Abstract
The prion protein occurs as a globular domain and a leading fragment whose structure is not well-defined. For the ovine species, all of the tryptophan residues are in the initial fragment, while the globular domain is rich in tyrosine residues. Using heme as a spectroscopic probe, we have studied the recombinant prion protein before and after a temperature-induced conformational change. As for most heme proteins, the absorption spectrum of heme-CO displays a red shift upon binding to the protein, and both the Y and W fluorescence are highly quenched. Flash photolysis kinetics of the PrP–heme-CO complex shows a low yield for the bimolecular phase, indicating a pocket around the hemes. By comparing the holoprotein and the truncated sequence corresponding to the globular domain, the stoichiometry was determined to be five hemes for the globular domain and two hemes for the leading fragment. At high temperature, the hemes are released; upon cooling, only two hemes bind, and only the tryptophan fluorescence is quenched; this would indicate that the globular domain has formed a more compact structure, which is inert with respect to the hydrophobic probe. The final state of polymerization is perturbed if the synthetic peptide “N3” (PrP residues 142–166, which include the first helix) is added to the prion protein solution; the temperature cycle no longer reduces the number of heme binding sites. This would indicate that the peptide may alter or inhibit the polymer formation.
Keywords: prion protein, heme, fluorescence energy transfer, protein polymerization
The spongiform encephalopathies or “prion diseases” are neurodegenerative pathologies affecting a growing number of animal species. Sheep scrapie, bovine encephalopathy, Kuru, Gerstmann-Sträussler-Scheinker syndrome, and Creutzfeldt-Jakob disease (CJD) belong to these diseases, which may be sporadic or transmitted. Several hypotheses have been advanced concerning the nature of the infectious agents, but the most widely admitted, the “protein only” hypothesis, proposes a proteic nature of the infectious agent (Bolton et al. 1982; Prusiner 1982). According to this hypothesis, the mechanism of the pathogenesis is the conversion of the cellular form of the prion protein (PrPc) of the host, into a pathogenic isoform (PrPsc) characterized by its insolubility, its high content in β-sheet, and its protease resistance. The difference between PrPsc and PrPc is only conformational, because no covalent modifications differentiate PrPsc from PrPc (Stahl et al. 1991).
The transmission of bovine spongiform encephalopathy could then occur via protein–protein interactions (Prusiner 1982; Caughey 2001). As for HbS in sickle cell disease, a nucleus of several proteins forms a template that can deplete the stock of remaining protein by forming a larger aggregate. Unlike HbS, the reaction for PrP is not reversible, the aggregate is highly resistant to degradation, and the pathogenic form is transmissible (Baudin-Chich et al. 1990; Lesecq et al. 1996, 1997). Thus, the protein conformation is of importance to understanding the pathology of this disease.
As for most complex reactions, the intermediate forms for the transition PrPc → PrPsc have not yet been isolated and analyzed. For the prion protein, even the end points are not well-described. For the nonpathologic form PrP, the structure of the globular domain (residues 104–256) was determined by Nuclear Magnetic Resonance, while the leading fragment (8–103) did not show a “stable” form (Riek et al. 1998; Lopez Garcia et al. 2000; Zahn et al. 2000). As for other protein aggregates, the pathological form PrPSc involves the formation of amyloid deposits whose molecular structure is difficult to determine. The initial events leading to deposit formation are thus of interest (Prusiner et al. 1983).
In this study we use heme as a probe of the initial conformation of the monomeric recombinant protein, and the temperature-induced polymeric form. Heme has proven to be a useful probe, because it allows detection by both absorption and fluorescence methods. Furthermore, these standard optical techniques allow a relatively simple and rapid test that is sensitive to protein conformation. In the case of calmodulin, heme binding occurs on a millisecond time scale, and heme-CO binds with high affinity only to the calcium bound conformation (Marden et al. 1990, 1994; Leclerc et al. 1993a).
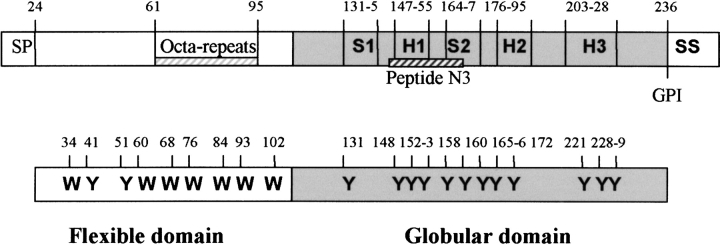
We have studied both the full sequence of ovine PrP, and the truncated form, corresponding to the globular domain. The ovine sequence has the advantage that all nine of the tryptophan residues are in the initial flexible domain, allowing a partial separation of the tyrosine fluorescence from the globular domain (shaded in Fig. 1 ▶). We show that heme binds to both domains of the PrP, but the heat-induced polymer releases heme from the globular domain.
Figure 1.
Schematic of the ovine PrP sequence. In addition to the full-length protein VRQ, we have expressed the truncated form Δ VRQ, which corresponds to the globular domain (shaded segment). Note that all the W residues are in the first “flexible” domain, which contains the glycine rich octa-repeats. N3 peptide (145–170) localization in the sequence is indicated as a hatched bar. SP = signal peptide, GPI = glyco phospholipid inositol fixation site, SS = signal sequence, H = αHelix, S = β-sheet.
Results
Interaction of heme-CO and the prion protein
Absorbance
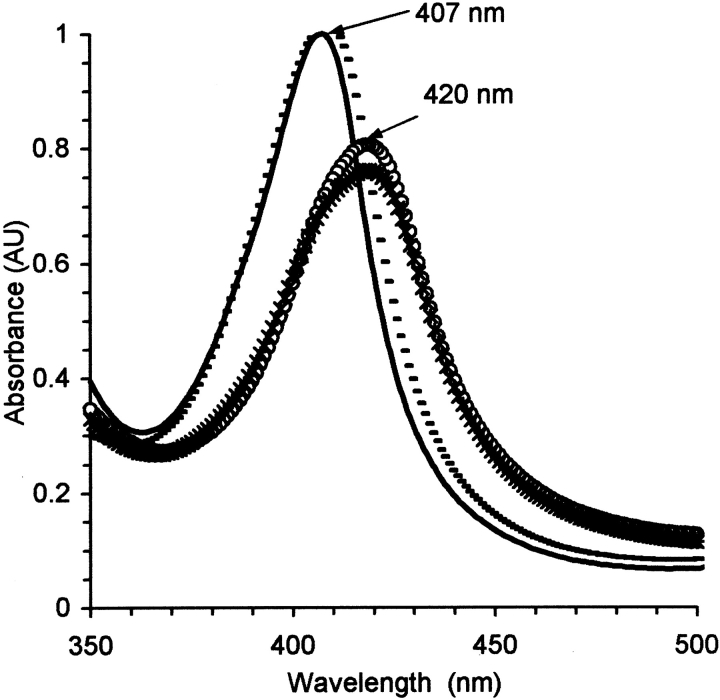
Heme-CO binds spontaneously to PrP, as evidenced by a redshift in the heme absorption spectrum (Fig. 2 ▶). Several hemes bind, probably with various affinities; the goal here was not to make a full mapping, but simply estimate the number bound at the concentration used. We used 20% glycerol in the solvent to eliminate sites of weak affinity. The fraction of hemes bound could be determined by analyzing the absorption spectrum as a sum of the bound and free spectra. Dilution studies to 0.2 μM did not show a significant release of the hemes, so the binding coefficients are <1 μM.
Figure 2.
Absorption spectra of heme-CO, without (—) and with 1.5 μM VRQ (empty circles) or 2 μM Δ VRQ (*) or 3.6 μM of the peptide N3 (- - - ). Solvent conditions were 10 μM in heme, in 20% glycerol with 20 mM MOPS buffer at pH 7.2.
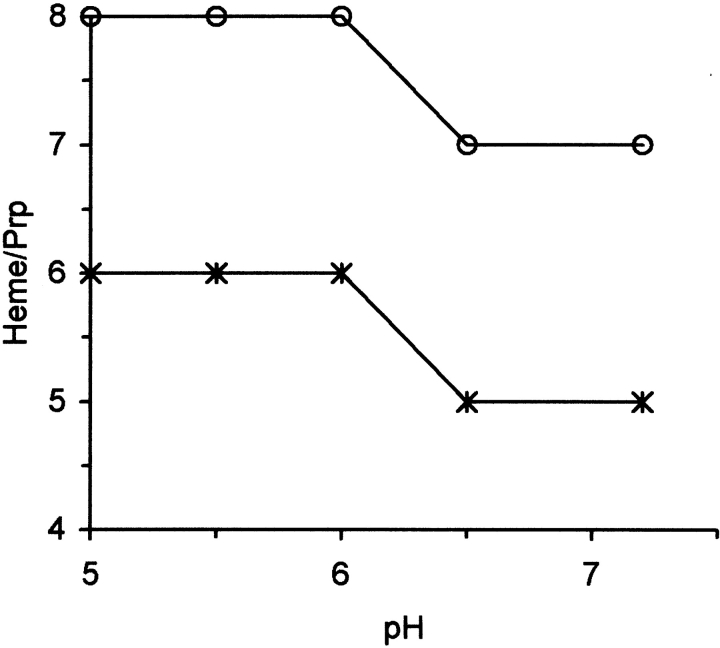
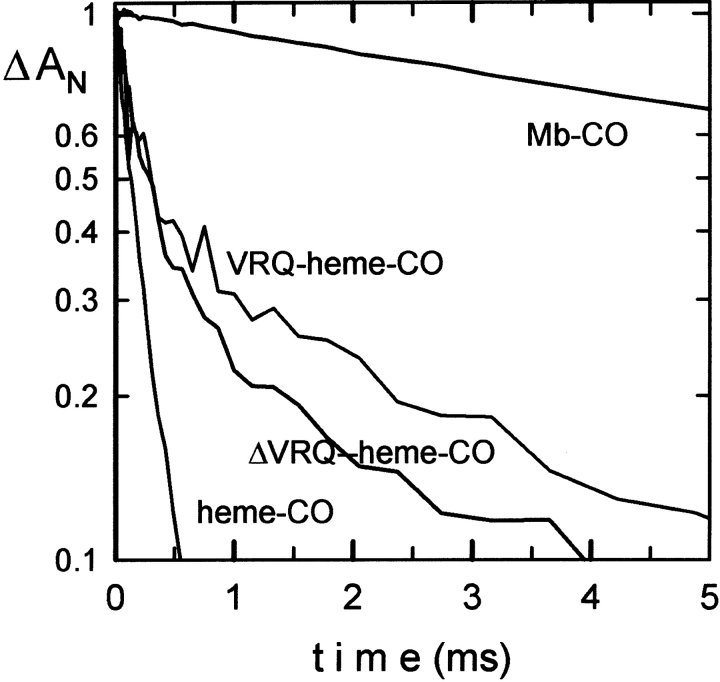
For the full-length protein a total of seven molecules of heme-CO can bind at pH 7.0 (Fig. 3 ▶). An additional molecule binds at lower pH values. The truncated protein ( Δ VRQ) binds five hemes at pH 7.0, indicating that only two hemes bind to the leading flexible fragment. The CO can be photolyzed from the heme group to determine whether the CO rebinding resembles that for free heme or a hemoprotein such as myoglobin. The flash photolysis kinetics displayed a low yield for the bimolecular phase, indicating the ligand does not escape easily. This suggests full protein pocket around the heme group (Fig. 4 ▶; Table 1).
Figure 3.
Effect of pH on the stoichiometry of the Heme-CO binding to the PrP holoprotein VRQ (empty circles) or the globular domain Δ VRQ (*). After a T-cycle, the values are 2 and 0, respectively, indicating that the globular domain no longer binds heme.
Figure 4.
CO-recombination kinetics of 10 μM Heme-CO before (empty circles) and after addition of full-length (VRQ) or truncated ( Δ VRQ) recombinant PrP protein at 2 μM. Data were collected at 436 nm and normalized to the observed amplitude at 4 μsec.
Table 1.
CO recombination kinetic constants for the complex Heme-CO with the full-length or truncated recombinant PrP protein
| Heme-CO | Heme-CO + VRQ | Heme-CO + Δ VRQ | Mb-CO | |
| Rapid rate (μM−1 sec−1) | 54 ± 3 | 43 | 40 | — |
| Slow rate (μM−1 sec−1) | — | 2.4 | 2.5 | 0.5 |
| Fraction unliganded (at 4 μsec) | 1 | 0.12 | 0.3 | 1 |
Effect of temperature cycle on the Heme-PrP interaction
Heating the sample to 85°C is sufficient to provoke a transition to the form with predominantly β-sheet secondary structure, as previously described (Rezaei et al. 2000). At the elevated temperature the absorption spectrum indicated free heme. Upon cooling, only two molecules of heme bind to the holoprotein, and the tyrosine fluorescence was recovered. Note that the same final state was obtained if the PrP sample was temperature cycled before addition of heme. This result indicates a strong dependence on the protein conformation for binding of the probe molecule.
In the absence of heme, the temperature cycle provokes an enhancement of the tyrosine fluorescence. There is thus less quenching of the tyrosine fluorescence in the globular domain by the solvent and/or the tryptophan residues of the nonstructured domain, suggesting a better domain separation. Because the intensity of the tyrosine fluorescence of PrP is not significantly different form free tyrosine, the enhanced yield of the heat cycled form is probably due to a change in local environment that decrease solvent access.
Interaction of hemin with the prion protein
Fluorescence
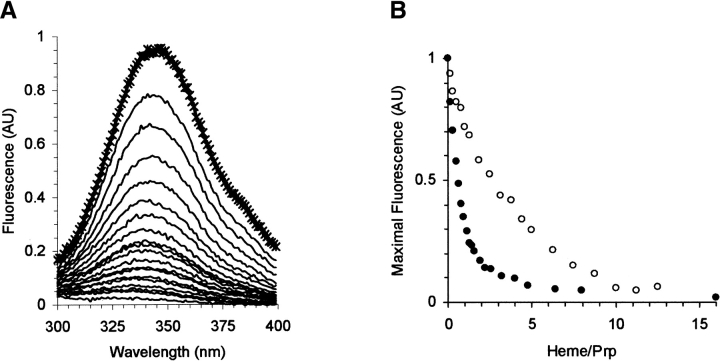
The protein fluorescence is quenched upon addition of heme, as expected, because heme is an excellent acceptor for energy transfer. This is the case for both the full-length and truncated protein, indicating that a heme is sufficiently close to all the W and Y residues (Fig. 5 ▶).
Figure 5.
(A) Fluorescence emission spectra of VRQ (0.6 μM), without (*) and with 0.1 to 9 μM heme (—). Data were corrected for the buffer spectrum. A similar result was obtained for the tyrosine fluorescence of truncated form Δ VRQ (data not shown). (B) Variation of the peak fluorescence intensity of VRQ (filled circles) and Δ VRQ (empty circles) vs. molar ratio heme/PrP. Data were normalized by the maximal fluorescence in the absence of heme.
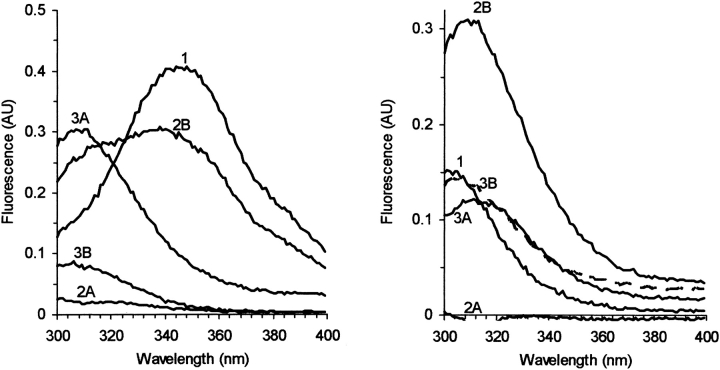
As observed in the absorption studies, the temperature cycle leads to a partial release of the hemes. Two pathways were employed, depending on whether hemin was added before (notation A) or after the temperature cycle (B). As shown in Figure 6 ▶ (curve 2A), addition of hemin can efficiently quench all the fluorescence. Note that the globular domain has Y but no W residues (Fig. 7 ▶). Because hemin can quench all the fluorescence, it apparently binds to both domains. The temperature cycle partially releases the hemes (Fig. 6 ▶, curve 3A), and one observes a recovery of the Y, but not the W, fluorescence.
Figure 6.
Fluorescence spectra of 0.6 μM holoprotein VRQ (left) and 0.8 μM of the truncated globular domain Δ VRQ, which has Y but no W residues (right). Two pathways were followed: 1) protein, 2A) after addition of 20 μM hemin, which efficiently quenches the fluorescence, 3A) after the temperature cycle to 85°C, which leads to a recovery of the tyrosine fluorescence. Alternatively the protein was first exposed to the temperature cycle 2B, which enhances the tyrosine signal, and then the hemin was added (3B), which quenches mainly the W signal. All spectra were measured at 25°C, in 20 mM MOPS buffer at pH 7.2.
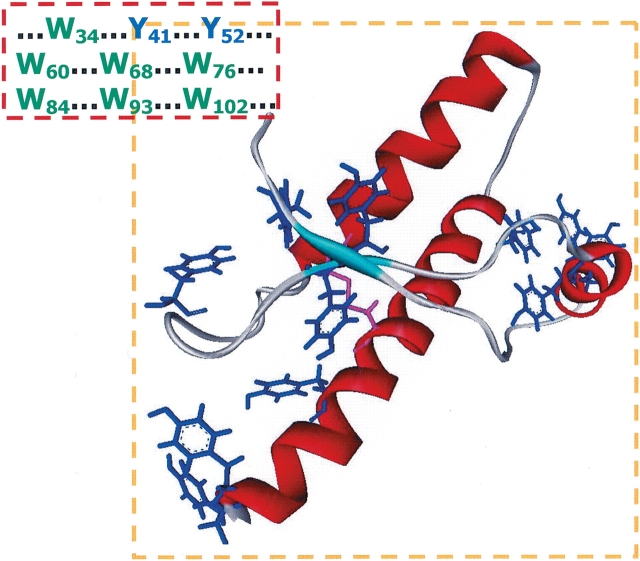
Figure 7.
Structure of the globular domain of PrP, determined by Nuclear Magnetic Resonance (Riek et al. 1998; PDB 1DX0), which has no W residues, but several Y (blue) clusters and a single SS bridge (magenta). The Y and W residues are listed for the N-terminal segment, whose structure is not determined.
If the temperature cycle is first performed on the PrP sample, an enhancement of tyrosine signal is observed (Fig. 6 ▶, curve 2B). Subsequent addition of hemin quenches mainly the tryptophan signal, without much change to the tyrosine fluorescence. The final states (Fig. 6 ▶, curve 3A or 3B) are similar for Δ VRQ, while the holoprotein showed some variability in the final properties: The W fluorescence was always quenched, but the final Y signal intensity was variable; this might be expected because the final state of the protein is a distribution of polymeric forms.
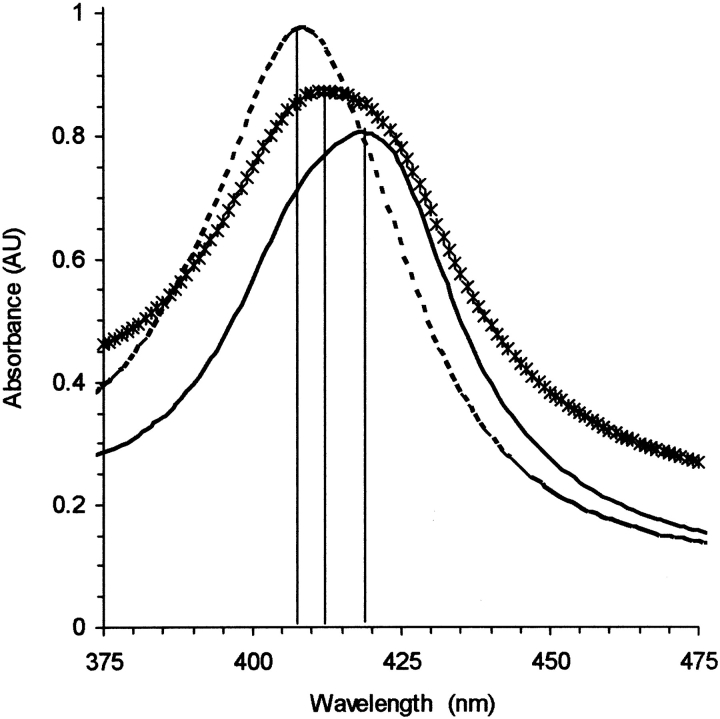
The absorption spectra for the various parts of the temperature cycle are shown in Figure 8 ▶. Addition of PrP to a heme-CO solution results in the transition from free to bound heme; however, heating the sample recovers the spectrum for free heme-CO. Returning to room temperature shows a bound form for heme-CO, but apparently different in the heme binding properties. The binding to the T-cycled (transconformed) species may thus involve different sites.
Figure 8.
Absorption spectra of 10 μM heme-CO with 2.4 μM of VRQ PrP. After mixing, the spectrum is typical of hemoproteins with a Soret peak near 420 nm (—); heating to 85°C recovers the spectrum for free heme-CO (- - -); subsequent cooling to 25°C results in the transconformed species of VRQ that binds less heme (*). Solvent conditions were 20% glycerol with 20 mM MOPS buffer at pH 7.2, 600 μM dithionite.
Complex of heme, PrP, and the peptide N3
Experiments were also made to study the combined interaction of the heme, PrP, and N3. The peptide N3 corresponds to the internal fragment of PrP, including the first α helix; it was initially studied as a template for the holoprotein, because the peptide undergoes a similar conformation change, and could thus serve as a model for the initiation of the transition (Kozin et al. 2001). The peptide does not show an interaction with heme.
The fluorescence measurements as described in Figure 7 ▶ were made with the peptide N3. Although no apparent interaction (based on absorption spectroscopy) occurs between the peptide and heme or the PrP, the temperature cycle in the presence of the peptide leads to a different final state (Fig. 9 ▶). Hemin is capable of quenching the fluorescence of both the protein and the peptide N3, indicating a ternary complex. The inert multimeric form of PrP that does not bind hemin is apparently no longer formed when N3 is present.
Figure 9.
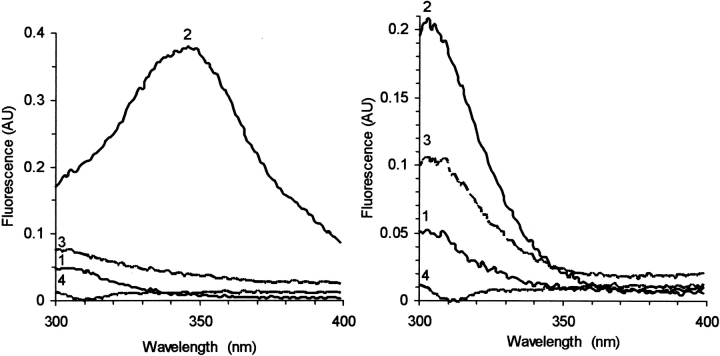
Interaction of the peptide N3 with holoprotein 0.6 μM VRQ (left) or 0.8 μM of the truncated Δ VRQ (right). Fluorescence spectra are for the peptide N3 (0.8 μM) without (1) and with (2) PrP, followed by the T-cycle (3), and finally addition of 20 μM Heme (4).
For the case of Δ VRQ, only Y residues contribute to the fluorescence, and addition of Δ VRQ to the peptide shows the expected sum of the intensities. The T-cycle shows some change in the intrinsic intensity (Fig. 9 ▶, curve 3), and subsequent addition of hemin shows a total quenching, as opposed to practically no effect of hemin on the protein alone.
Discussion
The present spectroscopic studies show a clear difference in the binding of the hydrophobic probe to the two domains of PrP. Five molecules of heme-CO can bind to the globular domain of PrP. This part of the protein is rich in tyrosine (12 residues), which forms three clusters. One possible binding mechanism is to insert the porphyrin between the tyrosine aromatic rings. This type of stacking has been proposed for the formation of heme dimers and higher polymers. As expected, the tyrosine fluorescence is highly quenched by the hemes.
The most surprising result is the release of the hemes after the temperature cycle. Previous studies have shown that the temperature cycle induces a transition from the α-helix form to a conformation with mainly β-sheet (Rezaei et al. 2002), as for the transition PrP → PrPSc. An unfolded or partially folded protein might expose additional hydrophobic surfaces that could bind the probe. However the T-cycled recombinant PrP showed no interaction of the globular domain with heme. In fact, the tyrosine fluorescence was higher than the initial form, indicating a modified environment and little quenching.
The heat-induced PrP transconformation was accompanied by a polymerization with an apparent molecular weight estimated to be 7 MD (Rezaei et al. 2002). The final form, therefore, has significant protein–protein interaction, and little space available for heme binding. A compact form, inert to external probes, would be compatible with the high resistance of the pathological form to degradation. However, it has not yet been shown that the temperature-induced aggregate has the pathogenic properties of PrPSc.
A different number of hemes bind to the two forms: before and after the T-cycle, which provokes the increase in the β-sheet conformation:
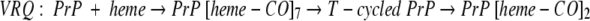
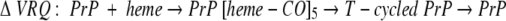
Heme as a probe is thus sensitive to the initial steps of the multimer formation, and the absorption spectrum of heme was used to detect the early steps of PrP polymer formation (Fig. 8 ▶).
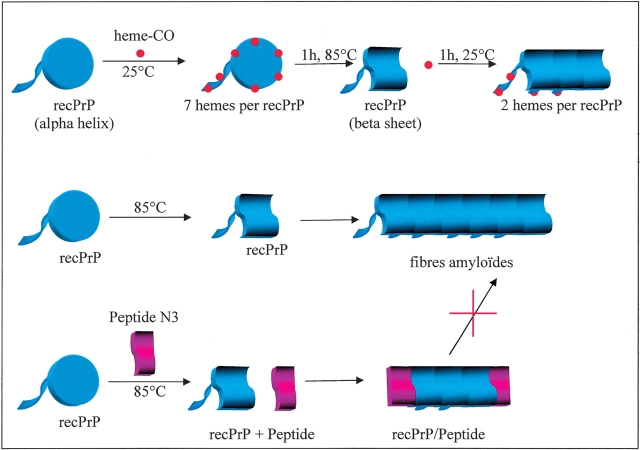
We also studied here whether the peptide N3 could participate in the temperature-induced formation of PrP polymers. We observed that addition of the peptide to the heme + PrP solution changed the properties of the final state. Without the peptide N3, the T-cycle provokes the releases of hemin, as evidenced by the recovery of the tyrosine fluorescence; with the peptide the PrP did not change its capacity to bind heme, as if the peptide prevented the formation of the form inert to hemin binding. A protein fragment such as N3 could therefore be used as an inhibitor. If it possesses the structural motif sufficient for insertion into the polymer, but lacks the complementary side to continue the polymer growth, the peptide would act as a cap to the fibre and inhibit further proteins from participating (Fig. 10 ▶). The peptide N3 appears to play this role, and could thus serve as a template to develop an inhibitor to the formation of PrPSc.
Figure 10.
Schematic diagram to summarize the conformational transitions of PrP (blue) in the presence of the probe heme and the peptide N3. The initial PrP is denoted as the two-domain form: disc for the globular domain and the flexible leader domain; this form binds a total of seven heme molecules (five to the globular domain); after the T-cycle the globular domain no longer binds heme. As previously described, the T-cycle leads to a conformation change and higher MW (middle scheme; Rezaei et al. 2002). The same pathway, but with the peptide N3 present, does not lead to the same final state. The form that does not bind heme is no longer produced; we hypothesize that the peptide acts as a cap to the polymer formation: It has a sufficient similarity to bind to the PrP protein to integrate in the nascent multimer, but does not provide the complementary surface to allow a propagation of the polymer growth.
Materials and methods
Solution preparation
Hemin (Sigma) was dissolved in 0.1 N NaOH at 10 mM. Sodium dithionite was dissolved at 100 mM in anaerobic 20 mM MOPS buffer at pH7.2. Heme-CO was prepared by reduction of hemin with the anaerobic solution of sodium dithionite in the presence of CO The heme-CO concentration was calculated from the Soret absorption at 407 nm (ɛ407 = 147,000 M−1 cm−1), and adjusted to a final concentration of 10 μM in the optical cuvette.
Sheep PrP proteins VRQ (full length) and Δ VRQ (without the N-terminal domain) were obtained by site directed mutagenesis following the method described by Rezaei et al. (2000). The concentrations of recombinant PrP proteins were determined by the bicinchoninic acid method; stock solutions of VRQ and Δ VRQ were at 63 μM and 80 μM, respectively, in 20 mM MOPS buffer at pH 7.2. The conformation transition for VRQ and Δ VRQ was induced by a temperature cycle of 25°C to 85°C and then back to 25°C; the length of time at the high temperature (at least several minutes) and maximum temperature (80°–95°C) did not effect the present results.
Peptide synthesis and purification
The N3 peptide (Fig. 1 ▶) was prepared on an Applied Biosystems 431A solid phase peptide synthesizer, using HMP-resin (4-hydroxymethyl phenoxymethyl-copoly(styrene-1% divinylbenzene)) and Fmoc (N-(9-fluorenyl)methoxycarbonyl) and 2-(H-benzotriazole-1-yl)1-1-3-3-tetramethyluronium hexafluoro-phosphate/N-hydroxybenzotriazole as coupling mediators (Kozin et al. 2001). After synthesis, peptide products were deprotected and cleaved from the support by treatment with trifluoroacetic acid. The peptide was purified by reverse-phase high-performance liquid chromatography (Waters) on a semipreparative column C4 eluted with linear gradient of water/acetonitrile/trifluoroacetic acid (10%–100%). Peptide fractions were pooled and lyophilized. To check the peptide purity, amino acid analysis and mass spectrometry were performed, and the results corresponded well to the expected sequences. The purity exceeded 99%.
Absorbance
The absorption spectra of heme-CO and hemin were measured at room temperature with a Varian Cary-400 spectrophotometer. Typical sample conditions were 10 μM in heme; in 20% glycerol with 20 mM MOPS buffer at pH 7.2. Recombinant sheep PrP proteins VRQ and Δ VRQ were added to the cuvette to a final concentration from 0.3 to 6 μM. Heme-CO absorption spectrum was recorded after each addition of protein. Spectra were corrected for the buffer and the dilution effect. Results versus pH were performed in the same conditions with the appropriate buffers: 20 mM sodium acetate for pH 5.0 and 5.5, 20 mM 2-(-Morpholino)-ethanesulfonic acid for pH 6.0 and 6.5, and 20 mM MOPS for pH 7.2. In these experiments, the number of heme-CO molecules fixed by one molecule of PrP protein was estimated by calculating the fraction of free heme-CO. The fraction bound was determined by simulating the observed absorption spectrum as a sum of the free and bound forms, where the bound form, probably not identical for all hemes, was measured under conditions for maximum interaction (high concentration and average ratio of PrP to heme).
Flash photolysis
CO-recombination kinetics of Heme-CO before and after addition of recombinant PrP proteins were measured after photodissociation of CO by 10-nsec pulses at 532 nm with a Quantel YG585 (France) laser. For these experiments sample concentrations were 10 μM for Heme-CO and 2 μM for VRQ and Δ VRQ prion proteins.
Fluorescence
Fluorescence spectra were recorded with a SLM-Aminco 8000 in 4 × 10-mm quartz cuvettes. The fluorescence emission of tryptophan and/or tyrosine residues of VRQ and Δ VRQ was recorded using 280 nm excitation in the presence and absence of heme. Sample concentrations were 0.6 μM and 0.8 μM, respectively, for VRQ and Δ VRQ in 20 mM MOPS buffer at pH7.2. The heme concentration varied from 0.1 at 10 μM. Spectra were corrected for the buffer, dilution, and inner filter effects (base on measurements of heme with free tryptophan and/or tyrosine). The affinity of a protein for heme can be estimated by successive dilution, to determine the concentration of the reactants were 50% of the heme-CO is bound (Leclerc et al. 1993b). In the case of the interaction of PrP with heme-CO, little change was observed over the range 10 μM to 0.2 μM, indicating a relatively high binding affinity. We thus treat the probe as an all-or-nothing reaction.
Acknowledgments
We thank Dr. T. Haertle and H. Rabesona for the generous gift of the N3 peptide. We thank also B. Doublet for help with the prion protein synthesis. This work was supported by funds from Inserm, Inra, and the GIS Prion program. C.P. was a recipient of the fellowship from the Gis Prion program.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
PrP, prion protein
PrPSc, scrapie form of PrP
recPrP, recombinant PrP
VRQ [valine 136, arginine 154, glutamine 171], variant of ovine PrP
Δ VRQ, fragment of VRQ recPrP comprising residues 103–34;
N3, fragment of human PrP comprising residues 142–63;
MOPS, 3-(-Morpholino)-propanesulfonic acid
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03409304.
References
- Baudin-Chich, V., Pagnier, J., Marden, M., Bohn, B., Lacaze, N., Kister, J., Schaad, O., Edelstein, S.J., and Poyart, C. 1990. Enhanced polymerization of recombinant human deoxyhemoglobin β 6 Glu → Ile. Proc. Natl. Acad. Sci. 87 1845–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton, D.C., McKinley, M.P., and Prusiner, S.B. 1982. Identification of a protein that purifies with the scrapie prion. Science 218 1309–1311. [DOI] [PubMed] [Google Scholar]
- Caughey, B. 2001. Interactions between prion protein isoforms: The kiss of death? Trends Biochem. Sci. 26 235–242. [DOI] [PubMed] [Google Scholar]
- Kozin, S.A., Bertho, G., Mazur, A.K., Rabesona, H., Girault, J.P., Haertle, T., Takahashi, M., Debey, P., and Hui Bon Hoa, G. 2001. Sheep prion protein synthetic peptide spanning helix 1 and β-strand 2 (residues 142–166) shows β-hairpin structure in solution. J. Biol. Chem. 276 46364–46370. [DOI] [PubMed] [Google Scholar]
- Leclerc, E., Leclerc, L., Cassoly, R., der Terrossian, E., Wajcman, H., Poyart, C., and Marden, M.C. 1993a. Heme binding to calmodulin, troponin C, and parvalbumin, as a probe of calcium-dependent conformational changes. Arch. Biochem. Biophys. 306163–168. [DOI] [PubMed] [Google Scholar]
- Leclerc, E., Leclerc, L., Poyart, C., and Marden, M.C. 1993b. Interaction of heme with amphiphilic peptides: Use of hemin-CN to probe the interaction of calmodulin with its target peptides. Arch. Biochem. Biophys. 306 158–162. [DOI] [PubMed] [Google Scholar]
- Lesecq, S., Baudin, V., Kister, J., Marden, M.C., Poyart, C., and Pagnier, J. 1996. Functional studies and polymerization of recombinant hemoglobin Glu-α2β26(A3) → Val/Glu-7(A4) → Ala. J. Biol. Chem. 271 17211–17214. [DOI] [PubMed] [Google Scholar]
- Lesecq, S., Baudin, V., Kister, J., Poyart, C., and Pagnier, J. 1997. Influence of the A helix structure on the polymerization of hemoglobin S. J. Biol. Chem. 272 15242–15246. [DOI] [PubMed] [Google Scholar]
- Lopez Garcia, F., Zahn, R., Riek, R., and Wuthrich, K. 2000. NMR structure of the bovine prion protein. Proc. Natl. Acad. Sci. 97 8334–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marden, M.C., Leclerc, L., and Poyart, C. 1990. Heme-CO as a probe of the conformational state of Calmodulin. FEBS Lett. 273 188–190. [DOI] [PubMed] [Google Scholar]
- Marden, M.C., Dufour, E., Christova, P., Huang, Y., Leclerc-L’Hostis, E., and Haertle, T. 1994. Binding of Heme-CO to bovine and porcine β-lactoglobulins. Arch. Biochem. Biophys. 311 258–262. [DOI] [PubMed] [Google Scholar]
- Prusiner, S.B. 1982. Novel proteinaceous infectious particles cause scrapie. Science 216 136–144. [DOI] [PubMed] [Google Scholar]
- Prusiner, S.B., McKinley, M.P., Bowman, K.A., Bolton, D.C., Bendheim, P.E., Groth, D.F., and Glenner, G.G. 1983. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell 35 349–358. [DOI] [PubMed] [Google Scholar]
- Rezaei, H., Marc, D., Choiset, Y., Takahashi, M., Hui Bon Hoa, G., Haertle, T., Grosclaude, J., and Debey, P. 2000. High yield purification and physicochemical properties of full-length recombinant allelic variants of sheep prion protein linked to scrapie susceptibility. Eur. J. Biochem. 267 2833–2839. [DOI] [PubMed] [Google Scholar]
- Rezaei, H., Choiset, Y., Eghiaian, F., Treguer, E., Mentre, P., Debey, P., Grosclaude, J., and Haertle, T. 2002. Amyloidogenic unfolding intermediates differentiate sheep prion protein variants. J. Mol. Biol. 322 799–814. [DOI] [PubMed] [Google Scholar]
- Riek, R., Wider, G., Billeter, M., Hornemann, S., Glockshuber, R., and Wuthrich, K. 1998. Prion protein NMR structure and familial human spongiform encephalopathies. Proc. Natl. Acad. Sci. 95 11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, N., Baldwin, M.A., and Prusiner, S.B. 1991. Electrospray mass spectrometry of the glycosylinositol phospholipids of the scrapie prion protein. Cell Biol. Int. Rep. 15 853–862. [DOI] [PubMed] [Google Scholar]
- Zahn, R., Liu, A., Luhrs, T., Riek, R., Von Schroetter, C., Lopez Garcia, F., Billeter, M., Calzolai, L. Wider, G., and Wuthrich, K. 2000. NMR solution structure of the human prion protein. Proc. Natl. Acad. Sci. 97 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]