Abstract
The Pointed (PNT) domain and an adjacent mitogen-activated protein (MAP) kinase phosphorylation site are defined by sequence conservation among a subset of ets transcription factors and are implicated in two regulatory strategies, protein interactions and posttranslational modifications, respectively. By using NMR, we have determined the structure of a 110-residue fragment of murine Ets-1 that includes the PNT domain and MAP kinase site. The Ets-1 PNT domain forms a monomeric five-helix bundle. The architecture is distinct from that of any known DNA- or protein-binding module, including the helix-loop-helix fold proposed for the PNT domain of the ets protein TEL. The MAP kinase site is in a highly flexible region of both the unphosphorylated and phosphorylated forms of the Ets-1 fragment. Phosphorylation alters neither the structure nor monomeric state of the PNT domain. These results suggest that the Ets-1 PNT domain functions in heterotypic protein interactions and support the possibility that target recognition is coupled to structuring of the MAP kinase site.
Transcription factor families are defined by highly conserved DNA-binding domains that display similar DNA recognition properties. The means by which individual family members control different genes therefore must be determined by regulatory mechanisms that enhance the specificity of DNA binding. In the ets gene family, which includes at least 18 members in the human genome, partnerships with additional transcription factors, as well as posttranslational modifications, help dictate specificity for distinct targets (1). These regulatory mechanisms converge on a highly conserved ≈80-aa region termed the Pointed (PNT) domain (2).
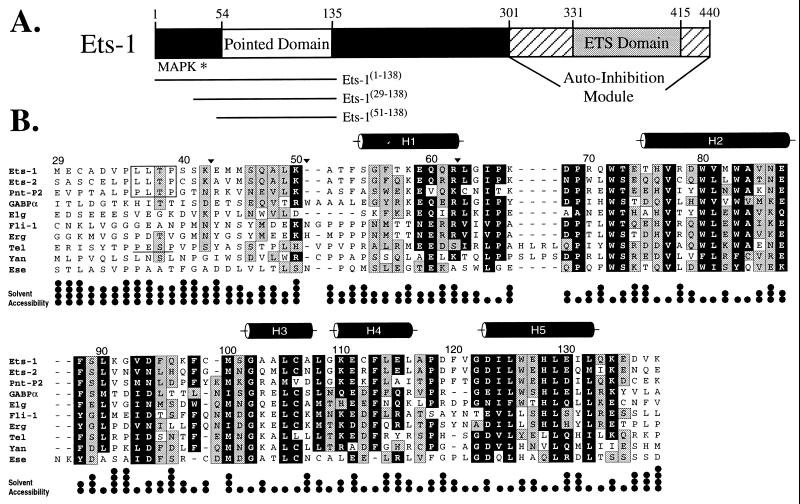
The PNT domain occurs in approximately one-third of the ets proteins, including Ets-1, Ets-2, GABPα, and TEL from vertebrates, and PNT-P2 and Yan from Drosophila (Fig. 1). This domain is proposed to mediate protein–protein interactions and to be regulated by ras-dependent signaling because of the presence of an adjacent mitogen-activated protein (MAP) kinase phosphorylation site (1). In particular, the PNT domain is implicated in the self-association of chimeric oncoproteins, identified in human leukemias, that result from chromosomal translocations of the gene encoding the ets protein TEL with segments of genes encoding several tyrosine kinases or the acute myeloid leukemia (AML)-1B transcription factor (3–9).
Figure 1.
The PNT domain is defined by a region of sequence conservation found in a subset of ets proteins. (A) Schematic diagram of the murine Ets-1 protein showing the locations of the PNT domain, MAP kinase phosphorylation site (Thr-38 ∗), and DNA-binding ETS domain with flanking autoinhibitory sequences (1). The central region of the protein contains putative transactivation domain(s). (B) Alignment of the sequences of PNT domains and preceding N-terminal regions from the ets family members murine Ets-1, Ets-2, GABPα, and Fli-1, human Erg, Tel, and Ese, and Drosophila PNT-P2, Elg, and Yan. The positions of highly conserved amino acids are highlighted in black (seven or more members having blosum62 substitution scores ≥1), and those of moderately conserved residues in gray (six or more members having blosum62 substitution scores ≥0). The MAP kinase phosphorylation sites identified in Ets-1, Ets-2, and PNT-P2 are boxed. Based on a consensus MAP kinase substrate sequence P-X-T/S-P, Tel also contains a potential phosphorylation site (underlined). Positions of tryptic cleavage in Ets-1(29–138) under conditions of partial proteolysis are denoted by ▾. The five α-helices (cylinders) in the Ets-1 PNT domain were identified by NMR methods. The fractional solvent accessibilities of the side chains in a low-energy structure of Ets-1(29–138) are illustrated (one • = 0–25%).
To date sequence conservation has defined the PNT domain, yet it has not been established that this region is a structural module that acts in a biological context. To create a framework for understanding the role of the PNT domain in the regulation of a variety of ets proteins and in the oncogenic potential of TEL fusion proteins, we have characterized structurally a fragment of Ets-1 that includes this domain and the adjacent MAP kinase phosphorylation site.
MATERIALS AND METHODS
Protein Samples.
DNA sequences encoding Ets-1(1–138), Ets-1(29–138), and Ets-1(51–138) were PCR-amplified from the full-length murine ets-1 cDNA and cloned into the pET28 (Ets-1(1–138)) or pET22b expression vectors (Invitrogen). The single substitutions of Leu-36 to Pro (L36P) or Thr-38 to Ala (T38A) were introduced into Ets-1(29–138) by PCR-based site-directed mutagenesis. Unlabeled and labeled Ets-1(29–138) and Ets-1(51–138) were prepared from Escherichia coli BL21(λDE3) grown in L-broth or minimal media, respectively, and purified by anion exchange (Mono Q; Pharmacia). Both proteins eluted with ≈200 mM NaCl in 50 mM Tris, pH 8.5. Ets-1(29–138) also was purified by reversed-phase chromatography using a Vydac C-18 column. Ets-1(1–138) was isolated by metal-chelation chromatography, followed by cleavage with thrombin to remove the N-terminal His6 tag. Ets-1 fragments were concentrated by lyophilization or with a micro-concentration device. Samples of ≈1.2 mM Ets-1(29–138) were prepared for NMR analysis by dialyzing the resuspended lyophilized protein against 2 mM NH4HCO3 and 5 mM DTT (pH 8.0), followed by 10 mM KCl, 10 mM KH2PO4, and 10 mM DTT (pH 6.5). D2O (10%) was added for a deuterium lock.
A 0.8 mM sample of uniformly 15N-labeled L36P Ets-1(29–138) was phosphorylated by incubating 5 mg of protein with 2.5 μg of sea star p44 MAP kinase (Upstate Biotechnology, Lake Placid, NY) and 10 mM ATP in buffer (12 mM Tris/12 mM β-glycerol phosphate/8 mM MgCl2/0.5 mM sodium vanadate/2 mM DTT, pH 7.2) at 16°C for ≈2 days (10). The reaction was monitored by electrospray ionization mass spectroscopy and, upon completion, the 31P,15N-labeled fragment was repurified by reversed-phase HPLC.
Protein Characterization.
Sedimentation equilibrium analysis of Ets-1(29–138) was performed by using a Beckman Model E ultracentrifuge operating at 22,000 rpm. The sample was 0.17 mM in 100 mM KCl, 25 mM KH2PO4, 10 mM DTT at pH 6.5 and 20°C. Size exclusion chromatography was carried out by using a HiLoad 16/60 Superdex 75 prep grade column (Pharmacia) with samples in 20 mM citrate, 150 mM KCl, and 10 mM DTT at pH 6.0 and 4°C. Protein stability was measured by using a Jasco (Easton, MD) J-720 spectropolarimeter to monitor the CD signal at 222 nm as a function of temperature. Samples were ≈20 μM protein (5 mM sodium phosphate, 5 mM sodium citrate, 5 mM sodium borate, 5 mM DTT, without or with 4 M urea at pH 7.0) in a 0.1-cm water-jacketed quartz cuvette. The urea was added to shift the unfolding transition to lower temperatures. Partial proteolysis of Ets-1(29–138) (2.75 μg) in 20 mM citrate, 150 mM KCl, and 1 mM DTT was carried out by incubation with 200 ng of trypsin for 2 min at pH 6.0 and 22.5°C, as described previously (11).
NMR Spectroscopy.
NMR experiments were carried out at 30°C on a Varian Unity 500 NMR spectrometer equipped with a pulsed-field gradient accessory. Data were processed by using nmrpipe (12) and pipp (13). Essentially complete 1H, 13C, and 15N spectral assignments of Ets-1(29–138) were obtained from an extensive set of heteronuclear NMR spectra (14). The diastereotopic methyl groups of valine and leucine were stereospecifically assigned by using biosynthetically directed 13C labeling (15). Resonances from the aromatic residues in Ets-1(29–138) were identified, and both histidines were shown to be in the neutral Nɛ2H tautomeric form at pH 6.5, as described elsewhere (16). Assignment of the amide resonances in the unmodified and phosphorylated forms of L36P Ets-1(29–138) were obtained by using 15N-total correlation spectroscopy (TOCSY)/nuclear Overhauser effect spectroscopy (NOESY)-heteronuclear single quantum correlation (HSQC) experiments. 15N T1, T2, and heteronuclear NOE relaxation data were recorded and analyzed as described by Farrow et al. (17).
Structure Determination.
An ensemble of 28 Ets-1(29–138) structures was computed from 1,568 distance (877 intraresidue, 309 sequential, 175 medium range, and 207 long range), 34 hydrogen-bonding, and 167 dihedral angle (65 φ, 70 ψ, and 32 χ1) restraints by using a simulated annealing protocol with x-plor version 3.8 (18). Interproton distance restraints were derived from three-dimensional (3D) 15N-NOESY-HSQC, 3D simultaneous 13C/15N-NOESY-HSQC, four-dimensional 13C,13C heteronuclear multiple quantum correlation (HMQC)-NOESY-HMQC spectra, and two-dimensional homonuclear NOESY experiments in D2O (for aromatic side chains), all recorded with τmix = 75 msec. Distances were calibrated as described previously (19). Hydrogen bonds were included as distance restraints for those amides remaining protonated 45 min after transfer of the protein to D2O buffer. Phi dihedral angles were restrained based on 3JHN-Hα coupling constants measured with the HNHA and HMQC-J experiments (20). Psi angles were restrained as described by Gagné et al. (21). Chi1 angles were restrained according to a staggered rotomer model using coupling patterns observed for stereospecifically assigned Hβ,β′ in 15N-TOCSY-HSQC and HNHB spectra and for the methyls of Thr, Ile, and Val in long-range 13Cγ-15N and -13C′ correlation spectra (20).
The x-plor energies for the ensemble of Ets-1(29–138) structures are: Etotal = 166.0 ± 5.3, Evdw = 7.0 ± 1.7, EL−J = −431.0 ± 22.8, Ecdih = 2.9 ± 0.6, and Enoe = 19.1 ± 1.6 kcal⋅mol−1. No distance or dihedral angle violation was greater than 0.25 Å or 5°, respectively, and the rms deviations from the idealized values are: bonds = 0.0040 ± 0.0001 Å, angles = 0.446o ± 0.006o, and improper angles = 0.283o ± 0.010o. Within this ensemble, 98.8% of residues have (φ, ψ) angles in the core or allowed regions of a Ramachandran plot, as determined by using procheck-nmr (22). All nonglycine residues in disallowed (φ, ψ) regions are located within the disordered termini of Ets-1(29–138).
RESULTS
Biophysical Characterization of Ets-1(29–138).
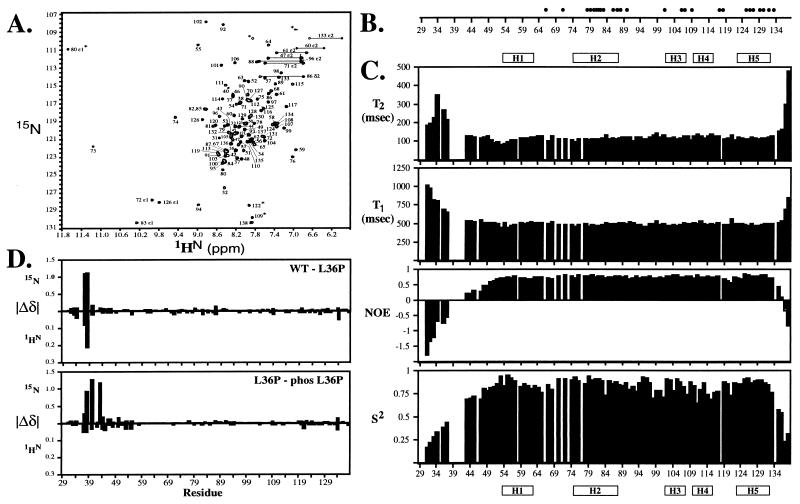
Ets-1(29–138) is a 110-residue polypeptide, encoded by exons III and IV of the ets-1 gene (23), which includes both the PNT domain and MAP kinase substrate site. The C-terminal three-quarters of this fragment, which corresponds to the PNT domain (Ser-54 to Glu-135), shares 30–65% sequence identity with nine other ets family members (Fig. 1). Ets-1(29–138), which was expressed as a soluble protein in E. coli, folds into a stable conformation with significant helical content as evidenced by a CD spectrum with pronounced minima at 208 and 222 nm ([θ]222 = −7,600 deg cm2⋅dmol−1), a well-dispersed NMR spectrum (Fig. 2A), and a reversible two-state unfolding transition with a midpoint temperature of ≈78°C at pH 7.0. Furthermore, Ets-1(29–138) is monomeric under a variety of experimental conditions as demonstrated by sedimentation equilibrium (MWapp = 11,400 Da; predicted = 12553.5 Da), 15N relaxation measurements, which revealed an overall rotational correlation time of 6.9 ns (Fig. 2C), and size exclusion chromatography, in which the protein eluted as a single peak near a 17-kDa marker. Taken together, these results establish that residues 29–138 from Ets-1 include an independent structural module.
Figure 2.
(A) The PNT domain (Ser-54 to Glu-135) is an independently folded structural module as evidenced by well-dispersed peaks in the 1H-15N HSQC spectrum of Ets-1(29–138). Residues N-terminal to this domain, including the MAP kinase substrate site, adopt a disordered conformation with 1HN chemical shifts that cluster near 8.2 ppm. Aliased peaks are identified by ∗. (B) Hydrogen-deuterium exchange studies identify amide protons that are protected from the solvent caused by hydrogen bonding and/or burial within Ets-1(29–138). • indicate residues with resolved 1H-15N HSQC cross peaks that have exchange rates >103 slower than expected for a random coil polypeptide. (C) NMR relaxation measurements provide information about the global tumbling and fast internal motions of Ets-1(29–138). Analysis of the amide 15N T1 and T2 lifetimes and heteronuclear 15N NOE values according to the model-free formalism (17) yields an overall rotational correlation time of 6.9 nsec for the protein and squared order parameters (S2) for each individual nonproline with a resolved cross peak. This correlation time is consistent with that expected for a monomeric protein of ≈12.5 kDa. Relatively uniform relaxation parameters, including NOEs > 0.5 and S2 values > 0.7, indicate that the residues comprising the PNT domain are generally well ordered. In contrast, residues 31–34, 36–37, 43–45, and 47–49 at the N terminus and 135–138 at the C terminus of Ets-1(29–138) are motionally disordered on a nano- to picosecond time scale as evident by NOEs < 0.5. Mutation of Leu-36 to Pro and phosphorylation of Thr-38 does not significantly change the relaxation properties of Ets-1(29–138) (not shown). (D) Chemical shift perturbations indicate that the effects of the Leu-36 to Pro mutation and the subsequent phosphorylation of Thr-38 are localized to the MAP kinase substrate site in the disordered N-terminal region of Ets-1(29–138). Shown are the absolute values of the changes in the amide 15N and 1HN chemical shifts (ppm) caused by the mutation and phosphorylation plotted versus residue number. The small changes observed for residues in the PNT domain reflect subtle differences in experimental conditions.
Structure Determination of Ets-1(29–138).
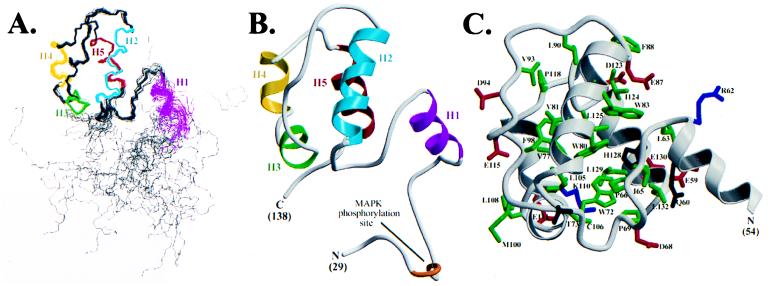
The structure of Ets-1(29–138) was determined by NMR spectroscopy (Fig. 3). The Ets-1 PNT domain shows a novel architecture consisting of five α-helices. The C-terminal portion of this domain, which includes helices H2–H5, folds into a well-defined bundle with rms distributions about the mean coordinate positions of 0.39 ± 0.06 Å for the backbone atoms and 0.85 ± 0.06 Å for all heavy atoms of residues 63–133. Helices H3 and H4 are arranged head to tail, yet are clearly separated by a bend centered at Gly-109 such that H3 is approximately perpendicular to H2, H4, and H5. Helix H5 is antiparallel to H4, and H2 lies across H3, H4, and H5. Consistent with this tertiary fold, helices H2, H4, and H5 are amphipathic, whereas the short H3 is nonpolar (Fig. 1B). Packing of these helices is mediated by several highly conserved hydrophobic and aromatic residues including Val-77, Trp-80, Val-81, Trp-83, and Ala-84 (H2), Leu-105 (H3), Phe-113 (H4), and Ile-124, Leu-125, His-128, Leu-129, and Leu-132 (H5) (Fig. 3C). Conserved residues in the loop between H2 and H3 (Leu-90, Val-93, and Phe-98) also contribute to the hydrophobic core of Ets-1(29–138).
Figure 3.
The tertiary structure of Ets-1(29–138) was determined by NMR methods. (A) Superimposition of the main chain atoms from 28 NMR-derived structures of Ets-1(29–138) aligned by using residues 63–133. The five α-helices in the PNT domain are colored (H1: residues 54–62; H2: 75–87; H3: 102–107; H4: 110–116; H5: 123–132), whereas the remainder of the main chain is shown in gray. The N and C termini of the molecule (residues 29–49 and 135–138) are disordered as evidenced by both high structural rms deviations and 15N NMR relaxation data. (B) Ribbon diagram (37) of a representative low-energy structure calculated for Ets-1(29–138). Only a single conformation is shown for the flexible N and C termini. (C) A low-energy structure of residues 26–132 of Ets-1(29–138) showing the positions of the side chains that are highly conserved (Fig. 1B) among the PNT domains of 10 ets proteins (green = hydrophobic, red = acidic, blue = basic, dark gray = polar).
Ets-1(29–138) contains a fifth helix (H1) that, although well defined by chemical shift, NOE, and J-coupling data, does not appear intimately associated with the core helical bundle (Fig. 3). The solvent exposure of helix H1 is consistent with its predominantly polar nature, its susceptibility to proteolysis at its C terminus (Arg-62) by trypsin, and its lack of protection against amide hydrogen exchange under the conditions examined (Figs. 1B and 2B). The position of this helix with respect to the remainder of the molecule is not precisely established, being determined by a small number of medium- and long-range NOE-derived distance restraints involving Leu-63, Ile-65, Pro-66, Pro-69, and Trp-72 in the loop between H1 and H2. Nevertheless, the helix itself is well defined locally, with an rms deviation of 0.53 ± 0.12 Å for the main chain atoms and 1.4 ± 0.15 Å for the heavy atoms of residues 54–62. 15N NMR relaxation measurements also indicate that the backbone of H1 is well ordered (Fig. 2C). Furthermore, CD and NMR spectroscopic measurements demonstrate that this helix unfolds cooperatively with the remainder of Ets-1(29–138) (not shown). These results indicate that helix H1, although exposed to the solvent, is an integral structural component of the Ets-1 PNT domain.
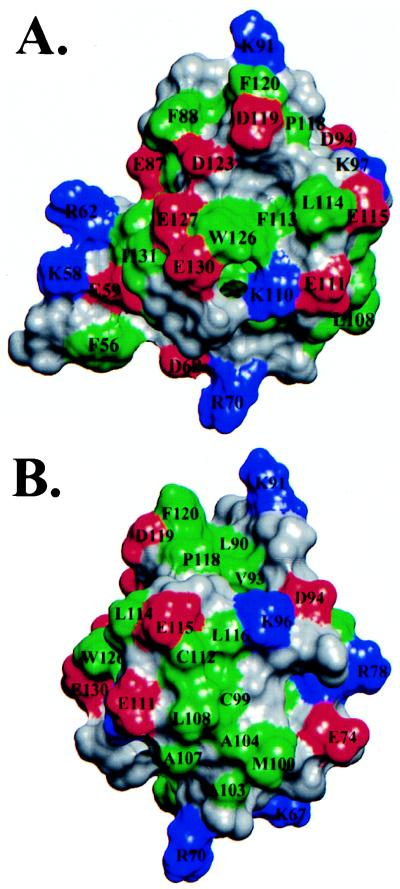
Inspection of the Ets-1 PNT domain structure reveals several potential protein binding sites. Protein–protein association results from both hydrophobic and electrostatic/hydrogen bonding interactions between interfaces composed of complementary nonpolar and charged/polar residues. A common type of interactive surface contains a hydrophobic patch surrounded by polar groups (24). One such surface in Ets-1(29–138), formed by helices H4 and H5, displays a hydrophobic region, centered around Trp-126, that is encircled by six acidic side chains (Fig. 4A). Similarly, the surface formed by helix H3 and the preceding loop from H2 contains several exposed hydrophobic residues that are surrounded by charged glutamates and lysines (Fig. 4B).
Figure 4.
Views of the van der Waals surface of the Ets-1 PNT domain (residues 54–135), illustrating potential protein–protein association interfaces centered on (A) helices H4 and H5, and (B) the loop connecting helices H2 and H3. The side chains are colored as green = hydrophobic, red = acidic, blue = basic, and gray = polar (and main chain).
The region of Ets-1(29–138) that precedes helix H1 is disordered. These amino acids display random coil amide 1HN and 15N chemical shifts (Fig. 2A) and very high rms deviations within the ensemble of calculated structures (Fig. 3A). Conformational mobility also is detected through 15N relaxation measurements (Fig. 2C). Consistent with these NMR results, the region is readily susceptible to tryptic cleavage, specifically at Lys-42 and Lys-50 (Fig. 1B). Furthermore, deletion of these residues to produce Ets-1(51–138) does not perturb the 1HN and 15N chemical shifts of the remaining amides that form helices H1-H5. Similar NMR measurements reveal that the first 28 residues of Ets-1(1–138) also are disordered (not shown). Together, these results demonstrate that the N-terminal ≈50 residues of Ets-1 are highly flexible in solution and structurally independent of the PNT domain, at least in the context of Ets-1(1–138) and Ets-1(29–138). The observation that this sequence is sensitive to trypsin cleavage in experiments performed on full-length Ets-1 (11) suggests that these residues also are disordered in the native protein.
Phosphorylation of Ets-1(29–138).
The function of Ets-1 in transcription assays is enhanced by ras-dependent signaling that requires a single MAP kinase substrate site, 36Leu-Leu-Thr-Pro39 (25–27). This site is located within the flexible, solvent-exposed N-terminal region of Ets-1(29–138) (Figs. 1 and 3). To provide a foundation for investigating the possible mechanisms by which phosphorylation regulates the function of ets proteins, we characterized the effects of this posttranslational modification on the structural and dynamic properties of Ets-1(29–138).
Ets-1(29–138) was phosphorylated by using activated sea star p44 MAP kinase. To increase the efficiency of this enzymatic reaction, Leu-36 was replaced with a proline in Ets-1(29–138). This substitution generated an optimized MAP kinase substrate sequence (36Pro-Leu-Thr-Pro39) (10, 28), which matched the site in Drosophila PNT-P2 (29), and led to the enhanced phosphorylation of Thr-38. The posttranslational modification was verified by mass spectroscopy, which showed the expected increase of 80 Da, and by 31P-NMR measurements. Mutation of Thr-38 to Ala completely prevented phosphorylation of Ets-1(29–138), confirming that Thr-38 is the only MAP kinase target (not shown).
The structural and dynamic properties of the unmodified and phosphorylated forms of L36P Ets-1(29–138) were compared with wild-type Ets-1(29–138) by three approaches. First, thermal denaturation experiments demonstrated that the midpoint unfolding temperatures of the proteins were essentially identical (≈57°C in 4 M urea at pH 7.0). Therefore, the modifications did not alter the stability of the Ets-1 fragment. Second, 15N relaxation studies revealed that the N-terminal regions of each protein, which includes the MAP kinase substrate site, remained motionally disordered (not shown). In addition, the rotational correlation times of Ets-1(29–138), derived from these relaxation measurements, did not change significantly upon mutation or phosphorylation. These dynamic measurements demonstrate that the modified proteins remained monomeric. Finally, for the highest resolution and most sensitive monitor of structural changes, the 1H-15N HSQC spectra of three Ets-1(29–138) variants were compared. As shown in Fig. 2D, the chemical shift perturbations caused by the modifications were highly localized to the amino acids immediately adjacent to Leu-36 and Thr-38. Thus, within the context of Ets-1(29–138), phosphorylation of Thr-38 does not significantly perturb the disordered character of the MAP kinase substrate site or its adjacent amino acids, nor alter the structure or oligomerization state of the PNT domain.
DISCUSSION
The Ets-1 PNT domain adopts a globular shape comprised of five α-helices, establishing that this region of conserved sequence is an autonomous structural domain. This fold is distinct from that of any known DNA-binding or protein-association module, as judged by the structural database comparison programs dali (30) and vast (31). The PNT domain frequently has been called the helix-loop-helix (HLH) region (3–7). This designation initially was proposed based on a secondary structure prediction that suggested an apparent resemblance to the dimerization domain found in the basic HLH transcription factors (32). Clearly, the structure of the Ets-1 PNT domain is not similar to that of the HLH motif. We therefore suggest strongly that this ets protein module be designated as the PNT (or Pointed) domain (2).
The PNT domain is proposed to mediate protein–protein interactions between members of the ets family of transcription factors. This hypothesis initially arose from studies indicating that the TEL PNT domain causes the self-association and constitutive activation of oncogenic fusion proteins formed by chromosomal translocations involving the tel locus with fragments of genes encoding the PDGFβ receptor, cABL, or JAK2 tyrosine kinases (3–6, 8, 9). Studies using qualitative techniques (i.e., two-hybrid screens and “glutathione S-transferase-pull down” and coimmunoprecipitation assays) have revealed homotypic as well as heterotypic interactions between some, but not all, ets family members. For example, fragments of TEL, including those corresponding to only helices H2–H5 of its PNT domain, self-associate; in contrast, the PNT domains of Ets-1, Erg, and GABPα do not (33, 34). In addition, segments of TEL and Fli-1, containing the PNT domains from each of these proteins, bind one another (34), whereas PNT domain-containing fragments of Ets-2 associate with a second ets protein, ERG (35).
Our biophysical studies demonstrate that Ets-1(29–138), both phosphorylated and unphosphorylated, is a monomer. Likewise, sedimentation equilibrium studies demonstrate that unphosphorylated full-length Ets-1 is monomeric in solution (J.J.S. and L. Joss, unpublished work). These results argue that the PNT domain from Ets-1 may bind to a heterotypic PNT domain(s) or may function through intra- or intermolecular interactions to unrelated protein-association modules. The structure of Ets-1(29–138) provides important clues for ongoing studies aimed at defining the potential targets for the PNT domains of ets family members.
Distinct patterns of protein–protein interactions involving specific ets transcription factors could arise from sequence variability among the PNT domains. As would be expected for a structural fold, the most highly conserved positions lie within the hydrophobic core (Figs. 1B and 3C). On the other hand, the most variable positions map to the surface of the domain, particularly on helices H1, H3, and H4 and in the extended loop linking H2 and H3, and to the disordered region including the MAP kinase phosphorylation site. The hydrophobic and charged groups forming the postulated association surfaces of Ets-1 (Fig. 4) are not strictly conserved among PNT domains, thus providing the potential for specific interactions with target proteins. Other possible sources of variation include a four-residue insertion between the predicted positions of helices H1 and H2 in the PNT domains from TEL and Yan (Fig. 1B). These inserted amino acids may alter or disrupt the packing of the exposed helix H1 against the core helical bundle or provide additional interprotein contacts. Finally, because of the low sequence conservation within the region corresponding to H1 in the Ets-1 fragment, it is plausible that this helix is not present in all ets PNT domains. The flexibility or absence of helix H1 could produce an additional hydrophobic interaction surface by exposing several conserved aromatic side chains corresponding to Trp-72, Trp-80, and Trp-83 in Ets-1 (Fig. 3C).
Phosphorylation of sequences adjacent to the PNT domain provides another potential source for specificity and regulation. Although phosphorylation enhances the transactivation function of Ets-1, Ets-2 and PNT-P2, the precise mechanism by which this occurs is unknown (25–27, 29). Our analysis of Ets-1(29–138) indicates that the MAP kinase substrate site lies within a flexible segment of this Ets-1 fragment (as well as that of Ets-1(1–138)), and that phosphorylation of Thr-38 does not change its structural or dynamic properties. These results imply that binding of this site by potential partner transcription factors, perhaps in conjunction with the PNT domain, is coupled to the ordering of these residues. This type of a regulated folding event is exemplified by the phosphorylation-dependent association of the KIX domain of the cAMP-regulated transcription factor CREB with its coactivator CREB binding protein (CBP). The KIX domain undergoes a random coil-to-helix transition that is induced by CBP binding, and not by phosphorylation alone (36).
In conclusion, the description of the Ets-1 PNT domain and its adjacent MAP kinase substrate site establishes this region of conserved sequence as a structural module, that in the case of Ets-1, is not affected by phosphorylation. The PNT domain, which is clearly unrelated to the helix-loop-helix motif, constitutes one of the few non-DNA binding domains of transcription factors whose structure has been characterized. The conserved and variable features of the PNT domain could accommodate either self-association or heterotypic interactions of specific ets family members.
Acknowledgments
We are grateful to Mr. Les Hicks and Dr. Cyril Kay for performing ultracentrifugation experiments, Ms. Isabelle Pot for assistance with cloning, Mr. Stéphane Gagné and Drs. Lewis Kay and Neil Farrow for providing computer programs, and Dr. Tom Alber for critical reading of this manuscript. This work was supported by grants from the National Cancer Institute of Canada with funds from the Canadian Cancer Society (L.P.M.), the National Institutes of Health GM 38663 and the Huntsman Cancer Institute (B.J.G.), the Leukemia Research Fund of Canada (to C.M.S.), the Jane Coffin Childs Memorial Fund for Medical Research (L.N.G.), the Natural Sciences and Engineering Research Council (C.D.M.), and the National Institutes of Health Training Grant CA09602 (J.J.S). Instrument support is provided by the Protein Engineering Network of Centres of Excellence (L.P.M.).
ABBREVIATIONS
- PNT
Pointed
- HSQC
heteronuclear single quantum correlation
- MAP
mitogen-activated protein
- NOESY
nuclear Overhauser effect spectroscopy
Footnotes
Data deposition: The coordinates, restraints, and chemical shifts for Ets-1(29–138) reported in this paper have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID codes 1bqv and r1bqvmr).
References
- 1. Graves B J, Petersen J M. In: Advances in Cancer Research. Vande Woude G, Klein G, editors. San Diego: Academic; 1998. pp. 1–55. [Google Scholar]
- 2.Klambt C. Development (Cambridge, UK) 1993;117:163–176. doi: 10.1242/dev.117.1.163. [DOI] [PubMed] [Google Scholar]
- 3.Golub T R, Barker G F, Lovett M, Gilliland D G. Cell. 1994;77:307–316. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]
- 4.Golub T R, Barker G F, Bohlander S K, Hiebert S W, Ward D C, Bray-Ward P, Morgan E, Raimondi S C, Rowley J D, Gilliland D G. Proc Natl Acad Sci USA. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golub T R, Goga A, Barker G F, Afar D E H, McLaughlin J, Bohlander S K, Rowley J D, Witte O N, Gilliland D G. Mol Cell Biol. 1996;16:4107–4116. doi: 10.1128/mcb.16.8.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll M, Tomasson M H, Barker G F, Golub T R, Gilliland D G. Proc Natl Acad Sci USA. 1996;93:14845–14850. doi: 10.1073/pnas.93.25.14845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiebert S W, Sun W, Davis J N, Golub T, Shurtleff S, Buijs A, Downing J R, Grosveld G, Roussel M F, Gilliland D G, et al. Mol Cell Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lacronique V, Boureux A, Della Valle V, Poirel H, Quang C T, Mauchauffe M, Berthou C, Lessard M, Berger R, Ghysdael J, Bernard O A. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- 9.Peeters P, Raynaud S D, Cools J, Wlodarska I, Grosgeorge J, Philip P, Monpoux F, Van Rompaey L, Baens M, Van den Berghe H, Marynen P. Blood. 1997;90:2535–2540. [PubMed] [Google Scholar]
- 10.Clark-Lewis I, Sanghear J, Pelech S L. J Biol Chem. 1991;266:15180–15184. [PubMed] [Google Scholar]
- 11.Jonsen M D, Petersen J M, Xu Q, Graves B J. Mol Cell Biol. 1996;16:2065–2073. doi: 10.1128/mcb.16.5.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delaglio F, Grzesiek S, Vuister G W, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 13.Garrett D S, Powers R, Gronenborn A M, Clore G M. J Magn Reson. 1991;95:214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Clore G M, Gronenborn A M. Protein Sci. 1994;3:372–390. doi: 10.1002/pro.5560030302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neri D, Szyperski T, Otting G, Senn H, Wüthrich K. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 16.Slupsky, C. M., Gentile, L. N. & McIntosh, L. P. (1998) Biochem. Cell Biol., in press. [DOI] [PubMed]
- 17.Farrow N A, Muhandiram R, Singer A U, Pascal S M, Kay C M, Gish G, Shoelson S E, Pawson T, Forman-Kay J D, Kay L E. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 18.Brünger A T. x-plor: A System for x-ray Crystallography and NMR. New Haven: Yale University Press; 1992. [Google Scholar]
- 19.Slupsky C M, Sykes B D. Biochemistry. 1995;34:15953–15964. doi: 10.1021/bi00049a010. [DOI] [PubMed] [Google Scholar]
- 20.Bax A, Vuister G W, Grzesiek S, Delaglio F, Wang A C, Tschudin R, Zhu G. Methods Enzymol. 1994;239:79–105. doi: 10.1016/s0076-6879(94)39004-5. [DOI] [PubMed] [Google Scholar]
- 21.Gagne S, Tsuda S, Li M X, Chandra M, Smillie L B, Sykes B D. Protein Sci. 1994;3:1961–1974. doi: 10.1002/pro.5560031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laskowski R A, Rullma J A C, MacArthur M W, Kaptein R, Thornton J M. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 23.Jorcyk C L, Watson D K, Mavrothalassitis G J, Papas T S. Oncogene. 1991;6:523–532. [PubMed] [Google Scholar]
- 24.Larsen T A, Olson A J, Goodsell D S. Structure (London) 1998;6:421–427. doi: 10.1016/s0969-2126(98)00044-6. [DOI] [PubMed] [Google Scholar]
- 25.Yang B-M, Hauser C A, Henkel G, Colman M S, van beveren C, Stacey K J, Hume D A, Maki R A, Ostrowski M C. Mol Cell Biol. 1996;16:538–547. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy S A, Chen D, Yang B-S, Garcia Ramirez J J, Cherwinski H, Chen X-R, Klagsbrun M, Hauser C A, Ostrowski M C, McMahon M. Mol Cell Biol. 1997;17:2401–2412. doi: 10.1128/mcb.17.5.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wasylyk C, Bradford A P, Gutierrez-Hartmann A, Wasylyk B. Oncogene. 1997;14:899–913. doi: 10.1038/sj.onc.1200914. [DOI] [PubMed] [Google Scholar]
- 28.Songyang Z, Lu K P, Kwon Y T, Tsai L-H, Filhol O, Cochet C, Brickley D A, Soderling T R, Bartleson C, Graves D J, et al. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Neill E, Rebay I, Tjian R, Rubin G M. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- 30.Holm L, Sander C. Nucleic Acids Res. 1997;25:231–234. doi: 10.1093/nar/25.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibrat J-F, Madej T, Bryant S H. Curr Opin Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 32.Seth A, Papas T S. Oncogene. 1990;5:1761–1767. [PubMed] [Google Scholar]
- 33.Jousset C, Carron C, Boureux A, Quang C T, Oury C, Dusanter-Fourt I, Charon M, Levin J, Bernard O, Ghysdael J. EMBO J. 1997;16:69–82. doi: 10.1093/emboj/16.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwiatkowski B W, Bastian L S, Bauer Jr T R, Schickwann T, Zielinska- Kwiatkowski A G, Hickstein D D. J Biol Chem. 1998;273:17525–17530. doi: 10.1074/jbc.273.28.17525. [DOI] [PubMed] [Google Scholar]
- 35.Basuyaux J P, Ferreira E, Stéhelin D, Butticè G. J Biol Chem. 1997;272:26188–26195. doi: 10.1074/jbc.272.42.26188. [DOI] [PubMed] [Google Scholar]
- 36.Radhakrishnan I, Perez-Alvarado G C, Parker D, Dyson H J, Montminy M R, Wright P E. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 37.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]