Abstract
Cdc14 phosphatases antagonize cyclin-dependent kinase–directed phosphorylation events and are involved in several facets of cell cycle control. We investigate the role of the fission yeast Cdc14 homologue Clp1/Flp1 in cytokinesis. We find that Clp1/Flp1 is tethered at the contractile ring (CR) through its association with anillin-related Mid1. Fluorescent recovery after photobleaching analyses indicate that Mid1, unlike other tested CR components, is anchored at the cell midzone, and this physical property is likely to account for its scaffolding role. By generating a mutation in mid1 that selectively disrupts Clp1/Flp1 tethering, we reveal the specific functional consequences of Clp1/Flp1 activity at the CR, including dephosphorylation of the essential CR component Cdc15, reductions in CR protein mobility, and CR resistance to mild perturbation. Our evidence indicates that Clp1/Flp1 must interact with the Mid1 scaffold to ensure the fidelity of Schizosaccharomyces pombe cytokinesis.
Introduction
In eukaryotes, progression through the cell cycle is directed by the activity of Cdks. Reversal of Cdk-dependent phosphorylation events involves the highly conserved Cdc14 family of phosphatases. First characterized for their essential role in facilitating mitotic exit in budding yeast, Cdc14 family members are now known to play numerous cell cycle roles, ranging from regulating centrosome duplication to controlling the terminal events of cytokinesis (Stegmeier and Amon, 2004). For example, cells lacking the fission yeast Cdc14 homologue Clp1/Flp1 (hereafter referred to as Clp1) exhibit defects in chromosome segregation (Trautmann et al., 2004), cytokinesis (Cueille et al., 2001; Trautmann et al., 2001), and mitotic exit (Esteban et al., 2004; Wolfe and Gould, 2004). Although the roles of Cdc14 phosphatases in cell division have been best characterized in yeast, cytokinesis defects are also observed in human cells after depletion of hCdc14A (Kaiser et al., 2002; Mailand et al., 2002). Thus, a regulatory role for Cdc14 phosphatases in mitotic exit and cytokinesis appears conserved in multiple organisms.
Cdc14 phosphatases localize dynamically to many subcellular compartments to carry out their functions (Stegmeier and Amon, 2004). For example, Clp1 is primarily nucleolar and at the spindle pole body during interphase (Cueille et al., 2001; Trautmann et al., 2001). During mitosis, Clp1 disperses throughout the nucleus and cytoplasm, concentrating at kinetochores (Trautmann et al., 2004), the mitotic spindle, and the contractile ring (CR), a highly dynamic structure that controls ingression of the division plane. Clp1 remains at the CR during its constriction and then reconcentrates in the nucleolus.
Consistent with its localization at the CR, several findings have established a regulatory role for Clp1 in cytokinesis. Cells lacking Clp1 display strong negative genetic interactions with many cytokinesis mutants, including those which disrupt CR assembly, septum formation, and cell wall synthesis (Trautmann et al., 2001; Mishra et al., 2004). In addition, genetic studies indicate that Clp1 stabilizes CRs when actin polymerization is inhibited by latrunculin A (lat A) treatment (Mishra et al., 2004). However, it is not known how Clp1 affects the process of cytokinesis nor whether Clp1's interaction with the CR is important for this function.
Proper assembly and localization of the CR for cell division in fission yeast requires Mid1, an anillin-related protein (Chang et al., 1996; Sohrmann et al., 1996). In early mitosis, Mid1 exits the nucleus and directly interacts with the cell cortex, anchoring the CR in the cell center (Celton-Morizur et al., 2004). In the absence of mid1, CR structures are frequently off centered and/or tilted leading to unequal nuclear and cellular division (Chang et al., 1996; Sohrmann et al., 1996). Once the division site has been established, several other proteins are recruited to this region to assemble the CR. One essential ring component that Mid1 recruits to the cortical region is myosin heavy chain Myo2, and mitotic dephosphorylation of the Myo2 C terminus has been implicated in regulating this interaction (Motegi et al., 2004).
To investigate the role of Clp1 at the division site, we used a proteomics approach to identify Mid1 as a Clp1-interacting protein and defined Mid1 association as the mechanism of Clp1 recruitment to the CR. By selectively disrupting Clp1–Mid1 interaction, we have been able to determine the functional consequence of Clp1 recruitment to the CR. We find that the phosphorylation status and dynamic properties of key CR components are altered in the absence of Clp1 activity and that otherwise silent mutations of CR components lead to cytokinetic failures in the absence of Clp1 CR localization. Our data offer a mechanistic explanation for Clp1's requirement in the fidelity of Schizosaccharomyces pombe cytokinesis.
Results
Clp1 and Mid1 interact in vivo
To identify proteins that might link Clp1 to the CR, we isolated tandem affinity purification (TAP) complexes from nda3(β-tubulin)-KM311–arrested cells producing C-terminally TAP-tagged Clp1. These cells arrest in a premetaphase-like state with a CR but lack a mitotic spindle (Hiraoka et al., 1984). The protein composition of the complexes was analyzed by 2D liquid chromatography tandem mass spectrometry. In addition to Clp1 at nearly 100% sequence coverage, this strategy identified Mid1 with 39.9% sequence coverage. Confirming the TAP analysis, reciprocal coimmunoprecipitation experiments demonstrated that Clp1 and Mid1 associate (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200709060/DC1). Yeast two-hybrid analysis and in vitro binding assays showed that an internal region of Mid1 (aa 331–534) interacted directly with the N-terminal catalytic domain of Clp1 (Fig. S1, B–E). Other proteins were identified in Clp1–TAP complexes and the validity of these hits is being investigated. No other CR components were recovered with such high coverage (unpublished data).
Clp1 requires mid1+ for localization to the CR
During early mitosis, both Clp1 and Mid1 localize to the CR (Fig. 1 A; Sohrmann et al., 1996; Cueille et al., 2001; Trautmann et al., 2001). Therefore, we examined whether the CR localization of either protein is dependent on the other. Live-cell imaging of Mid1-GFP cells demonstrated that Clp1 does not affect Mid1 ring localization (Fig. 1 A). In nda3-KM311–arrested cells, Clp1-GFP was observed at kinetochores and in rings (Fig. 1 A). In nda3-KM311 mid1Δ cells, however, Clp1-GFP rings were not detected, although Clp1-GFP was still observed at kinetochores (Fig. 1 A). Lack of Clp1-GFP rings in mid1Δ cells was not because of a change in Clp1 levels (Fig. 2 E and Fig. S1 F). To verify that CRs could be detected in mid1Δ cells, we examined the localization of Clp1-YFP together with Cdc15-CFP, which localizes to the CR independently of Mid1 (Sohrmann et al., 1996). In 97% (193/198) of nda3-KM311–arrested cells examined, Clp1-YFP and Cdc15-CFP colocalized in rings (Fig. 1 B). Although Cdc15-CFP formed rings in mid1Δ cells, Clp1-YFP could not be detected in these rings (0/102 cells). Time-lapse videomicroscopy of cells progressing through mitosis confirmed that Clp1-GFP localized to kinetochores and the mitotic spindle in mid1Δ cells with similar timing and intensity as in mid1+ cells but failed to form a ring (Fig. 1 C; Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200709060/DC1). These data indicate that Mid1 is necessary to recruit Clp1 to the CR.
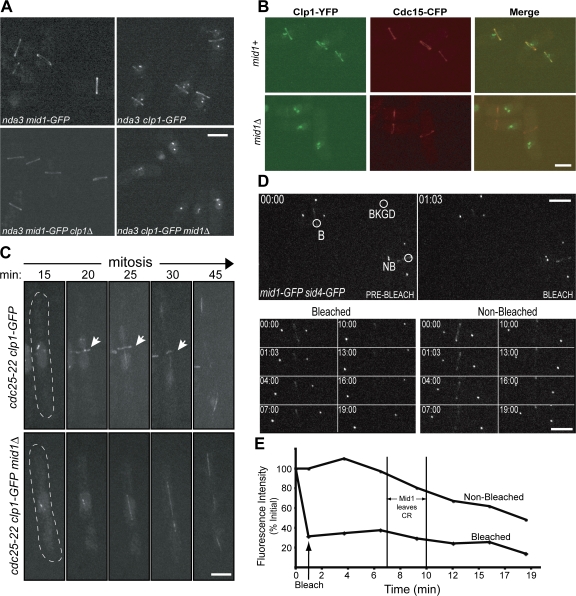
Figure 1.
Clp1 depends on Mid1 for localization to the CR. (A) Live-cell images of nda3-KM311 mid1-GFP, nda3-KM311 mid1-GFP clp1Δ, nda3-KM311 clp1-GFP, and nda3-KM311 clp1-GFP mid1Δ cells after incubation at 18°C for 7 h. (B) Live-cell images of nda3-KM311 clp1-YFP cdc15-CFP and nda3-KM311 clp1-YFP cdc15-CFP mid1Δ cells after arrest by incubation at 18°C for 7 h. Over 100 cells were examined for each strain. (C) Time-lapse confocal microscopy of live cdc25-22 clp1-GFP and cdc25-22 clp1-GFP mid1Δ cells during mitosis. Exponentially growing cells were incubated at the restrictive temperature (36°C) for 3.5 h, and then released to the permissive temperature (25°C) for 15 min. Indicated time points are from time of release. Arrows indicate Clp1 ring. Images are from Videos 1 and 2 (available at http://www.jcb.org/cgi/content/full/jcb.200709060/DC1). (D and E) Representative images (D) and fluorescence recovery curves (E) for Mid1-GFP FRAP in mid1-GFP sid4-GFP cells. Sid4-GFP signal determined mitotic stage and vertical lines indicate the window when Mid1 begins to dissociate from the CR. B, bleach region; NB, nonbleached region (used to correct for overall bleaching); BKGD, background region (used to correct for overall bleaching). Images are from Video 3 (available at http://www.jcb.org/cgi/content/full/jcb.200709060/DC1). Bars, 5 μm.
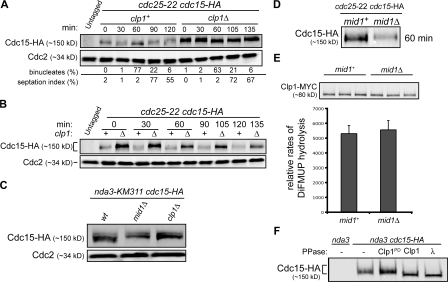
Figure 2.
Clp1 localization to the CR is required for Cdc15 dephosphorylation. (A and B) cdc25-22 cdc15-HA and cdc25-22 cdc15-HA clp1Δ cells were grown to mid log phase, shifted to 36°C for 4 h, and then released to permissive temperature (25°C). Samples were collected at the indicated times. Denatured protein extracts were analyzed by Western blot analysis with HA and Cdc2 antibodies. Completion of mitosis was determined by monitoring binucleate formation and septation index. Because of a delay in Cdc2 inactivation, septation peaks ∼15 min later in clp1Δ cells (Wolfe and Gould, 2004). (C) nda3-KM311 cdc15-HA, nda3-KM311 cdc15-HA mid1Δ, and nda3-KM311 cdc15-HA clp1Δ strains were arrested by incubation at 18°C for 7 h, and then samples were collected and denatured protein extracts normalized for Cdc15 levels were analyzed by Western blot analysis with HA antibodies. (D) Samples from cdc25-22 cdc15-HA and cdc25-22 cdc15-HA mid1Δ cells were prepared as in A and B. The 60-min time point is shown here. Western blot analysis of additional time points, binucleate formation, and septation index are included in Fig. S2 A (available at http://www.jcb.org/cgi/content/full/jcb.200709060/DC1). (E) clp1-MYC (mid1+) and clp1-MYC mid1Δ (mid1Δ) cells were grown at 32°C to log phase. Immunoprecipitated Clp1 phosphatase activity was determined by DiFMUP hydrolysis and Clp1 levels were determined by Western blot analysis. Reactions were performed in triplicate for standard error analysis. (F) Native protein extracts from nda3-KM311 and nda3-KM311 cdc15-HA cells were incubated with HA antibodies to immunoprecipitate Cdc15-HA. Immunoprecipitates were incubated with MBP (−), MBP-Clpl, MBP-Clp1PD, or λ phosphatase and analyzed by Western blot analysis with HA antibodies.
The S. pombe CR is a highly dynamic structure (Pelham and Chang, 2002; Wong et al., 2002). However, if Mid1 serves as a scaffold to link the cortex with CR components, such as Clp1, we reasoned that Mid1 might remain stably bound to the cortex. This was tested by FRAP analysis of Mid1-GFP. After spindle pole body separation, which was detected with Sid4-GFP (Chang and Gould, 2000), Mid1-GFP rings were bleached and recovery was monitored for the duration of Mid1 ring localization in a nonbleached cell (Fig. 1, D–E; and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200709060/DC1). Bleached Mid1-GFP rings showed little signal recovery after 15 min, indicating that Mid1 is very immobile while at the CR.
Clp1 regulates Cdc15 dephosphorylation at the CR
A plausible role for the Mid1–Clp1 interaction is to position Clp1 at the CR so that it can dephosphorylate CR components and thereby modify CR properties. Although several CR components are phosphoproteins (Wolfe and Gould, 2005) and might be Clp1 substrates, one essential CR component whose phosphorylation is known to vary during the cell cycle is Cdc15 (Fankhauser et al., 1995). Cdc15 is essential for cytokinesis (Fankhauser et al., 1995) and is responsible for recruiting F-actin nucleators to the division site (Fankhauser et al., 1995; Carnahan and Gould, 2003). To investigate whether Cdc15 dephosphorylation depends on Clp1, Cdc15 phosphorylation status was monitored in the presence and absence of clp1. As observed previously (Fankhauser et al., 1995), Cdc15 became progressively dephosphorylated as cells progressed through mitosis. Corresponding with the time of CR formation, Cdc15 was partially dephosphorylated (Fig. 2, A and B; Fankhauser et al., 1995). This Cdc15 phosphoisomer reproducibly accumulated in nda3-KM311–arrested cells (Fig. 2 C). Additional Cdc15 dephosphorylation followed coincident with anaphase resulting in a hypophosphorylated form (Fig. 2, A and B). Hyperphosphorylated Cdc15 was reestablished upon septation. In clp1Δ cells, the Cdc15 dephosphorylation observed at anaphase did not occur to the same extent as in wild-type cells (Fig. 2, A and B). Reduced Cdc15 dephosphorylation was also observed in cells expressing only the clp1-C286S phosphatase-dead allele (Wolfe and Gould, 2004) progressing through mitosis (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200709060/DC1). This mutant has been shown previously to localize to the CR during mitosis (Trautmann et al., 2004). Further supporting the idea that Cdc15 is a Clp1 target, recombinant Clp1 dephosphorylated the partially phosphorylated Cdc15 in immunocomplexes purified from nda3-KM311–arrested cells (Fig. 2 F) and a small amount of Clp1 specifically associated with Cdc15 immune complexes isolated from nda3-KM311–arrested cells (Fig. S2 C). These results indicate that Clp1 phosphatase activity is required for maximal Cdc15 dephosphorylation during mitosis.
We then reasoned that Clp1-dependent Cdc15 dephosphorylation should also be compromised in mid1Δ cells, in which Clp1 localization to the CR does not occur (Fig. 1). Indeed, Cdc15 in nda3-KM311–arrested mid1Δ cells was in the hyperphosphorylated form, comigrating with Cdc15 from nda3-KM311–arrested clp1Δ cells (Fig. 2 C). Furthermore, Clp1-dependent Cdc15 dephosphorylation was not detected in synchronized mid1Δ cells progressing through mitosis (Fig. 2 D; and Fig. S2 B). Clp1 phosphatase activity assays verified that the difference in Cdc15 phosphorylation was not a result of reduced Clp1 phosphatase activity in the absence of mid1 (Fig. 2 E). Collectively, our results indicate that Mid1 is required to properly localize Clp1 to the CR where Clp1 substrates, such as Cdc15, undergo Clp1-dependent dephosphorylation.
Clp1 phosphatase activity regulates Cdc15 and myosin ring stability
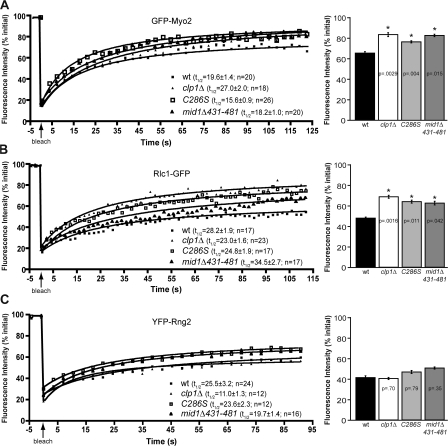
Given the previously established role for Clp1 in stabilizing CRs in response to mild perturbation (Mishra et al., 2004), we reasoned that even in the absence of CR disruption there might be an intrinsic difference in the dynamics of CR components in the absence of Clp1 function. To address this possibility, we examined the dynamics of four CR components involved in CR assembly and stability: Cdc15, the myosin II components, Myo2 (heavy chain) and Rlc1 (regulatory light chain), and the actin binding IQGAP, Rng2. Myosin II was of particular interest, as its recruitment to the CR is partially dependent on Mid1, and Myo2 dephosphorylation has been implicated in mediating a Mid1–Myo2 interaction (Motegi et al., 2004). In particular, a regulatory dephosphorylation event has been shown to occur in a C-terminal Myo2 fragment (Motegi et al., 2004) and Clp1 can dephosphorylate this fragment (Fig. S2 D), suggesting Myo2 as a possible second Clp1 target at the CR. After photobleaching, the half-times of GFP-Myo2 (19.6 ± 1.4 s; see Fig. 4 A), Rlc1-GFP (28.2 ± 1.9 s; see Fig. 4 B), and YFP-Rng2 (25.5 ± 3.2 s; see Fig. 4 C) recovery were not significantly different from each other but appeared slower than that of Cdc15-GFP (14.5 ± 0.6 s; Fig. 3 B; P = 0.1, 0.0055, 0.043, respectively). The half-times were not significantly altered in clp1Δ cells relative to the wild type. However, the absence of clp1 resulted in a significant increase to the mean mobile fraction of Cdc15-GFP (P = 0.0018; Fig. 3, B and C), GFP-Myo2 (P = 0.015; Fig. 4 A), and Rlc1-GFP (P = 0.0016; Fig. 4 B). To distinguish whether it was Clp1 activity or Clp1 presence at the CR that led to this difference, we examined CR dynamics in clp1-C286S cells, as this mutant localizes to the CR but lacks activity (Trautmann et al., 2004). As in clp1Δ cells, the half-times of Cdc15-GFP, GFP-Myo2, and Rlc1-GFP were not significantly altered in clp1-C286S cells relative to wild type (Fig. 3 B; and Fig. 4, A and B) but the mean mobile fraction was higher (Fig. 3, B and C; and Fig. 4, A and B). Interestingly, there was no difference in the half-time or mean mobile fraction of YFP-Rng2 in the absence of Clp1 activity (Fig. 4 C). GFP-Myo2, Rlc1-GFP, and YFP-Rng2 levels were similar in wild-type and clp1 mutant cells (Fig. S2 E). In addition, a significant increase in Cdc15 levels was not observed in clp1-C286S cells (Fig. S2 A), indicating that the observed differences in Cdc15 dynamics in clp1Δ cells are unlikely to result from the modest increase in overall Cdc15 levels in this strain. These results not only indicate that Clp1 influences CR component dynamics selectively to provide stability to the CR but provide the first evidence that the dynamics of individual CR components are different.
Figure 4.
Clp1 regulates myosin II dynamics at the CR. (A–C) Confocal FRAP measurements of GFP-Myo2 (A), Rlc1-GFP (B), and YFP-Rng2 (C) at the ring in wild-type, clp1Δ, clp1-C286S, and mid1Δ431-481 cells. Fluorescence recovery curves, including half-times with standard error, number of cells per strain analyzed, and mobile fractions are shown. Best-fit curves derived from the mean intensity values of n cells (Prism software) were used to calculate half-times and mobile fractions. R2 values range from 0.9552 to 0.9869. Asterisks indicate significant differences between wild-type and experimental cells as determined by student's t test. P-values and standard error are included.
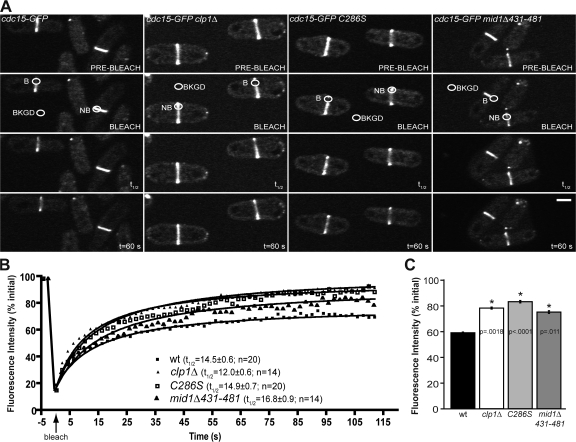
Figure 3.
Clp1 regulates Cdc15 dynamics at the CR. (A) Representative images from confocal FRAP measurements of Cdc15-GFP at the CR in wild type, clp1Δ, clp1-C286S, and mid1Δ431-481 cells. B, bleach region; NB, nonbleached region (used to correct for overall bleaching); BKGD, background region (used to correct for overall bleaching). Bar, 5 μm. (B) Fluorescence recovery curves including half-times with standard error and number of cells analyzed per strain. Best-fit curves derived from the mean intensity values of n cells (Prism software) were used to calculate half-times and mobile fractions. R2 values: wt, 0.9877; clp1Δ, 0.9802; clp1-C286S, 0.9819; and mid1Δ431-481, 0.9762. (C) Mobile fraction graphs from confocal FRAP measurements of Cdc15-GFP. Asterisks indicate significant differences between wild-type and experimental cells as determined by student's t test. P-values and standard error are included.
Clp1 at the CR regulates cytokinesis
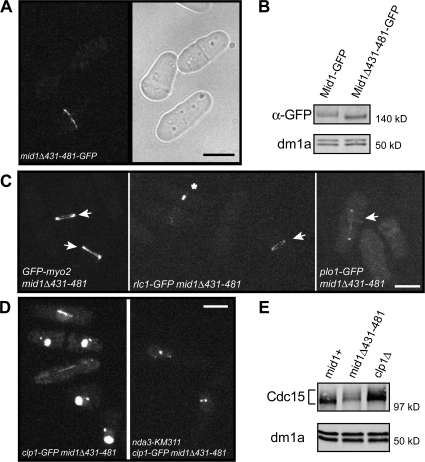
To determine if physical association of Clp1 with the CR is required for CR stability, we developed a strain expressing a Mid1 mutant that disrupts its interaction with Clp1. Yeast two-hybrid analysis was used to narrow the region within Mid1 necessary for Clp1 binding. Mid1 deleted of aa 431–481 failed to interact with Clp1 (unpublished data). When integrated at the endogenous mid1 locus and C-terminally tagged with GFP, mid1Δ431–481-GFP cells formed medial septa and Mid1Δ431–481-GFP localized at the CR (Fig. 5 A), which was centrally located and perpendicular to the cell length. In addition, protein levels of Mid1Δ431–481-GFP were similar to those of wild-type Mid1-GFP (Fig. 5 B). Myo2 and the polo-like kinase Plo1 both depend on Mid1 for proper CR localization and tight ring formation (Bahler et al., 1998a; Motegi et al., 2004). GFP-Myo2, Rlc1-GFP, and Plo1-GFP CR localization were unaffected by the deletion of residues 431–481 in Mid1 (Fig. 5 C). In contrast, Clp1-GFP was not detected at the CR but was visible at kinetochores in nda3-KM311–arrested mid1Δ431–481 cells (Fig. 5 D), just as we observed in mid1Δ cells (Fig. 1). Consistent with this, in nda3-KM311–arrested mid1Δ431–481 cells, Cdc15 remained in a hyperphosphorylated form, comigrating with Cdc15 from nda3-KM311–arrested clp1Δ cells (Fig. 5 E). As in cells lacking Clp1 activity, the half-times of Cdc15-GFP (16.8 ± 0.9 s; Fig. 3, A and B), GFP-Myo2 (18.2 ± 1 s; Fig. 4 A), Rlc1-GFP (34.5 ± 2.7 s; Fig. 4 B), and YFP-Rng2 (19.7 ± 1.4 s; Fig. 4 C) recovery after photobleaching were not significantly different from that of the wild type. However, the mean mobile fractions of Cdc15-GFP (P = 0.011; Fig. 3, A–C), GFP-Myo2 (P = 0.0029; Fig. 4 A), and Rlc1-GFP (P = 0.042; Fig. 4 B) at the ring were significantly increased in mid1Δ431–481 cells relative to the wild type. Clp1-GFP was also detected in the nucleolus and at the spindle pole body in interphase cells and the mitotic spindle (Fig. 5 D), confirming that Mid1 is responsible for recruiting Clp1 to the CR.
Figure 5.
Clp1 requires Mid1 association for CR localization. (A) Live-cell GFP and light images of mid1Δ431–481-GFP cells. (B) Denatured protein extracts were prepared from mid1-GFP and mid1Δ431–481-GFP cells and analyzed by immunoblotting with GFP and dm1a (α-tubulin) antibodies. (C) Live-cell imaging of GFP-Myo2, Rlc1-GFP, and Plo1-GFP in mid1Δ431–481 cells. Arrows indicate rings. Asterisk indicates constricting ring. (D) Live-cell imaging of clp1-GFP mid1Δ431–481 cells incubated at 32°C and nda3-KM311 clp1-GFP mid1Δ431–481 cells after incubation at 18°C for 7 h. Bars, 5 μm. (E) nda3-KM311 clp1-GFP (mid1+), nda3-KM311 clp1-GFP mid1Δ431–481, and nda3-KM311 clp1Δ strains were arrested by incubation at 18°C for 7 h, and then samples were collected and denatured protein extracts were analyzed by immunoblotting with Cdc15 and dm1a antibodies.
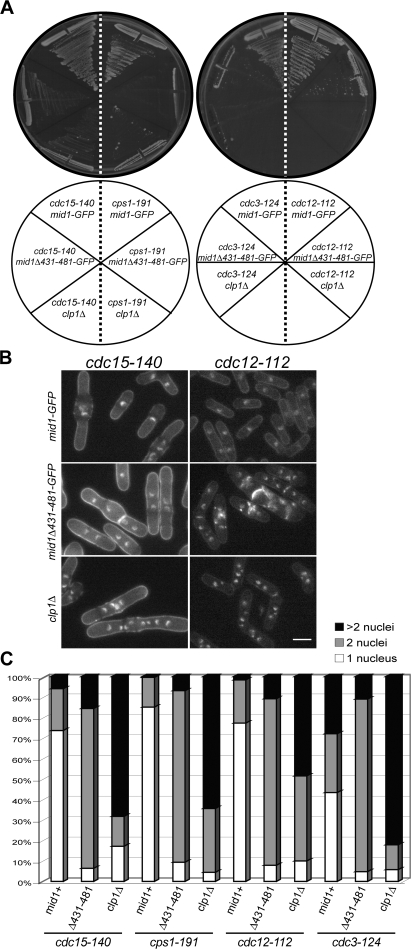
Cells lacking clp1 are predisposed to cytokinesis failure when the CR is challenged (Mishra et al., 2004). Indeed, strong genetic interactions were observed between mid1Δ431–481 and several mutations in CR components that also show negative genetic interactions with clp1Δ (Mishra et al., 2004). At the semipermissive temperature (32°C), cdc15-140 (Fankhauser et al., 1995), formin cdc12-112 (Chang et al., 1996), profilin cdc3-124 (Balasubramanian et al., 1994), and 1,3-β-glucan synthase cps1-191 (Liu et al., 1999) cells are viable and the majority of the population contains one nucleus, as is the case for wild-type cells (Fig. 6, A–C). However, under the same conditions, double mutants with mid1Δ431–481 did not form colonies (Fig. 6 A) and the majority of cells accumulated two or more nuclei (Fig. 6, B and C), which is indicative of cytokinetic failure. These results indicate that Clp1 activity is required at the CR for successful cytokinesis when specific CR or septum assembly components are compromised.
Figure 6.
Clp1 at the CR regulates cytokinesis. (A) Cells of the indicated genotype were assayed for colony formation on YE plates after 3 d at 32°C. (B and C) Cells of the indicated genotype were grown to mid log phase, shifted to 32°C for 5 h, ethanol fixed, and then stained with methyl blue and DAPI. At least 200 cells were scored for number of nuclei. Bar, 5 μm.
Discussion
In this paper, we investigate the function of Clp1 at the division site and pinpoint a phosphatase-dependent regulatory mechanism that influences CR dynamics and stability. We identify anillin-related Mid1 as the key targeting molecule recruiting Clp1 to the CR but not affecting Clp1's other localizations, making Mid1 the first reported cytoplasmic Cdc14 phosphatase tether. Once localized at the CR through Mid1 binding, Clp1 regulates the kinetics with which Cdc15 and myosin II are maintained at the CR. CR-associated Clp1 becomes essential for successful cytokinesis when specific components of the cytokinetic machinery are challenged, adding necessary robustness to the process of cell division.
Although we have a good understanding of the temporal assembly of the S. pombe CR (Wu et al., 2003), the mechanisms that regulate this ordered process are still unclear. Our FRAP results strongly support a scaffolding role for Mid1. Probably its stable association with the cortex, in marked contrast to the dynamic nature of other tested CR components (Pelham and Chang, 2002; Wong et al., 2002; this study), allows Mid1 to effectively recruit and organize other CR proteins into a focused ring structure and to secure the CR in place. Recruiting Clp1 is apparently one function of Mid1 in the CR assembly process.
The involvement of Clp1 phosphatase activity in regulating S. pombe CR dynamics is indicative of a phosphorylation/dephosphorylation-driven mechanism, and we have investigated Cdc15 as one of probably several Clp1 targets at the CR. Cdc15 phosphorylation is altered in clp1Δ, clp1-C286S, mid1Δ, and mid1Δ431–481 cells, and an alteration in Cdc15 phosphorylation status in clp1Δ cells was also reported by Wachtler et al. (2006). What is the function of Clp1-mediated Cdc15 dephosphorylation? Clp1 might regulate interactions between Cdc15 and other CR proteins or plasma membrane lipids. The F-BAR/EFC domain that is present in Cdc15 generally binds phospholipid bilayers (Itoh et al., 2005), and Cdc15 also uses this domain to bind the formin Cdc12 (Carnahan and Gould, 2003). In addition to Cdc15, other pombe cdc15 homology family members are phosphoproteins (Chitu and Stanley, 2007), and phosphoregulation may be a conserved mechanism to control their functions. Analysis of Cdc15 phosphosite mutants in vivo will be necessary to test the precise molecular mechanism of its regulation, and an investigation of other potential Clp1 CR targets will likely contribute to a more complete picture of the complexity of CR regulation.
Actin and myosin are initially recruited to the division site independently (Motegi et al., 2000). Several lines of evidence indicate that later mitotic events, including their turnover at the division site, require their cooperation. In mammalian cells, myosin II light chain influences the turnover of actin at the CR via a phosphorylation-dependent mechanism (Murthy and Wadsworth, 2005). In rat cells, inhibition of myosin II motor activity causes a twofold increase in the recovery time of actin, as assessed by FRAP, which leads to an accumulation of actin at the division site and eventual cytokinesis failure (Guha et al., 2005). We observed an approximate twofold increase in Cdc15 and myosin II mobility when Clp1 phosphatase activity was defective. In addition, we observed an increase in cytokinesis failure in cells lacking Clp1 activity at the CR when CR formation was disturbed. Given that Clp1 influences two major components that provide structural integrity to the CR and are among the most abundant proteins concentrated at the division site (Wu and Pollard, 2005), any alteration to their regulation is likely to disrupt the fidelity of cytokinesis. FRAP analysis indicated that Rng2 dynamics were unaffected by Clp1 and that the recovery of Cdc15 after photobleaching was measurably faster than myosin II or Rng2. These data are the first to indicate variable CR component dynamics.
Based on our results, Clp1 at the CR regulates the dynamic properties of specific CR components during an unperturbed mitosis, and this mechanism is likely to account in part for the observed cytokinesis failure in 3–5% of a clp1Δ population (Cueille et al., 2001; Trautmann et al., 2001). When the natural assembly of the CR or septum is challenged, Clp1 activity is specifically required at the CR to ensure CR integrity and completion of cytokinesis. In budding yeast, Cdc14 export from the nucleus to the cytoplasm is required for CR constriction (Bembenek et al., 2005), raising the possibility that Cdc14, like Clp1, needs to anchor with the CR for proper ring function. However, our data also indicate that Clp1 must be able to influence CR stability from other locations. Cell cycle progression in the presence of low doses of lat A requires septation initiation network (SIN) signaling and Clp1 activity (Mishra et al., 2004). Although Clp1 functions to maintain SIN activity, the SIN is required for nuclear exclusion of Clp1 until cytokinesis is complete (Trautmann et al., 2001; Mishra et al., 2004). We found that Clp1 activity at the CR is dispensable for Clp1-SIN signaling in response to low-dose lat A treatment (Fig. S3). This is consistent with the finding that Clp1 is dispersed throughout the cytoplasm rather than concentrated at the CR in the presence of low-dose lat A (Mishra et al., 2004). Our analyses of the mid1D431–481 mutant demonstrate that Clp1 utilizes at least two mechanisms to influence cytokinesis, one of which requires direct association with the CR and the other of which involves signaling to the cytokinetic machinery. The dual activities of Clp1 are further indications of the robustness of cytokinesis, a process protected by several redundant mechanisms. Cdk activity inhibits cytokinesis in both human and yeast cells (Murray, 2004; Wolfe and Gould, 2005) but the mechanism is unknown. Given that Clp1 dephosphorylates sites of Cdk phosphorylation, these studies reveal at least one likely mechanism by which Cdk activity antagonizes cytokinesis and suggest that through similar mechanisms other Cdc14 family members will be found to regulate CR dynamics.
Materials and methods
Strains, media, and molecular biology methods
The S. pombe strains used in this study (Table I) were grown in yeast extract (YE) as described in the figure legends. Induction of the nmt promoter (Maundrell, 1993) was achieved by growing cells in thiamine (repressing conditions) and then washing cells three times in medium lacking thiamine (inducing conditions). The clp1, mid1, and cdc15 ORFs were tagged at the 3′ end with 13XMYC:kanR, 3XHA:kanR, YFP:kanR, GFP:kanR, CFP:kanR, or TAP:kanR cassettes as previously described (Bahler et al., 1998b). The C terminus of myo2 encoding aa 1394–1526 was PCR amplified with oligos containing NdeI and BamHI sites on the 5′ and 3′ ends, respectively. The PCR product was NdeI–BamHI digested and cloned into the thiamine-repressible pREP41-HA vector to yield pKG3967. An NcoI site was introduced into mid1 at aa 431 and 481 by site-directed mutagenesis, and then standard molecular biology techniques were used to generate and integrate the mid1Δ431–481 allele at the endogenous mid1 locus. Yeast cells were transformed using a lithium acetate–based procedure (Keeney and Boeke, 1994). Proper integration of tags was confirmed by whole-cell PCR and Western blot or fluorescence microscopy, as appropriate. Tagged loci were transmitted to other strains by standard mating, sporulation, and tetrad dissection methods. The yeast two-hybrid system used in this study was described previously (James et al., 1996). All plasmids were generated by standard molecular biology techniques. cDNAs cloned into the bait plasmid pGBT9 or the prey plasmid pGAD424 (Clontech Laboratories, Inc.) were sequenced to ensure the absence of PCR-generated mutations and correct reading frame. To generate vectors for recombinant fusion protein production, gene fragments were PCR amplified, inserted into pGEX-2T or pMAL-2C, and then sequenced to verify the fidelity of the sequence.
Table I. Yeast strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| KGY246 | h− ade6-M210 leu1-32 ura4-D18 | Laboratory stock |
| KGY648 | h+ cdc3-124 clp1∷ura4 ade6-M21X leu1-32 ura4-D18 | Laboratory stock |
| KGY1278 | h+ rlc1-GFP:ura4 ade6-M210 leu1-32 ura4-D18 | Laboratory stock |
| KGY1985 | h+ cdc25-22 cdc15-HA:kanR clp1∷ura4 ade6-M210 leu1-32 ura4-D18 | This study |
| KGY2417 | h− mid1-GFP:kanR ade6-M210 leu1-32 ura4-D18 | Laboratory stock |
| KGY2882 | h− clp1-MYC:kanR ade6-M216 leu1-32 ura4-D18 | Trautmann et al. (2001) |
| KGY3019 | h− cdc15-GFP:kanR ade6-M210 leu1-32 ura4-D18 | Carnahan and Gould (2003) |
| KGY3078 | h− cdc15-140 mid1-GFP:kanR ade6-M21X leu1-32 ura4-D18 | Laboratory stock |
| KGY3079 | h− mid1-GFP:kanR sid4-GFP:kanR ade6-M21X leu1-32 ura4-D18 | Laboratory stock |
| KGY3155 | h− cps1-191 mid1-GFP:kanR ade6-M21X leu1-32 ura4-D18 lys1-151 | Laboratory stock |
| KGY3173 | h+ nda3-KM311 mid1-GFP:kanR ade6-M210 leu1-32 | This study |
| KGY3350 | h− cdc25-22 cdc15-HA:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY3381 | h− clp1∷ura4 ade6-M216 leu1-32 ura4-D18 | Trautmann et al. (2001) |
| KGY3382 | h− cdc25-22 clp1-GFP:kanR ade6-216 leu1-32 ura4-D18 | This study |
| KGY3388 | h+ nda3-KM311 clp1-MYC:kanR leu1-32 | This study |
| KGY3612 | h− nda3-KM311 ura4-D18 leu1-32 | Laboratory stock |
| KGY3783 | h+ nda3-KM311 clp1∷ura4 ade6-M21X leu1-32 ura4-D18 | Wolfe and Gould (2004) |
| KGY4410 | h− nda3-KM311 cdc15-HA:kanR ade6-M21X leu1-32 | This study |
| KGY4630 | h+ nda3-KM311 clp1-GFP:kanR leu1-32 ura4-D18 | This study |
| KGY4791 | h− cps1-191 clp1∷ura4 ade6-M21X leu1-32 ura4-D18 lys1-151 | Trautmann et al. (2001) |
| KGY4815 | h− nda3-KM311 mid1-MYC:ura4 leu1-32 ura4-D18 | This study |
| KGY4833 | h+ cdc15-140 clp1∷ura4 ade6-M21X leu1-32 ura4-D18 | Laboratory stock |
| KGY4881 | h− clp1-MYC:kanR mid1∷ura4 ade6-M216 leu1-32 ura4-D18 | This study |
| KGY5298 | h+ kanR:GFP-myo2 ade6-M210 leu1-32 ura4-D18 | Wu et al. (2003) |
| KGY5318 | h+ kanR:YFP-rng2 ade6-M210 leu1-32 ura4-D18 | Wu et al. (2003) |
| KGY5482 | h+ cdc25-22 cdc15-HA: kanR mid1∷ura4 ade6-M210 leu1-32 ura4-D18 | This study |
| KGY5594 | h− nda3-KM311 cdc15-HA:kanR clp1∷ura4 leu1-32 | This study |
| KGY5598 | h+ nda3-KM311 cdc15-HA:kanR mid1∷ura4 ade6-M21X ura4-D18 leu1-32 | This study |
| KGY5615 | h− cdc25-22 clp1-GFP:kanR mid1∷ura4 ade6-21X leu1-32 ura4-D18 | This study |
| KGY5616 | h− nda3-KM311 clp1-GFP:kanR mid1∷ura4 ade6-M21X leu1-32 ura4-D18 | This study |
| KGY5617 | h+ nda3-KM311 clp1∷ura4 mid1-GFP:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY5626 | h− nda3-KM311 cdc15-HA:kanR clp1-MYC:kanR ade6-M21X leu1-32 | This study |
| KGY5627 | h− cdc15-GFP:kanR clp1∷ura4 ade6-M21X leu1-32 ura4-D18 | This study |
| KGY5630 | h+ nda3-KM311 cdc15-CFP:kanR clp1-YFP:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY5631 | h+ nda3-KM311 clp1-GFP:kanR mid1-MYC:ura4 leu1-32 ura4-D18 | This study |
| KGY5667 | h+ nda3-KM311 cdc15-CFP:kanR clp1-YFP:kanR mid1∷ura4 ade6-M21X leu1-32 ura4-D18 | This study |
| KGY5812 | h+ nda3-KM311 clp1-MYC:kanR mid1∷ura4 ura4-D18 leu1-32 | This study |
| KGY5907 | h+ cdc15-GFP:kanR C286S-MYC:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY5924 | h− rlc1-GFP:ura4 clp1∷ura4 ade6-M21X leu1-32 ura4-D18 | This study |
| KGY5925 | h− rlc1-GFP:ura4 C286S-MYC:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY5944 | h− kanR:YFP-rng2 C286S-MYC:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY5946 | h− kanR:YFP-rng2 clp1∷ura4 ade6-M21X leu1-32 ura4-D18 | This study |
| KGY5956 | h− kanR:GFP-myo2 clp1∷ura4 ade6-M21X leu1-32 ura4-D18 | This study |
| KGY5958 | h+ kanR:GFP-myo2 C286S-MYC:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY6039 | h− cdc25-22 cdc15-HA:kanR C286S-MYC:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY6040 | h− cdc25-22 cdc15-HA:kanR clp1-MYC:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY6462 | h− mid1Δ431-481 ade6-M210 leu1-32 ura4-D18 | This study |
| KGY6485 | h− mid1Δ431-481 clp1-GFP:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY6486 | h− mid1Δ431-481-GFP:kanR ade6-M210 leu1-32 ura4-D18 | This study |
| KGY6487 | h− nda3-KM311 mid1Δ431-481 clp1-GFP:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY6549 | h− cps1-191 mid1Δ431-481-GFP:kanR ade6-M21X leu1-32 ura4-D18 lys1-151 | This study |
| KGY6597 | h− cdc15-140 mid1Δ431-481-GFP:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY6599 | h− cdc15-GFP:kanR mid1Δ431-481 ade6-M21X leu1-32 ura4-D18 | This study |
| KGY6605 | h− cdc12-112 clp1∷ura4 ade6-M21X leu1-32 ura4-D18 | This study |
| KGY6606 | h− cdc12-112 mid1Δ431-481-GFP:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY6607 | h− cdc12-112 mid1-GFP:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY6624 | h+ cdc3-124 mid1-GFP:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY6626 | h+ cdc3-124 mid1Δ431-481-GFP:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY6795 | h− mid1Δ431-481 plo1-GFP:kanR ade6-M21X leu1-32 ura4-D18 | This study |
| KGY6796 | h− mid1Δ431-481 kanR:GFP-myo2 ade6-M21X leu1-32 ura4-D18 | This study |
| KGY6797 | h− mid1Δ431-481 rlc1-GFP:ura4 ade6-M21X leu1-32 ura4-D18 | This study |
| KGY7202 | h− mid1Δ431-481:FLAG:kanR kanR:YFP-rng2 ade6-M21X leu1-32 ura4-D18 | This study |
Protein methods
Whole-cell lysates were prepared in NP-40 buffer either in native or denaturing conditions as previously described (Gould et al., 1991). Lysates were subjected to immunoprecipitation with anti-GFP (3E6; Invitrogen), anti-MYC (9E10), anti-HA (12CA5), or anti-Cdc2 (4711). Western blot analysis and λ phosphatase assays were performed as previously described (Tasto et al., 2003), except primary antibodies were detected with secondary antibodies coupled to Alexa Fluor 680 (Invitrogen) and then scanned on an Odyssey machine (LI-COR Biosciences). Quantification of protein intensities was performed using Odyssey version 1.2. Mass spectrometry analysis was performed as previously described (MacCoss et al., 2002) on TAP complexes, which were prepared from 8L cells as previously described (Tasto et al., 2001).
In vitro binding assays
All recombinant bacterially produced proteins were purified on either glutathione beads (GST) or amylose beads (MBP) as previously described (Carnahan and Gould, 2003), except binding buffer consisted of 20 mM Tris-HCl, pH 7.4 , 150 mM NaCl, 2 mM EDTA, and 0.1% Triton X-100. Proteins were resolved by SDS-PAGE, followed by Coomassie blue staining or Western blot analysis with anti-GST or anti-MBP to visualize proteins.
In vitro phosphatase assays
Immunoprecipitated Cdc15-HA and HA-Myo2-Ct were incubated in the presence of recombinant MBP, MBP-Clp1, or phosphatase-dead MBP-Clp1 (MBP-Clp1PD) at 30°C for 30–45 min in phosphatase assay buffer (50 mM imidazole, pH 6.9, 1 mM EDTA, and 1 mM DTT). Reactions were terminated by the addition of SDS sample buffer and by boiling for 5 min. Proteins were resolved by SDS-PAGE and detected by Western blot with anti-HA. Clp1-MYC phosphatase activity from cells was determined by DiFMUP (Invitrogen) continuous assays as previously described (Wolfe et al., 2006).
Microscopy
GFP, YFP, or CFP-tagged proteins were visualized using a spinning disk confocal microscope (Ultraview LCI; PerkinElmer) equipped with a 100×/1.40 Plan-Apochromat oil immersion objective. All images were acquired at 25°C. For time-lapse images, cells were placed on a hanging drop glass slide containing YE agar and covered with a coverslip. Images were captured with a charge-coupled device camera (Orca ER; Hamamatsu) using Metamorph software (7.1; MDS Analytical Technologies) or Ultraview LCI software (PerkinElmer) and then processed using Volocity software (version 3.5.1; Improvision). For time-lapse and static images, z-series optical sections were taken at 0.5-μm spacing. Time-lapse images were obtained at an interval of 60 s.
FRAP experiments in Figs. 3 and Figs.4 were performed on a confocal microscope (LSM 510; Carl Zeiss, Inc.) equipped with a 63×/1.40 Plan-Apochromat oil immersion objective. A 1.12-μm2 circular region of the ring was bleached with a sequence of 10 high-intensity laser iterations and then pictures were taken every 1.5 (Cdc15-GFP and Rlc1-GFP) or 3 s (GFP-Myo2 and YFP-Rng2). Fluorescence intensities were analyzed using 510 LSM software (Carl Zeiss, Inc.), and values were normalized to a region within the ring of a nonbleached cell and the background to correct for overall bleaching. Mid1-GFP FRAP was performed on a spinning disk confocal microscope (Ultraview LCI) equipped with a 100×/1.40 Plan-Apochromat oil immersion objective and Micropoint laser ablation system (Photonic Instruments). After bleach of a 2.84-μm2 circular region of the Mid1-GFP ring, z-series optical sections were captured with a charge-coupled-device camera (Orca ER) at 0.5-μm spacing every 3 min. Normalized data were plotted using Prism 4.0c software (Graphpad Software, Inc.). Mobile fractions and half-time values were calculated from the best-fit curve equation as the difference in final and bleach intensity and the time to reach half of the final intensity after photobleaching, respectively. Student's t test and standard error were calculated to determine significant differences. Differences reported as significant have p-values <0.05 and nonoverlapping standard error intervals.
Online supplemental material
Fig. S1 shows the interaction between Clp1 and Mid1 through coimmunoprecipitation, yeast 2-hybrid, and in vitro binding assays and Clp1 protein levels in the absence of Mid1. Fig. S2 shows Clp1-dependent dephosphorylation of Cdc15 and Myo2 C terminus, coimmunoprecipitation of Cdc15 with Clp1, and protein levels of GFP-Myo2, Rlc1-GFP, and YFP-Rng2 in the absence of Clp1 activity. Fig. S3 shows wild-type, clp1Δ, and mid1Δ431–481 cells treated with low-dose lat A. Video 1 is live-cell videomicroscopy of Clp1-GFP localization during mitosis. Video 2 is live-cell videomicroscopy of Clp1-GFP localization in mid1Δ cells during mitosis. Video 3 is live-cell videomicroscopy of Mid1-GFP FRAP, corresponding with Fig. 1 (D and E), in mid1-GFP sid4-GFP cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200709060/DC1.
Supplemental Material
Acknowledgments
We would like to thank Drs. Srinivas Venkatram, J. Shawn Goodwin, and Joseph Tasto for their technical assistance, Dr. Viesturs Simanis for plasmids used to generate materials for this study, Drs. Dannel McCollum and Ryoma Ohi for critically reading the manuscript, and other members of the Gould laboratory for helpful discussions.
We are thankful for the following support: D.M. Clifford, National Institutes of Health grants F32-GM076897 and T32-CA09582; B.A. Wolfe, National Institutes of Health grant GM068786; R.H. Roberts-Galbraith, National Science Foundation fellowship DGE-02387141; and J.R. Yates, National Institutes of Health grant P41RR11823. This work was supported by National Institutes of Health grant GM068786. K.L. Gould is an Investigator of the Howard Hughes Medical Institute.
Abbreviations used in this paper: CR, contractile ring; lat A, latrunculin A; SIN, septation initiation network; TAP, tandem affinity purification; YE, yeast extract.
References
- Bahler, J., A.B. Steever, S. Wheatley, Y. Wang, J.R. Pringle, K.L. Gould, and D. McCollum. 1998. a. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J. Cell Biol. 143:1603–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler, J., J.Q. Wu, M.S. Longtine, N.G. Shah, A. McKenzie III, A.B. Steever, A. Wach, P. Philippsen, and J.R. Pringle. 1998. b. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951. [DOI] [PubMed] [Google Scholar]
- Balasubramanian, M.K., B.R. Hirani, J.D. Burke, and K.L. Gould. 1994. The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J. Cell Biol. 125:1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembenek, J., J. Kang, C. Kurischko, B. Li, J.R. Raab, K.D. Belanger, F.C. Luca, and H. Yu. 2005. Crm1-mediated nuclear export of Cdc14 is required for the completion of cytokinesis in budding yeast. Cell Cycle. 4:961–971. [DOI] [PubMed] [Google Scholar]
- Carnahan, R.H., and K.L. Gould. 2003. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J. Cell Biol. 162:851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celton-Morizur, S., N. Bordes, V. Fraisier, P.T. Tran, and A. Paoletti. 2004. C-terminal anchoring of mid1p to membranes stabilizes cytokinetic ring position in early mitosis in fission yeast. Mol. Cell. Biol. 24:10621–10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, F., A. Woollard, and P. Nurse. 1996. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J. Cell Sci. 109:131–142. [DOI] [PubMed] [Google Scholar]
- Chang, L., and K.L. Gould. 2000. Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc. Natl. Acad. Sci. USA. 97:5249–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitu, V., and E.R. Stanley. 2007. Pombe Cdc15 homology (PCH) proteins: coordinators of membrane-cytoskeletal interactions. Trends Cell Biol. 17:145–156. [DOI] [PubMed] [Google Scholar]
- Cueille, N., E. Salimova, V. Esteban, M. Blanco, S. Moreno, A. Bueno, and V. Simanis. 2001. Flp1, a fission yeast orthologue of the s. cerevisiae CDC14 gene, is not required for cyclin degradation or rum1p stabilisation at the end of mitosis. J. Cell Sci. 114:2649–2664. [DOI] [PubMed] [Google Scholar]
- Esteban, V., M. Blanco, N. Cueille, V. Simanis, S. Moreno, and A. Bueno. 2004. A role for the Cdc14-family phosphatase Flp1p at the end of the cell cycle in controlling the rapid degradation of the mitotic inducer Cdc25p in fission yeast. J. Cell Sci. 117:2461–2468. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., A. Reymond, L. Cerutti, S. Utzig, K. Hofmann, and V. Simanis. 1995. The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell. 82:435–444. [DOI] [PubMed] [Google Scholar]
- Gould, K.L., S. Moreno, D.J. Owen, S. Sazer, and P. Nurse. 1991. Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 10:3297–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha, M., M. Zhou, and Y.L. Wang. 2005. Cortical actin turnover during cytokinesis requires myosin II. Curr. Biol. 15:732–736. [DOI] [PubMed] [Google Scholar]
- Hiraoka, Y., T. Toda, and M. Yanagida. 1984. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 39:349–358. [DOI] [PubMed] [Google Scholar]
- Itoh, T., K.S. Erdmann, A. Roux, B. Habermann, H. Werner, and P. De Camilli. 2005. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev. Cell. 9:791–804. [DOI] [PubMed] [Google Scholar]
- James, P., J. Halladay, and E.A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 144:1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, B.K., Z.A. Zimmerman, H. Charbonneau, and P.K. Jackson. 2002. Disruption of centrosome structure, chromosome segregation, and cytokinesis by misexpression of human Cdc14A phosphatase. Mol. Biol. Cell. 13:2289–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, J.B., and J.D. Boeke. 1994. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 136:849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., H. Wang, D. McCollum, and M.K. Balasubramanian. 1999. Drc1p/Cps1p, a 1,3-beta-glucan synthase subunit, is essential for division septum assembly in Schizosaccharomyces pombe. Genetics. 153:1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCoss, M.J., W.H. McDonald, A. Saraf, R. Sadygov, J.M. Clark, J.J. Tasto, K.L. Gould, D. Wolters, M. Washburn, A. Weiss, et al. 2002. Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl. Acad. Sci. USA. 99:7900–7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand, N., C. Lukas, B.K. Kaiser, P.K. Jackson, J. Bartek, and J. Lukas. 2002. Deregulated human Cdc14A phosphatase disrupts centrosome separation and chromosome segregation. Nat. Cell Biol. 4:318–322. [DOI] [PubMed] [Google Scholar]
- Maundrell, K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 123:127–130. [DOI] [PubMed] [Google Scholar]
- Mishra, M., J. Karagiannis, S. Trautmann, H. Wang, D. McCollum, and M.K. Balasubramanian. 2004. The Clp1p/Flp1p phosphatase ensures completion of cytokinesis in response to minor perturbation of the cell division machinery in Schizosaccharomyces pombe. J. Cell Sci. 117:3897–3910. [DOI] [PubMed] [Google Scholar]
- Motegi, F., K. Nakano, and I. Mabuchi. 2000. Molecular mechanism of myosin-II assembly at the division site in Schizosaccharomyces pombe. J. Cell Sci. 113:1813–1825. [DOI] [PubMed] [Google Scholar]
- Motegi, F., M. Mishra, M.K. Balasubramanian, and I. Mabuchi. 2004. Myosin-II reorganization during mitosis is controlled temporally by its dephosphorylation and spatially by Mid1 in fission yeast. J. Cell Biol. 165:685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, A.W. 2004. Recycling the cell cycle: cyclins revisited. Cell. 116:221–234. [DOI] [PubMed] [Google Scholar]
- Murthy, K., and P. Wadsworth. 2005. Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr. Biol. 15:724–731. [DOI] [PubMed] [Google Scholar]
- Pelham, R.J., and F. Chang. 2002. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature. 419:82–86. [DOI] [PubMed] [Google Scholar]
- Sohrmann, M., C. Fankhauser, C. Brodbeck, and V. Simanis. 1996. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 10:2707–2719. [DOI] [PubMed] [Google Scholar]
- Stegmeier, F., and A. Amon. 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38:203–232. [DOI] [PubMed] [Google Scholar]
- Tasto, J.J., R.H. Carnahan, W.H. McDonald, and K.L. Gould. 2001. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast. 18:657–662. [DOI] [PubMed] [Google Scholar]
- Tasto, J.J., J.L. Morrell, and K.L. Gould. 2003. An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J. Cell Biol. 160:1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann, S., B.A. Wolfe, P. Jorgensen, M. Tyers, K.L. Gould, and D. McCollum. 2001. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr. Biol. 11:931–940. [DOI] [PubMed] [Google Scholar]
- Trautmann, S., S. Rajagopalan, and D. McCollum. 2004. The S. pombe Cdc14-like phosphatase Clp1p regulates chromosome biorientation and interacts with Aurora kinase. Dev. Cell. 7:755–762. [DOI] [PubMed] [Google Scholar]
- Wachtler, V., Y. Huang, J. Karagiannis, and M.K. Balasubramanian. 2006. Cell cycle-dependent roles for the FCH-domain protein Cdc15p in formation of the actomyosin ring in Schizosaccharomyces pombe. Mol. Biol. Cell. 17:3254–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, B.A., and K.L. Gould. 2004. Fission yeast Clp1p phosphatase affects G(2)/M transition and mitotic exit through Cdc25p inactivation. EMBO J. 23:919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, B.A., and K.L. Gould. 2005. Split decisions: coordinating cytokinesis in yeast. Trends Cell Biol. 15:10–18. [DOI] [PubMed] [Google Scholar]
- Wolfe, B.A., W.H. McDonald, J.R. Yates III, and K.L. Gould. 2006. Phospho-regulation of the Cdc14/Clp1 phosphatase delays late mitotic events in S. pombe. Dev. Cell. 11:423–430. [DOI] [PubMed] [Google Scholar]
- Wong, K.C., V,M. D'Souza, N.I. Naqvi, F. Motegi, I. Mabuchi, and M.K. Balasubramanian. 2002. Importance of a myosin II-containing progenitor for actomyosin ring assembly in fission yeast. Curr. Biol. 12:724–729. [DOI] [PubMed] [Google Scholar]
- Wu, J.Q., and T.D. Pollard. 2005. Counting cytokinesis proteins globally and locally in fission yeast. Science. 310:310–314. [DOI] [PubMed] [Google Scholar]
- Wu, J.Q., J.R. Kuhn, D.R. Kovar, and T.D. Pollard. 2003. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell. 5:723–734. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.