Abstract
The coronavirus, mouse hepatitis virus strain JHM, causes acute and chronic neurological diseases in rodents. Here we demonstrate that two closely related virus variants, both of which cause acute encephalitis in susceptible strains of mice, cause markedly different diseases if mice are protected with a suboptimal amount of an anti-JHM neutralizing antibody. One strain, JHM.SD, caused acute encephalitis, while infection with JHM.IA resulted in no acute disease. Using recombinant virus technology, we found that the differences between the two viruses mapped to the spike (S) glycoprotein and that the two S proteins differed at four amino acids. By engineering viruses that differed by only one amino acid, we identified a serine-to-glycine change at position 310 of the S protein (S310G) that recapitulated the more neurovirulent phenotype. The increased neurovirulence mediated by the virus encoding glycine at position S310 was not associated with a different tropism within the central nervous system (CNS) but was associated with increased lateral spread in the CNS, leading to significantly higher brain viral titers. In vitro studies revealed that S310G was associated with decreased S1-S2 stability and with enhanced ability to mediate infection of cells lacking the primary receptor for JHM (“receptor-independent spread”). These enhanced fusogenic properties of viruses encoding a glycine at position 310 of the S protein may contribute to spread within the CNS, a tissue in which expression of conventional JHM receptors is low.
Mouse hepatitis virus (MHV) strain JHM is an enveloped, nonsegmented positive-strand RNA virus belonging to the Coronaviridae family within the Nidovirales order (7). JHM infection of experimental animals has been extensively used to study virus pathogenesis in the central nervous system (CNS). Susceptible mice develop an acute, fatal viral encephalomyelitis after intranasal or intracranial inoculation, with clinical signs that include hunching, ruffled fur, irritability, and lethargy. Viral antigen is detected in gray- and white-matter structures throughout the brain. Rodents chronically infected with MHV develop demyelinating diseases with many similarities to multiple sclerosis in humans (21, 55, 56). Chronic infection results if animals are infected with an attenuated virus or with a virulent virus in the presence of anti-JHM antibodies or T cells (5, 13, 26, 31, 39, 48, 57, 69). In one model of chronic infection, suckling mice were infected with JHM and nursed by dams previously immunized against the virus (48). Under these conditions, sufficient antibody was transmitted to suckling mice to prevent acute encephalitis but not to mediate virus clearance. Consequently, 40 to 90% of mice developed hindlimb paralysis and histological evidence of a demyelinating encephalomyelitis at 3 to 8 weeks postinfection (p.i.) (48). Since the inception of this maternal antibody model, we have used a specific strain of JHM, JHM.Iowa (JHM.IA), for all of our experiments.
A second virulent variant of JHM, JHM.San Diego (JHM.SD; formerly called MHV-4 [25]), is also studied in several laboratories. Both JHM.IA and JHM.SD cause a uniformly fatal encephalitis in naive 6-week-old C57BL/6 mice, with a 50% lethal dose of approximately 1 PFU (22, 25, 45, 48). Of note, disease onset was approximately 12 h earlier after intranasal inoculation with JHM.SD (unpublished data). Unexpectedly, however, the two viruses caused very different diseases after infection of suckling mice in the maternal antibody model. Infection with JHM.IA resulted in no acute disease in the presence of maternal antibody. By contrast, infection with JHM.SD resulted in a fulminant encephalitis in all maternal-antibody-protected suckling mice.
The genome of JHM encodes four structural proteins: the nucleocapsid (N) protein, the transmembrane (M) protein, the spike (S) glycoprotein, and the small envelope (E) protein (30). The JHM S protein is an oligomeric type I integral membrane glycoprotein, which is translated into a single protein and cleaved by a Golgi protease into S1 and S2 subunits of approximately 90 kDa each (54, 58). The S protein is responsible for receptor binding, fusion of viral and cell membranes, and cell-to-cell fusion (4, 10). Binding to the host cell receptor is mediated by the N-terminal 330 amino acids of the S1 subunit (28). Not surprisingly, the greatest amount of sequence heterogeneity among strains of MHV is present in the S1 subunit (11).
The cellular receptors for JHM are members of the carcinoembryonic antigen family of cell adhesion molecules (CEACAMs), with CEACAM1a identified as the principal host cell receptor (9, 23, 74). Binding to these proteins is believed to induce S1 release and conformational changes that expose a hydrophobic region within S2, promoting fusion between the viral envelope and the cell membrane (15, 27, 33, 63, 67, 73). Of note, the S1-S2 complex of JHM.SD is unstable; viruses with enhanced S1-S2 stability are selected by passage through tissue culture cells (17, 27). In addition, JHM.SD is a potent mediator of cellular fusion, even in the absence of binding to the specific host cell receptor for JHM (14, 41). Other viruses that are less neurovirulent, such as strain A59 of MHV (MHV-A59), do not mediate receptor-independent spread (14). Although JHM primarily infects the murine CNS, expression of CEACAM1a is low in the CNS, making its role in JHM infection of the CNS less certain (19, 20). Receptor-independent spread would therefore facilitate JHM spread in the CNS.
Data from multiple other studies further illustrate differences between S proteins from various isolates that are likely to contribute to differences in pathogenesis (4, 16, 18, 32, 42, 44, 73, 74). Here we postulated that the differences in pathogenesis shown by JHM.IA and JHM.SD also resulted from differences in the S protein. To investigate this possibility, we took advantage of recent advances in reverse genetics that have facilitated genetic manipulation of the JHM genome. Infectious clones of several coronaviruses have been described (1, 6, 65, 70, 71), but a JHM infectious clone has not yet been developed. An alternative approach, termed targeted recombination, is based on the high recombination rate exhibited by coronaviruses and is useful for introducing mutations into the 3′ end of the coronavirus genome (35). In one instance, selection of recombinant viruses takes advantage of the strong species specificity shown by the S protein (29). This method was used to demonstrate the important role that the S protein plays in coronavirus pathogenesis (51, 52). In this study, we investigated the pathogenic differences between JHM.SD and JHM.IA in greater detail, making use of targeted recombination to develop recombinant viruses that express the JHM.SD or JHM.IA S proteins in the context of the JHM.IA genome. These viruses were used to probe the relationship between neurovirulence and the fusogenic properties of the S protein measured in vitro.
MATERIALS AND METHODS
Virus and cells.
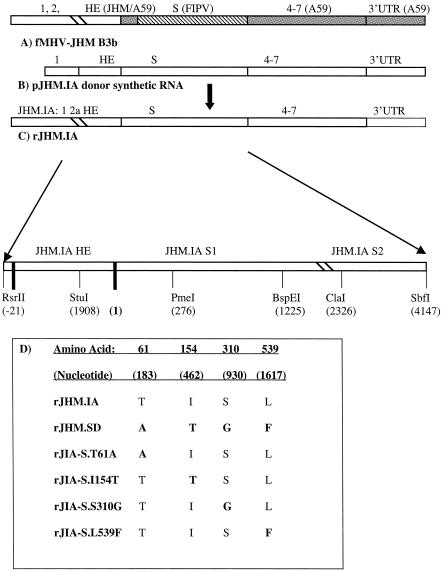
JHM was grown and titered as described previously (48). A chimeric recombinant virus encoding the feline S protein (designated fMHV-JHM clone B3b) was used as the recipient for targeted recombination (29, 45). This virus was propagated in FCWF (feline) cells. fMHV-JHM B3b contains genes 1 and 2a of JHM.IA, the 5′ end of the HE gene of JHM.IA followed by the 3′ end of the HE gene of MHV-A59, the ectodomain of the S gene of feline infectious peritonitis virus (FIPV), and the transmembrane domain and endodomain of the S gene of MHV-A59. The remaining 3′ end of fMHV-JHM was also derived from MHV-A59 (Fig. 1).
FIG. 1.
Schematic of recombinant viruses. (A) Interspecies chimeric virus fMHV-JHM B3b. fMHV-JHM carries the region from gene 1 through the 5′ end of HE from JHM.IA, the 3′ end of HE from MHV-A59, the ectodomain of FIPV gene 3, the transmembrane domain and endodomain of gene 3 from MHV-A59, and the region from gene 4 through the 3′ untranslated region (3′ UTR) from MHV-A59 (45). This chimeric virus infects only feline cells. (B) RNA transcribed from pJHM. As an example, construction of rJHM.IA is shown. Donor RNA (8 kb) was transcribed in vitro as described in Materials and Methods. pJHM.IA carries sequences entirely derived from JHM.IA. (C) Construction of rJHM.IA. fMHV-JHM B3b-infected feline cells were transfected with donor RNA transcribed from pJHM.IA to create rJHM.IA. (D) Donor RNA from pJHM.SD was used to create rJHM.SD. Point mutations were introduced into pJHM.IA to create the recombinant viruses listed. The sources of genomic or donor RNA segments are indicated by open rectangles for JHM.IA, hatched rectangles for FIPV, and shaded rectangles for MHV-A59. S1-S2 cleavage occurs between nt 2307 and 2308.
Mice.
Specific-pathogen-free 5- to 6-week-old B6 mice were obtained from the National Cancer Institute (Frederick, Md.) and Jackson Laboratories (Bar Harbor, Maine). In some experiments, female mice were immunized with infectious JHM prior to mating, as described previously (48). Alternatively, female mice were immunized passively with a 1:1 mixture of anti-S neutralizing antibodies (monoclonal antibody [MAb] 5A13.5 [neutralizing titer, 1:45,000] and MAb 5B19.2 [neutralizing titer, 1:2,700]; provided by Michael Buchmeier, The Scripps Research Institute). Sixty microliters of antibody in 500 μl of phosphate-buffered saline (PBS) was delivered by intraperitoneal inoculation at 7 days postpartum as described previously (50). Three days later, suckling mice were intranasally infected with 2 × 104 to 4 × 104 PFU of JHM or recombinant virus. To control for variability in the amount of protective antibody transmitted to the suckling mice, one-third of each litter was inoculated with a different virus. Each virus was analyzed in a minimum of three litters.
RNA sequence analysis.
To sequence the S gene of JHM.IA, RNA was harvested from infected 17Cl-1 cells by using TriReagent (Molecular Research Center, Inc., Cincinnati, Ohio) as described previously (49). PCR products were prepared from cDNA and inserted into a TOPO vector according to the manufacturer's instructions (Invitrogen Corporation, Carlsbad, Calif.). Individual clones were sequenced with an automated sequencer (ABI 373A Stretch Sequencer; Applied Biosystems, Foster City, Calif.).
Plasmids and PCR mutagenesis.
pJHM.SD (formerly designated pJHM) was derived as previously described (45). pJHM.SD contains the JHM.SD S gene fused to genes 2b, 4, 5a, E, M, and N derived from JHM.IA. pJHM.SD was used to transcribe RNA for the recombinant virus rJHM.SD. pJHM.IA is identical to pJHM.SD except that the sequence was modified to encode the S protein of JHM.IA. pJHM.IA was used to transcribe RNA for the recombinant virus rJHM.IA. Overlapping extension PCR and subcloning were used to create recombinant viruses in which the amino acids differing between rJHM.SD and rJHM.IA were individually introduced into rJHM.IA (Fig. 1).
To change Ile at position 154 to Thr, an upstream PCR product was synthesized using primer 5′S01 (nucleotides [nt] −6 to +12, CTAAACATGCTGTTCGTC) (nucleotide positions are relative to the first nucleotide of S protein) and downstream primer 3′IT.S (nt 446 to 467, GGTATATGTACAAACAGAAGC), introducing the I154T mutation. A downstream PCR product was constructed using primer [5′IT.S (nt 447 to 468, GCTTCTGTTTGTACATATACC)], complementary to the downstream primer of the first PCR product, and 3′ primer [3′S09 (nt 2061 to 2083, TGATATGCAGCAGAAACACGGC)]. The final PCR product was synthesized by using the two outer primers (5′S01 and 3′S09). The same process was repeated to introduce a glycine at residue 310 of the spike, using the unique inner primers [3′SG.S (nt 921 to 939, GGACCGTGTAACCGGATAG)] and 5′SG.S (nt 921 to 940, CTATCCGGTTACACGGTCC). I154T- or S310G-containing PCR products were then restricted with PmeI (nt 276) and BspEI (nt 1225) and inserted into p022. p022 encodes JHM.IA sequence from an RsrII site (introduced 22 nt upstream of gene 2b) through the 3′ end of the S gene (SbfI site [nt 4147]), resulting in plasmids p022.I154T and p022.S310G.
To introduce the T61A change into p022, a fragment from pJHM.SD containing the T61A difference was excised with RsrII and PmeI (p022.T61A). The Leu at position 539 was changed to Phe by restricting pJHM.SD with BspEI and ClaI (nt 2326) and subcloning this fragment into p022 (p022.L539F). p022 clones containing the single point mutations in the S gene were restricted with StuI (introduced at nt 1908 of gene 2b) and SbfI and inserted into pJHM.IA to create pJHM.IA.S.T61A, pJHM.IA.S.I154T, pJHM.IA.S.S310G, and pJHM.IA.S.L539F. These modified pJHM.IA constructs were subsequently used to transcribe RNA to create recombinants rJIA-S.T61A, rJIA-S.I154T, rJIA-S.S310G, and rJIA-S.L539F, respectively. Recombinant viruses containing point mutations were labeled as follows (taking rJIA-S.T61A as an example): rJIA for recombinant JHM strain IA, S for S protein, and T61A for the specific mutation within S. All PCR products were sequenced prior to further use.
Targeted recombination.
Targeted recombination was carried out as described previously (29, 45). Briefly, FCWF cells infected with fMHV-JHM were transfected with donor RNAs transcribed from one of the six plasmids described above, and recombinants were selected on 17Cl-1 murine cells. Recombinant viruses were plaque purified twice, amplified on 17Cl-1 cells, and titered on HeLa cells expressing the JHM receptor (HeLa-MHVR).
Donor RNA was prepared by using a mMESSAGE mMACHINE T7 kit (Ambion, Inc., Austin, Tex.) as recommended by the manufacturer except that GTP was added to a final concentration of 4.5 mM. To ensure that the desired mutations were not lost during propagation in tissue culture cells, RNA harvested from infected cells was purified and 500 bases within the S gene encompassing the introduced mutation were sequenced. For each recombinant virus, at least two recombinants derived from independent recombination events were characterized.
Virus neutralization assays.
Sequential dilutions of serum from dams immunized against JHM were mixed with equal volumes of virus containing 100 PFU and were incubated on ice for 30 min. Samples were then titered on HeLa-MHVR cells.
Histology and immunohistochemistry.
Brains were harvested from infected mice on day 5 p.i., fixed in Histochoice (Amresco, Solon, Ohio), and embedded in paraffin. Seven-micrometer-thick sections were baked for 1 h at 56°C and stained with hematoxylin. Viral antigen was detected by using anti-nucleocapsid (anti-N) MAb 5B188.2 (kindly provided by M. Buchmeier) as described previously (60).
Growth kinetics in tissue culture.
17Cl-1 cells were infected with virus at a multiplicity of 1 PFU per cell. Samples were harvested at the times indicated, and viral titers were determined on HeLa-MHVR cells.
pH stability and thermal inactivation.
Equal amounts of the indicated viruses were diluted 10-fold in PBS buffered to pH 6.0, 7.0,or 8.0, prepared as previously described (27). Viruses were then incubated at 37°C for the indicated times. Samples were subsequently titered on HeLa-MHVR cells.
Metabolic radiolabeling and immunoprecipitation of S proteins.
A series of 17Cl-1 cell cultures (106 cells in 10-cm2-diameter wells) were inoculated with rJHM viruses for 1 h at 37°C and then incubated until 50% of cells had fused into syncytia, typically observed by 12 h p.i. The medium was removed, and cells were rinsed three times with isotonic saline and then replenished with a 35S labeling medium (methionine- and cysteine-free Dulbecco's modified Eagle medium containing 0.1% dialyzed fetal calf serum; 1 ml per well) containing 50 μCi of Tran35S-label (ICN Biomedicals, Inc., Aurora, Ohio) per ml. After a 30-min pulse-labeling period, the medium was removed and saved (0-h chase). Parallel monolayers were rinsed twice with cold saline and then replenished with 1 ml of medium per well. At hourly intervals, the medium was removed from these cultures (1- and 2-h chase).
To remove debris, samples were centrifuged at 2,000 × g for 10 min at 5°C and then at 20,000 × g for 10 min at 5°C. Clarified medium was overlaid onto linear 3-ml 10 to 30% (wt/wt) sucrose gradients in HNB (25 mM sodium HEPES [pH 7.4], 100 mM NaCl, 0.1% bovine serum albumin). Centrifugation was performed at 5°C in an SW60 rotor for 1 h at 55,000 rpm. The top 1-ml volumes from each gradient, containing free S1 fragments, were collected and stored at −80°C. Gradient material remaining in each tube was decanted, and pellets containing intact virions were dissolved in 0.1 ml of sodium dodecyl sulfate (SDS) lysis buffer (62 mM Tris HCl [pH 6.8], 2% SDS, 5% 2-mercaptoethanol, 2.5% Ficoll, 0.01% bromphenol blue). The S1 fragments remaining at the tops of sucrose gradients were immunoprecipitated by adsorption to N-CEACAM-Fc, a soluble form of the CEACAM1a receptor (16). Immunoprecipitated proteins were dissolved in 0.1 ml of SDS lysis buffer. Equal volumes of 35S protein samples were subjected to SDS-polyacrylamide gel electrophoresis as described previously (27), and X-ray films exposed to dried gels were developed to visualize radioactive protein bands.
Receptor-independent spread.
Monolayers of baby hamster kidney (BHK) cells, negative for JHM receptors, or 17Cl-1 cells, positive for JHM receptors, were trypsinized, spun, and washed with PBS containing 0.1% bovine serum albumin (PBS-B). Cells were resuspended at 106/ml and labeled with 1 μM carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes, Eugene, Oreg.) in PBS-B for 10 min at 37°C. Cells were then washed three times and allowed to adhere to coverslips. In parallel, 17Cl-1 murine cells were infected with the indicated viruses at a multiplicity of 1 PFU per cell. After 30 min, cells were washed twice to remove nonadherent virus. After 4 h, the infected 17Cl-1 cells were trypsinized, diluted in 5% fetal bovine serum-Dulbecco's modified Eagle medium, and counted. Infected cells were then overlaid onto CFSE-labeled BHK cells at a ratio of 1:50. An equal number of infected 17Cl-1 cells was overlaid on uninfected 17Cl-1 cells as a positive control. After 18 h, cells were fixed. Viral antigen was detected by sequential treatments with an anti-N MAb and a Texas red-conjugated goat anti-mouse antibody (ICN). Cells were then examined by fluorescence microscopy. Receptor-independent spread was indicated by the presence of multinucleated cells on BHK cells that stained for both CFSE and viral antigen.
Statistical analysis.
Statistical analyses were performed using unpaired t tests.
RESULTS
The JHM.SD S protein mediates enhanced neurovirulence.
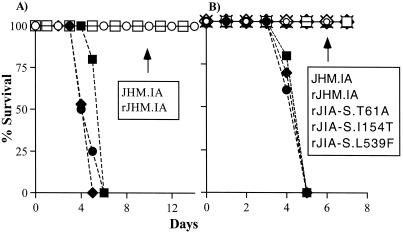
Our preliminary results suggested a striking difference in pathogenicity when suckling mice were inoculated intranasally with 2 × 104 to 4 × 104 PFU of JHM.IA or JHM.SD and nursed by dams immunized previously with infectious JHM.IA. Mice infected with JHM.SD uniformly died in the first 5 to 7 days p.i., whereas those infected with JHM.IA all survived the acute infection (Fig. 2A). The specific strain of JHM used for immunization did not matter, because dams actively immunized with live JHM.SD, as opposed to JHM.IA, were also able to protect pups infected with JHM.IA, but not pups infected with JHM.SD (data not shown). Inoculation of naive 6-week-old B6 mice with the same inoculum of virus resulted in acute encephalitis in all recipients, although there was a 12-h increase in the time until death in mice infected with JHM.IA (data not shown). These results collectively showed that JHM.SD was more virulent than JHM.IA and that this difference was accentuated in suckling mice in the presence of maternally derived anti-JHM antibody. To determine whether this difference in virulence could be attributed to a loss of recognition of one or more antibody epitopes that were crucial for protection in JHM.SD-infected mice, we mixed serum from actively immunized dams with aliquots of either JHM.SD or JHM.IA. We were unable to detect any difference in the ability of the sera to neutralize the two viruses (Fig. 3), suggesting that antibody escape did not contribute to the increased virulence exhibited by JHM.SD. Further, a panel of anti-S MAbs raised against JHM.SD recognized JHM.IA, consistent with the lack of antibody escape (47).
FIG. 2.
Virulence of spike variants. Suckling mice were infected intranasally with the indicated viruses and nursed by dams actively immunized with infectious JHM.IA (A) or passively immunized with a cocktail of two anti-JHM neutralizing antibodies (B) as described in Materials and Methods. All pups were monitored daily for signs consistent with acute encephalitis (hunching, ruffled fur, lethargy). The fraction surviving at each day p.i. is shown. Each virus was used to infect five to eight mice. Symbols: ○, JHM.IA; •, JHM.SD; □, rJHM.IA; ▪, rJHM.SD; ▴, rJIA-S.T61A; ◊, rJIA-S.I154T; ⧫, rJIA-S.S310G; ×, rJIA-S.L539F. Note that only JHM.IA, JHM.SD, rJHM.SD, rJHM.IA, and rJIA-S.S310G are analyzed in panel A.
FIG. 3.
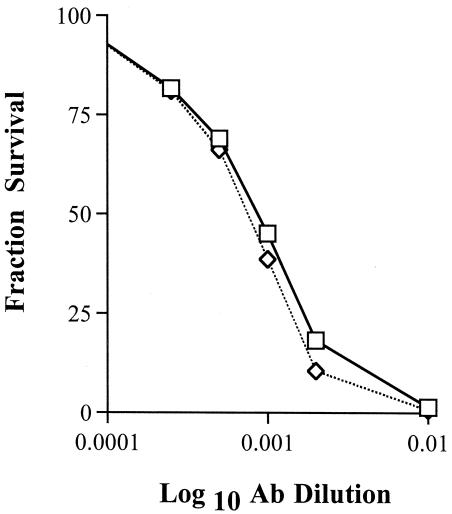
Neutralization of JHM.IA and JHM.SD by maternal serum. Dams were immunized three to five times with infectious JHM.IA. Sera were prepared 3 weeks after the last immunization and treated at 56°C for 30 min. Dilutions of sera were incubated with an equal volume of the indicated virus for 30 min on ice. Residual virus titers were determined by a plaque assay on HeLa-MHVR cells as described in Materials and Methods. Representative curves with serum from an individual dam are shown. Sera from five dams were analyzed in these experiments with similar results. Symbols: ⋄, JHM.IA; □, JHM.SD. Ab, antibody.
These experiments involved three immunizations of dams with infectious JHM in Freund's adjuvant. To simplify the protocol in subsequent experiments, dams were immunized passively with a mixture of two anti-JHM neutralizing antibodies 7 days after delivery (3 days before pups were inoculated with JHM). Using amounts of antibody that were previously shown to protect suckling mice from acute encephalitis caused by JHM.IA (50), we were not able to protect mice from JHM.SD-induced encephalitis (Fig. 2B). Of note, JHM.SD was not completely resistant to the anti-JHM antibody, since two- to threefold greater amounts of passively administered antibody protected mice from clinical disease caused by either virus.
Previous reports have demonstrated the key role that the S protein plays in MHV-induced pathogenesis (5, 17, 51). Therefore, targeted recombination was used to develop recombinant viruses that differed only in the source of the S protein. In all recombinant viruses, all genes except the S gene were identical to the JHM.IA sequence. For each experiment, at least two independent isolates were studied. Identical results were obtained in all cases, so only results from single isolates are shown below. As shown in Fig. 2, the recombinant viruses containing the two different S proteins exactly mirrored the parental strains in the clinical diseases they caused. Maternal antibody-protected suckling mice infected with the recombinant virus encoding the JHM.SD S protein (rJHM.SD) developed an acute, fatal encephalitis, whereas those infected with rJHM.IA, encoding the JHM.IA S protein, did not. We next compared the published sequence of the JHM.SD S protein (46) to that of the JHM.IA isolate. The two proteins differed in only four amino acids, all located within the S1 subunit (Table 1).
TABLE 1.
Nucleotide and amino acid differences between JHM.IA and JHM.SD spikes
| Nucleotide position and difference (JHM.IA to JHM.SD) | Amino acid position | Amino acid in:
|
|
|---|---|---|---|
| JHM.IA | JHM.SD | ||
| 181 (G to A) | 61 | Thr | Ala |
| 461 (T to C) | 154 | Ile | Thr |
| 928 (A to G) | 310 | Ser | Gly |
| 1617 (G to T) | 539 | Leu | Phe |
Gly at S310 is critical for enhanced JHM.SD neuropathogenicity.
To investigate which amino acids were responsible for the differences in pathogenesis that we observed, four additional recombinants were developed in which the unique amino acids from JHM.SD were introduced into the JHM.IA S protein (Fig. 1D). A single mutation in which a serine at position 310 was changed to a glycine dramatically increased the virulence of the JHM.IA recombinant (Fig. 2). None of the other three changes increased the virulence of JHM.IA, and in fact, as shown below, some of the variants were less neurovirulent than the rJHM.IA parental isolate. Sequence analysis showed that all four mutations were stable and that no second-site mutations were detected after passage in mice (data not shown).
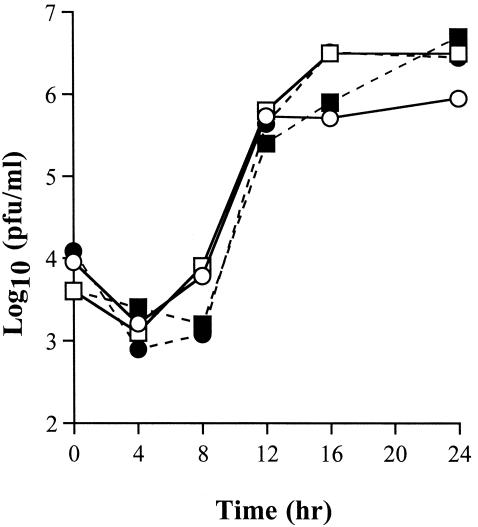
The differences in spike sequence did not cause any differences in the ability of the recombinant viruses to replicate in murine fibroblast 17Cl-1 tissue culture cells (Fig. 4). For these experiments, cells were infected with JHM.SD, JHM.IA, or one of the six recombinant viruses. The two wild-type nonrecombinant viruses (JHM.SD and JHM.IA) replicated with identical kinetics, as did the two parental wild-type recombinants (rJHM.IA and rJHM.SD). All of the single-amino-acid recombinants also replicated with the same kinetics as rJHM.IA and rJHM.SD (data not shown). Of note, all the recombinants exhibited a slight but consistent delay in virus growth at 8 h p.i. relative to the nonrecombinant viruses. The basis of this delay is not known, but it has been observed previously (45).
FIG. 4.
Kinetics of virus production in 17Cl-1 cells. Cells were infected with the indicated viruses at a multiplicity of 1 PFU per cell. Cells and supernatants were harvested at the indicated times and freeze-thawed. Titers were measured by plaque assay on HeLa-MHVR cells. Symbols: □, JHM.IA; ○, JHM.SD; ▪, rJHM.IA; •, rJHM.SD.
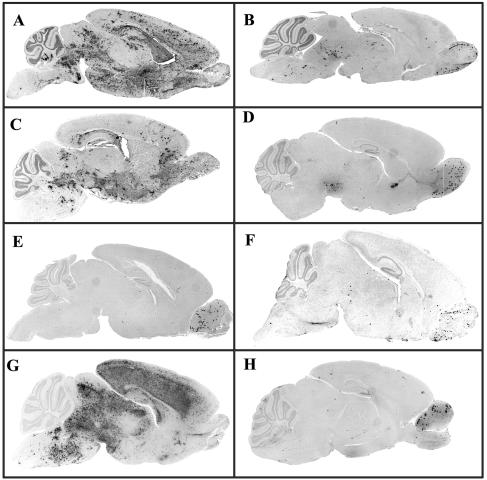
To assess the role of enhanced virus replication in the increased neurovirulence shown by JHM.SD, rJHM.SD, and rJIA-S.S310G, virus titers from the CNSs of infected mice were measured. Titers were 1 to 2 log units higher in the CNSs of mice infected with the more neurovirulent strains (Table 2). These increased virus titers correlated with higher levels of virus antigen in the infected CNS. Viral antigen was detected throughout the CNSs of mice infected with JHM.SD, rJHM.SD, or rJIA-S.S310G (Fig. 5A, C, and G, respectively). The pattern of antigen distribution was similar to that observed in suckling or adult mice infected with JHM.IA in the absence of protective antibody (3). Virus antigen was detected initially in the olfactory bulb, the site of initial infection in the CNS after intranasal inoculation, and in structures that are neuroanatomically connected to the bulb, such as the primary olfactory cortex, lateral hypothalamus, medial septal nucleus, and anterior amygdaloid area. Over the next 1 to 2 days, the virus spread to more distally connected structures, such as the medial parabrachial nucleus, and spread laterally from primary, secondary, and tertiary sites of infection, to involve most of the CNS. In striking contrast, viral antigen was confined to a few sites, all connected to the olfactory bulb, in mice infected with JHM.IA, rJHM.IA, rJIA-S.T61A, rJIA-S.I154T, or rJIA-S.L539F (Fig. 5B, D, E, F, and H). Since JHM.SD is very mildly hepatotropic, it was possible that hepatitis contributed to the rapid death observed after infection with the JHM.SD-like viruses. However, histological examination of livers from moribund pups showed no pathological changes (data not shown), demonstrating that the presence of maternally derived antibodies protected mice from liver disease.
TABLE 2.
JHM titers in the infected brain at day 5 p.i.
| Virus(no. of mice analyzed) | Viral titer (log10 PFU/g)a |
|---|---|
| JHM.IA (6) | 3.97 ± 0.53 |
| JHM.SD (8) | 5.92 ± 0.54 |
| rJHM.IA (6) | 3.87 ± 0.35 |
| rJHM.SD (7) | 5.58 ± 0.60 |
| rJIA-S.T61A (6) | 2.89 ± 0.36 |
| rJIA-S.I154T (9) | 3.54 ± 0.61 |
| rJIA-S.S310G (6) | 5.24 ± 0.68 |
| rJIA-S.L539F (8) | 3.10 ± 0.29 |
Boldfaced titers are significantly higher than all the others (P < 0.005).
FIG. 5.
Enhanced spread within the CNS by viruses encoding a Gly at position 310 of the S protein. Suckling mice were infected intranasally at the age of 10 days with either JHM.SD (A), JHM.IA (B), rJHM.SD (C), rJHM.IA (D), rJIA-S.T61A (E), rJIA-S.I154T (F), rJIA-S.S310G (G), or rJIA-S.L539F (H). Five days p.i., brains were harvested and fixed as described in Materials and Methods. Viral antigen was detected with the anti-N MAb 5B188.2 and counterstained with hematoxylin.
Gly at spike residue 310 diminishes S1-S2 stability.
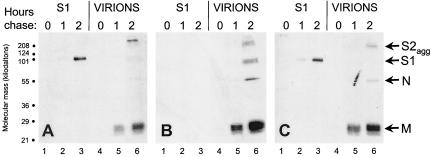
In infected tissue culture cells, about half of the spike proteins that successfully fold into native oligomers are incorporated into virions at intracellular budding sites and then are transported out into the medium. During transport, JHM spike proteins are quantitatively cleaved into noncovalently interacting S1 and S2 subunits. Once in tissue culture growth medium, these noncovalent S1-S2 associations are often poorly maintained, and particularly unstable associations are often evident in the most virulent virus strains (17, 27). To determine whether the Gly-to-Ser change at spike residue 310 changed JHM.IA S1-S2 heteromeric stability, infected cells were pulse-labeled for 30 min with 35S-labeled amino acids, and the fate of radioactive S proteins was monitored during a 2-h chase period. Electrophoretic analyses revealed that 35S-labeled virions, harvested from the chase medium by ultracentrifugation, contained S2, S1, M, and N proteins (Fig. 6). S2 was identified in all virion preparations as high-molecular-weight, SDS-resistant aggregates. These aggregates were not dissociated into monomeric, ∼95-kDa S2 because virions were not boiled in the SDS solubilizing buffer. This electrophoretic behavior permitted clear identification of S1 fragments electrophoresing as ∼100-kDa monomeric fragments. Notably, these ∼100-kDa S1 fragments were retained on rJHM.IA virions (Fig. 6B, lane 6) but were barely detectable on rJHM.SD or rJIA.S.S310G virions (Fig. 6A and C, lanes 6). The S1 fragments produced during rJHM.SD and rJ.IA.S310G infection were readily captured by immunoprecipitation from the supernatants remaining after virions had been pelleted (Fig. 6A and C, lanes 3), indicating that they had separated from S2. The levels of these free S1 proteins exceeded those found on complete rJHM.IA virions (Fig. 6B, lane 6), consistent with S1 shedding from both secreted virions and infected cell surfaces during the 2-h chase period. S1 shedding was undetectable in rJHM.IA-infected cultures (Fig. 6B, lane 3). These findings indicate that the labile JHM.SD spikes could be stabilized by the four JHM.IA-specific amino acid changes in S1 and that the S310G change in JHM.IA had a powerful destabilizing effect.
FIG. 6.
Effects of the JHM.IA mutations on S1 shedding from virions. 17Cl-1 cells infected with rJHM.SD (A), rJHM.IA (B), or rJ.IA.S.S310G (C) were pulse-labeled for 30 min with Tran35S-label and then chased in nonradioactive medium for the indicated times. Media were then collected, virions were pelleted by ultracentrifugation, and S1 fragments which remained soluble were immunoprecipitated onto NCEACAM-Fc beads (16). Radioactive proteins were separated by electrophoresis and visualized by fluorography.
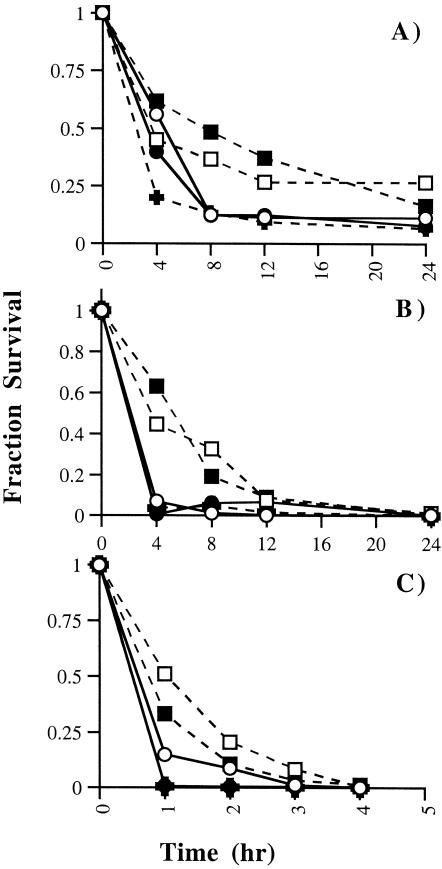
The relative abundance of virion-associated S1 correlated with virus infectivity. This was evident from a determination of the infectivities of cell-free viruses after exposures to various buffers at 37°C. At all pH values tested, the infectivity titers of JHM.SD, rJHM.SD, and rJIA-S.S310G declined rapidly in comparison with those of JHM.IA and rJHM.IA (Fig. 7). These findings indicated that the labile JHM.SD and rJIA.S.S310G viruses were poorly maintained in tissue culture environments.
FIG. 7.
Thermostability of spike variants. Cell-free virus was incubated in solutions at a pH of 6.0 (A), 7.0 (B), or 8.0 (C) at 37°C. Aliquots were removed at the indicated time points and titered on HeLa-MHVR cells. The fraction of virus surviving at each time point is shown. Symbols: (□), JHM-IA; ○, JHM-SD; ▪, rJHM.IA; •, rJHM.SD; +, rJIA-S.S310G. Data are representative of four independent experiments.
Gly at spike residue 310 confers CEACAM-independent membrane fusion activity.
Previous findings have correlated extremely labile interactions between S1 and S2 with the ability to infect cells lacking specific receptors for JHM (14). Hypotheses explaining this property suggest that S1-S2 complexes on infected cell surfaces release their S1 spontaneously, without any requirement for S1 to engage CEACAM glycoproteins on adjacent cells. This spontaneous reorganization reveals portions of the S proteins, most likely in S2, which are necessary for coalescing membranes.
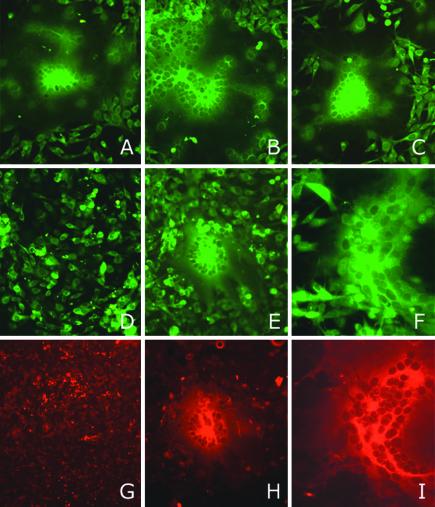
The striking differences in S1-S2 stability among the rJHM isolates (Fig. 6) warranted investigation of this CEACAM-independent membrane fusion activity. To this end, 17Cl-1 cells were infected with rJHM.IA or rJHM.SD and overlaid onto BHK cells (lacking murine CEACAM) previously labeled with CFSE. There were no differences in syncytium formation when cells infected with either virus were overlaid onto permissive 17Cl-1 cells (Fig. 8A and B). However, when the infected cells were overlaid onto BHK cells, striking differences were evident. Large syncytia (25 to 35 nuclei) resulted when rJHM.SD-infected cells were overlaid (Fig. 8E and H), whereas infection with rJHM.IA resulted, most commonly, in no syncytia (Fig. 8D and G). In one out of six experiments, infection with rJHM.IA resulted in a few small syncytia (3 to 5 nuclei). This absence of syncytia was also observed when the BHK cells were overlaid with 17Cl-1 cells infected with rJIA.S.A61T, rJIA.S.I154T, or rJIA.S.L539F (data not shown). However, large syncytia, similar in size to those observed after infection with rJHM.SD, were observed after cells infected with rJIA.S.S310G were overlaid onto either 17Cl-1 (Fig. 8C) or BHK (Fig. 8F and I) cells. Thus, the ability to infect cells lacking murine CEACAM receptors correlated with the stability of S1-S2 heteromers and also with neurovirulence.
FIG. 8.
Effect of a Gly at position 310 of the S protein on receptor-independent spread. 17Cl-1 cells (JHM permissive) infected with rJHM.IA (A, D, and G), rJHM.SD (B, E, and H), or rJIA.S.S310G (C, F, and I) were overlaid onto CFSE-labeled uninfected 17Cl-1 cells (A through C) or BHK cells (non-JHM permissive) (D through I) at a 1:50 ratio as described in Materials and Methods. At 20 h p.i., cells were fixed, and viral antigen was detected by sequential treatments with an anti-N antibody and a Texas red-conjugated goat anti-mouse antibody. Cells were visualized by fluorescent microscopy. Green fluorescence, CFSE labeling; red fluorescence, Texas red labeling. Only CSFE-labeled cells are shown in panels A through C.
DISCUSSION
Origins of JHM.IA and JHM.SD.
The precise origins of JHM.IA and JHM.SD are not known with certainty. All JHM strains were derived from a single mouse with hindlimb paralysis identified by Cheever et al. (8). With passage through the brains of suckling mice, the initial isolate became increasingly virulent, and it is likely that JHM.SD is one of these virulent isolates. JHM.IA, while also derived from the same stocks, may have mutated during the course of passage in tissue culture cells. JHM.SD does indeed undergo adaptive changes during passage in tissue culture, and the adaptive mutations are routinely fixed into S genes (18, 27). The environmental conditions accounting for these changes may be complex, but it is reasonable to suspect that the in vitro incubation temperature and the pH of the growth medium selectively eliminate the more labile virus particles. Extracellular JHM.SD particles are extremely temperature sensitive, while tissue culture variants including JHM.IA are relatively stable against thermal inactivation of infectivity, particularly at slightly basic pH conditions (Fig. 7C) (27, 66).
Stability of S1-S2 interaction.
As with the spike proteins of human immunodeficiency viruses (37), the receptor-binding subunits on JHM S proteins can detach from virions (59). Spontaneous S1 detachment was pronounced in the thermolabile JHM.SD strain but essentially absent in the more stable JHM.IA strain (Fig. 6). Similar patterns correlating thermostability to S1-S2 heteromeric stability have been observed and have been attributed to large deletion mutations in S1 (27) or to point mutations in a portion of S2 designated “heptad repeat 1” (27, 40). Relative to JHM.SD, JHM.IA contained four amino acid differences in S1, and a systematic analysis of single-site revertants clearly demonstrated that the JHM.IA S1 subunits detached from S2 when serine 310 was converted back to glycine. This finding adds to the spectrum of different spontaneous JHM mutations that stabilize S1-S2 associations, and while one cannot rule out effects that the S310G change might have on overall S protein conformation, this finding raises the possibility that S1 links directly to S2 at regions near residue 310. Residue 310 is within a portion of S1 that binds murine CEACAM, the primary MHV receptor (28), and functional interactions of this region with S2 have been proposed recently (36).
Several reports have documented that infection with JHM variants with more stable S1-S2 complexes result in less syncytial development than infection with the virulent JHM.SD strain (27, 36, 41). In a comparison of syncytial expansion of JHM.SD and JHM.IA on CEACAM-bearing cells, no differences were observed (Fig. 8A and B), suggesting that the previously hypothesized correlation between S1-S2 stability and fusion activity is not absolute. However, in parallel fusion assays using cells lacking murine CEACAMs, we identified clear differences among the variants. Here JHM.IA did not spread to cells lacking CEACAM, but the expected JHM.SD-induced fusion was observed (14, 41) (Fig. 8D and E, respectively). The introduced S310G reversion fully restored CEACAM-independent fusion to the level of that with JHM.SD (Fig. 8F). In preliminary experiments, we performed the reciprocal replacement of Gly310 in JHM.SD with Ser in the context of vaccinia virus constructs expressing the S protein. This change eliminated CEACAM-independent fusion (data not shown). Thus, we suggest that CEACAM-independent infection depends on Gly310 and also on labile S1-S2 interactions. Weak connections between S domains may be necessary to spontaneously reveal membrane fusion machinery in S2 and thus cause syncytia without any need for a CEACAM “trigger.”
Effect on neurovirulence.
In previous studies, mutations that compromised CEACAM-dependent cell fusion had profound effects on virulence (17). JHM.IA and JHM.SD mediated CEACAM-dependent cell fusion to similar extents (Fig. 8A and B), and consequently, the effect on neurovirulence was more subtle, since both viruses caused acute encephalitis in naïve mice with the same 50% lethal dose (22, 25, 45). However, the S1 mutations in JHM.IA specifically diminished only CEACAM-independent fusion (Fig. 8), and this unique feature of the IA variants correlated with decreased neurovirulence in the presence of suboptimal amounts of an anti-JHM antibody (Fig. 2). In the most striking example, the S310G reversion in JHM.IA that restored CEACAM-independent fusion concomitantly caused JHM.IA to revert to the JHM.SD neurovirulent phenotype. A Gly at position 310 is also present in several other strains of MHV, including strain A59, which are less neurovirulent than MHV-IA or MHV-SD and do not mediate receptor-independent spread, suggesting that this amino acid must interact with other MHV-SD-specific amino acids to mediate increased pathogenicity (14, 34).
We note that the three other amino acid differences between JHM.IA and JHM.SD may also contribute, more subtly, to the virulence of JHM.SD in the presence of the S310G change. Another highly neurovirulent isolate of JHM, cl-2, isolated after passage through rat brains, encodes the same four amino acids as JHM.SD, suggesting that these four amino acids are required for optimal virulence (62). Of particular interest are the T61A and I154T changes. Both of these changes are located within the first 330 amino acids of S, the region critical for binding to the host cell receptor (28). T61A is a conservative change but is adjacent to residue 62, which has previously been shown to be crucial for receptor binding (61). The third change, L539F, is located within the hypervariable region. This region of the S protein tolerates point mutations and deletion, making it less likely that this change contributes to pathogenesis. Notably, none of these point mutations affect a consensus N-glycosylation motif.
Previous analyses have revealed low expression of the primary receptor for JHM, CEACAM1a, in the murine CNS (19, 20), even though JHM infects the CNS primarily. These low levels of expression may be adequate to support JHM replication. Another possibility is that other members of the CEACAM family or a completely novel receptor is utilized in the CNS. Two other members of the CEACAM family, CEACAM2 and PSG16 (previously designated brain CEA), are both expressed in the CNS, although PSG16 does not function as a receptor for JHM (9, 43). Alternatively, JHM.SD may be able to compensate for low receptor density in the CNS by having a lower threshold for undergoing conformational changes necessary for fusion activation. This property would increase the ability of JHM.SD, rJHM.SD, and rJIA-S.S310G to initiate infection of cells by both receptor-dependent and receptor-independent mechanisms. After intranasal inoculation, this enhanced infectivity may directly increase the ability of the virus to spread laterally from sites neuroanatomically connected to the olfactory bulb.
Another possibility is that JHM.SD and its derivatives spread more rapidly within the CNS because the initial immune response to this strain was compromised and therefore was unable to control the infection. Infection of macrophages, important for initiating the immune response, has been documented in mice infected with MHV-2 or MHV-3, both of which are hepatotropic strains of MHV (2, 12). Activation of macrophages in mice infected with MHV-3 resulted in induction of prothrombinase with subsequent fulminant hepatitis (12). Differential infection of these cells or dendritic cells by JHM.IA and JHM.SD might contribute to the observed differences in neurovirulence. Precedents for this come from other viral infections. Infection of dendritic cells or monocytes may facilitate viral replication in humans infected with measles virus or dengue virus (24, 68). Infection of dendritic cells with immunosuppressive strains of lymphocytic choriomeningitis virus resulted in deletion or dysfunction of antiviral CD8 T cells (38, 53, 72).
Collectively, data from our study and those previously published suggest a critical role for S1-S2 instability in enhancing the replication of JHM in the murine CNS. The crystal structure of CEACAM1 has been published recently (64), but that of the JHM S protein is not yet known. Knowledge of both crystal structures will be essential for understanding the conformational changes of the S proteins of JHM.IA and JHM.SD that result in differences in neurovirulence.
Acknowledgments
This work was supported by grants from the NIH (NS 365092 and NS 40438 to S.P.; NS31616 to T.M.G.) and the National Multiple Sclerosis Society (to S.P.). E.O. was supported by an NIH predoctoral National Research Service Award (F31 Al10348).
We thank C. Martin Stoltzfus and Ajai Dandekar for helpful discussions and critical review of the manuscript and Lecia Pewe for valuable technical assistance.
REFERENCES
- 1.Almazan, F., J. M. Gonzalez, Z. Penzes, A. Izeta, E. Calvo, J. Plana-Duran, and L. Enjuanes. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 97:5516-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bang, F. B. 1978. Genetics of resistance of animals to viruses. I. Introduction and studies in mice. Adv. Virus Res. 1978:269-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, E., M. Cassell, and S. Perlman. 1993. Two neurotropic viruses, herpes simplex virus type 1 and mouse hepatitis virus, spread along different neural pathways from the main olfactory bulb. Neuroscience 57:1007-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle, J. F., D. G. Weismiller, and K. V. Holmes. 1987. Genetic resistance to mouse hepatitis virus correlates with absence of virus-binding activity on target tissues. J. Virol. 61:185-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchmeier, M. J., H. A. Lewicki, P. J. Talbot, and R. L. Knobler. 1984. Murine hepatitis virus-4 (strain JHM)-induced neurologic disease is modulated in vivo by monoclonal antibody. Virology 132:261-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casais, R., V. Thiel, S. G. Siddell, D. Cavanagh, and P. Britton. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 75:12359-12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 8.Cheever, F. S., J. B. Daniels, A. M. Pappenheimer, and O. T. Bailey. 1949. A murine virus (JHM) causing disseminated encephalomyelitis with extensive destruction of myelin. J. Exp. Med. 90:181-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, D., M. Asanaka, K. Yokomori, F. Wang, S. Hwang, H. Li, and M. M. C. Lai. 1995. A pregnancy-specific glycoprotein is expressed in the brain and serves as a receptor for mouse hepatitis virus. Proc. Natl. Acad. Sci. USA 92:12095-12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, A. R., R. L. Knobler, H. Powell, and M. J. Buchmeier. 1982. Monoclonal antibodies to murine hepatitis virus-4 (strain JHM) define the viral glycoprotein responsible for attachment and cell-cell fusion. Virology 119:358-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Groot, R. J., J. A. Lenstra, W. Luytjes, H. G. Niesters, M. C. Horzinek, B. A. van der Zeijst, and W. J. Spaan. 1987. Sequence and structure of the coronavirus peplomer protein. Adv. Exp. Med. Biol. 218:31-38. [DOI] [PubMed] [Google Scholar]
- 12.Fingerote, R. J., M. Abecassis, M. J. Phillips, Y. S. Rao, E. H. Cole, J. Leibowitz, and G. A. Levy. 1996. Loss of resistance to murine hepatitis virus strain 3 infection after treatment with corticosteroids is associated with induction of macrophage procoagulant activity. J. Virol. 70:4275-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleming, J. O., M. D. Trousdale, F. El-Zaatari, S. A. Stohlman, and L. P. Weiner. 1986. Pathogenicity of antigenic variants of murine coronavirus JHM selected with monoclonal antibodies. J. Virol. 58:869-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallagher, T., M. Buchmeier, and S. Perlman. 1992. Cell receptor-independent infection by a neurotropic murine coronavirus. Virology 191:517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher, T. M. 1996. Murine coronavirus membrane fusion is blocked by modification of thiols buried within the spike protein. J. Virol. 70:4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallagher, T. M. 1997. A role for naturally occurring variation of the murine coronavirus spike protein in stabilizing association with the cellular receptor. J. Virol. 71:3129-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallagher, T. M., and M. J. Buchmeier. 2001. Coronavirus spike proteins in viral entry and pathogenesis. Virology 279:371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallagher, T. M., C. Escarmis, and M. J. Buchmeier. 1991. Alteration of the pH dependence of coronavirus-induced cell fusion: effect of mutations in the spike glycoprotein. J. Virol. 65:1916-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godfraind, C., N. Havaux, K. V. Holmes, and J. P. Coutelier. 1997. Role of virus receptor-bearing endothelial cells of the blood-brain barrier in preventing the spread of mouse hepatitis virus-A59 into the central nervous system. J. Neurovirol. 3:428-434. [DOI] [PubMed] [Google Scholar]
- 20.Godfraind, C., S. G. Langreth, C. B. Cardellichio, R. Knobler, J. P. Coutelier, M. Dubois-Dalcq, and K. V. Holmes. 1995. Tissue and cellular distribution of an adhesion molecule in the carcinoembryonic antigen family that serves as a receptor for mouse hepatitis virus. Lab. Investig. 73:615-627. [PubMed] [Google Scholar]
- 21.Haring, J., and S. Perlman. 2001. Mouse hepatitis virus. Curr. Opin. Microbiol. 4:462-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haspel, M. V., P. W. Lampert, and M. B. A. Oldstone. 1978. Temperature-sensitive mutants of mouse hepatitis virus produce a high incidence of demyelination. Proc. Natl. Acad. Sci. USA 75:4033-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes, K. V., and G. S. Dveksler. 1994. Specificity of coronavirus/receptor interactions, p. 403-443. In E. Wimmer (ed.), Cell receptors for animal viruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Karp, C. L., M. Wysocka, L. M. Wahl, J. M. Ahearn, P. J. Cuomo, B. Sherry, G. Trinchieri, and D. E. Griffin. 1996. Mechanism of suppression of cell-mediated immunity by measles virus. Science 273:228-231. [DOI] [PubMed] [Google Scholar]
- 25.Knobler, R., M. Haspel, and M. B. A. Oldstone. 1981. Mouse hepatitis virus type-4 (JHM strain) induced fatal nervous system disease. Part I. Genetic control and the murine neuron as the susceptible site of disease. J. Exp. Med. 153:832-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Körner, H., A. Schliephake, J. Winter, F. Zimprich, H. Lassmann, J. Sedgwick, S. Siddell, and H. Wege. 1991. Nucleocapsid or spike protein-specific CD4+ T lymphocytes protect against coronavirus-induced encephalomyelitis in the absence of CD8+ T cells. J. Immunol. 147:2317-2323. [PubMed] [Google Scholar]
- 27.Krueger, D. K., S. M. Kelly, D. N. Lewicki, R. Ruffolo, and T. M. Gallagher. 2001. Variations in disparate regions of the murine coronavirus spike protein impact the initiation of membrane fusion. J. Virol. 75:2792-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubo, H., Y. K. Yamada, and F. Taguchi. 1994. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 68:5403-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai, M. M. C., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lecomte, J., V. Cainelli-Gebara, G. Mercier, S. Mansour, P. J. Talbot, G. Lussier, and D. Oth. 1987. Protection from mouse hepatitis virus type 3-induced acute disease by an anti-nucleoprotein monoclonal antibody. Arch. Virol. 97:123-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leparc-Goffart, I., S. T. Hingley, M. M. Chua, X. Jiang, E. Lavi, and S. R. Weiss. 1997. Altered pathogenesis of a mutant of the murine coronavirus MHV-A59 is associated with a Q159L amino acid substitution in the spike protein. Virology 239:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo, Z., and S. R. Weiss. 1998. Roles in cell-to-cell fusion of two conserved hydrophobic regions in the murine coronavirus spike protein. Virology 244:483-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luytjes, W., L. S. Sturman, P. J. Bredenbeek, J. Charite, B. A. M. van der Zeijst, M. C. Horzinek, and W. J. M. Spaan. 1987. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology 161:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masters, P. S. 1999. Reverse genetics of the largest RNA viruses. Adv. Virus Res. 53:245-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuyama, S., and F. Taguchi. 2002. Communication between S1N330 and a region in S2 of murine coronavirus spike protein is important for virus entry into cells expressing CEACAM1b receptor. Virology 295:160-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore, J. P., J. A. McKeating, R. A. Weiss, and Q. J. Sattentau. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139-1142. [DOI] [PubMed] [Google Scholar]
- 38.Moskophidis, D., F. Lechner, H. Pircher, and R. M. Zinkernagel. 1993. Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature 362:758-761. [DOI] [PubMed] [Google Scholar]
- 39.Nakanaga, K., K. Yamanouchi, and K. Fujiwara. 1986. Protective effect of monoclonal antibodies on lethal mouse hepatitis virus infection in mice. J. Virol. 59:168-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nash, T., and M. J. Buchmeier. 1997. Entry of mouse hepatitis virus into cells by endosomal and nonendosomal pathways. Virology 233:1-8. [DOI] [PubMed] [Google Scholar]
- 41.Nash, T., and M. J. Buchmeier. 1996. Spike glycoprotein-mediated fusion in biliary glycoprotein-independent cell-associated spread of mouse hepatitis virus infection. Virology 223:68-78. [DOI] [PubMed] [Google Scholar]
- 42.Navas, S., S. H. Seo, M. M. Chua, J. D. Sarma, E. Lavi, S. T. Hingley, and S. R. Weiss. 2001. Murine coronavirus spike protein determines the ability of the virus to replicate in the liver and cause hepatitis. J. Virol. 75:2452-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nedellec, P., G. S. Dveksler, E. Daniels, C. Turbide, B. Chow, A. A. Basile, K. V. Holmes, and N. Beauchemin. 1994. Bgp2, a new member of the carcinoembryonic antigen-related gene family, encodes an alternative receptor for mouse hepatitis viruses. J. Virol. 68:4525-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohtsuka, N., Y. K. Yamada, and F. Taguchi. 1996. Difference in virus-binding activity of two distinct receptor proteins for mouse hepatitis virus. J. Gen. Virol. 77:1683-1692. [DOI] [PubMed] [Google Scholar]
- 45.Ontiveros, E., L. Kuo, P. S. Masters, and S. Perlman. 2001. Inactivation of expression of gene 4 of mouse hepatitis virus strain JHM does not affect virulence in the murine CNS. Virology 290:230-238. [DOI] [PubMed] [Google Scholar]
- 46.Parker, S. E., T. M. Gallagher, and M. J. Buchmeier. 1989. Sequence analysis reveals extensive polymorphism and evidence of deletions within the E2 glycoprotein gene of several strains of murine hepatitis virus. Virology 173:664-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perlman, S., G. Jacobsen, A. L. Olson, and A. Afifi. 1990. Identification of the spinal cord as a major site of persistence during chronic infection with a murine coronavirus. Virology 175:418-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perlman, S., R. Schelper, E. Bolger, and D. Ries. 1987. Late onset, symptomatic, demyelinating encephalomyelitis in mice infected with MHV-JHM in the presence of maternal antibody. Microb. Pathog. 2:185-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pewe, L., G. Wu, E. M. Barnett, R. Castro, and S. Perlman. 1996. Cytotoxic T cell-resistant variants are selected in a virus-induced demyelinating disease. Immunity 5:253-262. [DOI] [PubMed] [Google Scholar]
- 50.Pewe, L., S. Xue, and S. Perlman. 1997. Cytotoxic T cell-resistant variants arise at early times after infection in C57BL/6 but not in SCID mice infected with a neurotropic coronavirus. J. Virol. 71:7640-7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phillips, J. J., M. M. Chua, E. Lavi, and S. R. Weiss. 1999. Pathogenesis of chimeric MHV4/MHV-A59 recombinant viruses: the murine coronavirus spike protein is a major determinant of neurovirulence. J. Virol. 73:7752-7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanchez, C. M., A. Izeta, J. M. Sanchez-Morgado, S. Alonso, I. Sola, M. Balasch, J. Plana-Duran, and L. Enjuanes. 1999. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 73:7607-7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sevilla, N., S. Kunz, A. Holz, H. Lewicki, D. Homann, H. Yamada, K. P. Campbell, J. C. de La Torre, and M. B. Oldstone. 2000. Immunosuppression and resultant viral persistence by specific viral targeting of dendritic cells. J. Exp. Med. 192:1249-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spaan, W., D. Cavanagh, and M. C. Horzinek. 1988. Coronaviruses: structure and genome expression. J. Gen. Virol. 69:2939-2952. [DOI] [PubMed] [Google Scholar]
- 55.Stohlman, S. A., C. C. Bergmann, and S. Perlman. 1998. Mouse hepatitis virus, p. 537-557. In R. Ahmed and I. Chen (ed.), Persistent viral infections. John Wiley & Sons, Ltd., New York, N.Y.
- 56.Stohlman, S. A., and D. R. Hinton. 2001. Viral induced demyelination. Brain Pathol. 11:92-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stohlman, S. A., G. K. Matsushima, N. Casteel, and L. P. Weiner. 1986. In vivo effects of coronavirus-specific T cell clones: DTH inducer cells prevent a lethal infection but do not inhibit virus replication. J. Immunol. 136:3052-3056. [PubMed] [Google Scholar]
- 58.Sturman, L., and K. Holmes. 1985. The novel glycoproteins of coronaviruses. Trends Biochem. Res. 10:17-20. [Google Scholar]
- 59.Sturman, L. S., C. S. Ricard, and K. V. Holmes. 1990. Conformational change of the coronavirus peplomer glycoprotein at pH 8.0 and 37 degrees C correlates with virus aggregation and virus-induced cell fusion. J. Virol. 64:3042-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun, N., D. Grzybicki, R. Castro, S. Murphy, and S. Perlman. 1995. Activation of astrocytes in the spinal cord of mice chronically infected with a neurotropic coronavirus. Virology 213:482-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suzuki, H., and F. Taguchi. 1996. Analysis of the receptor-binding site of murine coronavirus spike protein. J. Virol. 70:2632-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taguchi, F., T. Ikeda, and H. Shida. 1992. Molecular cloning and expression of a spike protein of neurovirulent murine coronavirus JHMV variant cl-2. J. Gen. Virol. 73:1065-1072. [DOI] [PubMed] [Google Scholar]
- 63.Taguchi, F., and Y. K. Shimazaki. 2000. Functional analysis of an epitope in the S2 subunit of the murine coronavirus spike protein: involvement in fusion activity. J. Gen. Virol. 81:2867-2871. [DOI] [PubMed] [Google Scholar]
- 64.Tan, K., B. D. Zelus, R. Meijers, J. H. Liu, J. M. Bergelson, N. Duke, R. Zhang, A. Joachimiak, K. V. Holmes, and J. H. Wang. 2002. Crystal structure of murine sCEACAM1a[1,4]: a coronavirus receptor in the CEA family. EMBO J. 21:2076-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thiel, V., J. Herold, B. Schelle, and S. G. Siddell. 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 82:1273-1281. [DOI] [PubMed] [Google Scholar]
- 66.Tsai, J. C., B. D. Zelus, K. V. Holmes, and S. R. Weiss. 2003. The N-terminal domain of the murine coronavirus spike glycoprotein determines the CEACAM1 receptor specificity of the virus strain. J. Virol. 77:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weismiller, D. G., L. S. Sturman, M. J. Buchmeier, J. O. Fleming, and K. V. Holmes. 1990. Monoclonal antibodies to the peplomer glycoprotein of coronavirus mouse hepatitis virus identify two subunits and detect a conformational change in the subunit released under mild alkaline conditions. J. Virol. 64:3051-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu, S. J., G. Grouard-Vogel, W. Sun, J. R. Mascola, E. Brachtel, R. Putvatana, M. K. Louder, L. Filgueira, M. A. Marovich, H. K. Wong, A. Blauvelt, G. S. Murphy, M. L. Robb, B. L. Innes, D. L. Birx, C. G. Hayes, and S. S. Frankel. 2000. Human skin Langerhans cells are targets of dengue virus infection. Nat. Med. 6:816-820. [DOI] [PubMed] [Google Scholar]
- 69.Yamaguchi, K., N. Goto, S. Kyuwa, M. Hayami, and Y. Toyoda. 1991. Protection of mice from a lethal coronavirus infection in the central nervous system by adoptive transfer of virus-specific T cell clones. J. Neuroimmunol. 32:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yount, B., K. M. Curtis, and R. S. Baric. 2000. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 74:10600-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zajac, A. J., J. N. Blattman, K. Murali-Krishna, D. Sourdive, M. Suresh, J. D. Altman, and R. Ahmed. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 188:2205-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zelus, B. D., J. H. Schickli, D. M. Blau, S. R. Weiss, and K. V. Holmes. 2003. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37 degrees C either by soluble murine CEACAM1 receptors or by pH 8. J. Virol. 77:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zelus, B. D., D. R. Wessner, R. K. Williams, M. N. Pensiero, F. T. Phibbs, M. de Souza, G. S. Dveksler, and K. V. Holmes. 1998. Purified, soluble recombinant mouse hepatitis virus receptor, Bgp1(b), and Bgp2 murine coronavirus receptors differ in mouse hepatitis virus binding and neutralizing activities. J. Virol. 72:7237-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]