Abstract
The human cytomegalovirus UL99-encoded pp28 is a myristylated phosphoprotein that is a constituent of the virion. The pp28 protein is positioned within the tegument of the virus particle, a protein structure that resides between the capsid and envelope. In the infected cell, pp28 is found in a cytoplasmic compartment derived from the Golgi apparatus, where the virus buds into vesicles to acquire its final membrane. We have constructed two mutants of human cytomegalovirus that fail to produce the pp28 protein, a substitution mutant (BADsubUL99) and a point mutant (BADpmUL99), and we have propagated them by complementation in pp28-expressing fibroblasts. Both mutant viruses are profoundly defective for growth in normal fibroblasts; no infectious virus could be detected after infection. Whereas normal levels of viral DNA and late proteins were observed in mutant virus-infected cells, large numbers of tegument-associated capsids accumulated in the cytoplasm that failed to acquire an envelope. We conclude that pp28 is required for the final envelopment of the human cytomegalovirus virion in the cytoplasm.
Human cytomegalovirus (HCMV) is the prototypical member of the betaherpesvirus family. Seroepidemiologic studies have shown that HCMV infection is widespread in the human population in both industrial and developing regions. In healthy individuals infection is generally asymptomatic, but the virus can cause serious disease in people with immature or compromised immune systems. It is the leading infectious disease cause of birth defects and a life-threatening adventitious agent in transplant recipients and AIDS patients (40, 43).
In virions, the double-stranded HCMV DNA resides within a capsid that is surrounded first by a tegument layer and then by an envelope. The tegument domain, which is unique to herpesvirus particles, contains approximately 30 virus-encoded proteins (1, 5, 14). Since they are components of virions, tegument proteins are delivered to cells at the very start of infection and they have the potential to function even before the viral genome is activated. For example, the UL83-encoded pp65 protein has been reported to block major histocompatibility complex class I presentation of a viral immediate-early protein (21), the UL47 protein acts during disassembly of the newly infecting virus particle (8), the UL82-encoded pp71 protein is a transcriptional activator (32) that helps to activate the immediate-early genes within infected cells (10), and the UL69 protein blocks cell cycle progression (23).
The pp28 protein of HCMV (31, 36) is a 190-amino-acid myristylated (51) phosphoprotein (39) that is expressed as a true late protein (28, 37), i.e., it is synthesized only after the onset of viral DNA replication. The pp28 protein is encoded by UL99, the last open reading frame positioned within a family of 3′-coterminal transcripts that share the same polyadenylation site (61) (Fig. 1A). It is one of the most abundant constituents of the tegument layer (5, 31) and is highly immunogenic (30, 39, 44).
FIG. 1.
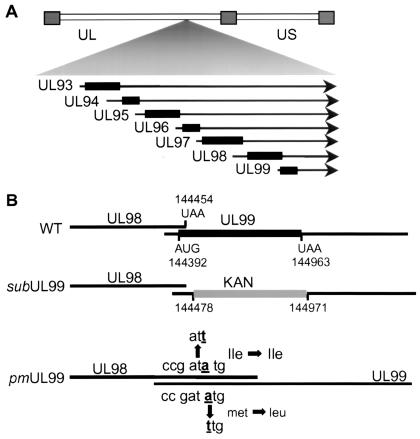
Diagrams of the HCMV genome, the region encoding UL99, and the mutations introduced into the UL99 coding region. (A) Map of the viral genome showing the location of UL93 to UL99 mRNAs. Repeated regions are represented by rectangles, unique regions (UL and US) are shown by a double line, and mRNAs are depicted by arrows. (B) Changes introduced into viral mutants. In BADsubUL99, a kanamycin-resistance marker is substituted for a substantial portion of the UL99 coding region, and in BADpmUL99, the UL99 ATG start codon is changed to TTG without altering the coding potential of the overlapping UL98 open reading frame, which is translated in a different reading frame.
Whereas some tegument proteins accumulate in the nuclei of infected cells (e.g., pp71) (25), pp28 resides in the cytoplasm (31), partially colocalizing with several other tegument proteins (pp150 and pp65) and envelope glycoproteins (gH, gB, and gp65) in a juxtanuclear site proposed to be the site of assembly of virions (50). This site coincides with markers for the trans-Golgi compartment, and the association of pp28 with membranes within this compartment requires that it be myristylated (50).
In this study, we have investigated the function of pp28 during viral replication. Using a bacterial artificial chromosome (BAC) carrying the HCMV genome (62), we constructed two pp28-null mutants, a substitution mutant, and a point mutant. The mutants can be propagated only in a pp28-expressing cell line. Characterization of their growth in normal fibroblasts revealed that pp28 is required at a late step in the HCMV replication cycle, during the envelopment of capsids in the cytoplasm.
MATERIALS AND METHODS
Cells and viruses.
Primary human foreskin fibroblasts (HFFs; passages 4 to 15) were cultured in Dulbecco's modified Eagle medium containing 10% fetal calf serum at 37°C in a 5% CO2 atmosphere. Complementing cells expressing pp28 and cells expressing pp28 with an N-terminal hemagglutinin (HA) epitope tag were constructed by using recombinant retroviruses that were prepared in Phoenix Ampho cells, as described by Kinsella and Nolan (29). The resulting retroviruses, Retropp28WT and RetroHA-pp28, were used to infect HFFs at passage 4 in the presence of Polybrene (15 μg/ml). In multiple experiments, the propagation of Retropp28WT in Phoenix Ampho cells produced low-titer retrovirus stocks, so either three or eight sequential infections were performed to yield HFFpp28-3x and HFFpp28-8x cells. RetroHA-pp28 grew to higher titers, and a single infection of HFFs produced HFFHApp28 cells expressing relatively high levels of the protein.
Mutant viruses unable to express pp28 and a revertant of one of the mutants were constructed by using the wild-type HCMV strain AD169 BAC(pAD12-29) (62). Alterations were made in pAD12-29 by allelic exchange with pGS284 (54). To alter the UL99 coding region in pAD12-29, three pGS284 derivatives were made: pGS284-KAN/UL99, pGS284-ATG-TTG/UL99, and pGS284-WT/UL99. pGS284-Kan/UL99 contains a kanamycin resistance gene cloned into the plasmid's BglII and NotI sites between PCR-amplified UL99-specific flanking sequences (nucleotides 143410 to 144478 and 144971 to 145996) (sequence numbers are from reference 13). pGS284-WT/UL99 carries an HCMV fragment containing the wild-type UL99 open reading frame (nucleotides 143782 to 144988). pGS284-ATG-TTG/UL99 has a UL99 allele that contains an A-to-T change at nucleotide 144392. This mutation changes the UL99 start codon from ATG to TTG, but it does not change the amino acid sequence of UL98, which overlaps UL99 by 61 bp (nucleotides 144393 to 144454) in a different reading frame. The integrity of the HCMV segment in each plasmid was confirmed by DNA sequence analysis. Allelic exchange was performed by homologous recombination in Escherichia coli GS500 cells as described previously (54, 62). BADsubUL99 (BAD is BAC-derived AD169) was made by recombination of pAD12-29 with pGS284-KAN/UL99 followed by selection and screening for kanamycin resistance. BADsubUL99 was used to make BADpmUL99 and BADrevUL99 by using pGS284-ATG-TTG/UL99 and pGS284WT/UL99, respectively, by screening for kanamycin sensitivity. The mutant BAC DNAs were analyzed by EcoRI and HindIII digestion and by Southern blot assay with probes to UL98, UL99, and the kanamycin resistance gene.
BAC DNAs (15 μg) were used to transfect HFFs and HFFpp28 cells in the presence of a plasmid expressing pp71 (1 μg) (27), which enhances the infectivity of HCMV DNA (4), and a cre-expressing plasmid (1 μg) to direct excision of the BAC sequences from the viral genome (54, 55) by electroporation. Virus stocks were prepared and infectious yields were determined by plaque assay on HFFpp28-8x cells. DNAs were prepared from infected cells and characterized by Southern blot analysis with probes for the UL82 and UL99 coding regions.
For analysis of mutant virus growth kinetics, HFFs and HFFpp28 cells were infected at a multiplicity of infection of 0.01 PFU/cell with BADwt (from the parental pAD12-29 BAC), BADsubUL99, BADpmUL99, or BADrevUL99 virus. At various times after infection, intracellular and extracellular viruses were collected by scraping the cells, disrupting them by sonication, pelleting the debris, and collecting the supernatant. Virus titers were determined by plaque assay on HFFpp28-8x cells.
The relative ratios of infectivity to number of virus particles was determined for BADwt and BADsubUL99 virions by using real-time PCR to quantify the amount of viral DNAs present in portions of virus stocks containing 103 PFU. The virus stocks were prepared by disrupting infected cells by sonication, pelleting the debris, and collecting the supernatant. The virus particles were incubated in lysis buffer (20 mM Tris-HCl, 50 mM EDTA, 200 mM NaCl, 1.2% sodium dodecyl sulfate [SDS], 200 μg of proteinase K/ml) for 2 h at 37°C, and the DNA was extracted with phenol-chloroform-isoamyl alcohol (24:24:1) and resuspended in TE buffer (pH 8.0) (10 mM Tris-HCl, 1 mM EDTA). Two dilutions from each viral DNA were amplified by using the iCicler iQ System (Bio-Rad), and the fluorescence from incorporated SYBR Green (Applied Biosystems) was measured at the end of each cycle. A portion of the UL10 open reading frame was arbitrarily chosen for amplification with the following primers: TGCGGCGACTGATTAACCATATC and AGGGTAACTGGATCACCTACTCTG.
Analysis of viral DNA and protein.
Viral DNA was analyzed by Southern blot assay. Cell lysates were prepared in a lysis buffer containing 100 mM NaCl, 10 mM Tris (pH 8.0), 25 mM EDTA, 0.5% SDS, and 0.1 mg of proteinase K/ml. After incubation for 3 h at 55°C, DNA was extracted twice with phenol-chloroform-isoamyl alcohol (24:24:1), precipitated with ethanol, and resuspended in TE buffer. After digestion with restriction enzymes, DNA was analyzed by Southern blotting with UL99- or UL82-specific [32P]dCTP-labeled DNA probes.
Viral DNA accumulation was monitored by slot blot assay. Fibroblasts were infected with BADwt, BADsubUL99, or BADpmUL99 at a multiplicity of infection of 0.01 PFU/cell. At various times after infection, total cellular DNA was prepared as described above, and aliquots (1 μg) were transferred to a nylon membrane by using a slot blot apparatus. The immobilized viral DNA was hybridized to a 32P-labeled pp71-specific DNA probe, and after washing, radioactive probe retained on the membrane was quantified by using a phosphorimager.
The accumulation of viral proteins in HCMV- and recombinant retrovirus-infected cells was assayed by Western blotting. At various times after infection, cells lysates were dissolved in RIPA lysis buffer (10 mM Tris [pH 7.4], 1% Triton X-100, 0.1% SDS, 1% deoxycholate, 160 mM NaCl, and a protease inhibitor cocktail; Roche). Protein was quantified in the lysate by Bradford assay, and aliquots of protein (40 μg) were mixed with a 2× sample buffer and separated in SDS-containing 10% polyacrylamide gels. The separated proteins were transferred to a nitrocellulose membrane and probed with antibodies specific for the following viral and cellular proteins: UL123-encoded IE1 (1B12 monoclonal antibody produced by immunization with a fusion protein containing the C-terminal domain of IE1) (A. Marchini, P. Robinson, and T. Shenk, unpublished data), pUL44 (ICP36; Virusys), UL82-encoded pp71 (10G11) (27), UL97 (gift from A. Marchini, Tribeca Pharmaceuticals), UL99-encoded pp28 (10B4-29 monoclonal antibody produced by immunization with disrupted virions) (M. Silva, P. Robinson, and T. Shenk, unpublished data), UL83-encoded pp65 protein (41), and tubulin (DM1A; Sigma). Anti-mouse immunoglobulin G antibody conjugated with horseradish peroxidase was used as the secondary antibody.
Immunofluorescence and electron microscopic observation of infected cells.
For immunofluorescence, fibroblasts grown on glass coverslips were infected with BADwt or BADsubUL99 at a multiplicity of infection of 0.01 PFU/cell. At 120 h postinfection, cells were washed twice with phosphate-buffered saline (PBS) at 37°C, fixed at room temperature with 2% paraformaldehyde in PBS for 20 min, and washed twice again with PBS. The fixed cells were permeabilized by treatment for 15 min at room temperature with PBS containing 0.1% Triton X-100, washed twice with PBS, and blocked by incubation for 30 min at room temperature with PBS containing 2% bovine serum albumin and 0.05% Tween 20. Cells were incubated at room temperature with primary antibody diluted in PBS containing 0.05% Tween 20 for 1 h, washed three times with buffer lacking the antibody, incubated with fluorochrome-conjugated anti-mouse or anti-rabbit secondary antibody for 1 h, and washed again three times. In some cases, after reacting with the secondary antibody, the Golgi compartment was stained by incubation of cells at room temperature with Alexa 488-conjugated lectin HPA (Molecular Probes) for 20 min. Coverslips were mounted in slow-fade solution (Molecular Probes) and analyzed by confocal microscopy. Primary antibodies used in these studies were UL32-encoded pp150 and UL83-encoded pp65 rabbit polyclonal antibodies (gift of M. Schrader, University of Marburg), UL99-encoded pp28 mouse monoclonal antibody (10B4-29), UL55-encoded gB (58-15 monoclonal antibody produced by immunization with disrupted virions) (P. Robinson and T. Shenk, unpublished data).
For electron microscopy, fibroblasts were infected with BADwt or BADsubUL99 at a multiplicity of infection of 0.01 PFU/cell. At 96 h postinfection, cells were washed with PBS and fixed with prewarmed 2.5% glutaraldehyde, 2 mM CaCl2, and 0.1 M sodium cacodylate (pH 7.4) for 2 h. The cells were rinsed in 0.1 M cacodylate buffer, collected from the culture plate in 1 ml of the same buffer, and centrifuged into a compact pellet. Cells were then treated with 1% osmium tetroxide, dehydrated with ascending concentrations of ethanol, embedded in LX-112 medium, and polymerized at 68°C for 72 h. Ultrathin sections (∼80 nm) were cut with a diamond knife, mounted on 200-mesh thin-bar copper grids, stained with uranyl acetate and lead citrate, and examined with a Philips CM-100 electron microscope operated at 60-kV accelerating voltage.
RESULTS
UL99-encoded pp28 is essential for HCMV replication.
To explore the requirement for pp28 during HCMV replication in cultured human fibroblasts, two pp28-null mutant viruses were constructed in the HCMV AD169 strain by recombination in E. coli, with a BAC system (62). The first pp28-null virus was BADsubUL99 (Fig. 1B). In this mutant virus the majority of the UL99 open reading frame, which encodes pp28, was deleted and replaced by a kanamycin resistance gene. The segment of the pp28 coding region that was retained was the 61-bp region that overlaps the C-terminal domain of the upstream UL98 open reading frame. A second mutant was produced to ensure that the insertion of the kanamycin resistance gene did not produce misleading results by altering the accumulation or function of mRNAs encoding UL93-UL98, which share a polyadenylation site with the UL99 mRNA (Fig. 1A). BADpmUL99 was produced by mutating the first ATG codon of UL99 to TTG (Fig. 1B). This changed the pp28 initiator methionine to leucine, thereby abolishing translation of UL99, but the mutation had no effect on the amino acid sequence of the UL98 protein. We also constructed a revertant for BADsubUL99, termed BADrevUL99, with the BAC system.
After construction of the BAC derivatives, they were shown to be free of gross rearrangements by digestion with HindIII, and the region containing the mutation was shown to have the predicted structure by Southern blot assay with probes for UL98, UL99, and, in the case of BADsubUL99, a probe for the kanamycin resistance gene (data not shown). The BAC DNAs were introduced into human fibroblasts by electroporation, and virus was recovered from BAC DNA containing the wild-type HCMV genome, BADwt, and the revertant, BADrevUL99. However, BAC DNAs containing the BADsubUL99 or BADpmUL99 genome did not produce infectious virus when introduced into human fibroblasts, suggesting that pp28 is required for HCMV replication.
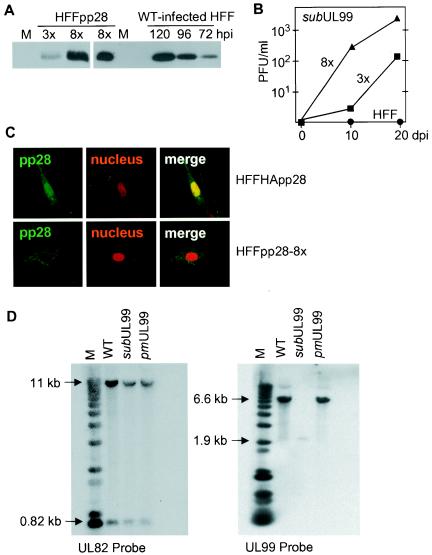
To rescue the growth of the mutant viruses, we prepared complementing cells that expressed pp28 from an integrated defective retrovirus. A recombinant retrovirus carrying the UL99 coding region (RetroUL99) grew poorly and produced relatively low-titer virus stocks (data not shown). Consequently, it was necessary to infect fibroblasts multiple times with RetroUL99 to increase the number of integrated recombinant retrovirus genomes in order to obtain sufficient expression of pp28 for complementation. The number of sequential infections correlated with the proportion of the fibroblast population expressing the protein. Three or eight sequential infections produced fibroblast populations termed HFFpp28-3x and HFFpp28-8x, in which about 30 and 80% of the cells, respectively, expressed pp28 at a level that could be detected by immunofluorescence (data not shown). We determined the levels of pp28 expressed and the growth efficiency of BADsubUL99 in these two cell populations. Western blot analysis demonstrated that HFFpp28-8x cultures expressed about five times more pp28 than HFFpp28-3x (Fig. 2A, left panel). The amount of the protein expressed by HFFpp28-8x cultures was comparable to that produced during the late phase of infection (120 h postinfection) in cultured fibroblasts, in which about 1% of the cells were infected with wild-type HCMV (Fig. 2A, right panel). Since about 80% of the cells in HFFpp28-8x cultures express pp28, as can be detected by immunofluorescence, the level of pp28 expression in each cell is substantially lower in HFFpp28-8x cells than in HCMV-infected cells. Nevertheless, the two pp28-deficient viruses, BADsubUL99 and BADpmUL99, were recovered from BAC DNAs following electroporation into HFFpp28-8x cells. BADsubUL99 grew to 10-fold-higher titers in HFFpp28-8x than in HFFpp28-3x (Fig. 2B).
FIG. 2.
Characterization of complementing cells and analysis of the structure of mutant DNAs derived from pp28-deficient virus produced in complementing cells. (A) Western blot analysis monitoring the expression of pp28 in mock-infected (M), HFFpp28-3x cells (3x), HFFpp28-8x cells (8x), and HCMV-infected fibroblasts (HFF) at different times (hours) postinfection (hpi). WT, wild type. (B) Growth of BADsubUL99 in HFFpp28-3x cells (3x), HFFpp28-8x cells (8x), and normal fibroblasts (HFF). Cells were infected, lysates of cells were prepared in the culture medium at the indicated times after infection, and virus was quantified by plaque assay on HFFpp28-8x cells. (C) Location of native pp28 and HA-tagged pp28 expressed from recombinant retroviruses in fibroblasts. Immunofluorescence was performed by using a pp28-specific antibody on HFFpp28-8x (bottom) cells and cells expressing HA-tagged pp28 (HFFHA-pp28, top). Three images are presented for each cell type: left, pp28 (green); middle, nuclear stain (red); right, multicolor merge. (D) Southern blot assay of viral DNAs. Total cell DNA was prepared from HFFpp28-8x cells infected with the indicated viruses, and DNA was cleaved with HindIII and analyzed by Southern blot assay with a UL82- and UL99-specific probe DNA. The sizes of labeled DNA fragments are indicated in kilobases relative to marker (M) DNA fragments.
We also prepared fibroblasts expressing pp28 tagged with the HA epitope at its N terminus (HFFHA-pp28). This modification would be expected to inhibit cleavage of the first methionine and subsequent myristylation of pp28. Although we did not directly test the myristylation state of the HA-tagged protein, we performed immunofluorescence experiments with a specific pp28 antibody and observed that most of the HA-tagged pp28 localized in the nucleus (Fig. 2C), as has been demonstrated when myristylation of pp28 was blocked by mutation (51). The growth of BADsubUL99 was compared in different cell types. It produced a yield of 2 × 104 PFU/ml in HFFpp28-8x cells, but it failed to generate detectable progeny in cells expressing the HA-tagged pp28, suggesting that myristylation is required for pp28 function.
To exclude the possibility of contamination with wild-type virus and to verify the changes in the substitution mutant and revertant virus genomes recovered from complementing cells, we performed Southern blot assays. HindIII-digested viral DNAs were analyzed with a UL99-specific probe (Fig. 2D, right panel) or, as a control, with a UL82-specific probe (Fig. 2D, left panel). The expected UL99-specific bands of 6.6 kb in BADwt and BADpmUL99 and 1.8 kb in BADsubUL99 were obtained. The structure of BADpmUL99 was confirmed by sequence analysis of the viral DNA in the region containing the point mutation (data not shown).
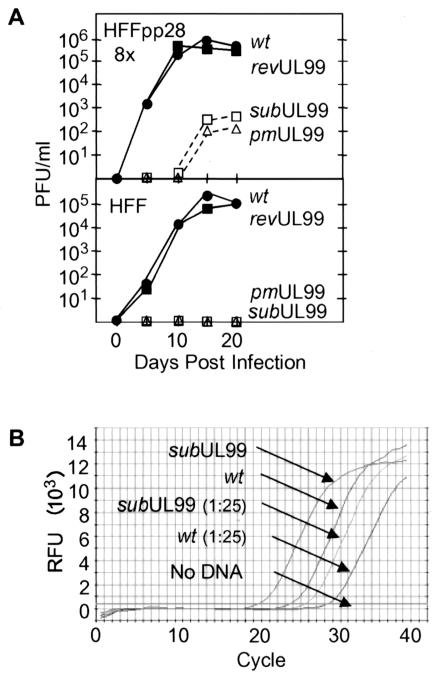
The growth kinetics of the mutant and revertant viruses were compared on normal human fibroblasts and HFFpp28-8x cells (Fig. 3A). Cells were infected at a multiplicity of infection of 0.01 PFU/cell, and virus yields were determined by plaque assay on HFFpp28-8x cells. BADrevUL99 grew to the same titers as BADwt in normal fibroblasts and in HFFpp28-8x cells, indicating that the defect in BADsubUL99 is caused by the specific changes made in the UL99 coding region and not by unintentional alterations in other regions of the viral genome. No infectious progeny virus was detected after BADsubUL99 or BADpmUL99 infection of human fibroblasts, which do not express pp28. Even in HFFpp28-8x cells, both mutants produced substantially reduced virus yields compared to wild-type or revertant viruses. Presumably, pp28 levels are limiting in the complementing cells, and as a result, the mutants do not produce wild-type yields. This interpretation is consistent with the observation that BADsubUL99 grows less well in HFFpp28-3x than in HFFpp28-8x cells (Fig. 2B).
FIG. 3.
Growth characteristics and relative particle-to-PFU ratios of wild-type (wt) and pp28-deficient viruses. (A) Production of infectious progeny in cultures of pp28-expressing fibroblasts (HFFpp28-8x) or normal fibroblasts (HFF). Cells were infected with BADwt (•), BADrevUL99 (▪), BADsubUL99 (□), or BADpmUL99 (▵) at a multiplicity of infection of 0.01 PFU/cell. Cells were lysed in culture medium at the indicated times after infection, and virus was quantified by plaque assay on HFFpp28-8x cells. (B) Relative amounts of DNA in BADsubUL99 compared to BADwt virions. DNA was prepared from a quantity of virus stock containing 103 PFU, and aliquots of undiluted DNA and diluted (1:25) DNA were quantified by real-time PCR with a primer pair that amplified a segment of the HCMV UL10 gene. Relative fluorescence units (RFU) are plotted as a function of cycle number.
It is possible that pp28-deficient viruses do not efficiently form plaques on HFFpp28-8x cells because the mutant growth defect is not completely complemented by the cells (Fig. 3A). Consequently, we employed a more sensitive infectivity assay to investigate the possibility that small quantities of virions are formed in normal fibroblasts that could not be detected by performing plaque assays on HFFpp28-8x cells. Normal fibroblasts and HFFHApp28 and HFFpp28-8x cells were infected with BADsubUL99, and 10 days later the cell lysates, produced by sonication of infected cells in fresh medium, as well as the medium from the infected cultures, were used to infect normal fibroblasts. After 24 h, the infected cells were fixed and immunofluorescence was performed with an antibody specific for the immediate-early IE1 protein. We readily detected IE1 expression in cells infected with the medium or cell lysate from BADsubUL99-infected HFFpp28-8x cells, but none was detected in the medium or cell lysates of mutant-infected normal fibroblasts or HFFHApp28 cells (data not shown). We conclude that no detectable infectious progeny are produced in mutant virus-infected fibroblasts that do not express pp28 or HA-tagged pp28, which is mislocalized (Fig. 2C) and presumably not myristylated.
Since it appears likely that the amount of pp28 is limiting in complementing cells, we investigated the possibility that large numbers of noninfectious particles, containing no pp28 or reduced amounts of pp28, were present in BADsubUL99 virus stocks. The number of infectious particles relative to the total number of particles, estimated from the amount of viral DNA, produced was analyzed by quantitative real-time PCR. We detected 2 to 5 times more DNA in aliquots of BADsubUL99 virus stocks that contained 103 PFU than in similar samples of BADwt (Fig. 3B). Although it appears that BADsubUL99 virus stocks contain somewhat more DNA-containing particles per unit of infectivity than BADwt stocks, the difference is modest relative to the growth defect observed for the mutant in normal fibroblasts. Indeed, it is possible that the mutant particles are as infectious as wild-type virions, because the titer of BADsubUL99 might be underestimated by plaque assay on HFFpp28-8x cells.
Mutant viruses lacking pp28 have a defect at a late stage of viral replication.
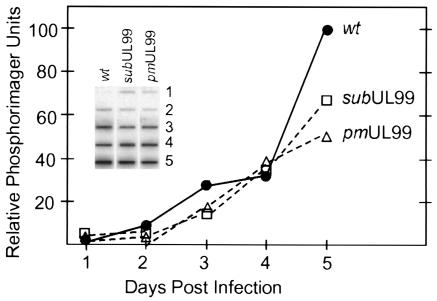
To determine whether the block to the growth of the pp28-deficient mutants was at an early or late stage of the viral replication cycle, we monitored the accumulation of viral DNA. Normal fibroblasts were infected at a multiplicity of infection of 0.01 PFU/cell, total DNA was prepared at intervals from days 1 to 5 after infection, and a slot blot assay was performed with an HCMV-specific probe. In two independent experiments, BADsubUL99 and BADpmUL99 DNAs accumulated at rates comparable to that observed for BADwt virus (Fig. 4). Although a modest difference between the mutant and wild-type viruses was evident on day 5 after infection, this difference can be explained by the fact that wild-type virus is completing its replication cycle and infecting new cells at this time, whereas the mutant viruses are not.
FIG. 4.
Accumulation of viral DNA after infection with wild-type (wt) and pp28-deficient viruses. Normal fibroblasts were infected with BADwt (•), BADsubUL99 (□), or BADpmUL99 (▵) at a multiplicity of infection of 0.01 PFU/cell, and total cell DNA was prepared at the indicated times after infection and assayed by slot blot with an HCMV-specific probe. Radioactivity was quantified by using a phosphorimager.
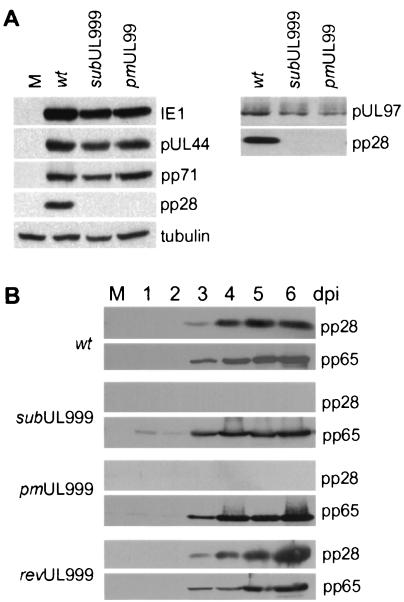
Next, we monitored the accumulation of representative viral proteins in normal fibroblasts infected with the pp28-deficient mutants. Cell lysates were prepared at 72 h postinfection and subjected to Western blotting with antibodies specific for the immediate-early IE1, early UL44 (DNA polymerase), and early-late pp71 proteins (Fig. 5A, left panel). As predicted by the DNA accumulation experiment, both mutant viruses expressed the immediate-early and early proteins, which are required for DNA replication. Both mutants also expressed the early-late protein, pp71, at normal levels, and in a control experiment, neither mutant produced a detectable amount of pp28.
FIG. 5.
Accumulation of viral proteins after infection with wild-type (wt) and pp28-deficient viruses. Normal fibroblasts were mock-infected (M) or infected with BADwt, BADsubUL99, or BADpmUL99 at a multiplicity of infection of 0.01 PFU/cell, and whole-cell extracts were prepared at 72 h (A) or at the indicated times after infection (days postinfection [dpi]) (B). Proteins were analyzed by Western blot analysis with antibodies specific for the UL123-encoded IE1, pUL97, UL83-encoded pp65, UL82-encoded pp71, pUL44, or UL99-encoded pp28.
We also examined the expression of pUL97, which, like pp71, is defined as an early-late protein (12). This protein was of special interest because the UL97 open reading frame resides upstream of UL99 in a family of 3′-coterminal transcripts that use the same polyadenylation site, which is located to the 3′ side of UL99. Consequently, it seemed possible that the substitution mutation in UL99 might influence expression of upstream open reading frames in the family. UL97 was expressed both in BADsubUL99 and BADpmUL99 (Fig. 5A, right panel), although at somewhat lower levels than seen for BADwt-infected cells. This difference in expression level can be explained again by the fact that cell lysates were prepared at 120 h after infection, and at this time postinfection, the wild-type virus is completing its replication cycle and spreading to new cells. Further, the point mutant, which would not be expected to have an effect on polyadenylation of mRNAs carrying upstream open reading frames, directed accumulation of pUL97 to the same level as seen for the substitution mutant.
Finally, we monitored expression of a true late protein (12), pp65, at 1 to 6 days after infection with the BADsubUL99, BADpmUL99, BADwt, or BADrevUL99 virus, and again, there was little or no difference in the accumulation of the viral protein among the viruses (Fig. 5B).
Envelopment of capsids within the cytoplasm requires pp28.
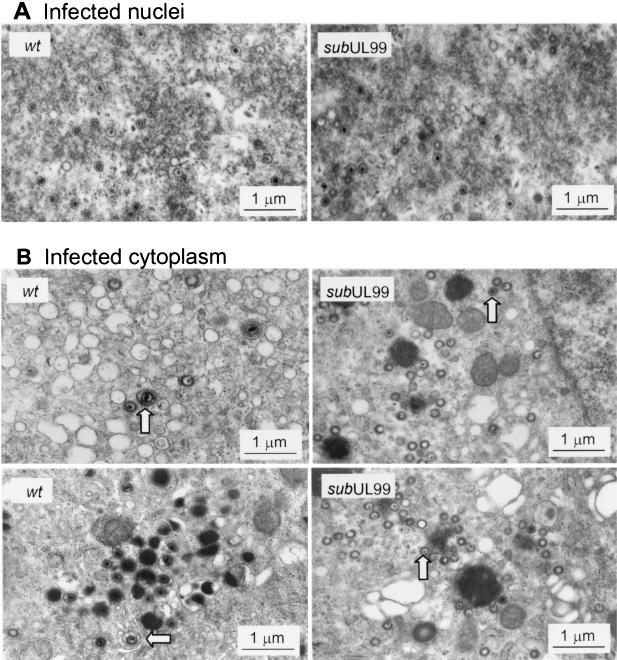
The late viral proteins (pp65, pp71, and UL97) that we examined proved to accumulate to normal or nearly normal levels after infection with pp28-deficient virus. Although we examined only a small subset of viral proteins, this result raised the possibility that the defect in mutant virus-infected cells might reside at the level of virus assembly. Consequently, we examined the morphogenesis of virions at 96 h after infection of normal fibroblasts with BADwt or BADsubUL99 by electron microscopy. All three types of capsids, A (empty), B (protein scaffold containing), and C (DNA containing), were evident in the nuclei of wild-type and mutant virus-infected cells (Fig. 6A). The number of each capsid type was counted in nuclear sections derived from different infected cells. Five sections from BADwt-infected nuclei contained 105 A, 241 B, and 106 C capsids, whereas five sections from BADsubUL99-infected nuclei contained 105 A, 226 B, and 186 C capsids. The pp28-deficient mutant and wild-type virus produced similar numbers and ratios (within twofold) of the three different capsids within the nucleus.
FIG. 6.
Accumulation of virus particles in normal fibroblasts infected with wild-type (wt) and pp28-deficient viruses. Cells were infected with BADwt or BADsubUL99 at a multiplicity of infection of 0.01 PFU/cell and processed for electron microscopy 96 h later. Images were printed at a final magnification of ×56,000. Representative infected nuclear (A) and cytoplasmic (B) regions are shown. Arrows identify enveloped virions in the cytoplasm of wild-type virus-infected cells and capsids in mutant-infected cytoplasm.
However, we observed a dramatic difference in the cytoplasmic sections (Fig. 6B). In BADwt-infected cells, type C capsids were observed that contacted the cytosolic face of membrane vesicles present in the Golgi area. Capsids were seen with the membrane beginning to invaginate and wrap around the particle to form the envelope of the complete virion. In contrast, an association between viral capsids and vesicle membranes was not observed in the cytoplasm of BADsubUL99-infected cells. None of the capsids within the cytosolic compartment acquired an envelope. However, BADsubUL99 cytoplasmic capsids contained a thickened electron-dense coating compared to nuclear capsids (Fig. 6). To quantify the increase in diameter, 50 nuclear and 50 cytoplasmic capsid particles were measured in sections of BADsubUL99-infected cells. The diameter of cytoplasmic particles increased by a factor of 1.18 ± 0.80, a change consistent with the increase in diameter from 1 to 1.3 nm reported for wild-type HCMV capsids (26). The increased diameter indicates that the mutant capsids associate with tegument proteins in the cytoplasm.
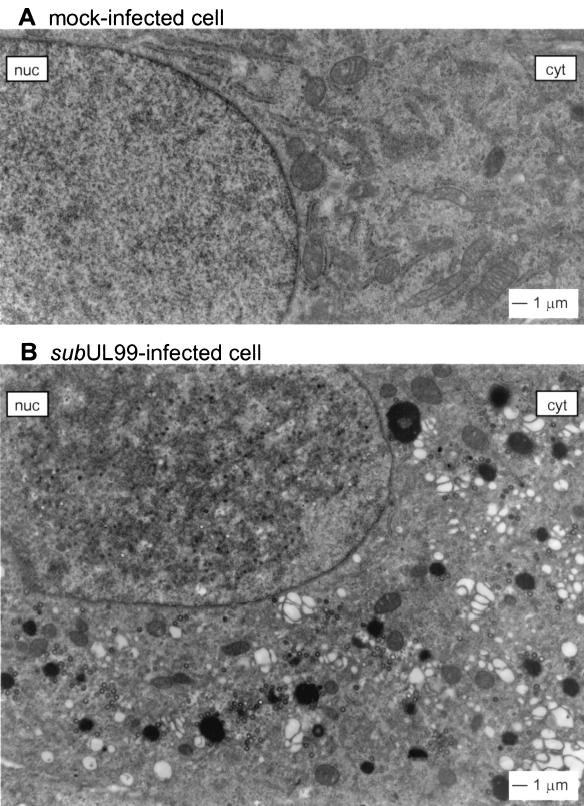
Mock-infected and mutant virus-infected cells are compared in Fig. 7. Approximately 240 cytoplasmic capsids were counted in the cytoplasmic section of the BADsubUL99-infected cell, but no enveloped particles were seen.
FIG. 7.
Comparison of mock-infected and pp28-deficient virus-infected cells. Cells were mock-infected (A) or infected with BADsubUL99 at a multiplicity of infection of 0.01 PFU/cell (B) and processed for electron microscopy at 96 h postinfection. Images were printed at a final magnification of ×13,600. The nucleus (nuc) and cytoplasm (cyt) are identified.
Localization of pp150, pp65, and gB is independent of pp28.
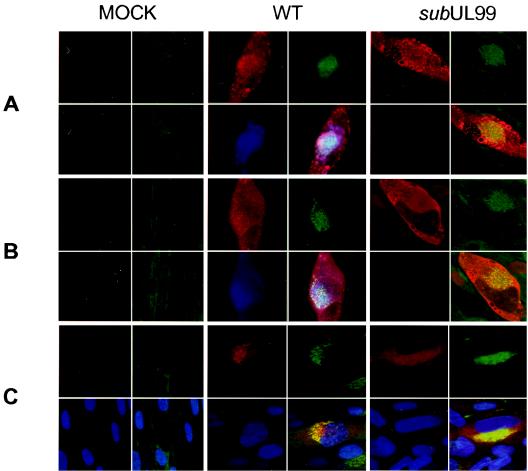
It has been reported that in HCMV-infected cells a series of tegument proteins (pp28, pp150, and pp65) and glycoproteins (gB, gH, and gp65) accumulate within a cytoplasmic region that partially colocalizes with the trans-Golgi network (50). This structure was proposed to be the site of final envelopment of tegument-coated capsids. Our data demonstrated that pp28 is required for the final envelopment of virions; consequently, we investigated the formation of this assembly region in the cytoplasm of cells infected with the pp28-deficient mutant by determining whether the location of p150, pp65, or gB was altered in the absence of pp28. Fibroblasts were infected with BADwt or BADsubUL99 for 120 h and analyzed by immunofluorescence with antibodies specific for tegument (pp150, pp65, and pp28) and envelope (gB) proteins (Fig. 8). The Golgi compartment was identified by using a fluorescent marker reagent, lectin HPA. In mutant and wild-type virus-infected cells, the fluorescence of the Golgi marker was substantially more intense than in mock-infected cells. The two tegument proteins, pp150 and pp65, as well as the envelope glycoprotein, gB, appeared to localize in the same region in BADwt- and BADsubUL99-infected cells. Therefore, we conclude that the structure near to the Golgi apparatus, which has been proposed to be the site of HCMV assembly, is formed in the absence of pp28; pp150, pp65, and gB do not depend on pp28 to accumulate in this region.
FIG. 8.
Localization of viral proteins expressed in normal fibroblasts infected with wild-type (WT) and pp28-deficient viruses. Cells were infected with BADwt or BADsubUL99 at a multiplicity of infection of 0.01 PFU/cell and processed for immunofluorescence at 120 h postinfection. Four images are presented for each cell. The upper-left quadrant shows UL32-encoded pp150 (A), UL83-encoded pp65 (B), or UL55-encoded gB (C) in red. The lower-left quadrants of panels A and B display pp28 in blue, whereas the lower-left panel of C contains blue-stained nuclei. In all panels, the upper-right quadrant displays Golgi in green and the lower-right panel shows a multicolored merged image.
DISCUSSION
Herpesvirus assembly is a multistage process that involves two major cellular compartments. Viral DNA is packaged into capsids in the nucleus, and several tegument proteins likely associate with the capsid in the nucleus. For example, the HCMV UL82-encoded pp71 (25) and the UL69-encoded ppUL69 tegument proteins (49) are localized to the nucleus during the late phase of infection. Whereas capsid assembly is relatively well understood, the mechanism for acquisition of the final envelope has been controversial. Two alternative pathways have been proposed for the envelopment of herpesvirus capsids (reviewed in reference 19). In one model, the capsid buds through the inner nuclear membrane and then travels through the secretory pathway, eventually exiting the cell with an envelope derived from the inner nuclear membrane. In the second model, the capsid buds through the inner nuclear membrane, acquiring an initial envelope that fuses with the outer nuclear membrane to release the capsid into the cytoplasm, and finally buds, together with its associated tegument proteins, through a membrane in the cytoplasm that is associated with the secretory system. Several lines of experimentation favor the second model (38). First, the phospholipid composition of the membrane enveloping extracellular herpes simplex virions is different than the composition of the nuclear membrane (59). Second, electron microscopic studies of herpes simplex virus morphogenesis have visualized the movement of its capsid from the nucleus to cytoplasm by sequential envelopment and de-envelopment as it passes through the inner and outer nuclear membranes (reference 53 and references therein), and it appears that a similar series of events relocates the HCMV capsid to the cytoplasm (reviewed in references 20 and 40). Third, the UL34 membrane protein of herpes simplex virus, which accumulates in the inner nuclear membrane, is present in perinuclear virions but not in extracellular virions (47). Fourth, a substantial portion of the tegument proteins are acquired by the maturing virion in the cytoplasm.
HCMV tegument proteins that accumulate in the cytoplasm late after infection include the UL32-encoded pp150 (24, 50), the UL99-encoded pp28 (31), the UL83-encoded pp65 (24, 50), and the UL25-encoded pUL25 (6). These, and presumably other tegument proteins, associate with the capsid in the cytoplasm, and then the complex acquires an envelope containing virus-specific glycoproteins. Electron microscopic studies demonstrate that tegument-coated particles acquire an envelope by budding into cytoplasmic vacuoles (20, 52, 56, 57). Immunofluorescence assays have suggested that the site of cytoplasmic envelopment is a juxtanuclear structure that includes viral tegument proteins as well as multiple virus-encoded envelope proteins (50). Using antibodies to cellular proteins, the cytoplasmic assembly center has been shown to partially overlap with a component of the trans-Golgi network (50), and we similarly observed that the location of viral proteins involved in the cytoplasmic assembly step partially overlapped with a Golgi marker (Fig. 8). Recently, Homman-Loudiyi et al. (26) have shown that the UL55-encoded gB, a major glycoprotein constituent of the HCMV envelope, colocalizes with several protein markers of Golgi-derived vacuoles that are destined for the plasma membrane. Consequently, it appears that the juxtanuclear HCMV assembly site is comprised of modified elements of the secretory apparatus. This view is reinforced by the ability of brefeldin A to block the envelopment of HCMV capsids (18), similar to the block it institutes in other herpesviruses (e.g., herpes simplex virus [15] and pseudorabies virus [60]).
In this report we have demonstrated that the pp28 protein plays a key role in the cytoplasmic phase of HCMV assembly. To study the function of pp28, we constructed two mutant viruses, BADsubUL99 and BADpmUL99 (Fig. 1B), which failed to produce pp28 (Fig. 5 and 8) and were defective for growth in normal fibroblasts (Fig. 3A). Viral DNA accumulated with wild-type kinetics reaching wild-type levels within mutant virus-infected cells (Fig. 4), several late proteins that were monitored by Western blot (Fig. 5) or immunofluorescence (Fig. 8) accumulated to wild-type levels, and numerous capsids accumulated in the nucleus of infected cells (Fig. 6A). However, once capsids reached the cytoplasm of mutant virus-infected cells, they did not associate with membranes of vesicles and, consequently, they did not acquire an envelope (Fig. 6B and 7). The phenotype of the pp28 mutants provides compelling support for the assembly model in which the HCMV capsid-tegument complex acquires its final envelope by budding through a secretory membrane within the cytoplasm.
BADsubUL99 capsids in the cytoplasm contained a thickened electron-dense coating compared to nuclear capsids (Fig. 6 and 7). This argues that, in the absence of pp28, capsids associate with tegument proteins but fail to bud through a membrane. It is interesting that Landini et al. (31) localized pp28 to the periphery of cytoplasmic capsids by electron microscopic examination of immunogold-labeled cells that were infected with HCMV. The label was not evident on vacuole membranes, where encapsidation is thought to occur (reference 26 and references therein), suggesting that pp28 might first interact with a capsid-tegument complex and then direct it to an appropriately modified membrane for envelopment. Further, pp28 forms a detergent-resistant association with capsids, suggesting it is more firmly associated with capsids than tegument proteins such as pp65 whose association is substantially disrupted by detergent treatment of virions (20).
Since we have monitored the expression of a limited subset of the late genes in mutant virus-infected cells, it is possible that pp28 influences the assembly process indirectly by stimulating the production of another HCMV late protein that was not assayed. However, pp28 is not known to modulate gene expression. We favor the view that pp28 acts directly to mediate the cytoplasmic envelopment of capsids for two reasons. First, pp28 is localized to the region within the cytoplasm at which assembly takes place in HCMV-infected cells (Fig. 8). Second, pp28 is a myristylated protein that interacts with cellular membranes (51). Studies of retrovirus assembly have revealed that myristylation of the gag protein is required for its targeting to cellular membranes with the concomitant budding of subviral particles to acquire an envelope (11, 46). Our results suggest that myristylation is required for pp28 function, since cells expressing HA-pp28, which cannot be myristylated, do not complement the defect of pp28-deficient viruses (Fig. 2).
It has been shown that, when expressed from a transfected plasmid in Cos7 cells, the pp28 viral tegument protein colocalized with a protein that resides in the endoplasmic reticulum-Golgi intermediate compartment (51), a compartment of the secretory pathway in which glycoproteins are sorted and transferred from the endoplasmic reticulum to the Golgi complex (reviewed in reference 22). However, in the context of viral infection, pp28 is relocated to the proposed assembly region in which other tegument proteins and glycoproteins are localized (51), indicating that interaction with other viral proteins or cellular proteins induced by infection is necessary for the proper localization of pp28. We tested the localization of other viral structural proteins to the cytoplasmic assembly site in the absence of pp28, but at the resolution of immunofluorescence, the pp150 and pp65 tegument proteins and the gB envelope glycoprotein were properly localized (Fig. 8). One might speculate that pp150, pp65, and other tegument proteins are directed to the assembly site by interacting, either directly or indirectly, with the cytoplasmic tails of virus-encoded envelope proteins like gB; it is also possible that these proteins interact with the capsid as well as the membrane assembly sites. Indeed, pp150 has been shown to bind directly to capsids (7, 58).
The BADsubUL99 and BADpmUL99 mutant viruses are extremely defective for growth in normal fibroblasts, producing no detectable infectious progeny (Fig. 2B and 3A). Since it is possible that plaque formation by the mutant viruses on the complementing cells is inefficient, we also assayed lysates of BADsubUL99-infected fibroblasts for the presence of virus able to infect cells and express the HCMV IE1 protein, but again, we obtained no evidence for the production of infectious progeny virus. The loss of pp28 leads to a complete block in the virus replication cycle. The strong dependence of viral growth on pp28 function is consistent with the observation that the sequence of the pp28 protein encoded by the Towne strain of HCMV is very similar to that of the AD169 protein (42) and with the presence of pp28 homologues in chimpanzee (17) and murine cytomegaloviruses (16, 45).
In addition to their failure to produce infectious progeny in normal human fibroblasts, the pp28-deficient mutants grew poorly in complementing cells (Fig. 3A). It is not likely that the mutant viruses contain a second site mutation because two different pp28-deficient mutants, BADsubUL99 and BADpmUL99, were generated, and both grew to the same limited extent in the complementing cells. Further, a revertant of BADsubUL99, termed BADrevUL99, grew like the wild-type virus, ruling out the possibility that the mutant phenotype resulted from a change at another location (Fig. 3A). The level of the pp28 protein is substantially reduced in the complementing cells compared to infected cells (Fig. 2A), and increasing amounts of pp28 (HFFpp28-3x versus HFFpp28-8x cells) supported the production of an increased yield of BADsubUL99 (Fig. 2B). Apparently, the poor growth of the mutant viruses in the complementing cells results from limiting amounts of pp28.
In some respects, the HCMV pp28 protein is functionally similar to the protein encoded by the herpes simplex type 1 UL11 gene. Both proteins are relatively small phosphoproteins (pp28, 190 amino acids; UL11, 96 amino acids), myristylated (34, 51), membrane associated (9, 35, 51), and tegument proteins (31, 51). The UL11 protein is also palmitylated (33), and it is not known if pp28 undergoes this modification. Further, as for the mutants described here (Fig. 6 and 7), a herpes simplex virus mutant unable to express the UL11 protein accumulated increased numbers of capsids in the nucleus and cytoplasm (3). However, there are significant differences between the two viral gene products. We have found that the growth phenotype of pp28-deficient mutants is extreme, i.e., no infectious virus can be detected after infection of normal fibroblasts (Fig. 3A). In contrast, UL11-deficient mutants grow poorly but nevertheless produce infectious progeny (3, 35). Further, whereas pp28 appears to be localized exclusively to the cytoplasm (51), UL11 is found in both the nuclear and cytoplasmic compartments of infected cells (2). Finally, HCMV can grow efficiently in pp28-expressing cells (Fig. 3A), but UL11-expressing cells are resistant to infection with herpes simplex virus, apparently at the point of fusion of the virion's envelope and the plasma membrane (48). It is possible that higher-level expression of pp28 might interfere with infection by wild-type virus, but at the level of expression we have achieved, we detect no inhibition. Additional experimentation will be needed to better delineate the functional similarities and differences between pp28 and the UL11 protein.
In conclusion, our mutant analysis has demonstrated that pp28 is required for the final cytoplasmic envelopment of tegument-associated HCMV capsids.
Acknowledgments
We thank D. Yu (Princeton University) for help with the AD169 BAC system, P. Robinson (Princeton University) for assistance in the production of monoclonal antibodies, J. Goodhouse and J. Schroer (Princeton University) for help with immunofluorescence, and M. Schrader (University of Marburg) and A. Marchini (Tribeca Pharmaceuticals) for generous gifts of antibodies.
This work was supported by a grant from the NIH (CA82396), and M.C.S. was supported by a predoctoral fellowship from the Brazilian government (Conselho Nacional de Desenvolvimento Científico e Tecnológico-CNPq).
REFERENCES
- 1.Adair, R., E. R. Douglas, J. B. Maclean, S. Y. Graham, J. D. Aitken, F. E. Jamieson, and D. J. Dargan. 2002. The products of human cytomegalovirus genes UL23, UL24, UL43 and US22 are tegument components. J. Gen. Virol. 83:1315-1324. [DOI] [PubMed] [Google Scholar]
- 2.Baines, J. D., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (UL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldick, C. J., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battista, M. C., G. Bergamini, M. C. Boccuni, F. Campanini, A. Ripalti, and M. P. Landini. 1999. Expression and characterization of a novel structural protein of human cytomegalovirus, pUL25. J. Virol. 73:3800-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baxter, M. K., and W. Gibson. 2001. Cytomegalovirus basic phosphoprotein (pUL32) binds to capsids in vitro through its amino one-third. J. Virol. 75:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bechtel, J. T., and T. Shenk. 2002. Human cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J. Virol. 76:1043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowzard, J. B., R. J. Visalli, C. B. Wilson, J. S. Loomis, E. M. Callahan, R. J. Courtney, and J. W. Wills. 2000. Membrane targeting properties of a herpesvirus tegument protein-retrovirus Gag chimera. J. Virol. 74:8692-8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bresnahan, W. A., and T. Shenk. 2000. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA 97:14506-14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers, J., A. Angulo, D. Amaratunga, H. Guo, Y. Jiang, J. S. Wan, A. Bittner, K. Frueh, M. R. Jackson, P. A. Peterson, M. G. Erlander, and P. Ghazal. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 73:5757-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 14.Chen, D. H., H. Jiang, M. Lee, F. Liu, and Z. H. Zhou. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10-16. [DOI] [PubMed] [Google Scholar]
- 15.Cheung, P., B. W. Banfield, and F. Tufaro. 1991. Brefeldin A arrests the maturation and egress of herpes simplex virus particles during infection. J. Virol. 65:1893-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cranmer, L. D., C. Clark, and D. H. Spector. 1994. Cloning, characterization, and expression of the murine cytomegalovirus homologue of the human cytomegalovirus 28-kDa matrix phosphoprotein (UL99). Virology 205:417-429. [DOI] [PubMed] [Google Scholar]
- 17.Davison, A. J., A. Dolan, P. Akter, C. Addison, D. J. Dargan, D. J. Alcendor, D. J. McGeoch, and G. S. Hayward. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17-28. [DOI] [PubMed] [Google Scholar]
- 18.Eggers, M., E. Bogner, B. Agricola, H. F. Kern, and K. Radsak. 1992. Inhibition of human cytomegalovirus maturation by brefeldin A. J. Gen. Virol. 73:2679-2692. [DOI] [PubMed] [Google Scholar]
- 19.Enquist, L. W., P. J. Husak, B. W. Banfield, and G. A. Smith. 1999. Infection and spread of alphaherpesviruses in the nervous system. Adv. Virus Res. 51:237-347. [DOI] [PubMed] [Google Scholar]
- 20.Gibson, W. 1996. Structure and assembly of the virion. Intervirology 39:389-400. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert, M. J., S. R. Riddell, B. Plachter, and P. D. Greenberg. 1996. Cytomegalovirus selectively blocks antigen processing and presentation of its immediate-early gene product. Nature 383:720-722. [DOI] [PubMed] [Google Scholar]
- 22.Hauri, H. P., F. Kappeler, H. Andersson, and C. Appenzeller. 2000. ERGIC-53 and traffic in the secretory pathway. J. Cell Sci. 113:587-596. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi, M. L., C. Blankenship, and T. Shenk. 2000. Human cytomegalovirus UL69 protein is required for the efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc. Natl. Acad. Sci. USA 97:2692-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hensel, G., H. Meyer, S. Gartner, G. Brand, and H. F. Kern. 1995. Nuclear localization of the human cytomegalovirus tegument protein pp150 (ppUL32). J. Gen. Virol. 76:1591-1601. [DOI] [PubMed] [Google Scholar]
- 25.Hensel, G. M., H. H. Meyer, I. Buchmann, D. Pommerehne, S. Schmolke, B. Plachter, K. Radsak, and H. F. Kern. 1996. Intracellular localization and expression of the human cytomegalovirus matrix phosphoprotein pp71 (ppUL82): evidence for its translocation into the nucleus. J. Gen. Virol. 77:3087-3097. [DOI] [PubMed] [Google Scholar]
- 26.Homman-Loudiyi, M., K. Hultenby, W. Britt, and C. Soderberg-Naucler. 2003. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-golgi network 46, and mannosidase II. J. Virol. 77:3191-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalejta, R. F., J. T. Bechtel, and T. Shenk. 2003. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol. Cell. Biol. 23:1885-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerry, J. A., M. A. Priddy, C. P. Kohler, T. L. Staley, D. Weber, T. R. Jones, and R. M. Stenberg. 1997. Translational regulation of the human cytomegalovirus pp28 (UL99) late gene. J. Virol. 71:981-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinsella, T. M., and G. P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405-1413. [DOI] [PubMed] [Google Scholar]
- 30.Landini, M. P., C. Re, G. Mirolo, B. Baldassarri, and M. La Placa. 1985. Human immune response to cytomegalovirus structural polypeptides studied by immunoblotting. J. Med. Virol. 17:303-311. [DOI] [PubMed] [Google Scholar]
- 31.Landini, M. P., B. Severi, G. Furlini, and B. de Giorgi. 1987. Human cytomegalovirus structural components: intracellular and intraviral localization of p28 and p65-69 by immunoelectron microscopy. Virus Res. 8:15-23. [DOI] [PubMed] [Google Scholar]
- 32.Liu, B., and M. F. Stinski. 1992. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J. Virol. 66:4434-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loomis, J. S., J. B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacLean, C. A., B. Clark, and D. J. McGeoch. 1989. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J. Gen. Virol. 70:3147-3157. [DOI] [PubMed] [Google Scholar]
- 35.MacLean, C. A., A. Dolan, F. E. Jamieson, and D. J. McGeoch. 1992. The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle. J. Gen. Virol. 73:539-547. [DOI] [PubMed] [Google Scholar]
- 36.Martinez, J., and S. C. St. Jeor. 1986. Molecular cloning and analysis of three cDNA clones homologous to human cytomegalovirus RNAs present during late infection. J. Virol. 60:531-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez, J., R. S. Lahijani, and S. C. St. Jeor. 1989. Analysis of a region of the human cytomegalovirus (AD169) genome coding for a 25-kilodalton virion protein. J. Virol. 63:233-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyer, H., A. T. Bankier, M. P. Landini, C. M. Brown, B. G. Barrell, B. Ruger, and M. Mach. 1988. Identification and prokaryotic expression of the gene coding for the highly immunogenic 28-kilodalton structural phosphoprotein (pp28) of human cytomegalovirus. J. Virol. 62:2243-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 41.Nowak, B., C. Sullivan, P. Sarnow, R. Thomas, F. Bricout, J. C. Nicolas, B. Fleckenstein, and A. J. Levine. 1984. Characterization of monoclonal antibodies and polyclonal immune sera directed against human cytomegalovirus virion proteins. Virology 132:325-338. [DOI] [PubMed] [Google Scholar]
- 42.Pande, H., K. Campo, B. Tanamachi, and J. A. Zaia. 1991. Human cytomegalovirus strain Towne pp28 gene: sequence comparison to pp28 of HCMV AD169 and stable expression in Chinese hamster ovary cells. Virology 184:762-767. [DOI] [PubMed] [Google Scholar]
- 43.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2705. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 44.Pereira, L., M. Hoffman, and N. Cremer. 1982. Electrophoretic analysis of polypeptides immune precipitated from cytomegalovirus-infected cell extracts by human sera. Infect. Immun. 36:933-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rein, A., M. R. McClure, N. R. Rice, R. B. Luftig, and A. M. Schultz. 1986. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc. Natl. Acad. Sci. USA 83:7246-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roller, R. J., and B. Roizman. 1994. A herpes simplex virus 1 US11-expressing cell line is resistant to herpes simplex virus infection at a step in viral entry mediated by glycoprotein D. J. Virol. 68:2830-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez, V., P. C. Angeletti, J. A. Engler, and W. J. Britt. 1998. Localization of human cytomegalovirus structural proteins to the nuclear matrix of infected human fibroblasts. J. Virol. 72:3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-golgi-intermediate compartment. J. Virol. 74:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Severi, B., M. P. Landini, and E. Govoni. 1988. Human cytomegalovirus morphogenesis: an ultrastructural study of the late cytoplasmic phases. Arch. Virol. 98:51-64. [DOI] [PubMed] [Google Scholar]
- 53.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment → deenvelopment → reenvelopment pathway. J. Virol. 75:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith, J. D., and E. De Harven. 1973. Herpes simplex virus and human cytomegalovirus replication in WI-38 cells. I. Sequence of viral replication. J. Virol. 12:919-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163-178. [PubMed] [Google Scholar]
- 58.Trus, B. L., W. Gibson, N. Cheng, and A. C. Steven. 1999. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J. Virol. 73:2181-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Genderen, I. L., R. Brandimarti, M. R. Torrisi, G. Campadelli, and G. van Meer. 1994. The phospholipid composition of extracellular herpes simplex virions differs from that of host cell nuclei. Virology 200:831-836. [DOI] [PubMed] [Google Scholar]
- 60.Whealy, M. E., J. P. Card, R. P. Meade, A. K. Robbins, and L. W. Enquist. 1991. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J. Virol. 65:1066-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wing, B. A., and E. S. Huang. 1995. Analysis and mapping of a family of 3′-coterminal transcripts containing coding sequences for human cytomegalovirus open reading frames UL93 through UL99. J. Virol. 69:1521-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu, D., G. A. Smith, L. W. Enquist, and T. Shenk. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]