Abstract
Hepatitis delta virus expresses two essential proteins, the small and large delta antigens, and both are required for viral propagation. Proper function of each protein depends on the presence of a common amino-terminal multimerization domain. A crystal structure, solved using a peptide fragment that contained residues 12 to 60, depicts the formation of an octameric ring composed of antiparallel coiled-coil dimers. Because this crystal structure was solved for only a fragment of the delta antigens, it is unknown whether octamers actually form in vivo at physiological protein concentrations and in the context of either intact delta antigen. To test the relevance of the octameric structure, we developed a new method to probe coiled-coil structures in vivo. We generated a panel of mutants containing cysteine substitutions at strategic locations within the predicted monomer-monomer interface and the dimer-dimer interface. Since the small delta antigen contains no cysteine residues, treatment of cell extracts with a mild oxidizing reagent was expected to induce disulfide bond formation only when the appropriate pairs of cysteine substitution mutants were coexpressed. We indeed found that, in vivo, both the small and large delta antigens assembled as antiparallel coiled-coil dimers. Likewise, we found that both proteins could assume an octameric quaternary structure in vivo. Finally, during the course of these experiments, we found that unprenylated large delta antigen molecules could be disulfide cross-linked via the sole cysteine residue located within the carboxy terminus. Therefore, in vivo, the C terminus likely provides an additional site of protein-protein interaction for the large delta antigen.
Hepatitis delta virus (HDV) is a subviral agent that requires the presence of the envelope proteins of the hepatitis B virus in order to produce infectious virions (26, 27). HDV possesses a 1.7-kb, minus-sense, closed circular RNA genome that folds into an unbranched rod-like structure (4, 10, 34). Although its genome contains several open reading frames, HDV requires the production of only two proteins for its propagation (35). One is a 195-amino-acid protein termed the small delta antigen (HDAg-S). The other is a 214-amino-acid isoform termed the large delta antigen (HDAg-L). These two proteins are encoded by the same coding sequences and are identical but for 19 additional amino acids present at the C terminus of HDAg-L (16, 35). Despite their similarities, HDAg-S and HDAg-L play fundamentally different roles in the HDV replicative cycle. Whereas HDAg-S is expressed both early and late and is required for viral replication (3, 11), HDAg-L is expressed only late in infection, is a potent inhibitor of replication, and is required for viral assembly (2, 3, 7).
HDV replication is thought to proceed via a double rolling circle mechanism that requires the presence of HDAg-S (3, 11) and a host RNA polymerase (5, 18, 19). In this process, transcription of the genomic template produces a complementary strand of RNA, termed the antigenome. The antigenome acts as a replication intermediate and serves as a template for a second round of rolling circle replication to produce additional copies of the genome (19, 20, 22). At a later stage in infection, some antigenomes undergo RNA editing. This process leads to the production of HDAg-L (16), which, through an unknown mechanism, acts as a potent inhibitor of HDV replication (3, 7). In addition, because of the additional 19 amino acids at its C terminus, HDAg-L is farnesylated and is able to associate with the envelope proteins of the hepatitis B virus (2, 29, 31). Hence, by virtue of its ability to interact with both HDAg-S and HDV genomic RNA, HDAg-L escorts HDV genomic ribonucleoprotein (RNP) complexes into assembling virions (29, 30, 32).
Based on various mutational studies, the region shared in common by the delta antigens can be divided into four distinct domains. These include an amino-terminal multimerization domain (13, 37), a bipartite nuclear localization signal between residues 67 and 88 (38), an RNA binding domain between residues 89 and 144 (14, 32), and a Pro/Gly-rich region between residues 145 and 214 (13). The multimerization domain is predicted to form a coiled-coil structure and is known to be critical for proper function of both HDAg-S and HDAg-L (13, 37). In HDAg-S, deletion or mutation of this domain leads to a loss of the ability to support HDV replication. Similarly, in HDAg-L, deletion or mutation of this domain leads to a loss of the ability both to inhibit replication and to copackage HDAg-S into virions (13, 37). A crystal structure has been solved for a peptide fragment that included residues 12 to 60 of the multimerization domain. In that structure, four antiparallel coiled-coil dimers assemble into an octameric ring. However, since this structure was solved from a synthetic peptide fragment, it is unclear whether octamers actually form in vivo at physiological protein concentrations and in the context of either entire delta antigen primary sequence (39). Studies using glutaraldehyde cross-linking had previously indicated that the delta antigens may assemble as multimers larger than a dimer (13, 28, 33, 37). However, because the mechanism by which glutaraldehyde cross-links proteins is not specific, it is uncertain whether multimers larger than a dimer formed from interactions that involved only the multimerization domain or whether interactions involving other regions of the protein were also involved.
Recently, Moraleda et al. (21) attempted to test the relevance of the crystal structure in relation to the natural proteins by using an alanine mutagenesis approach. In their study, residues predicted to be involved in stabilizing the octamer were converted to alanines and the resulting HDAg-S and HDAg-L mutants were tested for loss of function. Surprisingly, they found that, while some mutations abolished the ability of HDAg-S to support replication, those same mutations had no effect on the ability of HDAg-L to either inhibit replication or copackage HDAg-S into virus-like particles. Moreover, when they tested the ability of their mutants to interact with a FLAG-tagged version of HDAg-S, they found that nearly all of their mutants retained the ability to interact with this tagged protein (21).
Based on that study, it is difficult to establish a correlation between the function of delta antigens and their multimerization status. First, it is unclear whether the interaction with the FLAG-tagged version of HDAg-S was due to formation of a dimer or formation of a higher-ordered multimer. Likewise, it is unknown whether a domain, in addition to the coiled coil, was responsible for this observed interaction. Additionally, since all of the relevant mutants involved a loss of function, the possibility that the mutation inhibited a process other than multimerization or that it had an unintended effect on protein folding and/or stability could not be ruled out. To directly probe the multimerization status of either delta antigen, either the wild-type proteins or, alternatively, mutants that are phenotypically null would have to be assayed. Since null mutants maintain full function, by definition they cannot have an unintended effect on folding, stability, or other properties.
To more precisely probe the oligomeric structure of the delta antigens in vivo and to gain a better understanding of the type of multimerization required for HDAg-S and HDAg-L function, we generated a panel of cysteine substitution mutants and used a disulfide bond formation assay to probe for the presence of antiparallel coiled-coil dimers and octamers in vivo. In structural studies, this type of strategy has proven to be useful for precisely defining areas of close contact within sites of protein-protein interaction (1, 17, 23). Using the solved crystal structure, we searched for pairs of residues that were predicted to be in proximity to one another and that were located at either the monomer-monomer or the dimer-dimer interface. These residues were then mutated to cysteines, and HDAg mutants containing one, two, or three Cys substitutions were generated. Since wild-type HDAg-S contains no Cys residues, formation of a disulfide bond should occur only with selected mutants in which two substituted Cys residues are in very high proximity. Following treatment of cell extracts with a mild oxidizing reagent, the presence or absence of a resulting disulfide bond could be visualized by running samples on nonreducing denaturing gels. Thus, by using selected phenotypically null Cys substitution, we could directly test the relevance of the crystal structure in relation to the structure assumed by the complete protein in vivo.
We found that, in vivo, the delta antigens dimerize through an antiparallel coiled-coil interaction. Likewise, we found that both HDAg-S and HDAg-L can form octamers that likely assume the ring-like structure depicted in the crystal structure. These structures could be found both inside cells and inside virus-like particles. Finally, during the course of these experiments, we found that wild-type HDAg-L can efficiently form a disulfide bond via the interaction of the sole C-terminal unprenylated Cys residue from one monomer with that from another. From these results, we suggest that the C-terminal 19 amino acids unique to HDAg-L may be an additional site of protein-protein interaction.
MATERIALS AND METHODS
Construction of HDAg Cys substitution mutants.
In this study, mutations are designated by the following convention: single-letter abbreviation for the residue in the wild-type sequence, followed by the residue number, followed by the single-letter abbreviation for the substituted residue. For example, L30C indicates a cysteine substitution for leucine at position 30. Substitutions in the HDAg coiled coil were generated via PCR, with primers containing the desired codon changes. PCR products were inserted between the SacII and EcoRI sites of pDL456, which expresses 1.2× unit-length HDV antigenomic RNA from a simian virus 40 promoter. Mutations were confirmed by sequencing. pDL456 derivatives were subsequently used to clone the desired mutation into either pDL444 or pDL445, which expresses HDAg-S or HDAg-L from a simian virus 40 promoter. The following mutations are listed with the codon changes in parentheses: E14C (GAA→TGT), L17C (CTC→TGC), I16C (ATC→TGC), W20C (TGG→TGT), V21C (GTG→TGC), A22C (GCC→TGC), G23C (GGA→TGC), L27C (TTA→TGT), L30C (CTC→TGC), L34C (CTC→TGC), T37C (ACA→TGT), L41C (CTC→TGC), I44C (ATA→TGT), N48C (AAT→TGT), N53C (AAC→TGC), I54C (ATC→TGC), and I57C (ATT→TGT).
Transfections and sample harvest.
HEK293 and Huh-7 cells were grown in 35-mm-diameter wells in Dulbecco's modified Eagle's medium (Cellgro; with 4.5 g of l-glucose per liter without l-glutamine) supplemented with fetal bovine serum (10%), nonessential amino acids (100 μM), l-glutamine (584 μg/ml), penicillin (100 U/ml), streptomycin (100 μg/ml), and amphotericin (0.25 μg/ml). Cells were grown overnight to 80% confluency and transfected with 3 μg of DNA by calcium phosphate precipitation. All transfected DNA samples contained 0.3 μg of an expression vector for secreted alkaline phosphatase (SEAP; pSS15). At 3 days posttransfection, SEAP activity was measured by a colorimetric assay to determine transfection efficiencies. To minimize disruption of protein-protein interactions, cells were lysed with 300 μl of 1× phosphate-buffered saline (PBS) containing 0.5% Triton X-100 and immediately frozen at −80°C. Aliquots were subsequently used in a cross-linking assay or processed to isolate total RNA for Northern blotting. Cells which were transfected with expression vectors for HDAg-S and HDAg-L mutants were harvested at 4 days posttransfection while those transfected with vectors expressing 1.2× unit-length HDV mutants were harvested at 7 days posttransfection.
Isolation of viral particles.
For isolation of viral particles, Huh-7 cells were grown in 10-cm-diameter plates and were transfected with a total of 33 μg of DNA containing 3 μg of SEAP vector (pSS15), 20 μg of an expression vector for the HBV small surface antigen (HBsAg-S; pBOM14), and 10 μg of expression vectors for the various HDAg mutants. At 4 days posttransfection, cells were lysed with 1 ml of 1× PBS-0.5% Triton X-100. Culture media from transfected cells were also collected and subjected to centrifugation at 5,000 × g for 10 min to pellet cell debris. Culture media were then transferred to new tubes and subjected to ultracentrifugation in a Beckman SW41 rotor run at 35,000 rpm for 3 h at 4°C. The resulting pelleted viral particles were then resuspended in 200 μl of 1× PBS-0.5% Triton X-100. Cell and viral extracts were subsequently used in disulfide cross-linking assays.
Disulfide cross-linking.
Disulfide cross-linking experiments were carried out essentially as described by Bonner et al. with some modifications (1). A 1 M solution of CuSO4 (in H2O) and a 500 mM solution of 1,10-phenanthroline (in 95% ethanol) were used to make a 100× stock solution of 50 mM CuSO4-130 mM 1,10-phenanthroline (CuPh). To induce disulfide bond formation, 1 μl of CuPh was added to 100 μl of cell extract. Samples were briefly vortexed and incubated at room temperature for 5 min. Following this brief incubation period, 33 μl of 4× Laemmli buffer (12) containing 8% sodium dodecyl sulfate (SDS) and 40% glycerol was added. Samples were then boiled at 95°C for 5 min. and homogenized with a 26-gauge needle. For reduced samples, 1 μl of 14.25 M β-mercaptoethanol was used instead of CuPh.
Western blotting.
Protein samples were run on either 6 or 12% acrylamide gels with an unstained protein ladder (Benchmark; Invitrogen) as a molecular weight standard. Electrophoretic transfer to nitrocellulose was performed at 100 V (∼800 mA) for 2 h to ensure sufficient transfer of high-molecular-weight species. The quality of the transfer was monitored by Ponceau staining of membranes. HDAg was detected by overnight incubation with rabbit polyclonal antiserum raised against recombinant (His)6-tagged HDAg (1:2,000 dilution). The following day, membranes were washed and incubated for 1 h with 125I-recombinant protein A to detect the primary antibody (1:1,000 dilution). Membranes were again washed, exposed overnight on a phosphor screen, and visualized using a phosphorimager (Storm Imager; Molecular Dynamics). Blocking of membranes, as well as dilution for primary antibody and recombinant protein A, was done in 1× PBS containing 1% powdered milk. Washes were performed at room temperature with 1× PBS.
RNA isolation and Northern blotting.
Total RNA was isolated from cell extracts with an RNeasy minikit (Qiagen). Because total RNA was isolated from samples lysed in 1× PBS-0.1% Triton X-100, the protocol for isolation of total RNA from animal cells was modified so that 3 volumes of RLT lysis buffer (Qiagen) were used for every 1 volume of cell lysate. Isolated RNA was mixed with 10× loading buffer containing 5 mM Vanadyl ribonucleoside complex and loaded onto 1.5% agarose gels. Samples were then electrotransferred to nylon membranes (Zeta Probe; Bio-Rad) and UV cross-linked. The amount of RNA loaded for each sample was monitored by ethidium bromine staining of agarose gels. Membranes were probed with a 32P-labeled antigenomic-sense RNA. This was generated by in vitro transcription with T7 RNA polymerase incubated with pSS159 linearized with Ecl136II and [32P]UTP. Prehybridization of membranes was performed for 2 h at 55°C while hybridization was performed overnight at 55°C (NorthernMax Hyb/Prehyb buffer; Ambion). The following day, membranes were subjected to a primary wash at room temperature in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS followed by a secondary wash at 70°C in 0.1× SSC-0.1% SDS. Membranes were then exposed overnight on a phosphor screen and visualized on a phosphorimager (Storm Imager; Molecular Dynamics).
RESULTS
Experimental design.
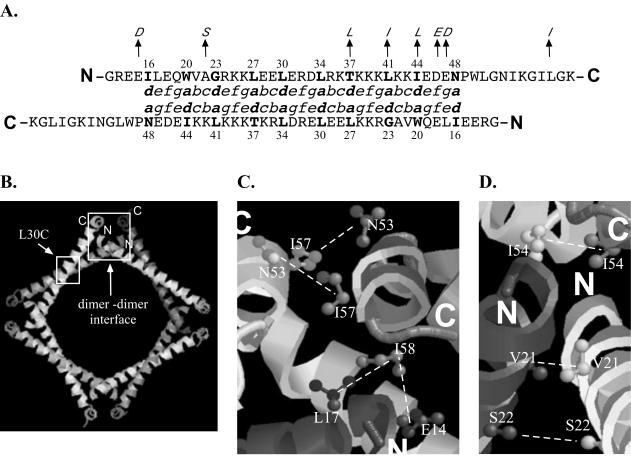
Based on the solved crystal structure, two regions of interaction appear to be important for assembly of octamers formed via the HDAg multimerization domain. The first region, which constitutes the monomer-monomer interface, spans residues 16 to 48 and involves an antiparallel coiled-coil interaction (39). As is characteristic of a coiled-coil domain, the primary sequence for this region consists of a heptad repeat in which hydrophobic amino acids are found at intervals of four and three residues. By convention, residues in the heptad repeat are designated as a to g, where a and d correspond to the hydrophobic repeats. These a and d positions are oriented on one side of an alpha helix and, by interacting with the a and d residues of a second alpha helix, form a hydrophobic core that stabilizes the coiled coil. In experiments performed with synthetic coiled coils, introduction of a single polar amino acid within the hydrophobic core can be tolerated and, in fact, influences the orientation assumed by the coiled coil (15, 24). We reasoned that introduction of Cys residues within the hydrophobic core of the delta antigen coiled coil may similarly be tolerated, and therefore we individually substituted Cys residues at all a and d positions throughout the delta antigen coiled coil.
For antiparallel coiled coils, a residues in one alpha helix are found to interact with d residues in the second alpha helix. Hence, if the HDAg coiled-coil domain assembles as an antiparallel coiled coil, disulfide cross-linking should be possible only when the appropriate combinations of a and d residue single mutants are coexpressed (Fig. 1A). In contrast, for parallel coiled coils, a given a residue in one helix is found to interact with the same a residues in the second helix and a given d residue in one helix interacts with the same d residue in the opposing helix. Hence, in this case, a single substitution mutant can give rise to a disulfide cross-linked species.
FIG. 1.
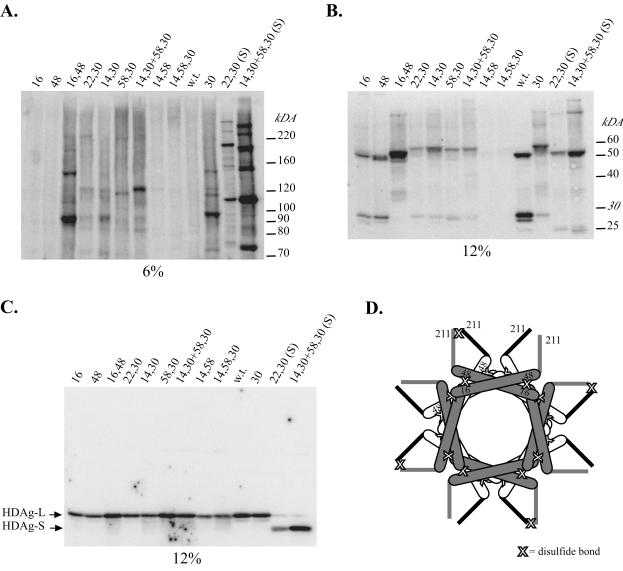
Cys substitutions at the monomer-monomer interface and the dimer-dimer interface. (A) Amino acid sequence for the multimerization domain of the Italian isolate used in this study. Residues in italics are those found in the American isolate peptide used for crystallization. Residues in boldface indicate a and d positions in the coiled coil which were targeted for Cys substitution. The sequences are aligned to indicate residues at the a and d positions that are predicted to interact with each other in an antiparallel coiled coil. (B) The two regions involved in stabilization of the octamer. Mutations designed to cross-link an octamer contained an L30C mutation (small box) and additional mutations at the dimer-dimer interface (large box). (C and D) Residues targeted for Cys substitution at the dimer-dimer interface. In the American isolate, residue 58 is an Ile while in the Italian isolate this residue is a Leu. The dashed lines indicate residues expected to form a disulfide cross-link when replaced by cysteines.
The crystal structure also demonstrates a second region involved in assembly of the octamer. This region consists of a four-helix bundle that brings together the ends of one dimer and those from another dimer. Stabilization of this region depends upon hydrophobic interactions involving both amino-terminal and carboxy-terminal residues of the peptide (39). To test for assembly of octamers in vivo, we first searched for pairs of residues within this domain that, when replaced with cysteines, were predicted to allow for disulfide bond formation involving monomers from two different dimers. For some mutations, the mutated residue within one monomer was predicted to be in proximity to that same residue within a second monomer. Hence, in this case, expression of a single mutant was predicted to yield a cross-linked dimer. For the remainder of the mutants, the mutated residue was predicted to be in proximity to a different residue that would also have to be mutated if a disulfide bond were to result. Hence, in these cases, either two different single mutants were coexpressed in cells or double mutants were assayed. To assay for octamer formation, mutants that formed disulfide bonds within the monomer-monomer interface were combined with those that formed disulfide bonds within the dimer-dimer interface (Fig. 1B).
In vivo assembly of HDAg-S dimers.
In this study, we used an isolate of HDV originally derived from an Italian patient, and as illustrated by Fig. 1A, the delta antigen primary sequence from this isolate differs from that of the peptide used to generate the crystal structure, which was derived from an American patient. However, since most of these differences in the multimerization domain involve conservative amino acid substitutions, this region of the delta antigen is assumed to be structurally similar in the two isolates. To test for the formation of an octameric structure in vivo, we first examined whether HDAg-S assembles as an antiparallel coiled coil. Expression vectors for HDAg-S mutants were constructed and used in various combinations to transfect either Huh-7 cells or HEK293 cells. A panel of mutants was generated that included all single Cys substitutions of the a and d positions in the predicted coiled-coil region. If the crystal structure was relevant to the structure assumed by HDAg-S in vivo, then only mixed transfections with the appropriate mutant combinations were expected to result in disulfide cross-linked HDAg-S dimers. Since the interior of a cell is a reductive environment where disulfide bonds rarely form, cell extracts were treated with CuPh, a reagent known to catalyze the air-mediated oxidation of free sulfhydryl groups (9). For example, a mixed transfection with the I16C mutant and the N48C mutant was expected to yield a cross-linked product. However, in single transfections with either mutant or in mixed transfections with the I16C mutant and any other mutant, no cross-linked product was expected (Fig. 1A).
When the various Cys substitution mutants were used in single transfections, all were expected to yield only HDAg-S monomers; however, with two mutants, species larger than monomers were observed (Fig. 2A). A larger species was observed with the W20C mutant; however, this species migrated significantly slower than a dimer. As shown in the figures below, the W20C mutant was not functional and was likely misfolded. We therefore hypothesized that the slower-migrating species resulted from formation of a disulfide bond that involved one misfolded W20C monomer and a host protein, perhaps a chaperone. In addition to the W20C mutant, a larger species was also observed with the L30C mutant. However, in this case, the migration of that species was consistent with that of a dimer. Upon close inspection of the crystal structure, this result was found to be consistent with the solved structure. Position 30 is centrally located within the antiparallel coiled coil and is adjacent to position 30 of the opposing monomer. To compare distances between residues targeted for Cys substitution, the distance between β-carbons in the crystal structure was measured. The distance between β-carbons for residue 30 and a second residue 30 in the dimer was found to be 5.7 Å. Hence, a β-carbon distance of 5.7 Å was sufficiently close to allow for disulfide bond formation. Although residue 34 is also located in the center of the coiled coil, the L34C single transfection did not yield a cross-linked dimer (Fig. 2A). However, in this case, the β-carbon distance was 6.4 Å. We concluded that the threshold β-carbon distance at which disulfide bond formation could occur was between 5.7 and 6.4 Å.
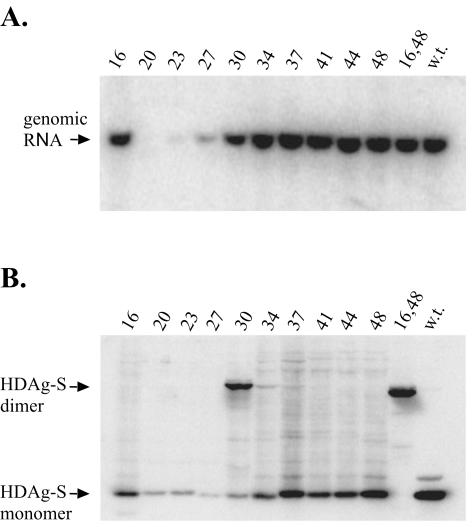
FIG. 2.
HDAg-S assembles as an antiparallel coiled coil. (A) Western blot for Cys substitution mutants transfected individually or in pairwise combinations into HEK293 cells. Disulfide cross-linking was performed on cell extracts as described in Materials and Methods. Samples were run on 12% acrylamide gels under nonreducing conditions. Lanes are numbered according to the residue targeted for Cys substitution. (B) Western blot for HEK293 cells cotransfected with the I16C mutant in pairwise combinations with additional Cys substitution mutants. Results were identical when Huh-7 cells were used. w.t., wild type.
As was predicted for an antiparallel coiled-coil interaction, when mutants that yielded monomers in single transfections were coexpressed with the appropriate partner mutant, cross-linked HDAg-S dimers could be detected. The extent of cross-linking in these mixed transfections exhibited some degree of variability, with the I16C and N48C pairing having the highest efficiency and the G23C and L41C pairing having the lowest efficiency (Fig. 2A). This variability in cross-linking likely reflected a partial disruption of HDAg-S structure with the W20C, G23C, and L27C mutants, since these mutants exhibited a loss of function (see below). Nevertheless, these results remain consistent with a coiled-coil structure that is antiparallel in orientation.
Finally, to test the specificity of the cross-linking assay, additional mixed transfections were performed in which the I16C mutant was paired in all possible combinations with the remaining Cys substitution mutants. Assuming that the crystal structure represents the actual structure assumed by the complete protein in vivo, the I16C mutant should be capable of cross-linking only to either an I44C mutant or an N48C mutant. As shown in Fig. 2B, this was indeed the case. In addition to the L30C-L30C cross-link that occurred when I16C was coexpressed with L30C, dimers were observed only when I16C was paired with I44C or N48C. These results indicated that the cross-linking assay is indeed specific.
In vivo assembly of HDAg-S dimers during HDV replication.
The previous experiments were performed with HDAg-S expression vectors, and hence, the intracellular concentration of delta antigen achieved is different from that which occurs during HDV replication. We next determined whether formation of antiparallel coiled-coil dimers also occurred when delta antigen was expressed as a consequence of HDV replication and in the presence of HDV RNA. In addition, since substitution of Cys residues for hydrophobic residues may have resulted in disruption of the HDAg-S structure, we wished to determine whether our various mutants were competent to support HDV replication. Therefore, the previously tested single Cys substitutions and an I16C,N48C double substitution were introduced into cDNA vectors that expressed circular antigenomic RNA. Although some delta antigen is expressed from messages transcribed directly from this vector, this level of expression represents less than 20% of that contributed by HDV replication at 7 days posttransfection (our unpublished results). HEK293 cells were transfected with these vectors and, at 7 days posttransfection, harvested for the disulfide cross-linking assay and Northern blot analysis. Since the transfected cDNA produces a transcript of antigenomic-sense RNA, in Northern blots the presence of genomic-sense RNA is indicative of HDV replication. As before, treatment of cell extracts with CuPh was expected to yield a disulfide cross-linked dimer only in certain transfections. In this case only the L30C single mutant and the I16C,N48C double mutant were expected to yield disulfide cross-linked dimers following single transfections.
From the Northern blots, we found that all but three mutants, W20C, G23C, and L27C, were able to support HDV replication near or at wild-type levels (Fig. 3A). Note that the three replication-defective mutants expressed much less delta antigen than did the other mutants (Fig. 3B), and this was consistent with the conclusion that, when replication occurs, it is responsible for the majority of delta antigen expression. As expected, of the mutants capable of supporting HDV replication, only an L30C mutant and an I16C,N48C double mutant yielded disulfide cross-linked dimers (Fig. 3B). In a mixed transfection with the I16C and N48C single mutants, an identical disulfide cross-linked dimer was also detected (data not shown). These results indicate that HDAg-S assembles as an antiparallel coiled coil during HDV replication and in the presence of HDV RNA. With the I16C,N48C double mutant, virtually 100% of the protein was resolved as a dimer. From this we concluded that, during HDV replication, essentially all delta antigen is present as dimers and/or higher-ordered multimers.
FIG. 3.
HDAg-S assembles as an antiparallel coiled coil during HDV replication. (A) Northern blot for HEK293 cells transfected with cDNA vectors expressing circular HDV antigenomic RNA containing Cys substitution mutations designed to probe for an antiparallel coiled-coil interaction. Cells were harvested 7 days posttransfection. (B) Western blot for CuPh-treated cell extracts obtained from cells transfected in the experiment shown for panel A. w.t., wild type.
We also observed that the W20C mutant, like the G23C and L27C mutants, produced lower levels of HDAg-S (Fig. 3B). However, unlike the G23C and L27C mutants, W20C does not appear to be capable of even partially supporting HDV replication. Based on the results from the mixed transfections, it appears that the W20C mutant is still able to assemble as a dimer (Fig. 2A, lane 20 + 44). When they were coexpressed with the appropriate Cys mutant, the extent of cross-linking for the G23C and L27C mutants correlated with the levels of replication for each mutant; however, this correlation did not hold true for the W20C mutant (Fig. 2A and 3A). In the crystal structure, W20 appears to stabilize the octamer by interacting with residues at both the monomer-monomer interface and the dimer-dimer interface (39). Thus, it is possible that the W20C mutation may have disrupted interactions at the dimer-dimer interface but not interactions at the monomer-monomer interface. We concluded that formation of HDAg-S dimers is necessary but not sufficient for the support of HDV replication.
In vivo assembly of HDAg-S octamers.
Since HDAg-S could assemble as an antiparallel coiled-coil dimer, we next wished to examine whether dimers could further assemble into octamers. Using a similar strategy, we substituted cysteines for residues located at the dimer-dimer interface such that formation of a disulfide bond was expected to cross-link a monomer from one dimer with a monomer from a second dimer. In addition, since the L30C mutation forms a cross-link at the monomer-monomer interface, double and triple mutants were designed that contained both the L30C substitution and Cys substitutions at the dimer-dimer interface. We anticipated that, by including this L30C mutation, octamers could be trapped via disulfide linkages at both the dimer-dimer interface and the monomer-monomer interface (Fig. 1B). These octamers could then be visualized by running samples under nonreducing conditions in 6% denaturing polyacrylamide gels. Depending on the location of the substituted cysteine at the dimer-dimer interface, either single or mixed transfections were expected to result in disulfide cross-linked products. For example, mutants with Cys substitutions at position 21, 22, or 54 were expected to yield disulfide cross-linked products in single transfections (Fig. 1D). In contrast, a mutant with a Cys substitution at position 14 was expected to yield cross-linked products only when it was coexpressed with a mutant containing a Cys substitution at position 58 (Fig. 1C).
As illustrated in Table 1, many of the mutants containing Cys substitutions at the dimer-dimer interface did not conform to the expected results. For example, from the crystal structure, the β-carbon distance from residue 21 to a second residue 21 was found to be 4.6 Å while the β-carbon distance from residue 22 to a second residue 22 was found to be 7.9 Å. The V21C mutant was therefore expected to cross-link more efficiently than the A22C mutant. Instead the A22C mutant yielded a cross-linked dimer (Fig. 4) while no cross-linked products could be detected for the V21C mutant. These results suggest that, in the context of intact HDAg-S, the actual interactions at the dimer-dimer interface may differ, at least slightly, from those observed in the crystal structure.
TABLE 1.
Mutations used for probing delta antigen multimerization
| Mutant | Replication | Dimers | Multimers | Expected products |
|---|---|---|---|---|
| E14C | + | +/−b | − | Monomers |
| L17C | + | − | − | Monomers |
| V21C | + | − | − | Dimers |
| A22C | + | + | − | Dimers |
| N53C | − | +/− | NDa | Monomers |
| I54C | − | − | − | Dimers |
| I57C | − | + | ND | Monomers |
| L58C | + | + | − | Monomers |
| E14C + L58C | + | + | ND | Dimers |
| L17C + L58C | + | + | ND | Dimers |
| E14C,L30C | + | + | + | Dimers |
| E14C,L58C | − | + | + | Multimers |
| L17C,L30C | + | + | +/− | Dimers |
| L17C,L58C | − | + | − | Multimers |
| V21C,L30C | +/− | + | − | Multimers |
| A22C,L30C | + | + | + | Multimers |
| N53C,L30C | − | + | + | Dimers |
| N53C,I57C | − | + | − | Dimers |
| I54C,L30C | − | − | − | Multimers |
| I57C,L30C | − | + | + | Dimers |
| L58C,L30C | + | + | + | Dimers |
| E14C,L30C + L58C,L30C | + | + | + | Multimers |
| L17C,L30C + L58C,L30C | + | + | + | Multimers |
| E14C,L58C,L30C | − | + | + | Multimers |
| L17C,L58C,L30C | − | + | − | Multimers |
| N53C,I57C,L30C | − | + | + | Multimers |
ND, not determined.
+/−, products formed with low efficiency.
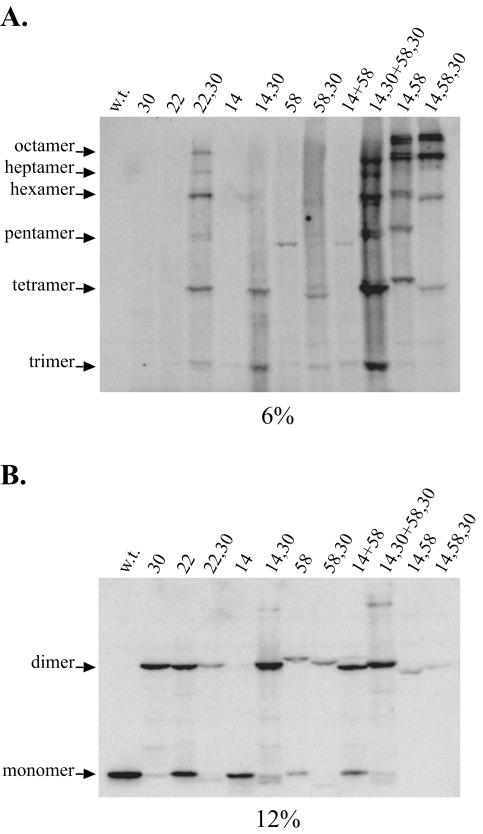
FIG. 4.
HDAg-S assembles as an octamer. The figure shows Western blots for CuPh-treated cell extracts obtained from HEK293 cells transfected with HDAg-S expression vectors containing Cys substitution mutations designed to probe for interactions at the dimer-dimer interface. w.t., wild type.
Additional evidence that might support this idea was obtained with the E14C and L58C mutants. Each of the E14C and L58C mutants was expected to yield only monomers in single transfections. However, these two mutants unexpectedly produced cross-linked dimers, albeit that dimer formation with the E14C mutant was very inefficient. When E14C and L58C were mixed, cross-linking was more efficient and the cross-linked species was resolved as a faster-migrating band (Fig. 4B, compare lanes 14, 58, and 14+58). Based on measurements of β-carbon distances in the crystal structure, the Cys58-Cys58 and the Cys14-Cys14 cross-links should not be possible. The β-carbon distance from residue 14 to a second residue 14 was found to be 17.1 Å while the β-carbon distance from residue 58 to a second residue 58 was found to be 13.3 Å. However, because the predominant dimeric species in the Cys14-plus-Cys58 mixed transfection is the Cys14-Cys58 cross-linked dimer, Cys58 must still be closer to Cys14 than it is to a second Cys58 residue.
Although the dimer-dimer interface in vivo might differ somewhat from that observed in the crystal structure, the cross-linking data nonetheless demonstrated that HDAg-S assembled as an octamer in vivo. Both the L30C mutant and the A22C mutant, in single transfections, yielded only cross-linked dimers. However, as expected, a double mutant containing these two mutations yielded cross-linked multimers as large as an octamer (Fig. 4). Additionally, because cross-linking at the dimer-dimer interface already occurs with either the E14C or the L58C single mutant, single transfections with either the E14C,L30C or the L58C,L30C double mutant yielded cross-linked multimers. Unlike the A22C,L30C double mutant, however, multimers larger than a tetramer were less readily detected (Fig. 4A). The relatively low abundance of cross-linked species larger than a tetramer resulted because the efficiency of cross-linking for the Cys14-Cys14 and the Cys58-Cys58 pairs was less than that for the Cys14-Cys58 pair. Mixed transfections involving the E14C,L30C and the L58C,L30C double mutants provided results consistent with this interpretation. In this transfection, the tetrameric band became more prominent and additional bands larger than a tetramer became more visible. In fact, by constructing an E14C,L58C double mutant and an E14C,L58C,L30C triple mutant, a greater proportion of HDAg-S could be trapped as multimers larger than a tetramer (Fig. 4A).
Assembly of HDAg-S octamers during HDV replication.
Because the previous experiments with Cys substitution mutants were performed with expression vectors for HDAg-S, we wished to determine whether octamers also form in the context of HDV replication. Likewise, we wished to determine whether mutants containing Cys substitutions at the dimer-dimer interface could still support HDV replication. Again, the various Cys substitution mutations were introduced into cDNA vectors expressing 1.2× unit-length antigenomic RNA. These were transfected into HEK293 cells, harvested at 7 days posttransfection, and assayed for disulfide cross-linking.
As shown in the Northern blots, several mutations resulted in disruption of HDAg-S function. Both an I54C single mutant and an I54C,L30C double mutant failed to support HDV replication (Fig. 5A). Based on the cross-linking data for these two mutants, the loss of ability to support HDV replication appears to be the result of a severe disruption of the coiled-coil domain. Assuming that the I54C mutation disrupted only interactions at the dimer-dimer interface, an I54C,L30C double mutant would be expected to produce dimers via cross-linking of Cys30 residues. However, an I54C,L30C double mutant actually failed to yield cross-linked dimers (Table 1). Thus, it appears that the I54C mutation disrupts HDAg-S function by abolishing interactions at both the dimer-dimer and monomer-monomer interfaces.
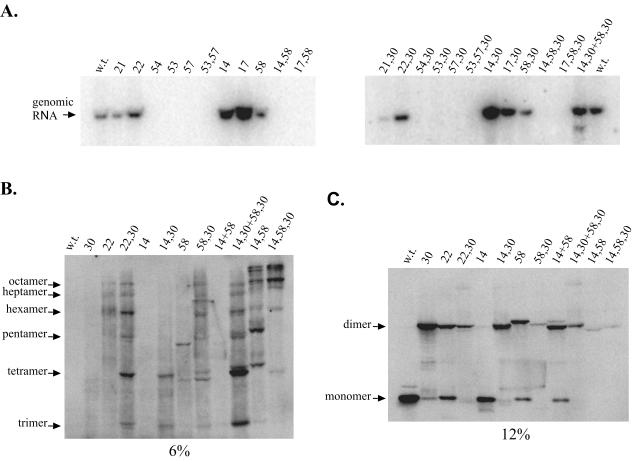
FIG. 5.
HDAg-S assembles as an octamer during HDV replication. (A) Northern blot for HEK293 cells transfected with cDNA vectors expressing 1.2× unit-length HDV antigenomic RNA containing Cys substitution mutations designed to probe for the dimer-dimer interaction. Cells were harvested 7 days posttransfection. (B and C) Western blots for a subset of CuPh-treated cell extracts obtained from cells transfected in the experiment shown in panel A. w.t., wild type.
Interestingly, other HDAg-S mutants that retained the ability to multimerize similarly failed to support HDV replication. For example, both the E14C,L58C double mutant and the E14C,L58C,L30C triple mutant produced octamers in cross-linking assays (Fig. 4A) but failed to support HDV replication (Fig. 5A). To ensure that the failure to support replication indeed was due to a defect in HDAg-S and did not result from a disruption in the HDV RNA structure, vectors that express these replication-defective mutant proteins were coexpressed in cells along with a form of HDV RNA that contains a frameshift and stop codon in the delta antigen open reading frame. This mutated HDV RNA does not produce any delta antigens but can undergo replication when functional HDAg-S is provided in trans. However, as before, the defective mutants were unable to support HDV replication in this assay (data not shown). Hence, the residues in question may participate in a process, in addition to multimerization, that is required for the ability of HDAg-S to support genome replication.
Finally, for mutants that retained the ability to support HDV replication, the results of cross-linking assays proved to be identical to those of experiments performed with HDAg-S expression vectors (Fig. 5B and C). Both an A22C single mutant and an A22C,L30C double mutant supported replication at near-wild-type levels (Fig. 5A). As before, the A22C single mutant produced disulfide-linked dimers while the A22C,L30C double mutant produced disulfide-linked multimers as large as an octamer. Similarly, the E14C single mutant, the L58C single mutant, the E14C,L30C double mutant, and the L58C,L30C double mutant were all also found to be capable of supporting HDV replication. As with the HDAg-S expression vectors, in single transfections, the E14C mutant produced mainly monomers while the L58C mutant produced a cross-linked dimer. When the two mutants were used in a mixed transfection, the dimer found in the L58C mutant was shifted to a faster-migrating band (Fig. 5C). Likewise, in single transfections, the E14C,L30C double mutant produced cross-linked species as large as tetramers while the L58C,L30C double mutant produced multimers as large as an octamer.
Two types of cross-linked tetramers are possible with the 58,30 double mutant. In type I, two dimers, each joined by a 58-58 cross-link, are joined to one another by a 30-30 cross-link. In type II, two dimers, each joined by a 30-30 cross-link, are joined to one another by a 58-58 cross-link. Since the 58-58 dimer ran more slowly than did the 30-30 dimer (Fig. 4B, compare 30 and 58 dimers) and since the type I tetramer has two 58-58 disulfides, while the type II tetramer has only one, we would expect that the type I tetramer should run more slowly than the type II tetramer. Furthermore, since disulfide bond formation was somewhat less efficient for the 58-58 dimer than for the 30-30 dimer (Fig. 4B, compare 30 and 58 dimers), we would expect that the type I tetramer should occur less frequently than the type II tetramer. Hence, a tetramer present as a doublet is predicted and the slower-migrating species in the doublet should be fainter than the faster-migrating species. This predicted pattern was actually observed both in Fig. 4A, lane 58,30, and in Fig. 5B, lane 58,30. Given that the 58,30 mutant was competent for replication, that it formed multimeric cross-linked species consistent with what were predicted, and that it assembled into octameric structures, we concluded that the L58C mutation did not significantly reduce delta antigen folding, multimerization, or function. Furthermore, in contrast to what is predicted by the crystal structure, we concluded that, in vivo, position 58 of one HDAg-S monomer is in proximity to position 58 of a second HDAg-S monomer present within a different dimer.
In a mixed transfection of E14C,L30C and L58C,L30C, multimers became more prominent than when either mutant was expressed individually (Fig. 5B). Hence, in vivo, as predicted in the crystal structure, residues 14 and 58 were in proximity. The sum of all results indicated that, during replication and in the presence of HDV RNA, HDAg-S indeed assembled as an octamer.
In vivo assembly of HDAg-L dimers and octamers.
In the previous cross-linking assays, the cell extracts used contained little or no HDAg-L. Therefore, to determine whether HDAg-L assembled into a structure similar to that of HDAg-S, we introduced the mutations used to probe the in vivo structure of HDAg-S into vectors that express HDAg-L. As before, HEK293 cells were transfected with these constructs and subsequently harvested for the disulfide cross-linking assays. Surprisingly, we found that wild-type HDAg-L can form a cross-linked dimer (Fig. 6B). HDAg-L contains a single Cys residue within its C-terminal 19 amino acids that is subject to posttranslational modification by prenylation (6, 25). The presence of a cross-linked dimer suggests that position C211 of HDAg-L, when it is not prenylated, is in proximity to C211 of a second molecule of HDAg-L that is also not prenylated. Given the specificity of this cross-linking assay, it is likely that the C-terminal interaction that was observed with the wild-type protein is a specific one, suggesting that the C-terminal 19 amino acids of HDAg-L may constitute a second site of protein-protein interaction.
FIG. 6.
HDAg-L assembles a structure similar to that of HDAg-S. (A and B) Western blots for CuPh-treated cell extracts obtained from HEK293 cells transfected with HDAg-L expression vectors containing a Cys substitution previously used to test the in vivo structure of HDAg-S. (C) Western blot for cell extracts treated with β-mercaptoethanol. Lanes labeled with “(S)” indicate HDAg-S Cys substitution mutants. w.t., wild type. (D) Location of CuPh-dependent disulfide cross-links in the I16C,N48C HDAg-L double mutant. Two stacked octameric rings, offset by 45°, are depicted. The bottom ring is shaded white and has thick black lines to denote the sequences downstream of position 60 that include the unprenylated C211 residue. The top ring is shaded gray and has thick gray lines to denote the sequences downstream of position 60 that include the unprenylated C211 residue. C211 cross-linking is proposed to result from interactions that occur between HDAg-L monomers present in different octamers. For simplicity, all C211 residues are depicted in the unprenylated form.
As a consequence of C211-mediated disulfide bond formation, all HDAg-L Cys substitution mutants tested produced at least dimeric species in the cross-linking assay. Hence, in this context, coiled-coil interactions could be observed only if a mutant(s) either caused an increase in the efficiency of dimer formation or caused the formation of higher-molecular-weight cross-linked products. Surprisingly, the I16C,N48C double mutant, which yielded monomer-monomer cross-linked dimers in the context of HDAg-S, instead yielded cross-linked multimers in the context of HDAg-L (Fig. 6A and B). The presence of multimers larger than a dimer indicates that the C211-C211 cross-link did not result from an interaction between two monomers in the same dimer but rather resulted from the interaction of two different dimers (Fig. 6D). In addition, this result indicated that, like HDAg-S, HDAg-L assembled as an antiparallel coiled coil. In a situation analogous to the one with the HDAg-S Cys substitution mutants, if HDAg-L assembled as a parallel coiled coil, multimers larger than a dimer would already be present in transfections with either the I16C or the N48C mutant alone. In contrast, only if HDAg-L assembled as an antiparallel coiled coil would multimers be absent with either the I16C or the N48C single mutant but present with the I16,N48C double mutant.
Finally, for mutants such as the E14C,L58C,L30C triple mutant and the E14C,L58C double mutant, which were expected to yield octamers, no species could be detected in either 6 or 12% polyacrylamide gels (Fig. 6A and B). However, upon treatment of cell extracts with a reducing agent, HDAg-L monomers could now be detected for these mutants (Fig. 6C). One interpretation of these results is that mutations designed to trap HDAg-L octamers were instead causing the formation of larger complexes that could no longer enter the gel. Additional evidence to support this hypothesis is provided by the A22C,L30C double mutant. In this case, the addition of the A22C mutation results in the disappearance of bands observed with the L30C single mutant (Fig. 6A and B). Despite this complication, the cross-linking results for HDAg-L follow the trends observed with the HDAg-S mutants. In single transfections with either the E14C,L30C double mutant or the L58C,L30C double mutant, only faint multimeric bands were detected. However, as with HDAg-S, in a mixed transfection with these two mutants, a more prominent band corresponding to a tetramer was detected (Fig. 6A). Similarly, in HDAg-S, the E14C,L58C double mutant and the E14C,L58C,L30C triple mutant proved to be the most efficient at cross-linking octamers. In HDAg-L, these two mutants may also have been the most efficient at cross-linking a multimeric complex too large to enter the gel, since no signal for HDAg-L could be detected in gels run under nonreducing conditions (Fig. 6A and B). Thus, it appeared that for both HDAg-S and HDAg-L, interactions at the monomer-monomer interface and the dimer-dimer interface were similar. We concluded that, like HDAg-S, HDAg-L assembles as an octamer in vivo.
Assembly of octamers in viral particles.
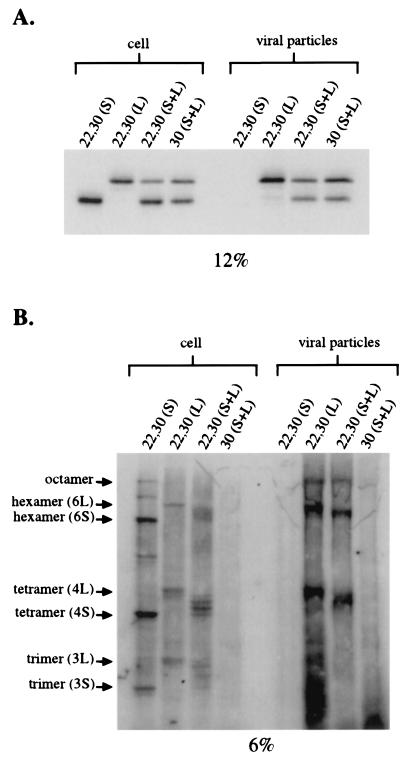
Although both HDAg-S and HDAg-L assembled into an octameric structure inside cells, it is unknown whether this same structure can be found inside virus-like particles. To test for the formation of octamers inside virus-like particles, Huh-7 cells were transfected with a cDNA vector expressing HBsAg-S along with cDNA vectors expressing either the A22C,L30C HDAg-S mutant alone, the A22C,L30C HDAg-L alone, a combination of both, or a combination of the L30C HDAg-S and L30C HDAg-L mutants. At 4 days posttransfection, cells and viral particles were harvested for cross-linking analysis. In order to determine the total amount of delta antigens expressed inside cells and inside viral particles, samples were also run under reducing conditions (Fig. 7A).
FIG. 7.
Assembly of delta antigen octamers inside viral particles. (A) Western blot for cell extracts and harvested viral particles treated with β-mercaptoethanol. Huh-7 cells were transfected with cDNA expressing HBsAg-S along with either the A22C,L30C HDAg-S mutant alone, the A22C,L30C HDAg-L mutant alone, both A22C,L30C HDAg-S and A22C,L30C HDAg-L mutants, or both L30C HDAg-S and L30C HDAg-L mutants. (B) Western blots for samples in panel A treated with CuPh. Compositions of multimeric bands are indicated in parentheses. 3S, three molecules of HDAg-S; 3L, three molecules of HDAg-L; 4S, four molecules of HDAg-S; 4L, four molecules of HDAg-L; 6S, six molecules of HDAg-L; 6L, six molecules of HDAg-L.
As expected, in the absence of any HDAg-L, the A22C,L30C HDAg-S mutant could not be packaged into virus-like particles (Fig. 7). In contrast, in a single transfection with the A22C,L30C HDAg-L mutant and in a cotransfection with the A22C,L30C HDAg-S and A22C,L30C HDAg-L mutants, multimeric species could be found both inside cells and inside viral particles (Fig. 7B). In a cotransfection with the L30C HDAg-S and L30C HDAg-L mutants, dimers could be similarly found both inside cells and inside viral particles (data not shown). These results indicated that the A22C,L30C double mutation did not disrupt the packaging and/or copackaging functions of either HDAg-S or HDAg-L. More importantly, these results indicate that, both inside cells and inside viral particles, the delta antigens assemble into a similar structure.
In the mixed transfection with the A22C,L30C HDAg-S and the A22C,L30C HDAg-L mutants, multimeric species designated as trimers, tetramers, and hexamers could be detected as multiple bands. These multiple bands likely represent the various combinations of HDAg-S and HDAg-L that can assemble into these higher-ordered structures (Fig. 7B). For example, for the species designated as tetramers, the fastest-migrating band likely represents four molecules of HDAg-S while the next higher migrating band likely represents three molecules of HDAg-S and one molecule of HDAg-L. Thus, it appears that the interaction of HDAg-S and HDAg-L via the coiled-coil domain occurs randomly, and there appears to be no preference for assembling either homomultimeric or heteromultimeric complexes.
DISCUSSION
In this study we found that, to a large extent, the structure solved from a crystallized delta antigen peptide is relevant to that actually assumed in vivo, when physiological levels of protein are expressed. We indeed found that both HDAg-S and HDAg-L assembled as antiparallel coiled coils and that each could further assemble into multimers as large as an octamer. These structures could be found both inside cells and inside viral particles. Moreover, these structures appear to have no preferred composition, since heteromultimers composed of various combinations of HDAg-S and HDAg-L could also be detected.
Interestingly, although all cross-linked mutant dimers possessed virtually identical molecular weights, subtle differences in their migration through denaturing gels were observed. A denatured cross-linked dimer can be envisioned as having four denatured peptide arms that emanate from the cross-link. Hence, when the I16C mutant was cross-linked to the N48C mutant, arm lengths of 16, 48, 147 (length of HDAg-S minus 48), and 179 (length of HDAg-S minus 16) amino acids resulted. We observed empirically that the length of the smallest arm dictated the mobility of the associated dimer. Hence, a 16-48 dimer ran slightly faster than a 20-44 dimer, which ran slightly faster than a 30-30 dimer, which ran slightly faster than a 58-58 dimer, which ran slower than a 14-58 dimer (Fig. 2A and 4B). The most dramatic difference was observed when the mobility of the wild-type HDAg-L dimer (211-211 cross-linked) was compared with that of the 30-30 cross-linked HDAg-L dimer (Fig. 6B, lanes w.t. and 30). In this case, the shortest arm length of the wild-type dimer was three amino acids, and it ran faster than the 30-30 dimer did. In the latter case, the shortest arm length was 30 amino acids.
In all figures, the bands labeled as monomer, dimer, trimer, etc., represented the predominant species present in the gel. However, in some cases, bands with weaker signals were observed migrating between the monomer and dimer positions. For example, in Fig. 4B, samples that have a strong dimer signal (lanes 30, 22, 14,30, 14 + 58, and 14,30 + 58,30) also have several weaker bands that are smaller than the dimer. We suspect that these bands are degradation products that resulted from boiling the sample in the presence of CuPh. Ascorbic acid added with CuPh is used as a reagent to nonspecifically cleave peptide bonds. In the absence of ascorbate but under a very high temperature, it is likely that CuPh can still promote limited proteolysis (36).
An unresolved question with respect to multimeric structures is their role in relation to delta antigen function. For example, what structure is required for the support of genome replication by HDAg-S? In disulfide cross-linking assays, the W20C mutant appeared to retain its ability to dimerize, and yet this mutant was unable to support genome replication. Hence, an HDAg-S dimer may not be able to support HDV replication, and perhaps octamers are required. However, additional mutants such as the E14C,L30C,L58C mutant were also incapable of supporting HDV replication yet appeared to retain the ability to form octamers. Perhaps at least some of these residues participate in a function, separate from octamer formation, which is also required for HDAg-S function. In the crystal structure, a ring was observed having a 50-Å pore that could potentially accommodate a molecule of genomic RNA (39). Perhaps HDAg-S octamers that are competent to support replication must contain a pore of the appropriate size and charge to properly accommodate HDV RNA. Alternatively, HDAg-S octamers may need to assume a very specific conformation in order to interact with host factors that are involved in HDV replication.
Prior to this study, it was uncertain whether the multimerization domain functions equivalently in the context of both HDAg-S and HDAg-L. HDAg-L functions as a dominant-negative inhibitor of replication, and for this property, a functional multimerization domain is required (13). Similarly, certain C-terminal deletion mutants of HDAg-S, in which only the first 81 and 88 amino acids are present, also act as dominant-negative inhibitors of replication (13). A potential model that could explain these observations is that sequences flanking the multimerization domain influence the way in which the multimerization domain interacts. Perhaps, for HDAg-S, flanking sequences can fold into a precise conformation so that a multimer capable of supporting HDV replication assembles. In contrast, for HDAg-L, the presence of the additional C-terminal 19 amino acids may result in a global change in protein structure such that sequences flanking the multimerization domain fold alternatively. Alternate folding of these flanking sequences and the subsequent interaction of HDAg-L with HDAg-S would then result in multimers with either defective conformations or incorrect numbers of HDAg molecules. Such a model would also explain the results observed with the 1-81 and 1-88 mutants. In this case, the absence of flanking sequences would prevent these truncated proteins from assuming an alternate conformation and, upon interaction with HDAg-S, would prevent assembly of a structure capable of supporting HDV replication. However, this model is not supported by the data obtained here, since the multimerization domains of HDAg-S and HDAg-L were found to function similarly. The similar function of the multimerization domain in the context of either HDAg-S or HDAg-L was also supported by our observation that, when coexpressed, the two proteins coassociate in multimers in a random fashion.
Surprisingly, we found that wild-type HDAg-L could be cross-linked via interactions between two unprenylated C211 residues. This interaction occurred between two monomers present in two different dimeric units. We also observed that mutants that were expected to yield octamers instead yielded larger complexes that failed to enter gels. A possible explanation for this result is that C211-mediated cross-linking occurred between molecules of HDAg-L that resided in different octamers. If, for instance, octamers were stacked on top of each other, formation of disulfide bonds exclusively via the coiled-coil domain would trap only HDAg-L octamers. However, if C211 present on different octamers could interact, cross-linking at both the coiled-coil domain and C211 could result in products composed of 16, 24, 32, or more molecules of HDAg-L. Such complexes would likely be too large to enter a 6% polyacrylamide gel. Based on the efficiency of C211 cross-linking, it appears that this region of HDAg-L constitutes a second domain involved in protein-protein interaction.
As a consequence of our efforts to probe the oligomeric structure of the delta antigens in vivo, we developed a methodology that has far broader applications. Here we demonstrated for the first time both that cysteine residues can be tolerated in the a and d positions of a coiled coil and that they can form disulfide bonds when so positioned. Thus, by using the methodology described here, it should be possible to test the validity in vivo of any coiled-coil prediction even in the complete absence of additional structural information. By simply individually mutating the predicted a and d positions to cysteine and then performing the disulfide assay, one could not only test the validity of the prediction but also discern whether the coiled-coil interaction is in a parallel or an antiparallel orientation. Large numbers of predicted coiled coils are presently being discovered as a consequence of genome sequencing projects. Using the method described here, it should now be possible to confirm these structures without the need for expensive and labor-intensive methods like X-ray crystallography.
In our assays, E14C HDAg-S and especially L58C HDAg-S were expected to yield only monomers but actually yielded disulfide cross-linked dimers. Furthermore, both E14C,L30C and L58C,L30C were expected to yield only dimers but actually yielded multimers as large as octamers. All of these mutants were able to support HDV replication, indicating that none of the mutations caused any major defects in protein folding, multimerization, or function. Hence, in the wild-type small delta antigen, it is highly likely that position 58 from one monomer is in proximity to that same position from a second monomer. The same is true, to a lesser extent, for position 14. Nowhere in the octameric crystal structure is position 58 found to be near position 58 in another monomer. For position 14, the residues are spaced even farther apart. How then can the crystal structure be reconciled with results obtained in this study? One obvious possibility is that, in vivo, the full-length HDAg-S protein assumes an octameric structure quite different from that assumed by the coiled-coil peptide in vitro. However, there is a second possibility that is consistent both with the crystal structure data and with all results presented here.
With several HDAg-L mutants, we obtained data consistent with the occurrence of an octamer-octamer interaction. When a crystal structure octamer is stacked on top of another, in planar fashion so that their pores are aligned, and is then rotated about the pore axis by approximately 18°, the position 58 residues from the bottom surface of one octamer are in very high proximity to the position 58 residues from the top surface of the second octamer. In this arrangement, position 14 residues are almost as proximal to one another as are the position 58 residues. Under such a condition, the crystal structure would predict formation of 58-58 and 14-14 cross-links, and this offset stacking of octamers could explain all of the unexpected cross-links observed here. We therefore favor this interpretation.
We have obtained indirect evidence in vivo to indicate that HDAg octameric rings stack on top of one another in an offset fashion. We have incorporated this finding into a new model for the structure of HDV RNPs. We propose that HDV RNPs possess helical symmetry that is imposed when octamers stack on top of one another in an offset manner. In the proposed structure, there are approximately 20 octameric units per helical turn. Previously, HDV genomic RNPs were estimated to contain roughly 200 HDAg monomers (8). If the model presented here and the prior estimates are both correct, then the HDV RNP is composed of stacked octamers that spiral for a little more than one helical turn. Interestingly, HDV is a negative-stranded RNA virus. For all other negative-strand RNA viruses, where the issue has been resolved, viral RNPs display helical symmetry. Further work will be needed to fully establish whether the HDV RNPs actually do possess helical symmetry.
Acknowledgments
We thank Jim Hogle for providing the expanded crystal structure coordinates that cover the complete octamer. We also thank John Coffin, Ralph Isberg, Claire Moore, and Cathy Squires for their helpful discussions and critical reading of the manuscript.
This work was supported by grant R01-AI40472 from the National Institutes of Health and by the Raymond and Beverly Sackler Research Foundation.
REFERENCES
- 1.Bonner, J. J., D. Chen, K. Storey, M. Tushan, and K. Lea. 2000. Structural analysis of yeast HSF by site-specific crosslinking. J. Mol. Biol. 302:581-592. [DOI] [PubMed] [Google Scholar]
- 2.Chang, F. L., P. J. Chen, S. J. Tu, C. J. Wang, and D. S. Chen. 1991. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 88:8490-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chao, M., S. Y. Hsieh, and J. Taylor. 1990. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 64:5066-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, P. J., G. Kalpana, J. Goldberg, W. Mason, B. Werner, J. Gerin, and J. Taylor. 1986. Structure and replication of the genome of the hepatitis delta virus. Proc. Natl. Acad. Sci. USA 83:8774-8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu, T. B., and J. Taylor. 1993. The RNAs of hepatitis delta virus are copied by RNA polymerase II in nuclear homogenates. J. Virol. 67:6965-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glenn, J. S., J. A. Watson, C. M. Havel, and J. M. White. 1992. Identification of a prenylation site in delta virus large antigen. Science 256:1331-1333. [DOI] [PubMed] [Google Scholar]
- 7.Glenn, J. S., and J. M. White. 1991. trans-Dominant inhibition of human hepatitis delta virus genome replication. J. Virol. 65:2357-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudima, S., J. Chang, G. Moraleda, A. Azvolinsky, and J. Taylor. 2002. Parameters of human hepatitis delta virus genome replication: the quantity, quality, and intracellular distribution of viral proteins and RNA. J. Virol. 76:3709-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobashi, K. 1968. Catalytic oxidation of sulfhydryl groups by o-phenanthroline copper complex. Biochim. Biophys. Acta 158:239-245. [DOI] [PubMed] [Google Scholar]
- 10.Kos, A., R. Dijkema, A. C. Arnberg, P. H. van der Meide, and H. Schellekens. 1986. The hepatitis delta (δ) virus possesses a circular RNA. Nature 323:558-560. [DOI] [PubMed] [Google Scholar]
- 11.Kuo, M. Y., M. Chao, and J. Taylor. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 63:1945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Lazinski, D. W., and J. M. Taylor. 1993. Relating structure to function in the hepatitis delta virus antigen. J. Virol. 67:2672-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, J. H., M. F. Chang, S. C. Baker, S. Govindarajan, and M. M. Lai. 1990. Characterization of hepatitis delta antigen: specific binding to hepatitis delta virus RNA. J. Virol. 64:4051-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lumb, K. J., and P. S. Kim. 1995. A buried polar interaction imparts structural uniqueness in a designed heterodimeric coiled coil. Biochemistry 34:8642-8648. [DOI] [PubMed] [Google Scholar]
- 16.Luo, G. X., M. Chao, S. Y. Hsieh, C. Sureau, K. Nishikura, and J. Taylor. 1990. A specific base transition occurs on replicating hepatitis delta virus RNA. J. Virol. 64:1021-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch, B. A., and D. E. Koshland, Jr. 1991. Disulfide cross-linking studies of the transmembrane regions of the aspartate sensory receptor of Escherichia coli. Proc. Natl. Acad. Sci. USA 88:10402-10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacNaughton, T. B., E. J. Gowans, S. P. McNamara, and C. J. Burrell. 1991. Hepatitis delta antigen is necessary for access of hepatitis delta virus RNA to the cell transcriptional machinery but is not part of the transcriptional complex. Virology 184:387-390. [DOI] [PubMed] [Google Scholar]
- 19.Macnaughton, T. B., S. T. Shi, L. E. Modahl, and M. M. Lai. 2002. Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J. Virol. 76:3920-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Modahl, L. E., T. B. Macnaughton, N. Zhu, D. L. Johnson, and M. M. Lai. 2000. RNA-dependent replication and transcription of hepatitis delta virus RNA involve distinct cellular RNA polymerases. Mol. Cell. Biol. 20:6030-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moraleda, G., K. Dingle, P. Biswas, J. Chang, H. Zuccola, J. Hogle, and J. Taylor. 2000. Interactions between hepatitis delta virus proteins. J. Virol. 74:5509-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moraleda, G., and J. Taylor. 2001. Host RNA polymerase requirements for transcription of the human hepatitis delta virus genome. J. Virol. 75:10161-10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nassal, M., A. Rieger, and O. Steinau. 1992. Topological analysis of the hepatitis B virus core particle by cysteine-cysteine cross-linking. J. Mol. Biol. 225:1013-1025. [DOI] [PubMed] [Google Scholar]
- 24.Oakley, M. G., and P. S. Kim. 1998. A buried polar interaction can direct the relative orientation of helices in a coiled coil. Biochemistry 37:12603-12610. [DOI] [PubMed] [Google Scholar]
- 25.Otto, J. C., and P. J. Casey. 1996. The hepatitis delta virus large antigen is farnesylated both in vitro and in animal cells. J. Biol. Chem. 271:4569-4572. [DOI] [PubMed] [Google Scholar]
- 26.Ponzetto, A., P. J. Cote, H. Popper, B. H. Hoyer, W. T. London, E. C. Ford, F. Bonino, R. H. Purcell, and J. L. Gerin. 1984. Transmission of the hepatitis B virus-associated delta agent to the eastern woodchuck. Proc. Natl. Acad. Sci. USA 81:2208-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizzetto, M., B. Hoyer, M. G. Canese, J. W. Shih, R. H. Purcell, and J. L. Gerin. 1980. Delta agent: association of delta antigen with hepatitis B surface antigen and RNA in serum of delta-infected chimpanzees. Proc. Natl. Acad. Sci. USA 77:6124-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozzelle, J. E., Jr., J. G. Wang, D. S. Wagner, B. W. Erickson, and S. M. Lemon. 1995. Self-association of a synthetic peptide from the N terminus of the hepatitis delta virus protein into an immunoreactive alpha-helical multimer. Proc. Natl. Acad. Sci. USA 92:382-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryu, W. S., M. Bayer, and J. Taylor. 1992. Assembly of hepatitis delta virus particles. J. Virol. 66:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryu, W. S., H. J. Netter, M. Bayer, and J. Taylor. 1993. Ribonucleoprotein complexes of hepatitis delta virus. J. Virol. 67:3281-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, C. J., P. J. Chen, J. C. Wu, D. Patel, and D. S. Chen. 1991. Small-form hepatitis B surface antigen is sufficient to help in the assembly of hepatitis delta virus-like particles. J. Virol. 65:6630-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, H. W., P. J. Chen, C. Z. Lee, H. L. Wu, and D. S. Chen. 1994. Packaging of hepatitis delta virus RNA via the RNA-binding domain of hepatitis delta antigens: different roles for the small and large delta antigens. J. Virol. 68:6363-6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, J. G., and S. M. Lemon. 1993. Hepatitis delta virus antigen forms dimers and multimeric complexes in vivo. J. Virol. 67:446-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, K. S., Q. L. Choo, A. J. Weiner, J. H. Ou, R. C. Najarian, R. M. Thayer, G. T. Mullenbach, K. J. Denniston, J. L. Gerin, and M. Houghton. 1986. Structure, sequence and expression of the hepatitis delta (δ) viral genome. Nature 323:508-514. [DOI] [PubMed] [Google Scholar]
- 35.Weiner, A. J., Q. L. Choo, K. S. Wang, S. Govindarajan, A. G. Redeker, J. L. Gerin, and M. Houghton. 1988. A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24δ and p27δ. J. Virol. 62:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, J., D. M. Perrin, D. S. Sigman, and H. R. Kaback. 1995. Helix packing of lactose permease in Escherichia coli studied by site-directed chemical cleavage. Proc. Natl. Acad. Sci. USA 92:9186-9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia, Y. P., and M. M. Lai. 1992. Oligomerization of hepatitis delta antigen is required for both the trans-activating and trans-dominant inhibitory activities of the delta antigen. J. Virol. 66:6641-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia, Y. P., C. T. Yeh, J. H. Ou, and M. M. Lai. 1992. Characterization of nuclear targeting signal of hepatitis delta antigen: nuclear transport as a protein complex. J. Virol. 66:914-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuccola, H. J., J. E. Rozzelle, S. M. Lemon, B. W. Erickson, and J. M. Hogle. 1998. Structural basis of the oligomerization of hepatitis delta antigen. Structure 6:821-830. [DOI] [PubMed] [Google Scholar]