Abstract
Heparin-like glycosaminoglycans, acidic complex polysaccharides present on cell surfaces and in the extracellular matrix, regulate important physiological processes such as anticoagulation and angiogenesis. Heparin-like glycosaminoglycan degrading enzymes or heparinases are powerful tools that have enabled the elucidation of important biological properties of heparin-like glycosaminoglycans in vitro and in vivo. With an overall goal of developing an approach to sequence heparin-like glycosaminoglycans using the heparinases, we recently have elaborated a mass spectrometry methodology to elucidate the mechanism of depolymerization of heparin-like glycosaminoglycans by heparinase I. In this study, we investigate the mechanism of depolymerization of heparin-like glycosaminoglycans by heparinase II, which possesses the broadest known substrate specificity of the heparinases. We show here that heparinase II cleaves heparin-like glycosaminoglycans endolytically in a nonrandom manner. In addition, we show that heparinase II has two distinct active sites and provide evidence that one of the active sites is heparinase I-like, cleaving at hexosamine–sulfated iduronate linkages, whereas the other is presumably heparinase III-like, cleaving at hexosamine–glucuronate linkages. Elucidation of the mechanism of depolymerization of heparin-like glycosaminoglycans by the heparinases and mutant heparinases could pave the way to the development of much needed methods to sequence heparin-like glycosaminoglycans.
Heparin-like glycosaminoglycans (HLGAGs) are one of the major components of the extracellular matrix and are present at the cell surface as part of proteoglycans (1, 2). HLGAGs are complex polysaccharides characterized by a disaccharide repeat unit of a uronic acid (either l-iduronic acid or d-glucuronic acid) which is linked 1–4 to a glucosamine (3). The modification of the functional groups of the sugar units (i.e., 2-O sulfate on the uronic acid and 3-O, 6-O, and N-sulfation of the hexosamine) (4), taken together with the variation in the chain length make HLGAGs the most acidic and heterogenous biopolymers. Together, these modifications allow for a wide array of HLGAG sequences and present a daunting challenge to understanding how certain sequences of HLGAGs elicit a biological response.
Of importance then is the development of molecular tools to study the in vivo roles of HLGAG sequences. One such strategy, being developed in our laboratories, is to use HLGAG degrading enzymes, or heparinases, to investigate the role and composition of biologically relevant HLGAG sequences. Three heparinases (I, II, and III) have been isolated from Flavobacterium heparinum; they differ from one another in terms of their size, molecular characteristics, and substrate specificities (5). By using these heparinases we have provided evidence for HLGAG involvement in fundamental biological processes such as angiogenesis (6) and development (7). While the heparinases have shown their use in delineating specific biological roles for HLGAG sequences, it is also possible that these enzymes can be used for the sequencing of HLGAG polymers when used in combination with a high precision analytical technique, such as matrix-assisted laser desorption ion mass spectrometry (MALDI MS), to identify enzyme-generated saccharide intermediates (8).
As a step toward the development of a sequencing protocol for HLGAGs, we recently have demonstrated that MALDI MS in combination with capillary electrophoresis can be used to successfully identify saccharide intermediates generated upon cleavage of defined oligosaccharides by heparinase I (9). We used this methodology to elucidate the mechanism of HLGAG depolymerization by heparinase I and discovered that it acts predominantly by an exolytic, processive mechanism, depolymerizing its substrate by binding HLGAG oligosaccharides and cleaving linkages starting from the nonreducing end of the polymer (10).
Heparinase II, another of the heparinases from F. heparinum, has the broadest substrate specificity of the three heparinases (11, 12). Heparinase II is able to cleave a wide range of disaccharide repeat units; indeed, linkages within HLGAG polysaccharides that are resistant to cleavage by heparinase I or III are cleaved by heparinase II (13–15). This suggested to us that heparinase II is an attractive candidate for developing a practical sequencing methodology for HLGAGs. To further develop heparinase II as an analytical tool for sequencing, we undertook an investigation of the mechanism of depolymerization of HLGAGs by heparinase II.
MATERIALS AND METHODS
Materials.
Substrate H1 was a gift from D. Tyrrell of Glycomed (Almeda, CA). Substrates O2, D3, and D4 were kindly provided by R. J. Linhardt after repurification by R. Hildeman (University of Iowa). P14 was a gift from Organon (P. Jacobs). Oligosaccharides were dissolved in deionized water at concentrations of 10–35 μM. Sucrose octasulfate was added as an internal standard where indicated. Heparinase II was diluted to a final concentration of 50 nM in a buffer containing 10 μM ovalbumin, 1 μM dextran sulfate, 50% glycerol, and 20 mM phosphate buffer (pH 7.0).
Enzymatic Purification.
Heparinase II from F. heparinum was purified as described (16). In addition, recombinant heparinase II and the heparinase mutant protein, C348A, where cysteine-348 is changed to alanine were expressed in Escherichia coli (17). Protein expression, isolation, and purification was carried out as described (17).
Digests.
Digests and mass spectral analyses were carried out as previously reported for heparinase I (9) with the exception that a calcium free buffer was used. Enzyme reactions using wild-type heparinase II derived from F. heparinum were performed by adding 1 μl of enzyme solution to 5 μl of substrate solution. Each time point was obtained by removing 0.5 μl of the reaction mixture and adding it to 4.5 μl of matrix solution. With E. coli-derived recombinant heparinase II (both wild-type and the C348A mutant), 1 μl of enzyme solution was added to 1 μl of substrate solution. In this case, the reaction was quenched after 60 min via the addition of 8 μl of matrix. All digests, whether with F. heparinum- or E. coli-derived recombinant heparinase II, were carried out at room temperature.
Derivatization.
Chemical derivatization of decasaccharide D3 was carried out by reacting 5 μl of oligosaccharide solution with 5 μl of 50 mM semicarbazide and 60 mM Tris/acetic acid (pH 5.0) for 18 hr at 30°C. Before digestion with the enzyme, the pH was adjusted to 7.0 by the addition of 1 μl of 50 mM Tris.
MS.
A saturated solution of caffeic acid (≈12 mg/ml in 30% acetonitrile/water) was used as the matrix solution. Seeded surfaces were prepared by using a modification (9) of the method developed by Xiang and Beavis (18). A 2-fold molar excess of peptide (arg-gly)19arg, isolated as the free base, was premixed with matrix before addition to the oligosaccharide solution (19). A 1-μl aliquot of sample/matrix mixture was deposited on the seeded stainless steel surface. After the formation of a polycrystalline layer, excess liquid was rinsed off with deionized water. MALDI MS spectra were acquired in the linear mode by using a Perseptive Biosystems (Framingham, MA) Voyager Elite reflectron time-of-flight instrument fitted with a 337-nm laser. Delayed extraction (20) was used to increase resolution (22 kV, grid at 93%, guide wire at 0.15%, pulse delay 150 ns, low mass gate at 1,000, 128 shots averaged). Mass spectra were calibrated externally by using signals for protonated (arg-gly)19arg and its complex with oligosaccharide H1.
RESULTS
Does Heparinase II Act Exolytically or Endolytically?
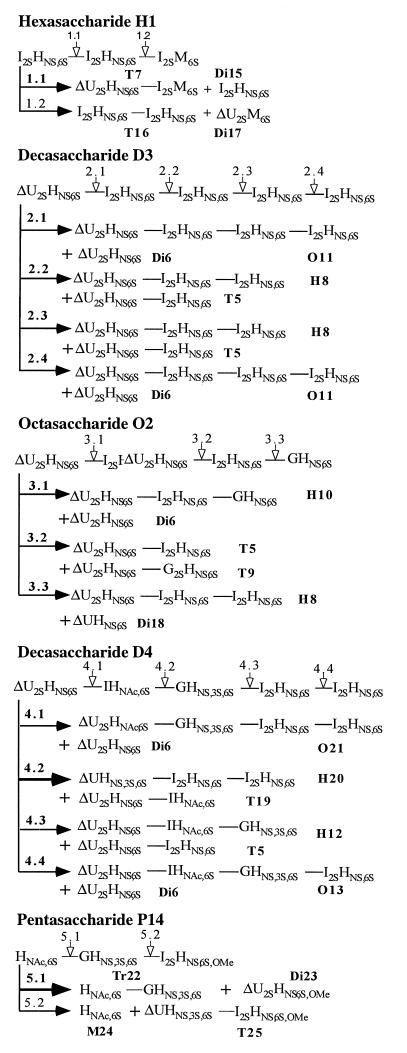
Table 1 lists the abbreviations, full sequences, and complexed [with (arg-gly)19arg] m/z values of all substrates and products that appear in this study. Fig. 1 lists the substrates used in this study and outlines the possible cleavage pathways of these substrates when acted on by heparinase II.
Table 1.
m/z of peptide/oligosaccharide complexes used in this study
| Substrates (1–4, 14) and products (5–13, 15–25) | Underivatized | Derivatized | |
|---|---|---|---|
| Monosaccharides | |||
| M24 | HNAc,6S | (4,742.19) | |
| Disaccharides | |||
| Di6 | ΔU2S–HNS,6S | 4,804.23 | 4,859.25 |
| Di15 | I2S–HNS,6S | 4,822.26 | |
| Di17 | ΔU2S–Man6S | (4,709.15) | |
| Di18 | ΔU–HNS,6S | (4,724.17) | |
| Di23 | ΔU2S–HNS,6S,OMe | 4,822.26 | |
| Trisaccharides | |||
| Tr22 | HNAc,6S–G–HNS,6S,3S | 5,143.49 | |
| Tetrasaccharides | |||
| T5 | ΔU2S–HNS,6S–I2S–HNS,6S | 5,381.72 | 5,437.79 |
| T7 | ΔU2S–HNS,6S–I2S–Man6S | 5,286.63 | |
| T9 | ΔU2S–HNS,6S–G–HNS,6S | 5,301.65 | |
| T16 | I2S–HNS,6S–I2S–HNS,6S | (5,399.74) | |
| T19 | ΔU2S–HNS,6S–I–HNAc,6S | 5,263.63 | |
| T25 | ΔU–HNS,6S,3S–I2S–HNS,6S,OMe | (5,410.72) | |
| Pentasaccharides | |||
| P14 | HNAc,6S–G–HNS,6S,3S–I2S–HNS,6S,OMe | 5,734.99 | |
| Hexasaccharides | |||
| H1 | I2S–HNS,6S–I2S–HNS,6S–I2S–Man6S | 5,882.13 | |
| H8 | ΔU2S–HNS,6S–[I2S–HNS,6S]2 | 5,959.20 | 6,015.27 |
| H10 | ΔU2S–HNS,6S–I2s–HNS,6S–G–HNS,6S | 5,879.13 | |
| H12 | ΔU2S–HNS,6S–I–HNAc,6S–HNS,6S,3S | 5,841.11 | |
| H20 | ΔU–HNS,6S,3S–I2S–HNS,6S–I2S–HNS,6S | 5,959.20 | |
| Octasaccharides | |||
| O2 | ΔU2S–HNS,6S–I2S–HNS,6S–I2S–HNS,6S–G–HNS,6S | 6,456.61 | |
| O11 | ΔU2S–HNS,6S–[I2S–HNS,6S]3 | 6,536.68 | |
| O13 | ΔU2S–HNS,6S–I–HNAc,6S–G–HNS,6S,3S–I2S–HNS,6S | 6,536.68 | 6,592.75 |
| O21 | ΔU–HNAc,6S–G–HNS,6S,3S–I2S–HNS,6S–I2S–HNS,6S | 6,536.69 | |
| Decasaccharides | |||
| D3 | ΔU2S–HNS,6S–[I2S–HNS,6S]4 | (6,996.07) | 7,052.14 |
| D4 | ΔU2S–HNS,6S–I–HNAc,6S–G–HNS,6S,3S–[I2S–HNS,6S]2 | (7,114.16) | |
2S, 3S, and 6S, 2-O, 3-O, or 6-O sulfation, (respectively); NS and NAc, N-sulfation and N-acetylation of the glucosamine.
Saccharides not observed are in parentheses. Oligosaccharides are abbreviated as follows: I, α-l-iduronic acid; G, β-d-glucuronic acid; ΔU, I or G with an unsaturated C4–C5 bond; H, α-d-2-deoxy,2-aminoglucose; and Man, anhydromannitol.
Figure 1.
Schematic representation of reactions of heparinase II digestion of the saccharides H1, D3(d), O2, D4, and P14. Substrates and products refer to Table 1.
Unlike either heparinase I or heparinase III, heparinase II has a broad substrate specificity, cleaving both sulfated and unsulfated substrates under nonforcing conditions (i.e., with enzyme-to-substrate ratios of 1:100 to 1:1,000). It was assumed that the relative susceptibility of scissile bonds should depend on the nature of residues flanking the glycosidic linkage. Therefore, two oligosaccharides, H1 and D3, containing only sulfated iduronate–hexosamine linkages, were used as substrates for the initial experiments. In this way, the mode of action of heparinase II (i.e., whether the enzyme acts exolytically or endolytically, processively or nonprocessively) could be investigated independently of any potential bias introduced by the chemical composition of the glycosidic linkage.
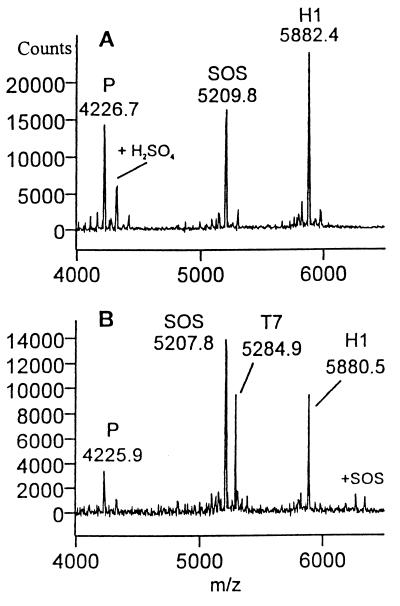
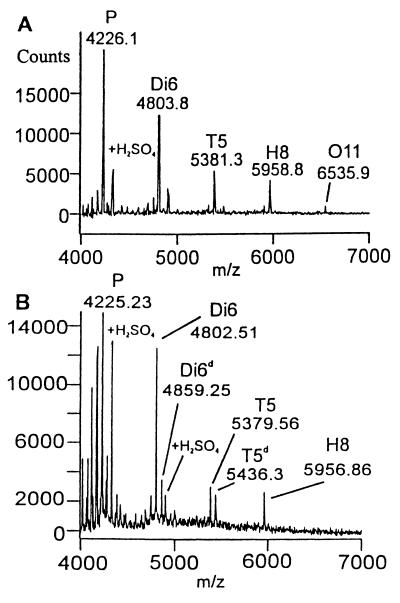
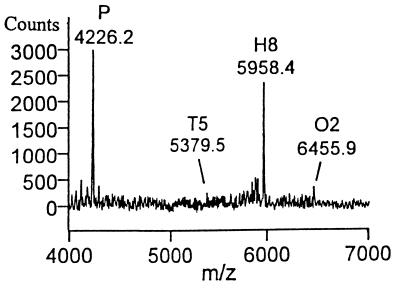
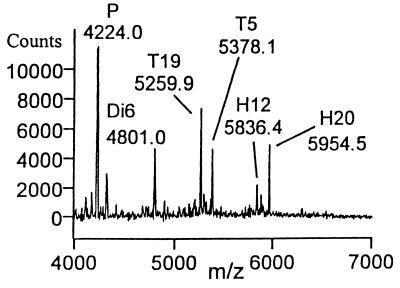
Upon heparinase II digestion of H1, tetrasaccharide T7 was observed concomitant with a decrease in the H1 signal, but T16 was not observed (Fig. 2). Thus, heparinase II cleaves H1 only at the scissile bond closest to the nonreducing end (Fig. 1, reaction 1.1). This result suggested that heparinase II might be exolytic; however, the absence of T16 (reaction pathway 1.2) also could be the result of an end group effect (i.e., the anhydromannitol moiety present at the reducing end could prevent the enzyme from cleaving linkage 1.2). Thus, to determine whether heparinase II acts exolytically, the decasaccharide D3 was treated with this enzyme. A number of intermediates were identified in the product profile (Fig. 3A). Octa- (O11), hexa- (H8), and tetrasaccharides (T5) were observed as intermediates. Furthermore, the amount of H8 produced was roughly equivalent to that of T5. Importantly, the amount of T5 remained relatively constant over time, indicating that the susceptibility of linkages to heparinase II is dependent on the size of the substrate. These results unambiguously show that heparinase II is an endolytic enzyme—i.e., for heparinase II, cleavage is possible at internal linkages (2.2 or 2.3 in D3) as well as external linkages (2.1 and 2.4 in D3).
Figure 2.
MALDI MS spectra of H1 digestion by heparinase II. P refers to the protonated peptide (arg-gly)19 arg. Sucrose octasulfate, H1, and T7 refer to the protonated complexes of peptide with sucrose octasulfate and with the oligosaccharide substrates and products (Table 1). (A) Mass spectrum profile of starting material H1. (B) Product profile after 60 min of digestion with heparinase II.
Figure 3.
(A) MALDI MS digestion profile of a 60-min heparinase II digestion of D3. Present in the product profile are O11, H8, T5, and Di6 (see Fig. 1 for reactions). (B) Heparinase II digestion of D3 after modification at the reducing end with semicarbazide (+57.06 mass units). Products with the mass tag are designated with a “d” (i.e., T5d and Di6d).
To confirm the results obtained with D3 and to establish whether heparinase II is a random, endolytic enzyme, D3 was derivatized with semicarbazide to introduce a mass tag at the reducing end. When derivatized D3 (D3d) was digested with heparinase II, only nonderivatized octa- (O11, but not O11d) and hexasaccharide (H8, but not H8d) were observed (Fig. 3B and Fig. 1, reaction pathways 2.3 and 2.4). In addition, derivatized tetrasaccharide (T5d) and both derivatized (Di6d) and nonderivatized (Di6) disaccharide were observed. In the case of D3d then, both H8 and O11 must be fragments derived from heparinase II cleavage and product release from the reducing end of D3d. Taken together, the presence of H8 and O11, coupled with the absence of H8d and O11d, indicate that the mechanism of action of heparinase II is not random (see Discussion for further interpretation). Thus, the digestion profiles of H1, D3, and D3d indicate that heparinase II is a nonrandom, endolytic enzyme.
Evidence for the Presence of Two Active Sites in Heparinase II.
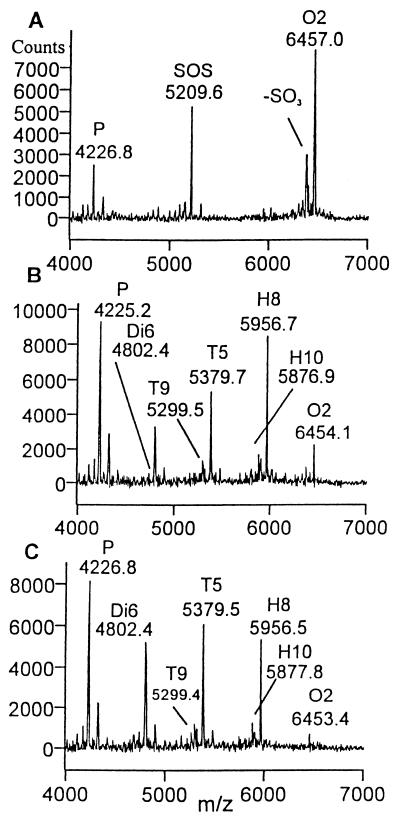
We sought further to corroborate the above findings with H1, D3, and D3d by using a substrate with more than one type of linkage. To this end, we investigated the mode of action of heparinase II on O2 that contains an unsulfated glucuronate at the reducing end. The dominant product observed in the initial time points of the digestion profile of O2 was H8 (data not shown), indicating cleavage at the unsulfated glucuronate (Fig. 1, reaction 3.3). This finding indicated that heparinase II was more efficient at cleaving linkages containing an unsulfated glucuronate than those containing a sulfated iduronate, even though this linkage is located next to the reducing terminus in O2 (Fig. 4). The substrate also was cleaved at sulfated iduronic acids, resulting in H10, T5, and T9 (reactions 3.1 and 3.2). Upon prolonged digestion the intermediate H8 was converted to T5 (reaction 3.3) and ultimately to Di6. This result further strengthens the notion that heparinase II, while endolytic, displays a nonrandom process of cleavage.
Figure 4.
MALDI MS spectra of heparinase II digestion of substrate O2. (A) Initial substrate profile before the introduction of heparinase II. Product profile after 60 min of digestion (B) and after 120 min of digestion (C).
The strong preference of heparinase II for the unsulfated glucuronate-containing linkage of O2 is consistent with the observation that heparinase II might contain two active sites, one of which is heparinase I-like, cleaving primarily heparin-like linkages, whereas the other site is heparinase III-like and cleaves primarily heparan sulfate-like linkages (17). In accordance with this hypothesis, we have identified a particular cysteine residue, cysteine 348, which is critical for the breakdown of heparin but not heparan sulfate by heparinase II (17). Therefore, we used a mutant heparinase II enzyme with this cysteine residue replaced by a nonfunctional alanine, hereafter referred to as the C348A mutant, to determine whether this dual active site model is correct.
Consistent with our hypothesis, H1 (which contains only sulfated iduronate linkages) was resistant to digestion by the C348A mutant (data not shown). In addition, the C348A mutant converted O2 almost exclusively to H8, resulting from cleavage at the unsulfated, glucuronic acid (Fig. 5 and Fig. 1, reaction 3.3); a small amount of T5 was also detected, which we interpret to be an impurity (data not shown). Furthermore, extending the digest time did not increase the amount of T5; thus, T9 could have arisen from heparinase II cleavage of a less sulfated impurity, present at ≈5% in the O2 substrate. Thus, the heparinase II C348A mutant does not appear to cleave glycosidic linkages flanked by sulfated iduronic acids. These results indicate that the activity of heparinase II or heparin-like and heparan sulfate-like regions of HLGAGs is distinct (see Discussion).
Figure 5.
MALDI MS spectrum of O2 digestion with the heparinase II C348A mutant. The figure represents the product profile after 120 min of digestion. A control of wild-type recombinant heparinase II was run at the same time. Its product profile at 120 min is identical to that in Fig. 4C.
Action of Heparinase II on 3-O-Sulfated Glucosamine.
In light of the above data regarding the size-dependent susceptibility of linkages to heparinase II digestion, we attempted to address the conflicting reports in the literature regarding the susceptibility of linkages proximate to a 3-O-sulfated glucosamine to heparinase II digestion. Previous reports suggest that heparinase II can cleave both scissile bonds flanking the disaccharide unit containing the 3-O-sulfated glucosamine (14). Later, Sugahara et al. (15) stated that a 3-O sulfate containing tetrasaccharide was resistant to heparinase II digestion, although most other tetrasaccharides were readily digested by this enzyme. To investigate this apparent discrepancy, we examined the heparinase II-treated digest profile of D4 and P14.
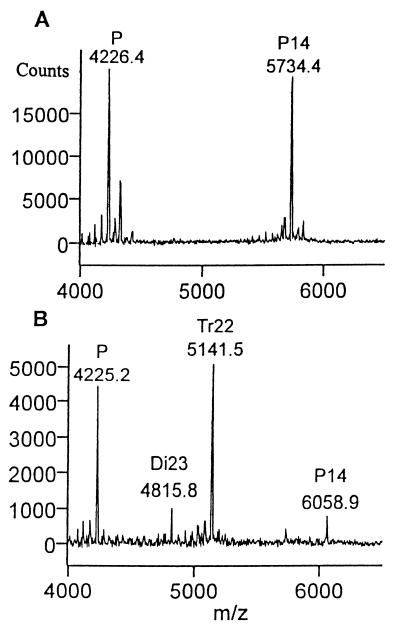
Substrate D4 presents a special case because it contains a 3-O-sulfated glucosamine in addition to an unsulfated glucuronate and an unsulfated iduronate linkage (see Table 1). The primary product of digestion of D4 with heparinase II was H20 (Fig. 6), indicating cleavage at the glycosidic linkage upstream of the 3-O-sulfated glucosamine (Fig. 1, reaction 4.2). Cleavage downstream of the 3-O-sulfated glucosamine (reaction 4.3) also took place, as H12 was observed in the mass spectrum. In addition, the tetrasaccharides T5 and T19 were detected. These results indicated that, within a decasaccharide length substrate, heparinase II was able to cleave linkages flanking a 3-O-sulfated glucosamine residue.
Figure 6.
MALDI MS spectrum of a heparinase II digestion of D4. The reaction was quenched at 60 min. Notation is similar to that of Fig. 3; chemical composition of products is outlined in Table 1.
To address the susceptibility of 3-O-sulfated glucosamine to cleavage by heparinase II within a shorter substrate, the product profile of pentasaccharide (P14) was investigated (Fig. 7). It showed that P14 was cleaved exclusively at the scissile bond upstream of the 3-O-sulfated glucosamine (Fig. 1, reaction 5.1). This finding is in contrast to the above results with D4, where linkages both upstream and downstream were cleaved. Together these results point to the fact that the susceptibility of the scissile bond downstream to a 3-O glucosamine depends on the length of the substrate; in decasacharide fragments or larger, the downstream linkage is susceptible to heparinase II action, whereas in shorter fragments this linkage is relatively resistant to cleavage.
Figure 7.
Analysis of the action of heparinase II on the pentasaccharide P14. (A) Profile of the pentasaccharide before introduction of heparinase II. (B) Product profile after 60 min of digestion.
DISCUSSION
Heparinase II Is an Endolytic, Nonrandom Enzyme.
The aim of this study was to elucidate the mode of HLGAG depolymerization by heparinase II using well-defined substrates. With sulfated substrates of the general composition (H6X,NS-I2S)x such as H1, D3, and D3d, heparinase II was found to cleave in an endolytic manner. In addition, the D3d results clearly show that heparinase II, while endolytic, cleaves sulfated substrates in a nonrandom manner.
No hexasaccharides or octasaccharides derived from cleavage at the nonreducing end (i.e., neither O11d nor H8d) were observed when D3d was digested with heparinase II. In addition, the smallest observed fragment derived from cleavage at the nonreducing end was the tetrasaccharide T5d. These results clearly indicate that the mode of action of heparinase II is not random, rather the enzyme possesses a bias for certain linkages. The nonrandom nature of heparinase II depolymerization seems to depend on the size of the substrate as well as the chemical composition of the linkages.
In a previous study, upon incubation of D3d with heparinase I the digestion profile of D3d showed only T5d and Di6 as products (9, 10). This result led us to conclude that heparinase I is an exolytic, processive enzyme that cleaves from the nonreducing to the reducing end. For heparinase II, the absence of derivatized hexa- and octasaccharide, plus the excess of Di6 to Di6d and the absence of H8d, indicates that heparinase II cleaves D3d in a manner similar to heparinase I. Thus, there may be some element of processivity to heparinase II. However, it is also possible that this product profile could be generated by nonrandom cleavage of D3d, followed by release of the product (which then would be a substrate for another round of cleavage). This interpretation is consistent with the roughly equivalent ratio of T5 to T5d in the product profile.
That heparinase I and heparinase II have a similar ability to cleave heparin-like regions of HLGAGs is not surprising considering their sequence similarity in functionally important regions. Both possess a Cardin–Weintraub consensus sequence. For heparinase II, the sequence reads 444FFKRTIAH451 (21), which is homologous to the heparin binding/active site region of heparinase I: 196IFKRNIAH203. In addition, evidence exists that His-451 plays a role in heparinase II similar to that which His-203 plays in heparinase I (21) for the breakdown of heparin. Thus, this sequence may represent part of the active site in heparinase II that is heparinase I-like. However, some fundamental aspects of the cleavage of heparin-like regions of HLGAGs by heparinase II must differ from heparinase I, because the enzymatic digest profile of substrates such as D3d are different for the two enzymes. In addition, heparinase II possesses the ability to cleave heparan sulfate-like regions of HLGAGs, which heparinase I cannot.
Heparinase II Possesses Two Active Sites.
A different mode of action was observed for heparinase II when an unsulfated glucuronate residue was present in the substrate. Heparinase II was found to exhibit a strong preference for unsulfated glucuronate linkages, which is consistent with the higher activity of heparinase II toward heparan sulfate, which is largely composed of unsulfated glucuronate as the uronate component. Cleavage at these sites appears to be independent of cleavage at sites containing sulfated iduronate linkages. Thus, it is proposed that heparinase II has a second active site domain, different from the one described above. The first site (site 1) cleaves heparin-like linkages (i.e., highly sulfated linkages flanked by iduronate moieties); the other site (site 2) cleaves primarily heparan sulfate-like regions (i.e., unsulfated linkages flanked by glucuronate moieties). Furthermore, we have identified a mutant of heparinase II, C348A, where the activity of site 1 has been greatly reduced or eliminated, whereas the activity associated with site 2 is intact. Because of the limited availability of substrates, we cannot at this time determine the mode of action of heparinase II at site 2.
The Activity of Heparinase II Depends on the Size of the Substrate.
With several of the substrates tested in this study it was apparent that the susceptibility of a glycosidic linkage depended on the size of the substrate. With D3d, the persistence of T5d indicates that the cleavage of oligosaccharides by heparinase II is slowed significantly when the substrate has been degraded to generate a tetrasaccharide, similar to what has been observed for heparinase I.
The size-dependent susceptibility of a linkage to heparinase II cleavage also was seen when we examined the ability of heparinase II to cleave saccharides containing a 3-O-sulfated glucosamine. Two substrates were used: one (D4), a decasaccharide; the other (P14), a pentasaccharide. We found that the susceptibility of linkages downstream of 3-O-sulfated glucosamine is influenced by the length of the oligosaccharide chain, with this linkage being susceptible when present in D4 but resistant when present in P14. Together, these results could resolve the controversy regarding the susceptibility of linkages downstream of a 3-O-sulfated glucosamine to heparinase II cleavage. In short, Linhardt et al. (14) were able to observe cleavage by heparinase II at sites both upstream and downstream of 3-O-sulfated glucosamine of longer oligosaccharides, whereas Sugahara and coworkers (15) did not observe cleavage at linkages downstream of 3-O sulfate in a tetrasaccharide. Our results clearly show that both observations are consistent with the mode of heparinase II activity as outlined in this study.
In summary, we have shown that heparinase II contains two active sites. Active site 1 is similar to the active site of heparinase I, except for the fact that it processes substrates endolytically, rather than exolytically. Active site 1 cleaves primarily sulfated linkages, whereas site 2 cleaves primarily unsulfated linkages. This study provides a framework for further investigations into the two active sites of heparinase II, exploiting it for the use of sequencing HLGAG oligosaccharides.
Acknowledgments
We thank Drs. R. Hileman, R. J. Linhardt, D. Tyrrell, and P. Jacobs for oligosaccharides. We also thank the National Institutes of Health (Grants GM57073 to R.S. and GM05472 to K.B.) and the Austrian–American Educational Commission (to A.J.R.) for funding.
ABBREVIATIONS
- HLGAGs
heparin-like glycosaminoglycans
- MALDI
matrix-assisted laser desorption ionization
- MS
mass spectrometry
References
- 1. Kjellen L, Lindahl U. Annu Rev Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- 2.Jackson R L, Busch S J, Cardin A D. Physiol Rev. 1991;71:481–539. doi: 10.1152/physrev.1991.71.2.481. [DOI] [PubMed] [Google Scholar]
- 3.Conrad H E. Ann NY Acad Sci. 1989;556:18–28. doi: 10.1111/j.1749-6632.1989.tb22486.x. [DOI] [PubMed] [Google Scholar]
- 4.Ernst S, Langer R, Cooney C L, Sasisekharan R. CRC Crit Rev Biochem Mol Biol. 1995;30:387–444. doi: 10.3109/10409239509083490. [DOI] [PubMed] [Google Scholar]
- 5.Linhardt R J, Rice K G, Kim Y S, Lohse D L, Wang H M, Loganathan D. Biochem J. 1988;254:781–787. doi: 10.1042/bj2540781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasisekharan R, Moses M A, Nugent M A, Cooney C L, Langer R. Proc Natl Acad Sci USA. 1994;91:1524–1528. doi: 10.1073/pnas.91.4.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binari R C, Staveley B E, Johnson W A, Godavarti R, Sasisekharan R, Manouikan A S. Development (Cambridge, UK) 1997;124:2623–2632. doi: 10.1242/dev.124.13.2623. [DOI] [PubMed] [Google Scholar]
- 8.Sutton C W, O’Neil J A, Cottrell J S. Anal Biochem. 1994;218:34–46. doi: 10.1006/abio.1994.1138. [DOI] [PubMed] [Google Scholar]
- 9.Rhomberg A J, Ernst S, Sasisekharan R, Biemann K. Proc Natl Acad Sci USA. 1998;95:4176–4181. doi: 10.1073/pnas.95.8.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst S, Rhomberg A J, Biemann K, Sasisekharan R. Proc Natl Acad Sci USA. 1998;95:4182–4187. doi: 10.1073/pnas.95.8.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nader H B. J Biol Chem. 1990;265:16807–16813. [PubMed] [Google Scholar]
- 12.Moffat C F, McLean M W, Long W F, Williamson F B. Eur J Biochem. 1991;202:531–541. doi: 10.1111/j.1432-1033.1991.tb16405.x. [DOI] [PubMed] [Google Scholar]
- 13.Desai U R, Wang H-M, Linhardt R J. Arch Biochem Biophys. 1993;306:461–468. doi: 10.1006/abbi.1993.1538. [DOI] [PubMed] [Google Scholar]
- 14.Linhardt R J, Turnbull J E, Wang H M, Loganathan D, Gallagher J T. Biochemistry. 1990;29:2611–2617. doi: 10.1021/bi00462a026. [DOI] [PubMed] [Google Scholar]
- 15.Sugahara K, Tohno-oko R, Yamada S, Khoo K H, Morris H R, Dell A. Glycobiology. 1994;4:535–544. doi: 10.1093/glycob/4.4.535. [DOI] [PubMed] [Google Scholar]
- 16.Godavarti R, Sasisekharan R. Biochem Biophys Res Commun. 1996;229:770–777. doi: 10.1006/bbrc.1996.1879. [DOI] [PubMed] [Google Scholar]
- 17.Shriver Z, Hu Y, Pojasek K, Sasisekharan R. J Biol Chem. 1998;273:22904–22912. doi: 10.1074/jbc.273.36.22904. [DOI] [PubMed] [Google Scholar]
- 18.Xiang F, Beavis R C. Rapid Commun Mass Spectrom. 1994;8:199–204. [Google Scholar]
- 19.Juhasz P, Biemann K. Carbohydr Res. 1994;270:131–147. doi: 10.1016/0008-6215(94)00012-5. [DOI] [PubMed] [Google Scholar]
- 20.Vestal M L, Juhasz P, Martin S A. Rapid Commun Mass Spectrom. 1995;9:1044–1050. [Google Scholar]
- 21.Shriver Z, Hu Y, Sasisekharan R. J Biol Chem. 1998;273:10160–10167. doi: 10.1074/jbc.273.17.10160. [DOI] [PubMed] [Google Scholar]