Abstract
Murine gammaherpesvirus 68 (MHV-68 [also referred to as γHV68]) is phylogenetically related to Kaposi's sarcoma-associated herpesvirus (KSHV [also referred to as HHV-8]) and Epstein-Barr virus (EBV). However, unlike KSHV or EBV, MHV-68 readily infects fibroblast and epithelial cell lines derived from several mammalian species, providing a system to study productive and latent infections as well as reactivation of gammaherpesviruses in vivo and in vitro. To carry out rapid genome-wide analysis of MHV-68 gene expression, we made DNA arrays containing nearly all of the known and predicted open reading frames (ORFs) of the virus. RNA obtained from an MHV-68 latently infected cell line, from cells lytically infected with MHV-68 in culture, and from the lung tissue of infected mice was used to probe the MHV-68 arrays. Using a tightly latent B-cell line (S11E), the MHV-68 latent transcription program was quantitatively described. Using BHK-21 cells and infected mice, we demonstrated that latent genes are transcribed during lytic replication and are relatively independent of de novo protein synthesis. We determined that the transcription profiles at the peak of lytic gene expression are similar in cultured fibroblast and in the lung of infected mice. Finally, the MHV-68 DNA arrays were used to examine the gene expression profile of a recombinant virus that overexpresses replication and transcription activator (RTA), C-RTA/MHV-68, during lytic replication in cell culture. The recombinant virus replicates faster then the parental strain and the DNA arrays revealed that nearly every MHV-68 ORF examined was activated by RTA overexpression. Examination of the gene expression patterns of C-RTA/MHV-68 over a time course led to the finding that the M3 promoter is RTA responsive in the absence of other viral factors.
Gammaherpesviruses are known to establish latency in lymphocytes and are associated with tumorigenesis (5-7, 10, 48). Two important human pathogens in the gammaherpesvirus subfamily of herpesviruses are Kaposi's sarcoma-associated herpesvirus (KSHV [also referred to as HHV-8]) and Epstein-Barr virus (EBV). KSHV and EBV are associated with several malignancies, including B-cell lymphomas, nasopharyngeal carcinoma, and Kaposi's sarcoma (22, 23, 27, 30, 32). Studies of KSHV and EBV are limited by the lack of cell lines able to support efficient productive infection and by the restricted host ranges of the viruses (11, 33). Murine gammaherpesvirus 68 (MHV-68 [also referred to as γHV68]) is another member of the gammaherpesvirus subfamily. However, in vitro cell culture systems are available to study productive de novo infection by MHV-68, as well as latency and reactivation (34, 40). MHV-68 forms plaques on monolayers of many cell lines, making it possible to genetically manipulate the viral genome. MHV-68 establishes lytic and latent infections in laboratory mice (47), providing a system for examining host-virus interactions (24, 25, 36, 42, 43). These characteristics of MHV-68 make it possible to examine the functions of individual viral genes at various points during the viral life cycle, including de novo infection. De novo infection analyses have not been possible for other gammaherpesviruses such as EBV and KSHV.
Herpesviruses have two distinct life cycle phases, latency and lytic replication. Reactivation from latency to lytic replication is essential for transmission of the virus from host to host and thus is one important aspect of herpesvirus biology. A viral protein, replication and transcription activator (RTA) is primarily encoded by open reading frame (ORF) 50, which is well conserved among gammaherpesviruses. RTA is necessary and sufficient to reactivate MHV-68 and drive the lytic cycle to completion in latently infected B cells (14, 19, 54, 55). Similarly, KSHV RTA has been shown to be sufficient to reactivate the virus from latently infected B cells derived from KSHV-associated lymphomas (20, 46). Although two EBV proteins, RTA and ZEBRA, function in a cooperative manner to reactivate the viral lytic cycle (3, 18, 21), RTA alone can disrupt latency in some latently infected cell lines (31, 56). These studies indicate that RTA of gammaherpesviruses plays a conserved role in virus reactivation.
We have constructed custom membrane arrays representing nearly all of the known and predicted MHV-68 ORFs to explore the patterns of viral gene expression. To illustrate the value of genome-wide transcription analysis, we used the MHV-68 DNA arrays to identify a novel regulatory element for a specific gene, to identify latency-associated transcripts not previously recognized, and to define the genome-wide effects of a specific genetic manipulation.
MATERIALS AND METHODS
Viruses and cells.
MHV-68 was originally obtained from the American Type Culture Collection (VR1465). The virus referred to as the parental virus is a recombinant EGFP/MHV-68 (tw25) constructed by insertion of the human cytomegalovirus (HCMV) promoter-driven EGFP cassette at the left of the MHV-68 genome (54). The C-RTA/MHV-68 virus was constructed by replacing the HCMV promoter-driven enhanced green fluorescent protein (EGFP) cassette with an HCMV promoter-driven RTA cassette (Tammy Rickabaugh, manuscript in progress). Viral stocks of wild-type (wt) MHV-68, EGFP/MHV-68, and C-RTA/MHV-68 were prepared as previously described (55). To infect BHK-21 cells, the viral inoculum in Dulbecco modified Eagle medium was incubated with cells for 1 h with occasional swirling. The inoculum was removed and replaced with fresh Dulbecco modified Eagle medium plus 10% fetal bovine serum. For the experiments involving cycloheximide (CHX; Sigma, St. Louis, Mo.), cells were treated at a concentration of 200 or 400 μg/ml 1 h prior to, during, and after viral inoculation until RNA was harvested. For the experiments with phosphonoacetic acid (PAA; Sigma), cells were treated at a concentration of 200 or 400 μg/ml after viral inoculation until RNA was harvested. For the experiments of infected lung tissue, BALB/c mice were inoculated intranasally with 5 × 105 PFU. Total RNA was harvested from the lungs of two mice at 1, 3, 5, and 7 days postinfection (dpi).
DNA array construction and probe hybridization.
Primers were designed to amplify ∼1-kb regions of DNA sequence from the 5′ end of known MHV-68 ORFs. Similarly, primers were designed to amplify ∼1-kb regions from the 3′ end of MHV-68 ORFs that are >2 kb in length (ORF 6, ORF 8, ORF 9, ORF 25, ORF 44, ORF 56, ORF 64, ORF 75a, ORF 75b, and ORF 75c). Moreover, primers were chosen to amplify a sequence from the murine cellular GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene which was used as a control. Each array element was cloned into pCRII by using the TA cloning kit (Stratagene, La Jolla, Calif.) according to the manufacturer's protocol. Common vector primers were designed to amplify the cloned array elements. The PCR products were isopropanol precipitated and resuspended in water at 100 ng/μl. Approximately 20 ng of DNA was spotted in quadruplicate on Hybond-N membrane (Amersham, Piscataway, N.J.) by using a replication system (Nalge Nunc International, Rochester, N.Y.). The DNA on the arrays was denatured for 5 min (0.5 M NaOH, 1.5 M NaCl), neutralized for 5 min (0.5 M Tris [pH 7.5], 1.5 M NaCl), submerged in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 5 min, and UV cross-linked to the membrane (Stratalinker). The quality of array element spotting was determined by the hybridization of a 32P-labeled oligonucleotide probe specific for the common primer sequence present at the ends of the PCR products. The oligonucleotide probe was end labeled with [γ-32P]ATP (Perkin-Elmer Life Sciences, Boston, Mass.) by using T4 polynucleotide kinase (Promega, Madison, Wis.) and purified by using a G25 spin column. The arrays were preincubated for 4 h in hybridization buffer (0.1 M K2HPO4 [pH 6.8], 7% sodium dodecyl sulfate [SDS], 1% bovine serum albumin, 1 mM EDTA) at 50°C. The labeled oligonucleotide was hybridized to the arrays for 15 h at 50°C in 10 ml of hybridization buffer. The arrays were washed twice for 15 min each time at 50°C in wash buffer I (40 mM Na2HPO4 [pH 6.8], 5% SDS 0.5% bovine serum albumin, 1 mM EDTA), with a final 15-min wash in wash buffer II (0.1× SSC, 0.1% SDS). Bound probe was detected by using a phosphor screen (Molecular Dynamics), and the signals were quantitated by using ImageQuant software (Molecular Dynamics). To determine the viral gene expression level, cells or mice were infected as described above, and the total RNA was harvested by using Tri-Reagent (Molecular Research Center, Inc., Cincinnati, Ohio) according to the manufacturer's protocol. Total cellular RNA (2 μg) was used in each reverse transcription (RT) reaction. The labeled cDNA probe synthesis was prepared by using a Strip-EZ RT kit (Ambion, Austin, Tex.) with [α-32P]dATP (Perkin-Elmer). cDNA was separated from unincorporated nucleotides by using a G25 spin column. The labeled probe was denatured at 95°C. The prehybridization, hybridization, and wash steps were carried out as described above except at a temperature of 65°C. The signal was quantitated as described above.
Data processing.
Local background for each array element was subtracted, and the mean phosphorimager (PI) signal from the two median spots was calculated, yielding one datum point for each viral gene. The PI units were expressed as a percentage of the average PI units of the housekeeping gene GAPDH. For the analysis of the temporal changes in gene expression patterns we controlled for differences in the amount of RNA by sorting datum points based on relative intensity and rank values assigned to each datum point. The ranked data set was then imported into the program Cluster (9). The images were generated by deselecting the cluster gene and cluster array options in the Cluster program (2). The results were visualized with the software TreeView (9). The resistance to CHX and PAA treatment was quantitated by expressing GAPDH-normalized values from the treated array as a percentage of the GAPDH-normalized value in the untreated array. Changes in gene expression were quantified by using a paired t test on four replicate spots for each ORF in untreated samples versus those treated with CHX or PAA. The Spearman rank correlation coefficient was used to assess the relationship between the magnitude of genes expressed in cell culture and in the lung of infected mice at the peak of viral gene expression.
Dual luciferase assay.
Two reporter plasmids were constructed by inserting two regions of the M3 promoter (1,200 and 600 bp upstream of the M3 TATA box) into the pGL3-Basic plasmid (Promega) containing the firefly luciferase coding sequence. The reporter plasmids were cotransfected into BHK-21, 293T, and NIH 3T3 cells with either pCMV-FLAG/RTA or pCMV-FLAG (Kodak). pRLSV40, which contains the coding sequence for Renilla luciferase under the control of a constitutively active human simian virus 40 (SV40) promoter, was included in each transfection and served as an internal control for transfection efficiency. At 24 h posttransfection, cell lysates were assayed for both firefly and Renilla luciferase activity. The dual luciferase reporter assay system (Promega) was used to test promoter activity. Fold activation was calculated by comparing the normalized firefly luciferase activity of pCMV-FLAG/RTA-transfected cells to that of pCMV-FLAG-transfected cells. Transfected BHK-21, 293T, and NIH 3T3 cells in a 12-well plate were washed with 1× phosphate-buffered saline and incubated with 100 μl of 1× passive lysis buffer provided by the manufacturer. Lysates were frozen and thawed three times and then centrifuged at top speed in a microcentrifuge for 5 min. Supernatants were diluted to 1/100 to obtain a reading in a linear range and assayed by using an Optocomp I Luminometer (MGM Instruments, Hamden, Conn.). The assays were carried out according to the manufacturer's protocol for the dual luciferase reporter assay system (Promega).
RESULTS
MHV-68 membrane array.
To characterize the gene expression patterns of MHV-68, custom DNA arrays were constructed representing nearly all of the known and predicted MHV-68 ORFs (52). PCR primers were designed based on published sequence data to amplify 83 regions of the viral genome representing 73 ORFs (Table 1). Seven MHV-68 ORFs are not represented on the arrays. Six of these (M10a, M10b, M10c, M12, M13, and M14) fall partially or completely within the G+C-rich 100-bp repeat or the G+C-rich terminal repeat regions of the genome, and it is not clear whether these putative ORFs indeed encode proteins. In addition, the short sequence representing ORF 38 partially overlaps with ORF 37 and is in extremely close proximity to ORF 39, making it problematic to clone while avoiding the neighboring genes. Of the 73 remaining ORFs, 10 (ORFs 6, 8, 9, 25, 44, ORF 56, 64, 75a, 75b, and 75c) are relatively long sequences and are therefore represented by two array elements: one near the 5′ end and one near the 3′ end of the gene.
TABLE 1.
MHV-68 primers used for array element synthesis
| Array element | Forward primer (5′) sequence | Reverse primer (3′) sequence | Location (nt positions) | Product size (bp) |
|---|---|---|---|---|
| vtRNA | ACCATTCGATGCAAATG | CTACACATGAAAATCCTGTGAG | 53-1891 | 1,833 |
| M1 | CATGCAGCTGGCCACCTTA | CCACGCAGTATTGTAGCGG | 2022-3028 | 1,005 |
| M2 | ATGAGGTTTCGTTTTCAGGT | AGCACCTTCACTGTTACTCC | 4025-4627 | 602 |
| M3 | CAGCCATGGCCTTCCTATCC | GAGTATCAATGATCCCCAAA | 6051-7282 | 1,231 |
| M4 | TGGGCACCCAGCCTAGATT | AGTGGCCACACACAACCGT | 8413-9611 | 1,198 |
| ORF 4 | GTCAAAAGTGGCACCACCC | GGTGGTACCGTCTGAGTGA | 9993-10918 | 925 |
| ORF 6-5′ | GGACCCGCTGGTTACATCT | CATGCAGGAATCTGGCACG | 11251-12378 | 1,127 |
| ORF 6-3′ | CTGTCTGCCAGCTTCGTGA | GGAGCAGCAGTGAACTGAG | 13381-14480 | 1,099 |
| ORF 7 | CCTCAGCCTGGTACCATAC | ACTCTGTAGGGAGGCGAAC | 14579-15614 | 1,035 |
| ORF 8-5′ | TCTGCTGTGCCACACGCAT | TCTGGTCACTGACGCAGCT | 16567-17609 | 1,042 |
| ORF 8-3′ | CAGAGACACCCTCATGTGG | GGCCACTAGAGGGAAGTTG | 17962-19060 | 1,098 |
| ORF 9-5′ | GTTCAGATTCCAGGCGGAC | CAGTAGGTCCAGGACCAAC | 19561-20719 | 1,158 |
| ORF 9-3′ | GTAGTGGACTTTGCCAGCC | GGTGCTCTGCCAAGTCAGA | 20951-22070 | 1,119 |
| ORF 10 | CTCTGGACCTACCACTGAC | AGTATCTGTCGCCTCGTGG | 22390-23384 | 994 |
| ORF 11 | GCCAACTATGCCTGGAAGC | ACTCAGCTCACCACCTGGA | 23551-24618 | 1,067 |
| K3 | TTGGATCTGCCACCAGCCA | CACCCCAGTCTACAACAGG | 24752-25312 | 560 |
| M5 | TTCTAGCCTGCTCATTGGCC | GGGATTTCCAGGTAGAGGG | 26290-26500 | 210 |
| ORF 17 | GATCATGACAGGCTCCTGC | TTCCTGCCCTCGCTTCATC | 28832-29879 | 1,047 |
| ORF 18 | AATGTCTCCTGGCCTGCAG | ATCTGGTGGGAGGGAAAGG | 29988-30669 | 681 |
| ORF 19 | GCATTCTGCGGCTTTGACC | TGGAAAGAGGGCGGACATG | 31041-32094 | 1,053 |
| ORF 20 | AGGAGCCAAGCAACTGCTC | ATGGTGCCTCCTGCTTCCA | 32272-32869 | 597 |
| ORF 21 | CTATGCAGAGCCAGAGGAG | AAACTCAGCAGCAGCCTCC | 32941-33926 | 985 |
| ORF 22 | TGTTTGTCTGCCCGGTTGG | CGTGAGCTGACTAGTCTGC | 35164-36410 | 1,246 |
| ORF 23 | GTCATCAAGGCCCCATCCT | GCGCATATTTGCGCAGCAC | 37081-38089 | 1,008 |
| ORF 24 | GAAGACTGCTCCTGGGCAT | GGCAGCCCATAGCAAAGAG | 38792-40069 | 1,277 |
| ORF 25-5′ | GAATCTGCAGCAGAGGGAC | GTCCCTCTGCTGCAGATTC | 40335-41490 | 1,155 |
| ORF 25-3′ | TCCTTTTGGCCAGCGACCT | CCACACATCCTGTTCGTCC | 42841-43869 | 1,028 |
| ORF 26 | ATCATGGCATCCAACAGGAA | GAGCTAGAGCCAACAGGTTCCT | 44420-45325 | 905 |
| ORF 27 | GGACCCTACCACTGTTCTG | TAAACATCCCTGGCTGCCG | 45392-45924 | 532 |
| ORF 29b | GCAATCACTGCCTGCCCTT | GGTCACTAGACAGCAGGAC | 46571-47279 | 708 |
| ORF 30 | AAATGTCTGCCCCCGGATG | TTGGCCGCGCTTGCATCTT | 47505-47736 | 231 |
| ORF 31 | GCGGCCTCTTTCTAACCCT | CAGATCGACCAGGAGCTCA | 47751-48359 | 608 |
| ORF 32 | TGAGCTCCTGGTCGATCTG | GGAGAAAGTCCCTCGTGAC | 48341-49420 | 1,079 |
| ORF 33 | CTTTACATGCACCACGTCC | GGTCAGATAGCCTCACGAG | 49677-50509 | 332 |
| ORF 29a | CCGCAGCACAGTTTCCAGT | GTGGCACACATCAAGGTGC | 50577-51425 | 348 |
| ORF 34 | CCGAAGCAGATCCTGTGCT | AATGCTGTCGTGGGTGGCA | 51490-52370 | 380 |
| ORF 35 | GACACGAAACTGCTGGCCA | CCATCGAGCCTCCAACAAG | 52426-52850 | 424 |
| ORF 36 | AGGATGGAGAGGACACTGC | TGGAGTGAACTGCTGCCGT | 52881-54046 | 1,165 |
| ORF 37 | GTTCTGGGAAACATGGCCC | AGGTACTCGTGCAGCACAG | 54180-55496 | 1,316 |
| ORF 39 | TGCCTGCCCTTAAAGTGCC | TTTGCTCTGGGGGGTTGTG | 55839-56950 | 1,111 |
| ORF 40 | GTGCTTCTGCCGCTGGTAA | AAGTCCTTTGGGCCGGATG | 57026-58686 | 1,660 |
| ORF 42 | CATCCTTCCCAGGCTGATG | CATACATCAACCCTGCGCG | 58899-59599 | 700 |
| ORF 43 | GTTTACCAGCTGGGCCAAC | TGGTGGCAGCACCTCTACT | 59634-61118 | 1,484 |
| ORF 44-5′ | AATCACAGGGACTGCTGGG | GGATGGCTGAGGTCTGAGT | 61488-62029 | 541 |
| ORF 44-3′ | GCTGGACTCGCCTCTTTGT | GTTCACTGCTCCTCATGGG | 62303-63559 | 1,256 |
| ORF 45 | CCCGATCTCTCGACCAATA | TCAATCCAACCATGGCCAG | 63652-64225 | 573 |
| ORF 46 | GCCTGGTAACCATGGACAC | GAGGGTTTGTTGTGCTGCG | 64291-65014 | 723 |
| ORF 47 | CCTGGGGCATAGTTCTTTG | GTGTCCATGGTTACCAGGC | 65013-65580 | 567 |
| ORF 48 | AAACCCGTGAAGGTGGTGG | TTCTGCCCAAGGACCATGG | 65636-66560 | 924 |
| ORF 49 | TCTCTGGAAAGCGTGGTCC | TTCTGCAGCGATGGCCTCT | 66750-67619 | 869 |
| ORF 50 | CAGGCATCCATGTGGGTAC | CAGCAGTGTCCTGGTTTGC | 67951-69339 | 1,388 |
| M7 | GCCGAGACTGTAGAGGGTA | GGTCGTATCCAAAGCAGGG | 69571-70841 | 1,270 |
| ORF 52 | CGATGGCGTCCAAAAAGCC | CTGTTTGAGAGGATGGGCC | 71013-71366 | 353 |
| ORF 53 | AATGCAGGTCCTGGTCGTG | AGGTTGTCTCCAGGGCACT | 71402-71702 | 300 |
| ORF 54 | CTCCTTTGTGCCCAAGCAC | TCTGCAGCTTCGTGCGGAA | 71820-72602 | 782 |
| ORF 55 | TGGGAAACCCGGTGTACCT | GTTTGGGTCAAGTGGGGTG | 72681-73287 | 606 |
| ORF 56-5′ | ACACGAGGGCGCAACTGAA | CAACCGTGTTGCTCGTGCA | 73393-74720 | 1,327 |
| ORF 56-3′ | TGCACGAGCAACACGGTTG | CCTCTACACTCGAGCATGC | 74702-75799 | 1,097 |
| M8 | TCCACCAGTTGAGGAGCCA | TCTGGGCGCAGTTTACCAG | 76040-76456 | 416 |
| ORF 57 | GTGCTGACCCACATGCTAG | CCTTGGTGCGGTGTGTTTG | 76650-77139 | 489 |
| ORF 58 | TCAGACGTGTTCCTCCCAC | TCCAGAAAGCAGCCAGCAG | 77323-78239 | 916 |
| ORF 59 | CTTGACACTGAGCGCCTCT | GTAGCTTGGGGCGCTTAGT | 78275-79424 | 1,149 |
| ORF 60 | ACCACGCTGGGTTCCTTGA | GGGTGGAATGAGAGCACAC | 79519-80445 | 926 |
| ORF 61 | CCAAGAAGGCCGAAGTCCT | GTGCAGACAAGAAGCACCC | 80523-81907 | 1,384 |
| ORF 62 | TGCATGCTGGGACTGTCAG | ACTGCCACGGTCCTCTAGA | 82867-84013 | 1,146 |
| ORF 63 | TGCACCTATGCACCCTGGA | CTTGGCTGGCAGCAGACTT | 84061-85104 | 1,043 |
| ORF 64-5′ | TGGCTGGGTTTCGCATCGA | TGGACATGGTCTGCGAGGT | 86586-87978 | 1,392 |
| ORF 64-3′ | TGCACGTGGAAACCCACCA | GTCATGGGTGACACCAGGA | 92721-93790 | 1,069 |
| ORF 65 (M9) | CAGGGTCAAAGCTCCAGCT | GGTCGATTCCCCCAAGTTC | 93992-94508 | 516 |
| ORF 66 | TCTATGGCCACCTCAGTGG | CTCCCTAAATGTCGCCCTC | 94581-95671 | 1,090 |
| ORF 67 | GACGGACATGGCTAACCAG | CTAGTTTACTGGCCAGCGC | 95743-96422 | 679 |
| ORF 68 | CATTGCCCCCCATGACTCA | AGAGCCTGTTGAGCGCATG | 96662-98078 | 1,416 |
| ORF 69 | CATGCGCTCAACAGGCTCT | AGCAGACATTATCGGCCGG | 98060-98850 | 790 |
| ORF 72 | TACACCTGCGACCTCCATG | TGGCAACGAGGAAAGGGCA | 102401-103197 | 796 |
| M11 | TGTTGTGCGTGCGCAGCTA | TTCGCCAGGGCATGCAACA | 103325-103890 | 565 |
| ORF 73 | CACTTGACCCACACCCTTC | GAGACCCTTGTCCCTGTTG | 103932-104930 | 998 |
| ORF 74 | AGCCACGATGCTTGTCCTG | CAAACTGCCCTCCTGACAG | 105050-106054 | 1,004 |
| ORF 75c-5′ | CAAGAACTGGTCCGTCTCG | GTCTTGTGTGGAGAGGCGA | 108896-110020 | 1,124 |
| ORF 75c-3′ | TGACGTGAGCGATGGTGGT | TGCCACTGCCAAGGGAAGA | 106152-107369 | 1,217 |
| ORF 75b-5′ | TCCAAGTCTATGCCAGGGC | CTTGGATGGAGCACCTGAC | 112671-113879 | 1,208 |
| ORF 75b-3′ | CAATGACTCCACAGACGCC | CCATCGGTGGATAGTGTCC | 110081-111129 | 1,048 |
| ORF 75a-5′ | CTCCGAGGTGGATGACAGA | TAACGATGGCTCCAGGACC | 116638-117740 | 1,102 |
| ORF 75a-3′ | CTTGGCCTCGCATGTACCA | TGCTGCCAGTGCCCATAAG | 114041-115420 | 1,379 |
Cloned array elements were spotted onto nylon membranes in quadruplicate. As a hybridization control, the mouse GAPDH gene was also spotted in quadruplicate in each of the four corners of the arrays. To test for spotting consistency between the quadruplicate spots, arrays were probed with a 32P-labeled common primer that was used to amplify the array elements. Quantitation of these signals showed the average variation between spots representing individual array elements to be ±5%. To eliminate small variations introduced by spotting inconsistency, all calculations were performed with an average of the two median spot values for each array element. To test the specificity of the arrays, cDNA probe was generated from total RNA from uninfected BHK-21 cells and hybridized to the DNA arrays. cDNA probes generated from uninfected BHK-21 cells did not hybridize to any MHV-68 ORFs but did hybridize to the mouse GAPDH probe (data not shown).
MHV-68 gene expression during latency.
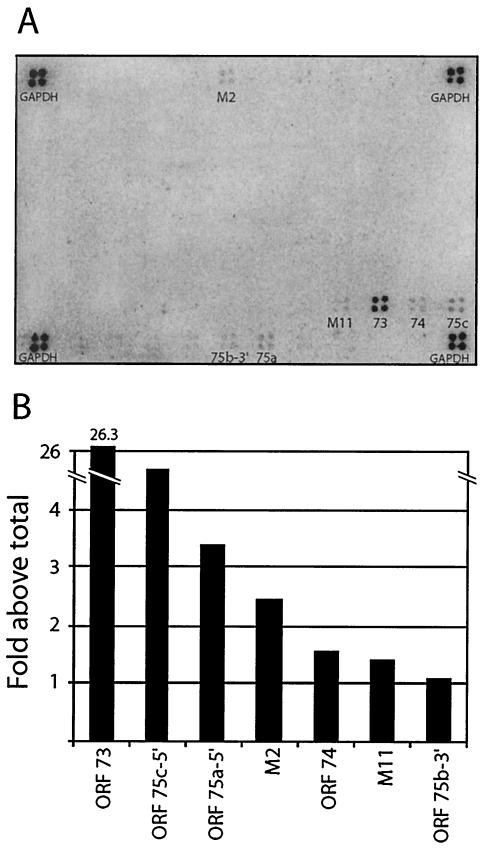
In addition to establishing and maintaining latency, latent viral proteins of gammaherpesviruses may play important roles in cell immortalization and transformation (49). Latent MHV-68 gene expression has previously been investigated by multiple groups (15, 41, 53). These studies utilized techniques such as Northern blot analysis, in situ hybridization, and real-time PCR in an attempt to identify latently associated genes. We used DNA arrays to simultaneously and quantitatively assess the relative levels of viral gene expression across the entire genome in a MHV-68 latently infected cell line (S11E). S11 is a cell line derived from a B-cell lymphoma in an MHV-68-infected mouse (50). The tightly latent S11E cell line was subcloned from S11 on the basis of a low spontaneous reactivation frequency (15). cDNA probe was generated from poly(A)-selected RNA from S11E cells and hybridized to membrane arrays (Fig. 1A). Consistent with previous reports, transcripts from M2, M11, ORF 73, and ORF 74 were detected in the latently infected cells (15, 53). In addition to the candidate latency genes, we also detected expression of regions of ORFs 75a, 75b, and 75c. To quantitate the levels of expression of the individual transcripts relative to one another, the signals for the detectable array elements were normalized to the total signal on the array. The results are shown graphically (Fig. 1B). The LANA homologue, ORF 73, was the most abundant transcript, being expressed ∼26-fold above the average array PI units. LANA is a well-established latent gene of KSHV and is thought to play an important role during latent viral persistence (12, 13, 17). These results confirm the expression of candidate latent genes (ORFs M2, M11, 73, and 74) and also identify potential new regions of the genome (ORFs 75a-5′, 75b-3′, and 75c-5′) that are transcriptionally active during latency.
FIG. 1.
Latent gene expression profile in S11E cells. (A) MHV-68 membrane arrays were probed with 32P-labeled cDNA generated by RT of poly(A)-selected RNA isolated from latently infected S11E cells. (B) Intensity of signal as a percentage of the average signal across the entire array. The experiment was repeated, and similar results were obtained.
Gene expression during productive replication of MHV-68 infection in vitro.
Many cell lines are highly permissive for MHV-68 replication, providing a system for examination of viral gene expression during de novo infection. To examine the temporal pattern of MHV-68 gene expression, we infected BHK-21 cells at 5 PFU/cell and harvested RNA at 4, 8, 12, 24, and 32 h postinfection (hpi) for array analysis. The robust nature of MHV-68 replication in cultured cells was reflected by abundant gene expression of most viral genes. RNA expression levels are shown in a tabular form (Fig. 2), as described by Chu et al. (2), with rows corresponding to the individual genes and columns corresponding to the successive time points at which RNA was harvested during de novo infection. RNA expression levels are represented in the matrix by a color scale based on numerical values quantitated from the DNA arrays, with increases in expression represented as graded shades of red. The genes were then sorted based on the overall abundance of each transcript over the course of the experiment and displayed in order of ascending abundance (Fig. 2A). There was an initial lag in detectable gene expression; however, by 8 hpi, expression from 100% of the array elements was detectable, including those corresponding to latency genes. Consistent with our current knowledge that herpesviruses express structural genes to high levels, 11 of the most abundant transcripts encode capsid proteins or glycoproteins. Of the 40 least-abundant transcripts, 12 encode proteins that function in nucleotide metabolism or DNA replication, a finding consistent with a lower level of expression of enzymes. All candidate latency genes (15, 53) were expressed during MHV-68 lytic replication in BHK-21 cells.
FIG.2.
Lytic gene expression profile. RNA samples from a time course postinfection of BHK-21 cells were analyzed by membrane array analysis. The data are graphically displayed with color to represent the quantitative changes in RNA abundance. Increases of RNA are shown as deeper shades of red. ORFs are color coded based on functional groups, with purple corresponding to nucleotide synthesis and DNA replication proteins, green corresponding to structural proteins, red corresponding to assembly proteins, blue corresponding to homologues of cellular signaling proteins, and black corresponding to unknown or other proteins. (A) The genes were sorted based on the overall abundance level throughout the course of the experiment. (B) The genes were ordered based on the time at which peak expression is reached and then sorted based on the relative abundance level at the peak of expression. The data are graphically displayed with color to represent the quantitative changes in RNA abundance. Increases of RNA are shown as deeper shades of red. M9, ORF 65.
Different viral genes reached peak expression levels at different time points. To determine whether the timing of expression was consistent with the function of each gene in the virus replication cycle, we grouped the genes based on the time at which peak expression is reached and then sorted the genes based on the relative abundance levels at the peak of expression (Fig. 2B). ORF 50 (RTA) and ORF 57, both important regulators of the gene expression cascade, peaked early at 8 hpi. Similarly, most ORFs involved in DNA replication (52), including ORF 6 (single-stranded DNA [ssDNA]-binding protein), ORF 9 (DNA polymerase), ORF 21 (thymidine kinase), and ORF 54 (dUTPase), ORF 59 (processivity factor), as well as ORFs 60 and 61 (small and large subunits of ribonucleotide reductase), peaked in expression at 8 to 12 hpi, indicating preparation for viral DNA replication. Interestingly, expression of the transcripts encoding components of the helicase-primase complex (ORFs 40, 44, and 56) peaked at later time points (24 to 32 hpi).
In general, genes encoding structural proteins and DNA packaging proteins peaked at later times postinfection. Most transcripts encoding structural proteins—including ORFs 8, 22, 39, and M7 (glycoproteins); ORFs 17, 25, 26, 43, and 65 (capsid proteins); and ORFs 63, 64, and 75a, 75b, and 75c (tegument proteins)—peaked at 12 to 24 hpi. However, one glycoprotein (ORF 47) and one tegument protein (ORF 19) peaked relatively early at 8 hpi and remained high. ORFs involved in DNA processing, although expressed early, also peaked later or continued to increase throughout the course of the infection. ORF 37 (alkaline exonuclease), ORF 62 (assembly protein), ORF 46 (uracil DNA glycosylase), and ORFs 29b, 67, and 68 (packaging proteins) all peaked in expression at 24 to 32 hpi, a finding consistent with their putative roles during the later phases of the replication process (35, 39). These arrays have shown that immediate-early proteins and proteins involved in DNA replication reach peak levels of expression first, followed by structural proteins and finally packaging proteins, a finding consistent with a classical virus gene expression cascade.
Sensitivity of MHV-68 transcription to inhibition of protein synthesis during de novo infection.
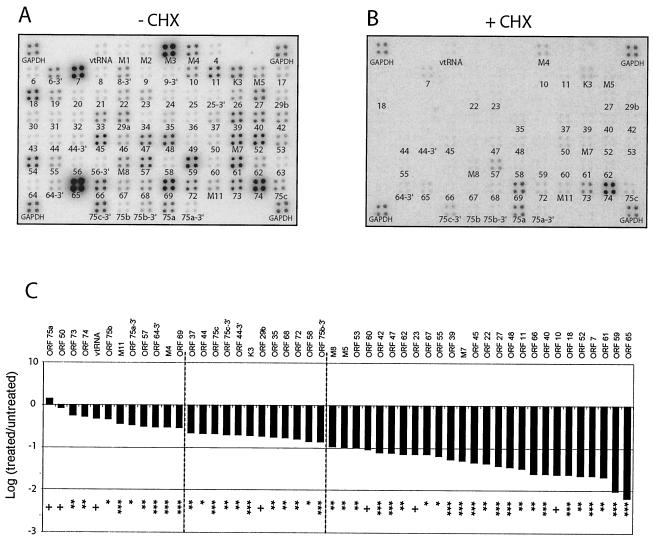
Herpesvirus immediate-early or alpha genes are defined as those that are expressed in the presence of CHX, a de novo protein synthesis inhibitor (35). To identify immediate-early transcripts of MHV-68, BHK-21 cells were infected at 5 PFU/cell in the absence (Fig. 3A) or presence (Fig. 3B) of CHX and RNA was harvested at 8 hpi for membrane array analysis. Expression levels from multiple regions of the genome were noticeably decreased compared to those from untreated cells. In the presence of CHX, the levels of classic early transcripts (ORFs 6, 9, 21, 40, 44, 56, 59, 60, and 61) and classic late transcripts (ORFs 8, 17, 22, 25, 26, 39, 43, 47, M7, and 65) were severely reduced or completely undetectable. In contrast, the expression level of the classic immediate-early transcript (ORF 50) was not affected by CHX treatment (Fig. 3B) (19, 55). Nevertheless, transcription from 50 regions of the genome remained detectable above background levels, although limited, in the presence of CHX. Among the regions of the genome that were transcriptionally active in CHX, five encode glycoproteins (ORFs 22, 39, 47, M7, and 68), five encode tegument proteins (ORFs 64, 67, and 75a, 75b, and 75c), two encode capsid proteins (ORFs 62 and 65), two encode DNA packaging proteins (ORFs 29b and 37), and 18 have no currently recognized function. The maximal reduction was >100-fold for ORF 65 (M9), indicating that drug inhibition was functional. To verify that CHX was effective under these conditions, a similar experiment was carried out by using twice the amount of CHX and similar results were obtained (data not show).
FIG. 3.
MHV-68 gene expression in the absence of de novo protein synthesis. MHV-68 membrane arrays were hybridized with oligo(dT)-primed cDNA synthesized from BHK-21 cells at 8 hpi with MHV-68 at 5 PFU/cell in the absence (A) or presence (B) of CHX. (C) Fold expression of observed transcripts in treated versus untreated samples. A Storm phosphorimager and the ImageQuant system were used to quantitate the signal from the array elements corresponding to 73 known and predicted MHV-68 ORFs. GAPDH-normalized values from the +CHX array (B) were divided by the corresponding GAPDH-normalized values from the untreated (−CHX) array (A) to derive the fold inhibition of gene expression relative to the untreated level for each array element. These values and their corresponding MHV-68 ORFs are ordered in the bar graph based on increasing fold inhibition of gene expression relative to the untreated level. Statistical significance of differences in expression is assessed by paired t test (+, P > 0.05; ✽, P < 0.05; ✽✽, P < 0.01; ✽✽✽, P < 0.001). Dashed lines were placed at the 0.5-log reduction and the 1-log reduction of expression in the presence of CHX.
To explore the notion that contamination from the viral inoculum contributed to the detection of 50 regions of the genome in the presence of CHX, we performed an additional experiment examining viral gene expression at earlier times postinfection. We infected BHK-21 cells at a multiplicity of infection of 5 and harvested RNA at 2 and 4 h postinfection for array analysis (data not shown). No viral transcripts were detectable at 2 hpi and only a few were detectable at 4 hpi. These results confirm that levels of viral RNA from the inoculum, if there is any, are below our detection limit and therefore could not have contributed to the observations in the CHX experiment.
To quantitate the levels of expression of the individual transcripts, we calculated the average of the PI counts for each array element with detectable levels above background. The PI units were normalized to the housekeeping gene GAPDH (Table 2). We assessed the level of resistance of each transcript to CHX by expressing the GAPDH-normalized value from the treated cells as a fraction of the GAPDH-normalized value in the untreated cells. These data are shown graphically to compare each ORF's level of resistance to CHX (Fig. 3C). The results showed that the previously described immediate-early gene ORF 50 (RTA) and those genes expressed during latency (ORFs M11, 73, and 74 and regions of 75a, 75b, and 75c) were weakly affected by CHX (<0.5 log). Moreover, a statistical analyses of the CHX data revealed that, although several regions of the genome were expressed to detectable levels in the presence of CHX, only two regions (ORFs 50 and 75a) were not affected by statistically significant amounts (P > 0.05).
TABLE 2.
Transcript abundance during MHV-68 infection
| Probe | Function | Transcript abundancea |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Normalized data at: |
Rank at: |
Normalized data |
Normalized data |
||||||||||||||
| 4 h | 8 h | 12 h | 24 h | 32 h | Lung 5d | 4 h | 8 h | 12 h | 24 h | 32 h | Lung 5d | +CHX | −CHX | +PAA | −PAA | ||
| vtRNA | vtRNA | ND | 0.52 | 1.47 | 1.47 | 3.98 | 0.35 | 50 | 58 | 59 | 60 | 5 | 0.07 | 0.13 | 0.87 | 4.40 | |
| M1 | Serpin | ND | 0.15 | 0.36 | 0.33 | 0.70 | 0.59 | 6 | 13 | 15 | 9 | 15 | ND | 0.07 | 0.99 | 1.21 | |
| M2 | Unknown | ND | 0.30 | 0.57 | 0.45 | 1.38 | 1.32 | 27 | 30 | 21 | 31 | 49 | ND | 0.12 | 2.23 | 2.24 | |
| M3 | Chemokine binding protein | 0.18 | 5.53 | 11.73 | 10.79 | 16.29 | 13.71 | 5 | 83 | 80 | 79 | 75 | 81 | ND | 10.79 | 68.30 | 44.42 |
| M4 | Unknown | ND | 0.53 | 0.96 | 0.53 | 1.43 | 0.74 | 53 | 50 | 28 | 33 | 22 | 0.32 | 1.06 | 11.26 | 1.81 | |
| ORF 4 | Complement regulatory protein | ND | 0.29 | 0.75 | 0.82 | 1.27 | 1.01 | 26 | 40 | 45 | 29 | 40 | ND | 0.10 | 0.81 | 1.09 | |
| ORF 6-5′ | ssDNA-binding protein | ND | 0.40 | 0.49 | 0.60 | 1.18 | 0.58 | 40 | 25 | 31 | 26 | 14 | ND | 0.14 | 0.78 | 1.24 | |
| ORF 6-3′ | ssDNA-binding protein | ND | 0.38 | 0.63 | 0.51 | 0.95 | 0.54 | 36 | 34 | 26 | 20 | 11 | ND | 0.74 | 2.17 | 2.29 | |
| ORF 7 | Transport protein | 0.14 | 2.89 | 7.01 | 5.65 | 16.53 | 8.43 | 4 | 76 | 77 | 75 | 76 | 78 | 0.16 | 7.10 | 6.92 | 9.96 |
| ORF 8-5′ | Glycoprotein B | ND | 0.26 | 0.77 | 0.64 | 1.54 | 0.69 | 19 | 43 | 35 | 35 | 21 | ND | 0.10 | 1.21 | 1.43 | |
| ORF 8-3′ | Glycoprotein B | ND | 0.71 | 1.95 | 1.37 | 3.10 | 1.76 | 61 | 63 | 58 | 57 | 60 | ND | 0.56 | 2.54 | 3.97 | |
| ORF 9-5′ | DNA polymerase | ND | 0.13 | 0.33 | 0.16 | 0.50 | 0.62 | 4 | 11 | 1 | 3 | 17 | ND | 0.06 | 0.87 | 1.16 | |
| ORF 9-3′ | DNA polymerase | ND | 0.06 | 0.32 | 0.27 | 0.48 | 0.56 | 1 | 8 | 7 | 2 | 13 | ND | 0.06 | 1.09 | 1.14 | |
| ORF 10 | Unknown | ND | 0.57 | 1.06 | 0.48 | 1.20 | 0.85 | 54 | 51 | 23 | 27 | 31 | 0.01 | 0.59 | 2.90 | 2.31 | |
| ORF 11 | Unknown | ND | 1.32 | 3.28 | 1.97 | 3.05 | 1.68 | 72 | 70 | 63 | 56 | 57 | 0.04 | 1.27 | 3.86 | 5.19 | |
| K3 | Modulator of immune response | ND | 1.42 | 4.59 | 5.81 | 8.97 | 4.91 | 74 | 73 | 76 | 71 | 73 | 0.21 | 1.08 | 6.96 | 11.64 | |
| M5 | Unknown | ND | 0.65 | 2.17 | 3.45 | 5.90 | 2.78 | 58 | 64 | 69 | 67 | 66 | 0.05 | 0.45 | 5.76 | 11.33 | |
| ORF 17 | Capsid protein | ND | 0.45 | 1.56 | 1.90 | 2.13 | 0.84 | 42 | 59 | 62 | 44 | 30 | ND | 0.28 | 2.85 | 1.65 | |
| ORF 18 | Unknown | ND | 1.10 | 1.67 | 1.33 | 1.62 | 1.03 | 68 | 61 | 57 | 39 | 41 | 0.04 | 1.67 | 30.58 | 5.78 | |
| ORF 19 | Tegument protein | ND | 0.83 | 1.46 | 0.80 | 2.18 | 1.11 | 64 | 57 | 44 | 46 | 46 | ND | 0.46 | 1.23 | 2.38 | |
| ORF 20 | Unknown | ND | 0.29 | 0.45 | 0.27 | 1.00 | 1.10 | 25 | 20 | 10 | 22 | 44 | ND | 0.13 | 1.54 | 1.93 | |
| ORF 21 | Thymidine kinase | ND | 0.29 | 0.43 | 0.22 | 0.79 | 0.55 | 24 | 17 | 4 | 13 | 12 | ND | 0.13 | 1.39 | 1.49 | |
| ORF 22 | Glycoprotein H | ND | 0.48 | 1.41 | 0.48 | 1.96 | 1.63 | 47 | 56 | 22 | 42 | 54 | 0.02 | 0.38 | 3.09 | 2.92 | |
| ORF 23 | Unknown | ND | 0.52 | 1.35 | 0.62 | 2.46 | 1.87 | 51 | 55 | 34 | 51 | 61 | 0.03 | 0.38 | 1.52 | 2.81 | |
| ORF 24 | Unknown | ND | 0.18 | 0.41 | 0.27 | 0.73 | 0.65 | 11 | 16 | 8 | 11 | 18 | ND | 0.08 | 0.94 | 1.35 | |
| ORF 25-5′ | Major capsid protein | ND | 0.13 | 0.23 | 0.31 | 0.58 | 0.32 | 3 | 2 | 13 | 4 | 3 | ND | 0.05 | 0.36 | 0.77 | |
| ORF 25-3′ | Major capsid protein | ND | 0.19 | 0.43 | 0.76 | 0.70 | 0.61 | 13 | 18 | 41 | 8 | 16 | ND | 0.04 | 1.00 | 1.53 | |
| ORF 26 | Capsid protein | ND | 0.84 | 3.21 | 2.96 | 6.17 | 2.54 | 65 | 69 | 67 | 68 | 64 | ND | 0.38 | 3.17 | 6.13 | |
| ORF 27 | Unknown | ND | 0.49 | 2.20 | 5.46 | 6.26 | 2.70 | 48 | 65 | 74 | 69 | 65 | 0.04 | 0.99 | 5.15 | 11.67 | |
| ORF 29b | Packaging protein | ND | 0.32 | 0.69 | 1.10 | 1.88 | 0.95 | 32 | 35 | 55 | 41 | 37 | 0.04 | 0.21 | 2.13 | 3.14 | |
| ORF 30 | Unknown | ND | 0.28 | 0.56 | 0.34 | 1.57 | 0.85 | 21 | 27 | 17 | 38 | 32 | ND | 0.20 | 3.47 | 4.92 | |
| ORF 31 | Unknown | ND | 0.25 | 0.34 | 0.33 | 1.00 | 0.67 | 18 | 12 | 16 | 21 | 20 | ND | 0.15 | 1.59 | 2.28 | |
| ORF 32 | Unknown | ND | 0.23 | 0.32 | 0.32 | 0.79 | 0.76 | 17 | 7 | 14 | 12 | 24 | ND | 0.11 | 0.99 | 1.55 | |
| ORF 33 | Unknown | ND | 0.50 | 0.69 | 0.78 | 2.26 | 0.88 | 49 | 37 | 43 | 48 | 36 | ND | 0.61 | 4.59 | 6.09 | |
| ORF 29a | Packaging protein | ND | 1.31 | 2.21 | 1.09 | 5.43 | 3.09 | 71 | 66 | 54 | 64 | 69 | ND | 0.79 | 4.58 | 7.68 | |
| ORF 34 | Unknown | ND | 0.16 | 0.24 | 0.25 | 0.79 | 0.65 | 8 | 4 | 5 | 14 | 19 | ND | 0.07 | 1.05 | 1.04 | |
| ORF 35 | Unknown | ND | 0.43 | 0.75 | 0.90 | 2.90 | 1.37 | 41 | 41 | 47 | 54 | 51 | 0.04 | 0.24 | 4.25 | 7.03 | |
| ORF 36 | Kinase | ND | 0.16 | 0.33 | 0.40 | 0.81 | 0.52 | 7 | 10 | 19 | 15 | 10 | ND | 0.08 | 0.97 | 1.47 | |
| ORF 37 | Alkaline exonuclease | ND | 0.32 | 0.47 | 0.73 | 0.89 | ND | 31 | 23 | 40 | 17 | 1 | 0.06 | 0.28 | 2.04 | 2.09 | |
| ORF 39 | Glycoprotein M | ND | 1.78 | 7.31 | 11.14 | 18.82 | 7.16 | 75 | 78 | 80 | 77 | 76 | 0.07 | 1.21 | 8.78 | 14.03 | |
| ORF 40 | Helicase-primase | ND | 1.11 | 6.41 | 10.60 | 19.90 | 8.29 | 69 | 75 | 78 | 78 | 77 | 0.02 | 0.98 | 7.48 | 13.93 | |
| ORF 42 | Tegument | ND | 0.52 | 1.21 | 1.78 | 3.02 | 1.05 | 52 | 53 | 61 | 55 | 43 | 0.03 | 0.51 | 4.63 | 4.82 | |
| ORF 43 | Capsid protein | ND | 0.13 | 0.23 | 0.27 | 0.66 | 0.45 | 5 | 3 | 9 | 6 | 6 | ND | 0.07 | 0.80 | 1.27 | |
| ORF 44-5′ | Helicase-primase | ND | 0.22 | 0.46 | 0.65 | 1.57 | 1.13 | 16 | 22 | 36 | 37 | 47 | 0.03 | 0.13 | 1.60 | 2.62 | |
| ORF 44-3′ | Helicase-primase | ND | 0.19 | 0.24 | 0.48 | 0.72 | 0.35 | 12 | 5 | 24 | 10 | 4 | 0.03 | 0.15 | 1.02 | 1.91 | |
| ORF 45 | Unknown | ND | 3.57 | 5.35 | 5.11 | 21.38 | 10.70 | 81 | 74 | 73 | 79 | 79 | 0.07 | 1.63 | 9.53 | 14.02 | |
| ORF 46 | Uracil DNA glycosylase | ND | 0.45 | 0.71 | 0.71 | 2.18 | 0.84 | 44 | 38 | 38 | 45 | 29 | ND | 0.36 | 4.91 | 4.15 | |
| ORF 47 | Glycoprotein L | ND | 0.65 | 0.87 | 0.61 | 2.03 | 1.41 | 59 | 46 | 32 | 43 | 52 | 0.08 | 1.04 | 7.56 | 2.63 | |
| ORF 48 | Unknown | ND | 0.37 | 0.53 | 0.67 | 1.43 | 0.77 | 35 | 26 | 37 | 34 | 25 | 0.05 | 1.38 | 13.58 | 3.30 | |
| ORF 49 | Unknown | ND | 0.16 | 0.27 | 0.39 | 0.65 | 0.46 | 9 | 6 | 18 | 5 | 7 | ND | 0.10 | 1.59 | 1.42 | |
| ORF 50 | Transcriptional activator | ND | 0.20 | 0.37 | 0.27 | 1.03 | 0.50 | 14 | 14 | 11 | 23 | 8 | 0.16 | 0.19 | 1.77 | 1.34 | |
| M7 | Glycoprotein 150 | ND | 3.10 | 12.17 | 13.72 | 29.93 | 11.63 | 78 | 81 | 81 | 81 | 80 | 0.09 | 1.73 | 11.63 | 19.40 | |
| ORF 52 | Unknown | ND | 3.19 | 15.60 | 26.93 | 53.79 | 17.20 | 79 | 83 | 82 | 82 | 82 | 0.05 | 1.97 | 14.46 | 21.60 | |
| ORF 53 | Unknown | ND | 0.48 | 1.64 | 2.47 | 5.54 | 0.88 | 46 | 60 | 64 | 65 | 35 | 0.05 | 0.44 | 4.88 | 5.23 | |
| ORF 54 | dUTPase | 0.13 | 0.45 | 0.69 | 0.20 | 0.95 | 1.04 | 3 | 43 | 36 | 3 | 18 | 42 | ND | 1.48 | 3.48 | 2.75 |
| ORF 55 | Unknown | ND | 0.36 | 0.57 | 0.98 | 1.64 | 1.42 | 34 | 28 | 49 | 40 | 53 | 0.02 | 0.38 | 1.86 | 3.00 | |
| ORF 56-5′ | Helicase-primase | ND | 0.08 | 0.09 | 0.16 | 0.30 | 0.30 | 2 | 1 | 2 | 1 | 2 | ND | 0.05 | 0.38 | 0.93 | |
| ORF 56-3′ | Helicase-primase | ND | 0.17 | 0.32 | 0.28 | 0.87 | 0.84 | 10 | 9 | 12 | 16 | 28 | ND | 0.08 | 1.06 | 1.43 | |
| M8 | Unknown | ND | 0.72 | 0.94 | 0.57 | 2.61 | 2.79 | 62 | 48 | 30 | 52 | 67 | 0.07 | 0.63 | 5.74 | 6.17 | |
| ORF 57 | Posttranscriptional regulator | 0.12 | 0.78 | 0.94 | 0.61 | 3.49 | 1.75 | 2 | 63 | 49 | 33 | 58 | 59 | 0.44 | 1.39 | 5.45 | 3.87 |
| ORF 58 | Unknown | ND | 0.30 | 0.46 | 0.78 | 1.54 | 0.99 | 29 | 21 | 42 | 36 | 39 | 0.02 | 0.11 | 2.16 | 2.84 | |
| ORF 59 | Processivity factor | ND | 0.69 | 0.74 | 0.85 | 1.12 | 0.87 | 60 | 39 | 46 | 25 | 34 | 0.03 | 2.67 | 13.79 | 2.61 | |
| ORF 60 | Ribonucleotide reductase (small) | ND | 0.30 | 0.47 | 0.54 | 1.36 | 0.96 | 28 | 24 | 29 | 30 | 38 | 0.01 | 0.10 | 1.72 | 1.81 | |
| ORF 61 | Ribonucleotide reductase (large) | 0.28 | 3.23 | 6.48 | 5.05 | 16.07 | 6.50 | 8 | 80 | 76 | 72 | 74 | 74 | 0.05 | 2.33 | 5.26 | 9.63 |
| ORF 62 | Assembly/DNA maturation | ND | 0.63 | 2.43 | 3.41 | 7.41 | 3.13 | 57 | 67 | 68 | 70 | 70 | 0.03 | 0.40 | 2.42 | 5.75/PICK> | |
| ORF 63 | Tegument protein | ND | 0.34 | 0.90 | 1.04 | 2.32 | 0.87 | 33 | 47 | 52 | 49 | 33 | ND | 0.09 | 1.07 | 1.26 | |
| ORF 64-5′ | Tegument protein | 0.38 | 0.30 | 0.59 | 0.49 | 0.69 | 1.10 | 10 | 30 | 32 | 25 | 7 | 45 | ND | 0.07 | 1.19 | 1.42 |
| ORF 64-3′ | Tegument protein | ND | 0.27 | 0.63 | 1.01 | 1.41 | 1.16 | 20 | 33 | 50 | 32 | 48 | ND | 0.29 | 1.75 | 3.64 | |
| ORF 65 (M9) | Capsid protein | ND | 5.14 | 13.96 | 28.05 | 64.10 | 22.55 | 82 | 82 | 83 | 83 | 83 | 0.15 | 22.19 | 154.13 | 268.20 | |
| ORF 66 | Unknown | ND | 0.85 | 1.92 | 4.48 | 13.83 | 2.21 | 66 | 62 | 70 | 73 | 63 | 0.03 | 1.29 | 16.44 | 26.19 | |
| ORF 67 | Tegument protein | ND | 0.39 | 0.79 | 0.98 | 4.45 | 1.67 | 38 | 44 | 48 | 62 | 56 | 0.03 | 0.40 | 2.50 | 6.23 | |
| ORF 68 | Glycoprotein | ND | 0.57 | 1.24 | 1.09 | 4.41 | 2.91 | 55 | 54 | 53 | 61 | 68 | 0.09 | 0.54 | 4.21 | 4.02 | |
| ORF 69 | Unknown | ND | 1.27 | 3.82 | 2.73 | 4.70 | 1.68 | 70 | 71 | 65 | 63 | 58 | 1.24 | 4.18 | 174.18 | 18.02 | |
| ORF 72 | v-Cyclin | ND | 0.39 | 0.76 | 0.73 | 2.74 | 1.33 | 39 | 42 | 39 | 53 | 50 | 0.15 | 0.94 | 121.95 | 14.89 | |
| M11 | vBCL-2 | ND | 0.28 | 0.58 | 0.42 | 2.40 | 2.13 | 22 | 31 | 20 | 50 | 62 | 0.14 | 0.39 | 52.88 | 6.43 | |
| ORF 73 | HHV-8 LANA homologue | 0.24 | 1.38 | 4.55 | 4.57 | 12.26 | 4.35 | 7 | 73 | 72 | 71 | 72 | 72 | 0.66 | 1.18 | 22.83 | 17.29 |
| ORF 74 | vIL8 receptor | 0.35 | 2.98 | 8.70 | 10.46 | 29.76 | 6.69 | 9 | 77 | 79 | 77 | 80 | 75 | 4.01 | 7.47 | 26.61 | 67.56 |
| ORF 75c-5′ | Tegument protein/FGARAT | 0.22 | 0.85 | 2.49 | 2.90 | 5.68 | 3.82 | 6 | 67 | 68 | 66 | 66 | 71 | 0.12 | 0.57 | 2.60 | 2.78 |
| ORF 75c-3′ | Tegument protein/FGARAT | ND | 0.61 | 1.11 | 1.78 | 3.55 | 0.84 | 56 | 52 | 60 | 59 | 27 | 0.25 | 1.22 | 8.47 | 12.81 | |
| ORF 75b-5′ | Tegument protein/FGARAT | ND | 0.28 | 0.43 | 0.26 | 0.95 | 0.74 | 23 | 19 | 6 | 19 | 23 | 0.05 | 0.11 | 0.74 | 1.03 | |
| ORF 75b-3′ | Tegument protein/FGARAT | 0.11 | 0.38 | 0.57 | 1.03 | 1.24 | 0.52 | 1 | 37 | 29 | 51 | 28 | 9 | 0.05 | 0.34 | 0.78 | 0.89 |
| ORF 75a-5′ | Tegument protein/FGARAT | 0.40 | 0.46 | 0.83 | 1.18 | 2.21 | 1.63 | 11 | 45 | 45 | 56 | 47 | 55 | 1.00 | 0.71 | 5.88 | 4.17 |
| ORF 75a-3′ | Tegument protein/FGARAT | ND | 0.21 | 0.40 | 0.52 | 1.09 | 0.82 | 15 | 15 | 27 | 24 | 26 | 0.04 | 0.13 | 1.31 | 1.52 | |
ND, not determined; Lung 5d, GAPDH-normalized PI units from the lung of infected mice at 5 dpi; +CHX and −CHX, with and without added CHX, respectively; +PAA and −PAA, with and without added PAA, respectively.
Sensitivity of MHV-68 transcription to inhibition of DNA replication during de novo infection.
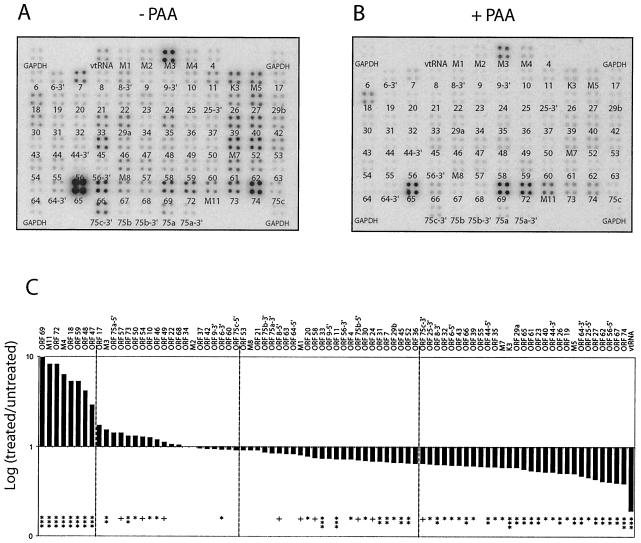
Herpesvirus early or beta genes are classified by the dependence of their expression on viral protein synthesis, and late or gamma genes are classified by the dependence of their expression on viral DNA replication. Therefore, inhibition of viral DNA synthesis with PAA, an inhibitor of viral DNA polymerase (29), blocks the expression of late genes, allowing them to be distinguished from immediate-early and E transcripts. MHV-68 transcript levels were examined by DNA array analysis in the presence of PAA. BHK-21 cells were infected at 5 PFU/cell in the absence (Fig. 4A) and presence (Fig. 4B) of PAA and total RNA was harvested at 24 h p.i. The expression of many genes was strongly inhibited by PAA. The levels of classic late transcripts (ORFs 25, 26, 39, 43, M7, 62, and 65) were severely reduced in the presence of PAA. In contrast, the expression levels of classic early transcripts (ORFs 6, 9, 54, and 59) were relatively unaffected by PAA treatment or detected at higher levels compared to the untreated array. Nevertheless, we observed that many MHV-68 ORFs were expressed to detectable levels by 24 hpi regardless of PAA treatment. To quantitate the levels of expression of the individual transcripts, we calculated the GAPDH-normalized value for each transcript (Table 2). We assessed the level of resistance of each transcript to PAA by expressing the GAPDH-normalized value from the treated cells as a fraction of the GAPDH-normalized value in the untreated cells. These data are shown graphically to compare each ORF's level of resistance to PAA (Fig. 4C). We performed a statistical analysis of the PAA data and found that many of the ORFs expressed to detectable levels in the presence of PAA were reduced by statistically significant levels by PAA treatment.
FIG. 4.
MHV-68 gene expression in the absence viral DNA synthesis. MHV-68 membrane arrays were hybridized with oligo(dT)-primed cDNA synthesized from BHK-21 cells at 24 hpi with MHV-68 at 5 PFU/cell in the absence (A) or presence (B) of PAA. (C) Fold expression of transcripts in treated versus untreated samples. A Storm phosphorimager and the ImageQuant system were used to quantitate the signal from the array elements corresponding to 73 known and predicted MHV-68 ORFs. GAPDH-normalized values from the +PAA array (B) were divided by the corresponding GAPDH-normalized values from the untreated array (−PAA) (A) to derive the fold inhibition of gene expression relative to the untreated level for each array element. These values and their corresponding MHV-68 ORFs are ordered in the bar graph based on increasing fold inhibition of gene expression relative to the untreated level. Statistical significance of differences in expression is assessed by paired t test (+, P > 0.05; ✽, P < 0.05; ✽✽, P < 0.01; ✽✽✽, P < 0.001). Dashed lines were placed at 20% reduction and at 30% reduction of expression in the presence of PAA.
The levels of some transcripts were significantly increased in the presence of PAA, suggesting their expression is negatively regulated by late gene expression. The most affected transcript was ORF 69 (unknown function) with a GAPDH-normalized value of >9-fold higher than the untreated signal. Other ORFs that exhibited increased transcript levels in the presence of PAA were ORF 47 (glycoprotein L), ORF 59 (processivity factor), ORF 72 (v-cyclin), M11 (vBCL-2), and ORFs 48, 18, and M4, encoding proteins of unknown function. Many transcript levels were unaffected by PAA treatment. Among the 20 transcripts least affected, six encode proteins involved in DNA replication or processing (ORFs 6, 9, 37, 46, 54, and 60), three encode structural proteins (ORFs 17, 22, and 68), and eight have unknown functions (ORFs 10, 34, 42, 49, and 69). Of the 30 transcripts whose expression levels are the most severely inhibited by PAA treatment, five encode capsid proteins (ORFs 25, 26, 43, 62, and 65), three encode tegument proteins (ORFs 19, 64, and 75c), two encode glycoproteins (ORFs 39 and M7), six have no currently recognized function (ORFs M5, 23, 27, 35, 55, and 66), and four encode proteins involved in DNA replication (ORFs 40, 44, 56, and 61). Twenty-two transcripts of the thirty most severely affected by PAA treatment reached peak levels of expression at later times (24 to 32 hpi) during productive replication (Fig. 2B), a finding consistent with their classification as late gene products. To verify that PAA was effective under these conditions, a similar experiment was carried out with twice the amount of PAA, and similar results were obtained (data not show).
MHV-68 gene expression during the natural course of infection in the lungs of mice.
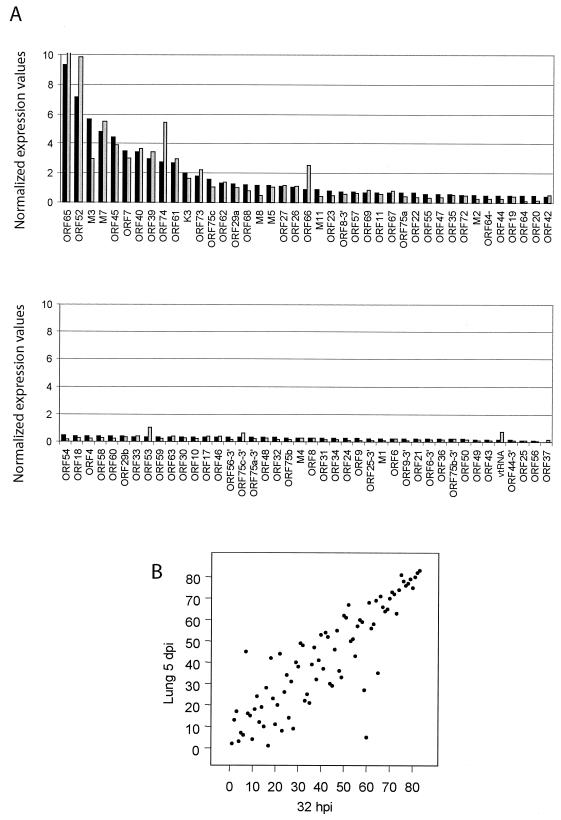
To determine the expression level of MHV-68 transcripts in vivo, we infected BALB/c mice by intranasal infection with 5 × 105 PFU. Total RNA was harvested from the lungs of two mice at 1, 3, 5, and 7 dpi. As was observed in cell culture, both latent and lytic transcripts were detected in the lungs during productive MHV-68 infection, a finding consistent with previous reports (34). Array analysis showed that viral gene expression in the lung peaked at 5 dpi. These data are consistent with previous observations that in the lung of mice infected at 5 × 105 PFU viral titers peak at 5 dpi (T. Wu, unpublished data). To compare the in vivo and in vitro gene expression patterns of MHV-68, the levels of each transcript during the peak of infection in the lungs were compared to those observed in infected BHK-21 cells. For both the tissue culture cell and infected lung arrays, 2 μg of total RNA was used for the probe synthesis reactions. We quantitated the GAPDH-normalized units for each transcript and ranked them based on the relative abundance level (Table 2). The transcripts most abundantly expressed in vitro are also abundantly expressed in vivo. Although there is a close match between the relative transcript levels in the lung and in cell culture at 32 hpi, the absolute viral transcript levels in the lung at the peak of viral gene expression were approximately two- to fivefold lower per microgram of total RNA. This may be due to the fact that the lung is composed of a heterogeneous population of cells, which will have various levels of susceptibility to MHV-68 infection and differentially support MHV-68 replication. To control for the relatively higher expression in culture, the signal for each array element was normalized to the total signal on the corresponding array. When the relative levels of gene expression between the two samples were compared, mRNA levels differed by <2-fold for 71 of the array elements (85%), and the largest difference was only 2.7-fold (Fig. 5A). The relative levels of transcripts encoding capsid proteins (ORFs 43, 62, and 65), glycoproteins (M7 and ORF 68), and the DNA packaging protein ORF 29a were equal or comparable in the lung and in cell culture during the later stages of infection (32 h time points). Rank correlation analysis showed that the relative expression of various MHV-68 gene products at 32 h after in vitro infection corresponded quite closely to those observed in the lungs of mice at 5 dpi (Spearman r = 0.86, P < 0.0001) (Fig. 5B). Similar results were observed when in vitro expression profiles at 24 hpi were compared to those observed in vivo (Spearman r = 0.73, P < 0.0001) (data not shown). In general, the BHK-21 model appears to faithfully mirror viral gene expression during in vivo infection.
FIG. 5.
Comparison of the MHV-68 transcription program at the peak of gene expression in vivo and in vitro. Total RNA was harvested from BHK-21 cells at 32 hpi or from the lungs of BALB/c mice at 5 dpi, and labeled cDNA probe was generated for hybridization to MHV-68 membrane arrays. The GAPDH-normalized phosphorimager units for each array element were divided by the total counts across the entire corresponding array. (A) These values and their corresponding MHV-68 ORFs are ordered in the bar graph based on decreasing gene expression levels in the lung (▪) and show the corresponding value in BHK-21 (░⃞). For ease of viewing, the values are illustrated in two graphs. (B) The Spearman rank correlation coefficient was used to assess the relationship between the magnitude of genes expressed in cell culture and in the lung of infected mice at the peak of viral gene expression.
Viral gene expression of an MHV-68 recombinant virus that overexpresses RTA.
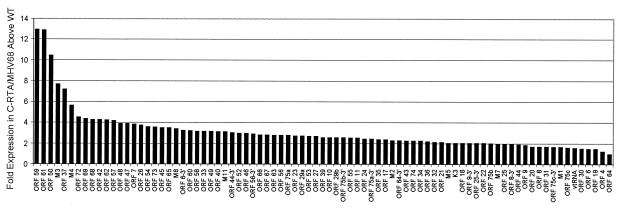
Our MHV-68 DNA arrays are also being used to investigate changes in viral gene expression in recombinant viruses. A recombinant virus (C-RTA/MHV-68) has been generated that overexpresses the viral transcription activator RTA (Tammy Rickabaugh, unpublished data). C-RTA/MHV-68 was constructed by inserting RTA under the CMV immediate-early promoter at the left end of the MHV-68 genome. BHK-21 cells were infected with C-RTA/MHV-68 or the parental virus, and RNA was harvested at 4, 8, and 12 hpi for array analysis. Cells infected with C-RTA/MHV-68 showed a marked enhancement of viral gene expression compared to the parental strain at all time points examined. GAPDH-normalized values from C-RTA/MHV-68 at 4 hpi were divided by the corresponding GAPDH-normalized values from the parental strain to derive the fold induction for each ORF (Fig. 6). These analyses revealed that all of the MHV-68 array elements detectable by 4 h postinfection were upregulated in the C-RTA/MHV-68 infection, although to different degrees, with a fold induction ranging from 12-fold (ORF 59) to 1.3-fold (ORF 64). Among the most highly activated genes were the functionally early proteins encoded by ORF 59 (processivity factor) and ORF 61 (ribonucleotide reductase large), both expressed in the C-RTA virus at 12-fold-higher levels than in the parental virus. M3 (soluble chemokine-binding protein) is expressed in the C-RTA virus at 7-fold-higher levels than in the parental strain. By 8 hpi, >80% of the ORFs examined were expressed in the C-RTA virus at fivefold-higher levels than in the parental virus (data not shown). Among the five most highly upregulated ORFs, two encode capsid proteins ORF 65 and ORF 26 at 30- and 22-fold above levels in the parental strain, respectively, and three encode proteins of unknown function (ORFs 66, 45, and M8). At 12 hpi, >57% of the ORFs examined were expressed in the C-RTA virus at fivefold-higher levels than in the parental virus (data not shown).
FIG. 6.
Gene expression profile of a recombinant MHV-68 overexpressing RTA compared to the parental strain. RNA was harvested from BHK-21 cells at 4 hpi with C-RTA/MHV-68 or the parental strain and labeled cDNA probe was generated for hybridization to MHV-68 membrane arrays. The GAPDH-normalized phosphorimager units for each array element from the recombinant virus array were compared to the corresponding value from the parental strain array. These data are shown graphically to compare each ORF's level of sensitivity to RTA overexpression.
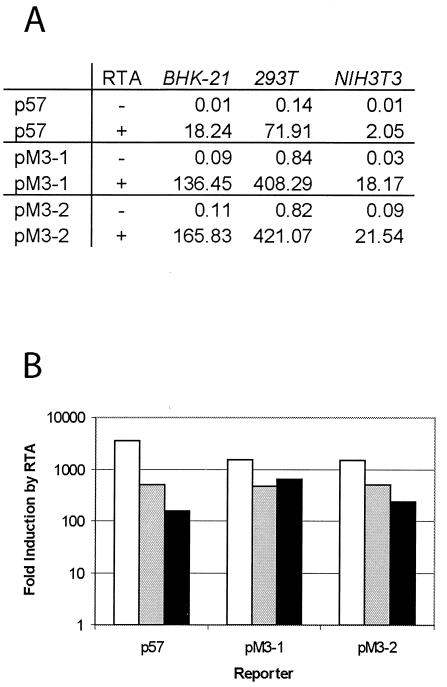
It is reasonable to assume that the genes whose expression is increased the most compared to the parental strain may be direct targets of the RTA protein. Since M3 is highly upregulated in the C-RTA virus, we considered it a potential target of RTA. To examine whether the M3 promoter is responsive to RTA, we constructed two reporter plasmids by inserting two regions of the M3 promoter (1,200 and 600 bp upstream of the M3 TATA box) into the pGL3-Basic plasmid (Promega) containing the firefly luciferase coding sequence. The ORF 57 promoter (p57) was used as a positive control for RTA responsiveness. This viral promoter has been shown to be responsive to wild-type MHV-68 RTA (19). In all three cell types tested, the M3 promoter constructs and p57 were responsive to RTA (Fig. 7). However, both of the pM3 reporter constructs yielded significantly higher luciferase activity than p57 in the presence of RTA. The highest level of luciferase activity in the presence of RTA was obtained for pM3-2, which contains 600 bp of sequence upstream of the M3 TATA box. The pM3-2 promoter was fivefold stronger than the p57 promoter in 293T cells and 10-fold stronger in NIH 3T3 cells. These results indicate that a region within 600 bp upstream of the M3 TATA box contains a RTA responsive element and suggests that M3 expression is activated by RTA in the absence of other viral factors. These data therefore support our conclusion from the membrane array that M3 is regulated by RTA.
FIG. 7.
Analysis of the M3 promoter responsivness to RTA by using reporter assays in BHK-21, 293T, and NIH 3T3 cells. Two regions of the M3 promoter were cloned into a pGL3-Basic vector to drive the expression of firefly luciferase as a reporter. (A) Luciferase activities of reporters cotransfected with pCMV-FLAG (−) or pCMV-FLAG/RTA (+). The M3 promoter constructs were cotransfected into BHK-21, 293T, and NIH 3T3 cells with pCMV-FLAG or pCMV-FLAG/RTA in the presence of a control vector, pRLSV40, that constitutively expresses Renilla luciferase driven by the SV40 promoter. At 24 h posttransfection, cells were harvested and dual luciferase assays were performed. Firefly luciferase activity in reporter constructs was normalized to the corresponding Renilla luciferase activity. (B) Activation of the M3 promoters by RTA in BHK-21, 293T, and NIH 3T3 cells. Fold activation of the reporter by RTA was obtained by comparing the normalized firefly luciferase activity of pCMV-FLAG/RTA-transfected cells to that of pCMV-FLAG-transfected cells. The data are an average of two separate experiments. Bars: □, BHK; ░⃞, 293T; ▪, 3T3.
DISCUSSION
MHV-68 is a model for the investigation of gammaherpesvirus infection. There has been much progress in deciphering the gene expression program during reactivation of KSHV by using DNA arrays (16, 28). However, the available KSHV lytic replication systems may not encompass all of the critical aspects of viral gene expression during de novo infection. MHV-68 elicits a productive infection both in cell culture and in its natural host, providing an ideal small-animal model for examining the function of viral genes during primary lytic infection. The gene expression patterns of a small subset of MHV-68 ORFs (M2, M3, 8, 9, K3, 50, 65, M11, 73, and 74) have been previously characterized individually by using different experimental backgrounds and analytical techniques (34, 44, 51, 53-55). More recently, the global gene expression program of MHV-68 has been evaluated by DNA array (1, 8). Genome-wide comparisons of MHV-68 gene expression during latency and productive infection in vivo and in vitro have not previously been done. We describe here custom MHV-68 DNA arrays constructed to analyze MHV-68 gene expression during latency and during lytic replication after de novo infection in vitro and primary infection in vivo. Analysis of the relative levels and temporal patterns of viral gene expression correlate well with expected patterns based on known and putative function of previously characterized genes. Our results both confirm previous studies on MHV-68 viral gene kinetics and highlight several distinctive ORFs whose expression pattern does not fit the expected kinetic class based on putative function.
The tight latency of the S11E cell line (15) facilitated the analysis of the MHV-68 latent transcription program. MHV-68 DNA array analysis revealed that regions of M2 (function unknown), M11 (vBCL-2), ORF 73 (LANA homologue), ORF 74 (vIL8 receptor), and ORFs 75a, 75b, and 75c are expressed in S11E cells. M2 expression was previously detected by Northern blot in the parental cell line, S11, and by real-time PCR in splenocytes of latently infected mice (15). Moreover, Virgin, et al. detected expression of ORFs M2, M11, 73, and 74 in mice by using RT-PCR (53). These reports, in addition to our own, suggest that there are similarities in the MHV-68 latency-associated gene expression program in cell culture and in infected tissue. However, there is a lack of consensus as to whether the candidate latency-associated genes (ORFs M2, M11, 73, and 74) are also expressed during lytic replication (34, 53). Our results indicate that the latency-associated genes are expressed during primary lytic infection of BALB/c mice (Table 2, lung 5d) and BHK-21 cells (Table 2). Moreover, the expression of the candidate latency-associated genes is highly resistant to CHX (Fig. 3C) treatment, suggesting that their transcription is primarily mediated by the existing transcription machinery. We therefore propose that latency may be the default gene expression program.
Previous studies have focused on candidate latency-associated genes based on sequence and positional homology with the latency genes of other gammaherpesviruses. However, the advantage of examining viral gene expression by array technology is the ability to assess gene expression across the entire genome. This benefit is evident based on our finding that a previously undetected region of the MHV-68 genome (ORF 75) is transcriptionally active during latency. Although there are no previous reports of ORF 75 expression during latency, this specific region of the MHV-68 genome may not have been analyzed thoroughly. The expression of ORF 75 in latently infected B cells was unambiguously detected in our membrane array (Fig. 1B). The fact that we did not detect expression from any known lytic transcript such as ORF 65 (small capsid protein), which is highly expressed during lytic replication, suggests that our cell line is tightly latent and that the expression of ORF 75 is latency associated. Moreover, a region of KSHV containing the positional homologue ORF 75 may also be transcriptionally active during latency (37).
In contrast to previous studies (15, 41, 53), the expression of M3 and ORF 65 was not detected in the latently infected S11E B cells (Fig. 1B). This may be due to tissue- and cell type-specific differences in MHV-68 gene expression in the individual latency models. However, the expression of latency-associated transcripts (ORFs M2, M11, 73, and 74) detected by membrane array analysis correlated well with data from previous studies. An alternative explanation is that the expression of M3 and ORF 65 during latency may actually represent lytic genes abundantly expressed by a small subset of reactivating cells. This idea is supported by our results showing that M3 and ORF 65 are consistently among the most highly expressed genes during the natural course of MHV-68 infection both in vivo and in vitro.
The gene products of many MHV-68 ORFs have been assigned putative functions based on sequence similarity to previously characterized herpes simplex virus (HSV) and KSHV genes. By using array technology, we have shown that most MHV-68 genes exhibit expression patterns that correlate with their presumed functions. Transcripts encoding proteins involved in DNA replication (ORFs 6, 9, 21, 54, 59, and 61) were expressed early during the course of infection (8 to 12 hpi) and were relatively resistant to PAA. In contrast, transcripts encoding virion structural proteins (ORFs 8, 25, 26, 39, 43, M7, 58, 63, 64, and 65) peaked later (12 to 24 hpi) and were sensitive to PAA. Transcripts encoding DNA packaging and virion assembly proteins (ORFs 29b, 62, and 67) were also sensitive to PAA and peaked after most structural genes (24 to 32 hpi).
Studies on HSV identified seven genes (UL 5, 8, 9, 29, 30, 42, and 52) required for viral origin-dependent DNA synthesis (26). All seven genes are expressed in a manner consistent with early gene kinetics. Three of these gene products form a complex which functions as a helicase-primase (4). MHV-68 encodes functional homologues of at least six of these DNA replication proteins. Three of the six (ssDNAbp, DNA Pol, and the processivity factor) reached peak levels of expression early (8 to 12 hpi) and were relatively resistant to PAA treatment, a finding consistent with early gene kinetics. However, the expression of the other three genes encoding the components of the helicase-primase complex reached peak levels much later (24 to 32 hpi) and are relatively sensitive to PAA treatment, suggesting late expression kinetics. KSHV also encodes homologues of the three helicase-primase components (38). Interestingly, the KSHV genes reach peak levels of expression at later times after reactivation than the other genes involved in DNA replication. This may indicate a previously undefined role for the helicase-primase complex during the later phases of the gammaherpesvirus life cycle.
Of the 73 ORFs represented on the DNA arrays, 10 (ORFs 6, 8, 9, 25, 44, 56, 64, 75a, 75b, and 75c) are relatively long sequences and were therefore represented by two array elements, one designed near the 5′ end and one near the 3′ end of the gene. Our rationale for representing these ORFs with two array elements was to control for the possibility of splicing events that may incorporate part but not the entire putative gene. In addition, representing these regions with two array elements allows the detection of previously undefined transcripts that read through part of the gene. In general, both 5′ and 3′ array elements detected each transcript with equal intensity. In some cases, the 3′ array element detected the transcript at slightly higher levels, which is consistent with the higher efficiency of reverse transcriptase oligo(dT) priming close to the poly A site. Notable exceptions were the higher intensity of the 5′ regions of ORFs 75a and 75c, a finding which is consistent with the possibility that this region of the genome may be alternatively spliced. The two array elements representing ORF 44 were also detected to different levels with the 5′ region higher. This suggests that the 5′ region of ORF 44 may encode a previously undefined gene.
While the expression of few viral genes was independent of de novo protein synthesis (RTA and latency-associated genes), there were a few early viral genes expressed to detectable levels in the presence of CHX. To verify that the drug was effective under these conditions, a similar experiment was carried out by using twice the amount of CHX, and similar results were obtained (data not show). The results suggest that the expression of some “early” genes is not absolutely dependent on immediate-early protein synthesis. One explanation for these results is that we are detecting basal transcription of viral ORFs from promoters containing sequences recognized by cellular transcription factors. Transcription in the presence of CHX may be possible owing to the abundance of cellular transcription machinery in the rapidly propagating BHK-21 cell line. Experiments by others using oligonucleotide-based microarrays of RNA from HSV-infected HeLa cells similarly detected low levels of early gene expression in the presence of CHX (45).
The utility of the MHV-68 membrane arrays in examining the gene expression patterns of recombinant viruses was evident in the analysis of C-RTA/MHV-68 infections. Using this technique we were able to look at nearly all MHV-68 ORFs in one experiment. This analysis revealed that RTA not only activates early genes but has a strong effect on the entire MHV-68 transcription program. By 4 hpi all ORFs tested were expressed to higher levels in C-RTA/MHV-68 than in the parental strain. Among the four ORFs that showed the greatest increase three (ORFs 59, 61, and 37) encode functionally early proteins (processivity factor, ribonucleotide reductase large, and alkaline exonuclease) and are therefore likely to be RTA responsive. The fourth, M3, which encodes a soluble chemokine-binding protein, has not previously been described as RTA responsive. These findings emphasize the value of genome-wide screening in defining targets of regulatory proteins on herpesvirus gene expression. Here we have shown that the M3 promoter is activated by RTA in the absence of other viral proteins. Our results demonstrating that M3 is responsive to RTA suggest the possibility that other highly upregulated ORFs identified in our array analysis are activated by RTA.
Although DNA arrays have proven invaluable in examining global gene expression patterns in organisms ranging from viruses to humans, it is recognized that the approach has some inherent technical limitations. One example of these limitations is that herpesviruses have a general underrepresentation of polyadenylation signal sequences that is likely to yield structurally polycistronic messages. The potential for misinterpretation of data has been reduced by examining the change in transcript levels over time or upon drug treatment rather than the absolute level of transcripts. Moreover, because the MHV-68 DNA array was spotted with PCR-amplified DNA and therefore double stranded, we cannot definitively identify the detected transcript as that labeled on the array. Because each array element is double stranded, transcripts encoded in either direction that pass through that region of the genome will hybridize to the array element. This problem is potentially avoided by using oligonucleotide-based DNA arrays However, cDNA arrays offer higher sensitivity then single-stranded oligonucleotide arrays. As is the case for all DNA array analyses, the information presented here provides a starting point for further detailed analyses of individual genes.
The membrane arrays described here make it possible to simultaneously study the expression of almost every MHV-68 ORF in a single experiment. With this powerful tool, we have assessed the transcription profile of genes with known or putative functions, as well as genes that have no currently recognized function. These data will provide a foundation for future work examining genes of both known and unknown functions. In addition, the MHV-68 DNA arrays have the potential to aid other types of analyses. The membrane arrays will facilitate the characterization of recombinant MHV-68 gene expression and assessment of the effect of mutations on the expression of other viral genes. In addition, the MHV-68 membrane arrays can be used to examine the effects of drugs on viral replication and gene expression and may therefore facilitate the identification of new antigammaherpesvirus drugs.
Acknowledgments
We thank Tonia Symensma for editing the manuscript.
This work was supported by NIH grants CA83525, CA91791, DE14153, the Stop Cancer Foundation (R.S.), and a special fellowship from the Leukemia and Lymphoma Foundation (T.-T.W.).
REFERENCES
- 1.Ahn, J. W., K. L. Powell, P. Kellam, and D. G. Alber. 2002. Gammaherpesvirus lytic gene expression as characterized by DNA array. J. Virol. 76:6244-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 3.Countryman, J., and G. Miller. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. USA 82:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crute, J. J., T. Tsurumi, L. A. Zhu, S. K. Weller, P. D. Olivo, M. D. Challberg, E. S. Mocarski, and I. R. Lehman. 1989. Herpes simplex virus 1 helicase-primase: a complex of three herpes-encoded gene products. Proc. Natl. Acad. Sci. USA 86:2186-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Decker, L. L., L. D. Klaman, and D. A. Thorley-Lawson. 1996. Detection of the latent form of Epstein-Barr virus DNA in the peripheral blood of healthy individuals. J. Virol. 70:3286-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decker, L. L., P. Shankar, G. Khan, R. B. Freeman, B. J. Dezube, J. Lieberman, and D. A. Thorley-Lawson. 1996. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J. Exp. Med. 184:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. van Marck, D. Salmon, I. Gorin, J. P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebrahimi, B., B. M. Dutia, K. L. Roberts, J. J. Garcia-Ramirez, P. Dickinson, J. P. Stewart, P. Ghazal, D. J. Roy, and A. A. Nash. 2003. Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J. Gen. Virol. 84:99-109. [DOI] [PubMed] [Google Scholar]
- 9.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flano, E., S. M. Husain, J. T. Sample, D. L. Woodland, and M. A. Blackman. 2000. Latent murine gammaherpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J. Immunol. 165:1074-1081. [DOI] [PubMed] [Google Scholar]
- 11.Ganem, D. 1997. KSHV and Kaposi's sarcoma: the end of the beginning? Cell 91:157-160. [DOI] [PubMed] [Google Scholar]
- 12.Gao, S. J., L. Kingsley, D. R. Hoover, T. J. Spira, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, P. Parry, Y. Chang, and P. S. Moore. 1996. Seroconversion to antibodies against Kaposi's sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi's sarcoma. N. Engl. J. Med. 335:233-241. [DOI] [PubMed] [Google Scholar]
- 13.Gao, S. J., L. Kingsley, M. Li, W. Zheng, C. Parravicini, J. Ziegler, R. Newton, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, Y. Chang, and P. S. Moore. 1996. KSHV antibodies among Americans, Italians, and Ugandans with or without Kaposi's sarcoma. Nat. Med. 2:925-928. [DOI] [PubMed] [Google Scholar]
- 14.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Husain, S. M., E. J. Usherwood, H. Dyson, C. Coleclough, M. A. Coppola, D. L. Woodland, M. A. Blackman, J. P. Stewart, and J. T. Sample. 1999. Murine gammaherpesvirus M2 gene is latency-associated and its protein a target for CD8+ T lymphocytes. Proc. Natl. Acad. Sci. USA 96:7508-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kedes, D. H., E. Operskalski, M. Busch, R. Kohn, J. Flood, and D. Ganem. 1996. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat. Med. 2:918-924. [DOI] [PubMed] [Google Scholar]
- 18.Lieberman, P. M., J. M. Hardwick, J. Sample, G. S. Hayward, and S. D. Hayward. 1990. The zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, S., I. V. Pavlova, H. W. T. Virgin, and S. H. Speck. 2000. Characterization of gammaherpesvirus 68 gene 50 transcription. J. Virol. 74:2029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 21.Manet, E., H. Gruffat, B. M. C. Trescol, N. Moreno, P. Chambard, J. F. Giot, and A. Sergeant. 1989. Epstein-Barr virus bicistronic mRNAs generated by facultative splicing code for two transcriptional trans-activators. EMBO J. 8:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonagh, D. P., J. Liu, M. J. Gaffey, L. J. Layfield, N. Azumi, and S. T. Traweek. 1996. Detection of Kaposi's sarcoma-associated herpesvirus-like DNA sequence in angiosarcoma. Am. J. Pathol. 149:1363-1368. [PMC free article] [PubMed] [Google Scholar]
- 23.Memar, O. M., P. L. Rady, and S. K. Tyring. 1995. Human herpesvirus-8: detection of novel herpesvirus-like DNA sequences in Kaposi's sarcoma and other lesions. J. Mol. Med. 73:603-609. [DOI] [PubMed] [Google Scholar]
- 24.Nash, A. A., and N. P. Sunil-Chandra. 1994. Interactions of the murine gammaherpesvirus with the immune system. Curr. Opin. Immunol. 6:560-563. [DOI] [PubMed] [Google Scholar]
- 25.Nash, A. A., E. J. Usherwood, and J. P. Stewart. 1996. Immunological features of murine gammaherpesvirus infection. Semin. Virol. 7:125-130. [Google Scholar]
- 26.Olivo, P. D., N. J. Nelson, and M. D. Challberg. 1989. Herpes simplex virus type 1 gene products required for DNA replication: identification and overexpression. J. Virol. 63:196-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pastore, C., A. Gloghini, G. Volpe, J. Nomdedeu, E. Leonardo, U. Mazza, G. Saglio, A. Carbone, and G. Gaidano. 1995. Distribution of Kaposi's sarcoma herpesvirus sequences among lymphoid malignancies in Italy and Spain. Br. J. Haematol. 91:918-920. [DOI] [PubMed] [Google Scholar]
- 28.Paulose-Murphy, M., N. K. Ha, C. Xiang, Y. Chen, L. Gillim, R. Yarchoan, P. Meltzer, M. Bittner, J. Trent, and S. Zeichner. 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purifoy, D. J., and K. L. Powell. 1977. Herpes simplex virus DNA polymerase as the site of phosphonoacetate sensitivity: temperature-sensitive mutants. J. Virol. 24:470-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rady, P. L., A. Yen, R. W. r. Martin, I. Nedelcu, T. K. Hughes, and S. K. Tyring. 1995. Herpesvirus-like DNA sequences in classic Kaposi's sarcomas. J. Med. Virol. 47:179-183. [DOI] [PubMed] [Google Scholar]
- 31.Ragoczy, T., L. Heston, and G. Miller. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72:7978-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 33.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 34.Rochford, R., M. L. Lutzke, R. S. Alfinito, A. Clavo, and R. D. Cardin. 2001. Kinetics of murine gammaherpesvirus 68 gene expression following infection of murine cells in culture and in mice. J. Virol. 75:4955-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2460. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 36.Sarawar, S. R., R. D. Cardin, J. W. Brooks, M. Mehrpooya, R. A. Tripp, and P. C. Doherty. 1996. Cytokine production in the immune response to murine gammaherpesvirus 68. J. Virol. 70:3264-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarid, R., T. Sato, R. A. Bohenzky, J. J. Russo, and Y. Chang. 1997. Kaposi's sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat. Med. 3:293-298. [DOI] [PubMed] [Google Scholar]
- 39.Shao, L., L. M. Rapp, and S. K. Weller. 1993. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology 196:146-162. [DOI] [PubMed] [Google Scholar]
- 40.Simas, J. P., and S. Efstathiou. 1998. Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol. 6:276-282. [DOI] [PubMed] [Google Scholar]
- 41.Simas, J. P., D. Swann, R. Bowden, and S. Efstathiou. 1999. Analysis of murine gammaherpesvirus-68 transcription during lytic and latent infection. J. Gen. Virol. 80:75-82. [DOI] [PubMed] [Google Scholar]
- 42.Speck, S. H., and H. W. Virgin. 1999. Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Curr. Opin. Microbiol. 2:403-409. [DOI] [PubMed] [Google Scholar]
- 43.Stevenson, P. G., and P. C. Doherty. 1998. Kinetic analysis of the specific host response to a murine gammaherpesvirus. J. Virol. 72:943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart, J. P., N. J. Janjua, N. P. Sunil-Chandra, A. A. Nash, and J. R. Arrand. 1994. Characterization of murine gammaherpesvirus 68 glycoprotein B (gB) homolog: similarity to Epstein-Barr virus gB (gp110). J. Virol. 68:6496-6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stingley, S. W., J. J. Ramirez, S. A. Aguilar, K. Simmen, R. M. Sandri-Goldin, P. Ghazal, and E. K. Wagner. 2000. Global analysis of herpes simplex virus type 1 transcription using an oligonucleotide-based DNA microarray. J. Virol. 74:9916-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sunil-Chandra, N. P., S. Efstathiou, J. Arno, and A. A. Nash. 1992. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J. Gen. Virol. 73:2347-2356. [DOI] [PubMed] [Google Scholar]
- 48.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. Gen. Virol. 73:3275-3279. [DOI] [PubMed] [Google Scholar]
- 49.Szekely, L., F. Chen, N. Teramoto, B. Ehlinhenriksson, K. Pokrovskaja, A. Szeles, A. Manneborgsandlund, M. Lowbeer, E. T. Lennette, and G. Klein. 1998. Restricted expression of Epstein-Barr virus (EBV)-encoded, growth transformation-associated antigens in an EBV- and human herpesvirus type 8-carrying body cavity lymphoma line. J. Gen. Virol. 79:1445-1452. [DOI] [PubMed] [Google Scholar]
- 50.Usherwood, E. J., J. P. Stewart, and A. A. Nash. 1996. Characterization of tumor cell lines derived from murine gammaherpesvirus-68-infected mice. J. Virol. 70:6516-6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Berkel, V., K. Preiter, H. W. T. Virgin, and S. H. Speck. 1999. Identification and initial characterization of the murine gammaherpesvirus 68 gene M3, encoding an abundantly secreted protein. J. Virol. 73:4524-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Virgin, H. W. t., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Virgin, H. W. T., R. M. Presti, X. Y. Li, C. Liu, and S. H. Speck. 1999. Three distinct regions of the murine gammaherpesvirus 68 genome are transcriptionally active in latently infected mice. J. Virol. 73:2321-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu, T. T., L. Tong, T. Rickabaugh, S. Speck, and R. Sun. 2001. Function of Rta is essential for lytic replication of murine gammaherpesvirus 68. J. Virol. 75:9262-9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, T. T., E. J. Usherwood, J. P. Stewart, A. A. Nash, and R. Sun. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol. 74:3659-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]