998;95:1933–1937. doi: 10.1073/pnas.95.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cook R J, Sitton J W, Haglund W A. Phytopathology. 1987;77:1192–1198. [Google Scholar]
- 32.Cook R J. Can J Plant Pathol. 1992;14:76–85. [Google Scholar]
- 33.Wilhelm S. Phytopathology. 1965;55:1016–1020. [Google Scholar]
- 34.Cook R J, Thomashow L S, Weller D M, Fujimoto D, Mazzola M, Bangera G, Kim D. Proc Natl Acad Sci USA. 1995;92:4197–4201. doi: 10.1073/pnas.92.10.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]