Abstract
Transcriptional profiling of embryogenic callus produced from Medicago truncatula mesophyll protoplasts indicated up-regulation of ethylene biosynthesis and ethylene response genes. Using inhibitors of ethylene biosynthesis and perception, it was shown that ethylene was necessary for somatic embryogenesis (SE) in this model legume. We chose several genes involved in ethylene biosynthesis and response for subsequent molecular analyses. One of these genes is a gene encoding a transcription factor that belongs to the AP2/ERF superfamily and ERF subfamily of transcription factors. We demonstrate that this gene, designated M. truncatula SOMATIC EMBRYO RELATED FACTOR1 (MtSERF1), is induced by ethylene and is expressed in embryogenic calli. MtSERF1 is strongly expressed in the globular somatic embryo and there is high expression in a small group of cells in the developing shoot meristem of the heart-stage embryo. RNA interference knockdown of this gene causes strong inhibition of SE. We also provide evidence that MtSERF1 is expressed in zygotic embryos. MtSERF1 appears to be essential for SE and may enable a connection between stress and development.
There have been numerous studies concerning the hormonal induction of somatic embryogenesis (SE) in a wide range of species. In almost all cases, auxin has a critical role and cytokinins are frequently involved (Fehér et al., 2003; Rose, 2004). Stress is also a factor that has been increasingly recognized as having an important role in the induction of SE (Touraev et al., 1997; Fehér et al., 2003; Nolan et al., 2006). Hormones and stress collectively induce dedifferentiation of differentiated cells and the initiation of an embryogenic program (Fehér et al., 2003; Ikeda-Iwai et al., 2003; Rose and Nolan, 2006).
The molecular mechanisms involved in the induction of SE from cultured tissue are not well understood. There has, however, been progress in identifying the involvement of the SOMATIC EMBRYO RECEPTOR KINASE (SERK) and a number of transcription factors. Arabidopsis (Arabidopsis thaliana) transformed with the AtSERK1 gene under the control of the cauliflower mosaic virus 35S promoter showed a marked increase in SE compared to wild-type cultures (Hecht et al., 2001). Ectopic expression of the transcription factors LEAFY COTYLEDON1 (LEC1; Lotan et al., 1998), LEC2 (Stone et al., 2001), BABY BOOM (Boutilier et al., 2002), and WUSCHEL (Zuo et al., 2002) in Arabidopsis cause spontaneous formation of somatic embryos on intact plants or explants. AGL15 is another transcription factor that promotes SE in Arabidopsis (Harding et al., 2003). In addition, many other genes are specifically expressed in SE (Imin et al., 2005).
In Medicago truncatula, high rates of somatic embryo formation can be induced in the Jemalong genotype 2HA (Rose et al., 1999) by application of the hormones auxin and cytokinin (Nolan et al., 2003). The 2HA genotype is a super embryogenic mutant that is 500-fold more embryogenic than wild-type Jemalong (Nolan et al., 1989; Rose et al., 1999; Rose and Nolan, 2006). M. truncatula is a model legume (Cook, 1999) with the sequencing of the gene-rich euchromatin nearing completion (Young and Shoemaker, 2006). Mutant resources (Tadege et al., 2005), numerous ESTs, microarray chips, proteomic tools, and physical and genetic maps are available for M. truncatula (VandenBosch and Stacey, 2003). The 2HA genotype coupled with the genomic and molecular genetics tools makes M. truncatula an attractive system to investigate the molecular genetics of SE (Nolan et al., 2003; Imin et al., 2005; Rose and Nolan, 2006).
In addition to the application of hormones to induce SE, there is the stress component, induced by the excision and culture of the explant, to consider (Nolan et al., 2006). In M. truncatula, there are many stress-related proteins associated with SE (Imin et al., 2004). A number of these proteins are differentially expressed between 2HA and Jemalong (Imin et al., 2005). Synthesis of the growth regulator ethylene can be rapidly evoked in response to a variety of biotic and abiotic stresses, including wounding (Kende and Zeevaart, 1997; Wang et al., 2002). Here, microarray studies on the induction of SE in M. truncatula identified genes predicted to encode ethylene biosynthesis and ethylene response proteins that are differentially expressed in SE. More detailed analysis of the role of ethylene in SE showed that a transcription factor of the AP2/ERF superfamily and ERF gene subfamily, designated SOMATIC EMBRYO RELATED FACTOR1 (MtSERF1), which is dependent on ethylene biosynthesis and perception for its expression, is required for SE in M. truncatula. MtSERF1 may enable a connection between stress and development.
RESULTS
Microarray Analysis
The use of mesophyll protoplasts was valuable for the microarray analysis because cultures are derived from one cell type and should identify critical gene expression changes more clearly than leaf explants. Leaf explants in addition to mesophyll cells contain cells of the vasculature, stomates, and epidermis. Trends in gene expression from 40- to 80-d-old 2HA cultures were profiled using a 16K oligonucleotide array and Cy3 and Cy5 fluorescent labels. At 40 d, the cultures are at the cell proliferation stage, at 60 d globular embryos are forming, and at 80 d heart- and later-stage embryos are forming (Fig. 1; Supplemental Fig. S1). We made direct comparisons between 40- and 60-d-old cultures, 60- and 80-d-old cultures, and 40- and 80-d-old cultures. The determination of up- and down-regulated genes was determined statistically using the strategies described in “Materials and Methods.” The statistical test is very important because the developing embryos are diluted among the proliferating cells and the fold change may be relatively small. Further, whereas there is a degree of synchrony in the production of embryos from protoplasts, embryo development is not perfectly synchronized. At 80 d of culture, embryo development in many cases has reached the heart stage, but synchronicity starts to be lost. Vascular tissue has also started to form in the callus at 80 d. We have grouped genes into functional classes to assist in the interpretation. These are the first transcriptional profiling data obtained from differentiating single protoplasts using large-scale microarrays.
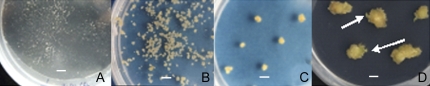
Figure 1.
Main stages of embryogenic callus development starting from single protoplasts. A to D, Microcalli (A); proliferating stage, 40 d of culture (B); appearance of embryos, globular stage, 60 d of culture (C); callus with embryos at heart and later stages of development, 80 d of culture (D). Arrows indicate embryos. Bars = 5 mm.
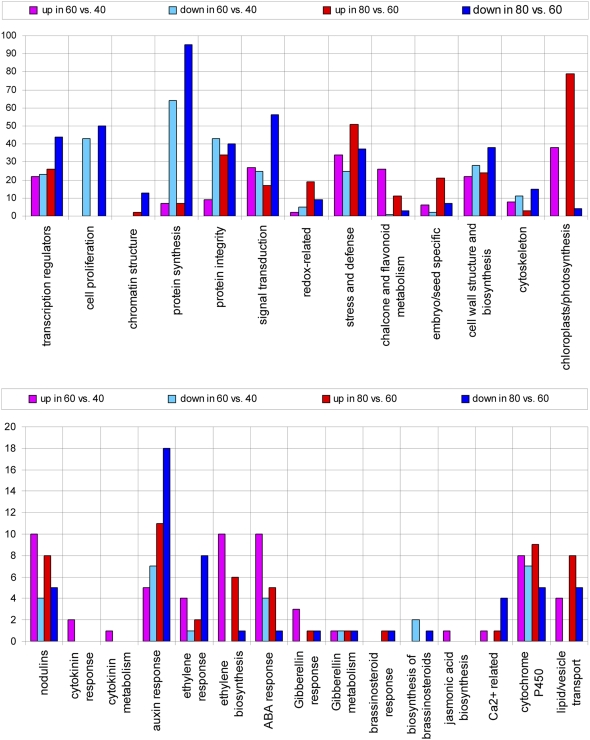
In Figure 2, we show the distribution of the number of genes associated with different functional classes that are up- or down-regulated for 60 d compared to 40 and 80 d. By including all genes that show statistically significant changes in expression, transcriptional changes occurring in only small numbers of cells will be included (see Supplemental Table S1). Our main interest is the time point where the cell culture (Supplemental Fig. S1) switches to SE formation (60 d) from proliferation (40 d). Statistically significant changes in expression were found for more than 1,500 genes at 60 d compared to 40 d: 883 and 823 genes were up- or down-regulated, respectively. Comparison of 80 and 60 d of culture revealed about 2,000 genes differentially expressed from which 889 were up-regulated and 1,089 down-regulated.
Figure 2.
Distribution of number of genes of different functional classes that are up- or down-regulated, for 60 d versus 40 d and 80 d versus 60 d of culture from single protoplasts. Adapted from Supplemental Table S1, which contains the significantly up- and down-regulated genes.
Development-Related Genes
Figure 2 shows the number of genes whose expression was up- or down-regulated within 27 functional groups. There is down-regulation of cell proliferation and protein synthesis genes (histones, DNA replication factors, ribosomal and a number of other translation associated proteins) as cells switch into SE. Two cyclin-dependent kinases, cdc2Ms1 and cdcMsF, which are actively expressed during the G2-to-M phase in alfalfa (Medicago sativa) cells (Magyar et al., 1997), are down-regulated at 60 d. These data are consistent with that of Thibaud-Nissen et al. (2003) in soybean (Glycine max), where the most rapid cell division occurs in early callus formation. Our data also demonstrate changes in expression of a number of cell wall-modifying enzymes as well as cell wall proteins. It is known that cells undergoing SE as well as zygotic embryogenesis show changes in cell wall polysaccharides and proteoglycans (Majewska-Sawka and Nothnagel, 2000). There is up-regulation of embryo-specific genes as the somatic embryos within the callus develop. There is also increased expression of chloroplast- and photosynthesis-related genes, reflecting plastid changes associated with the development of the embryogenic callus in low light. The developmental changes are consistent with the morphological development of the embryogenic callus and support the reliability of the arrays.
Stress-Related Genes
There is up-regulation of genes involved in biosynthesis of flavonoids, redox, P450, and other stress-related genes that could be related to general stress that is a part of cell culture and an important component in the induction of SE (Nishiwaki et al., 2000; Ikeda-Iwai et al., 2003; Belmonte and Yeung, 2004; Stasolla et al., 2004). Most of the enzymes from the isoflavonoid biosynthetic pathway are up-regulated at 60 d (chalcone reductase and chalcone synthase, isoflavone 2′-hydroxylase, isoflavone reductase) and their product can be involved in defense or nodulation processes.
Hormone- and Regulatory-Related Genes
We categorized a number of functional groups that were likely to have regulatory roles and provide a useful overview of the potential contributors to the regulatory networks involved in somatic embryo induction and development. Transcriptional regulators, signal transduction and hormone biosynthesis, and hormone response genes are represented. Auxin and cytokinin are the hormones supplied so it might be expected that there would be changes in gene expression for many genes directly related to these hormones. This was the case for the auxin response genes, but less so for the cytokinin response genes. What was of particular interest was the up-regulation of ethylene biosynthesis genes at both time points and ethylene response genes in the SE transition period.
To obtain a view of the major transcriptional changes involved in the induction of SE, we focused on a selection of genes from Supplemental Table S1 showing a 2-fold or greater change (e.g. see Hass et al., 2004) for 60 d compared to 40 d. The data in Figure 2 are derived from all genes that are statistically up- or down-regulated and are found in Supplemental Table S1. These data reinforce the view that stress and hormone responses are well represented. There are genes responsive to ethylene, abscisic acid (ABA), and GA, which are hormones not present in the culture medium, in addition to the auxin and cytokinin response genes.
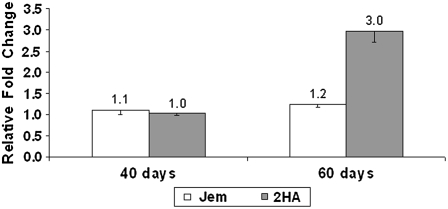
We were interested in the contribution of stress responses to successful SE. Therefore, we focused on the ethylene biosynthesis genes and an ethylene response transcription factor, the APETALA2/ETHYLENE RESPONSE ELEMENT BINDING PROTEIN (AP2/EREBP) homolog TC102138. The AP2/EREBP homolog was of more interest than other ethylene response genes because of its pattern of expression in quantitative reverse transcription (qRT)-PCR studies (detailed below); it showed a near 2-fold (1.94, included in Table I) increase and was a transcription factor. In a separate protoplast experiment, the increase in expression in AP2/EREBP occurred in the highly embryogenic 2HA at 60 d, but not in the near nonembryogenic Jemalong (Fig. 3). We designated the ethylene-responsive AP2/EREBP homolog MtSERF1.
Table I.
Genes up-regulated ≥2 times at 60 d of protoplast culture versus 40 d of culture
| Fold Change | IDs | Similarity to Known Proteins |
|---|---|---|
| Chalcone and flavonoid metabolism | ||
| 2.8–3.5 | TC100398, TC100400, TC100398 | Chalcone reductase |
| 2.0–2.3 | TC106536, TC106544 | Chalcone synthase |
| 3.4 | TC107720 | UDP-glycose:flavonoid glycosyltransferase |
| 3.2–3.4 | TC94281, TC96312 | Isoflavone reductase |
| 2.1 | TC100787 | Isoflavone 2′-hydroxylase |
| Transcription factors | ||
| 2.5–3.6 | TC107549, TC94651, TC1015293 | MYB |
| 2.3–2.8 | TC97324, TC101761 | WRKY |
| 2.1 | TC100528, TC96130 | No apical meristem (NAM) |
| Nodulins | ||
| 2.5–3.2 | TC111031, AL383966 | NOD-like membrane protein |
| 2.5–3.6 | TC100836, TC94419, TC107353 | Nodule specific |
| 2.2 | TC103767 | ENOD18 |
| 2.2 | TC95616 | N21-like protein |
| Cytokinin response | ||
| 2.2 | TC94601 | Ser/Thr protein kinase |
| Auxin response | ||
| 2.6 | TC95234 | Auxin-induced protein homolog F8F16.140 (Arabidopsis) |
| 2.6 | TC94351 | Zea mays IN2-2 protein |
| Ethylene biosynthesis | ||
| 2.8–6.9 | TC106654, TC106655 | ACO |
| 2.3 | TC95406 | ACS |
| Ethylene response | ||
| 2.8–3.3 | TC43436, TC105017 | Ethylene-induced esterase/lipase (Citrus sinensis) |
| 1.94 | TC102138 | MtSERF1 |
| ABA response | ||
| 2.2–3.9 | TC95327, TC106638 | ABA and environmental stress-inducible protein |
| GA response | ||
| 5 | TC100404 | LTCOR11, |
| 3.8 | TC94215 | GAST-like gene product (Fragaria × ananassa) |
| 2.2 | TC95411 | Snakin-1 (Solanum tuberosum) |
| GA metabolism | ||
| 2.9 | TC103730 | GA 2-oxidase (Pisum sativum) |
| Jasmonic acid biosynthesis | ||
| 2.2 | TC107322 | Allene-oxide cyclase |
| Stress and defense | ||
| 2.3 | TC106640 | Ferritin |
| 2.1–4.7 | TC106639, TC106641, TC100143 | Cold- and drought-regulated protein |
| 2.2 | TC108259 | Dehydration stress-induced protein (Brassica napus) |
| 2.8 | TC101709 | Putative esterase |
| 2.1–7 | TC101688, TC95383, TC94626, TC94626 | Pathogenesis-related protein |
| 2.4 | TC94759, TC100686 | Putative disease resistance protein |
| 2.2–5 | TC108315, TC107261, TC106851, TC95164, TC100746 | Peroxidase |
| 4.7 | TC100966 | Environmental stress-induced protein |
| 2 | TC102781 | Temperature stress-induced lipocalin, partial (87%) |
| 2.2 | TC97485 | β-Glucan-elicitor receptor (soybean) |
| Signal transduction | ||
| 2.7 | TC100498 | Protein kinase MMK4, cold- and drought-induced (alfalfa), complete |
| 2.2 | TC109312 | Putative LRR receptor-like kinase |
| 2–2.5 | TC102218, TC94601, TC94008 | Ser/Thr-specific protein kinase |
| 2.5 | TC103069 | Similarity to calmodulin, partial (46%) |
| Ca2+ related | ||
| 2.3 | TC108816 | Ca2+/H+-exchanging protein |
| Chloroplasts/photosynthesis | ||
| 2–2.7 | TC106570, TC93920, TC94106 | Rubisco small subunit |
| 2–2.2 | TC100390, TC100390, TC106432 | Chlorophyll a/b-binding protein |
| Cytochrome P450 | ||
| 2.1–2.3 | TC100502, TC100504, BE941365 | Cytochrome P450 |
| Lipid transport | ||
| 2.1–4.5 | TC94445, TC95002, TC94143, TC94138, TC93922 | Nonspecific lipid-transfer protein precursor (LTP) |
| Protein integrity | ||
| 3.5 | TC106781 | Kunitz proteinase inhibitor |
| Redox-related | ||
| 2 | TC104047 | Thioredoxin 3 |
| Cell wall structure and biosynthesis | ||
| 2.3–3.1 | TC106576, TC106582 | Extensin-like protein |
| 2.5–2.7 | TC94063, TC94968 | Cyanogenic β-glucosidase |
| 2.7 | TC100486 | Xyloglucan endotransglycosylase, brassinosteroid-regulated protein BRU1 (soybean) |
| 2.1–2.4 | TC101143, TC94366 | Pectinesterase-like protein |
| 2.2 | TC100597 | Haloacid dehalogenase-like hydrolase, putative ripening-related protein (Vitis vinifera) |
| 2.1 | TC100580 | Pro-rich cell wall protein |
| 2–2.5 | TC94670, TC93935 | Glucan endo-1,3-β-d-glucosidase |
Figure 3.
MtSERF1 (TC102138) expression in embryogenic 2HA and nonembryogenic Jemalong after 40 and 60 d of protoplast culture using qRT-PCR. sem indicated.
Table II.
Genes down-regulated ≥2 times at 60 d of protoplast culture versus 40 d of culture
| Fold Change | No. of Genes | Similarity to Known Proteins |
|---|---|---|
| Cell proliferation | ||
| 2.4 | TC99694 | DNA topoisomerase II |
| 2 | TC109376 | Histone H3 |
| 2–2.1 | TC95372, TC100976 | Histone H2A |
| Transcription regulators | ||
| 2 | TC103975 | TGACG-motif binding protein (Vicia fava) |
| Auxin response | ||
| 5 | TC106520 | Pro-rich protein auxin-induced (alfalfa) |
| 2.7 | TC102735 | Auxin-induced protein AUX28 |
| 2.2 | TC110388 | Auxin-induced protein 22E (indole-3-acetic acid-induced protein ARG14) |
| ABA response | ||
| 6.7 | TC106508 | Abscisic stress-ripening protein homolog (Prunus armeniaca) |
| Ethylene biosynthesis | ||
| 2.4 | TC104552 | ACS |
| Nodulins | ||
| 4.8 | TC106522 | ENOD12 (Vicia sativa) |
| 2.7 | TC109877 | Nodulin protein (Arabidopsis) |
| Stress and defense | ||
| 2–2.3 | TC106484, TC95489 | Peroxidase precursor |
| 2.3 | TC94562 | Pathogenesis-related protein PR-1 precursor (M. truncatula) |
| 2 | TC107866 | Defensin AMP1 (Dahlia merckii) |
| Aquaporins | ||
| 2.3 | TC100851 | Multifunctional aquaporin (M. truncatula) |
| Signal transduction | ||
| 2.2–2.4 | TC108015, TC103969 | Cdc2-like protein kinase (alfalfa) |
| 2.3 | TC109033 | Ser/Thr-specific protein kinase homolog (Arabidopsis) |
| 2.3 | TC95992 | DNA-binding protein phosphatase 2C (N. tabacum) |
| Cytoskeleton | ||
| 2.1 | TC100408 | Tubulin α-1 chain (Pisum sativum) |
| Protein integrity | ||
| 2.1 | TC102787 | Ubiquitin-conjugating enzyme (Arabidopsis) |
| 2 | TC95356 | Ser proteinase (Arabidopsis) |
| Redox-related | ||
| 2 | TC95190 | l-Ascorbate oxidase |
| Cell wall structure and biosynthesis | ||
| 2.3–4.8 | TC100994, TC94484, TC100993, TC94613 | α-Expansin |
| 2.2–3.3 | TC112360, TC107321 | Arabinogalactan protein |
| 2–2.5 | TC94066, TC96847, TC96066 | β-1,3-Glucanase |
| 3 | TC98773 | Cellulase precursor (P. sativum) |
| 3.2 | TC104377 | Cellulose synthase (Arabidopsis) |
| 2.4 | TC94609 | Endo-β-1,4-glucanase (Fragaria × ananassa) |
| 2.1–3.1 | TC108758, TC110405 | Extensin-like protein |
| 3.2 | TC101945 | Mannan endo-1,4-β-mannosidase (Arabidopsis) |
| 2.3–3.5 | TC100685, TC109691 | Pectate lyase |
Gene Expression Analysis Using qRT-PCR
Measurements of gene expression using qRT-PCR were carried out for both the ethylene biosynthesis genes and the ethylene response gene on leaf explants to see whether these genes were similarly up-regulated as they were using mesophyll protoplasts. Leaf explants are experimentally simpler than using isolated single protoplasts and are commonly used to produce embryogenic callus for legume transformation experiments (Wang et al., 1996; Chabaud et al., 2003). Experiments can be turned over more quickly using leaf explants because they produce embryos about 40 d earlier than protoplasts These experiments were carried out with both the highly embryogenic 2HA and the near nonembryogenic wild-type Jemalong.
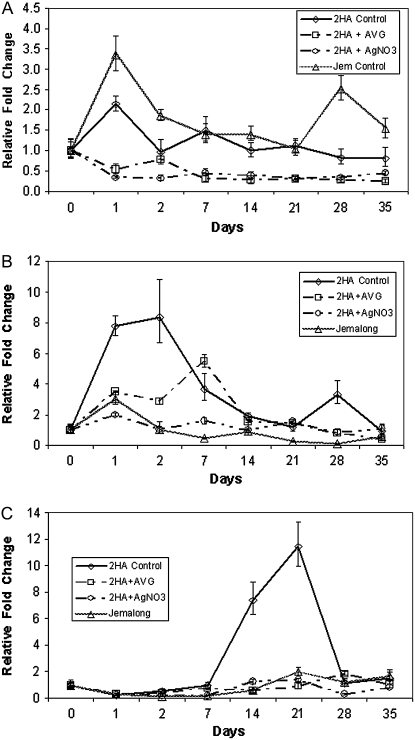
The ethylene biosynthesis genes are expressed quite early and expression continues throughout the culture period in the embryogenic 2HA. The expression pattern of the ethylene biosynthesis genes and MtSERF1, the ethylene response gene, is shown in Figure 4. The peak of expression in 2HA for ACC SYNTHASE (ACS) transcription is day 1 of culture and day 1 to 2 in the case of ACC OXIDASE (ACO) transcription. When the expression of MtSERF1 was measured, it first showed an increase in expression between day 7 and day 14 and peaked at day 21 when embryos are starting to form in a partially synchronous fashion, the transition period between day 40 and 60 in the protoplast experiments. Expression then declines, but continues as more embryos are formed; the amount of expression after day 21 varies according to the amount of embryogenesis. In all four biological repeats, the same inductive pattern was evident. Gene expression was also measured with the nonembryogenic line Jemalong, which showed high expression of ACS, but little expression of ACO and MtSERF1. The data overall indicate that inhibitors of ethylene perception (Ag+) and biosynthesis (aminoethoxyvinylglycine [AVG]) inhibit the expression of all three genes in embryogenic tissue. In the case of ACO expression, the peak of expression is delayed and clearly reduced in the AVG treatment.
Figure 4.
Relative expression of ACS (TC95406; A), ACO (TC106655; B), and MtSERF1 (TC102138; C) during the development of embryogenic cultures from leaf explants of 2HA or nonembryogenic Jemalong using qRT-PCR. The 2HA cultures were also treated with the ethylene biosynthesis inhibitor AVG at 10 μm or the ethylene perception inhibitor Ag+ at 10 μm. All data for each time point are derived from the same cDNA. The fold change is normalized to the starting leaf tissue. sem indicated.
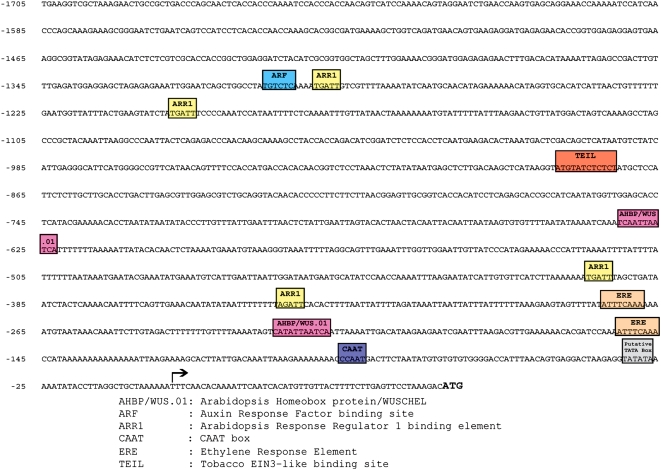
As MtSERF1 is a member of the AP2/ERF family of transcription factors, the promoter region was examined for an ethylene response element (ERE). A 1,758-bp region upstream from the transcription start site was isolated, cloned, and sequenced. In addition to the TATA and CAAT boxes, in silico analysis indicated that the promoter region contained a number of potential regulatory elements (Fig. 5). Two ERE elements were present, as well as two WUSCHEL-binding sites, four ARABIDOPSIS RESPONSE REGULATOR1 (ARR1) elements that are associated with cytokinin signaling, an AUXIN RESPONSE FACTOR (ARF) element, and a TOBACCO EIN3-LIKE (TEIL) element.
Figure 5.
Core and responsive element motifs in a 1,758-bp region upstream from the transcription start site of MtSERF1. In addition to the TATA and CAAT boxes, in silico analysis indicated that the promoter region contained a number of potential regulatory elements indicated in the figure.
Somatic Embryo Induction
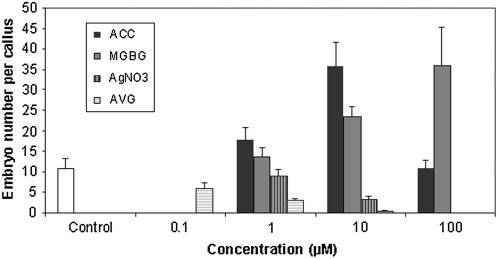
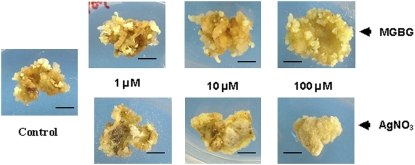
To further investigate the role of ethylene in SE, experiments were carried out with leaf explants using stimulators of ethylene biosynthesis and inhibitors of ethylene biosynthesis and perception. These data are shown in Figure 6 and results clearly indicate a marked influence of ethylene on SE. The addition of the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC), the substrate for ACO (Pierik et al., 2006), caused a marked increase of SE at 10 μm. Similarly, methylglyoxal bis(guanylhydrazone) (MGBG), which increases the availability of the ethylene precursor S-adenosyl-Met (SAM) by inhibiting polyamine synthesis, which utilizes the same precursor, stimulates ethylene synthesis (Lee et al., 1997). MGBG at 100 μm stimulated SE to the same extent as 10 μm ACC. Conversely, the ethylene biosynthesis inhibitor AVG, which inhibits the conversion of SAM to ACC (Yu and Yang, 1979), and Ag+, an inhibitor of ethylene perception (Bleecker, 1999), strongly inhibited SE at 1 and 10 μm, respectively, and there were no embryos at 10 and 100 μm, respectively. Representative photographs of some treatments shown in Figure 7 also illustrate the influence of ethylene on callus development from leaf explants. Stimulators of ethylene biosynthesis, using MGBG as the example, have small increases in callus development and inhibitors of ethylene action, such as Ag+, cause small decreases, but still allow callus development to occur. This is clearly shown at 100 μm Ag+, where there is complete inhibition of embryo formation but callusing of the leaf explant has still occurred.
Figure 6.
The effects of ethylene biosynthesis stimulators (ACC and MGBG) and inhibitors of biosynthesis and perception (AVG and Ag+) on embryo number in 2HA. To modify the levels of ethylene production, the following compounds were added to the medium: (1) the ethylene precursor ACC, which stimulates ethylene biosynthesis; (2) the stimulator of ethylene biosynthesis MGBG; (3) the inhibitor of ethylene perception silver nitrate (AgNO3); and (4) the inhibitor of ethylene biosynthesis AVG. Three different concentrations (i.e. 1, 10, and 100 μm) were employed for ACC, MGBG, and AgNO3, whereas AVG was employed at 0.1, 1, and 10 μm. Embryo numbers were counted after 11 weeks of culture. sem indicated.
Figure 7.
The effect of the ethylene biosynthesis stimulator (MGBG) and inhibitor of perception (Ag+) on the development of embryogenic callus from 2HA leaf explants. Embryos have developed in the dark for 11 weeks. Note large numbers of embryos at 100 μm MGBG and callus development without embryos in the case of 100 μm Ag+. Arrows point to embryos. Bars = 8 mm. In parallel experiments with Jemalong, only two embryos were produced across all treatments. This confirms that wild-type Jemalong rarely produces embryos.
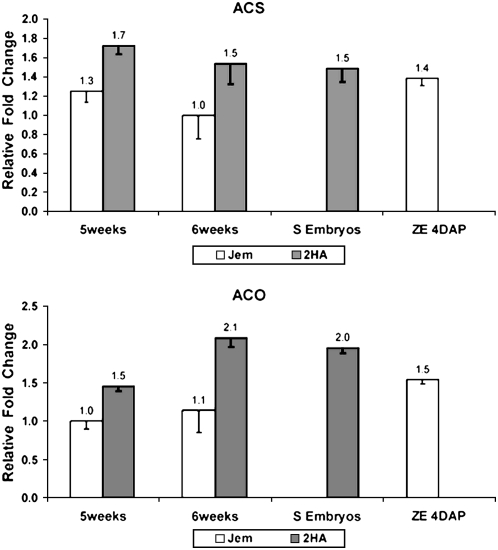
Given that embryogenic cultures are a mixture of embryos and callus cells, to establish a stronger connection with ethylene biosynthesis and embryo formation, we directly compared nonembryogenic callus with embryogenic callus, somatic embryos, and ovules. The ACS and ACO genes are consistently expressed at higher levels in embryogenic tissue, somatic embryos, and ovules with globular-stage embryos compared to nonembryogenic callus (Fig. 8).
Figure 8.
Relative expression of ACS (TC95406) and ACO (TC106655) in nonembryogenic callus (Jem) compared to embryogenic callus (2HA) obtained from leaf explants, somatic embryos (S embryos), and ovules with globular-stage embryos 4 dap. The SE data are shown only for the embryogenic 2HA, and zygotic embryogenesis (ZE) is shown only for Jem.
Localization of MtSERF1 Expression and Requirement for SE
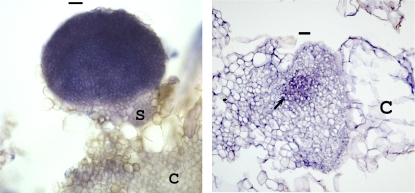
To localize MtSERF1 expression, we carried out in situ hybridization. The MtSERF1-specific probe was a 376-bp fragment from the 3′ region. As shown in Figure 9, MtSERF1 is strongly expressed in embryos. In the globular embryo, MtSERF1 is expressed throughout the embryo. This is very clearly shown in thick sections of fresh tissue where the callus cells show little, if any, hybridization signal. Later in the heart-stage embryo, using thinner sections of paraffin-embedded tissue, we were able to show that hybridization predominates in a small group of cells in the developing shoot meristem.
Figure 9.
In situ hybridization of MtSERF1 RNA probe. The globular-stage embryo shows expression over the whole embryo (left), whereas in the heart-stage embryo (right), expression is localized to a small group of cells (arrow) in the developing shoot meristem just below the apical notch (arrow). S, Suspensor-like structure; C, callus cells. Bars = 40 μm.
Given the MtSERF1 expression in somatic embryos, we investigated whether similar expression was present in zygotic embryos. Ovules at different times after pollination were collected and MTSERF1 expression measured using qRT-PCR. Expression increased during ovule development and then declined (Supplemental Fig. S2), similar to the pattern in embryogenic callus (Fig. 4C). The peak of ovule expression corresponded to the globular stage and had clearly declined at the torpedo stage (7 d after pollination [dap]). In situ hybridization studies showed expression was present in the embryo, but not in the ovule wall (F.R. Mantiri, S. Kurdyukov, X.-D. Wang, and R.J. Rose, unpublished data).
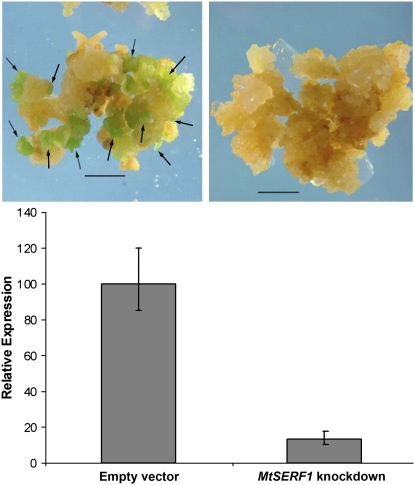
To examine whether MtSERF1 expression is required for SE, we used an RNA interference (RNAi) approach. As shown in Figure 10, transgenic MtSERF1 knockdown calli produced no somatic embryos when compared to their empty vector counterparts. For the empty vector control, 90% of 28 transformed calli produced embryos (with an average of 13.6 embryos/callus). To confirm the effects of RNA knockdown, we performed qRT-PCR on the calli. Results showed that the level of the transcripts in knockout calli was only 15% of that of empty vector calli. We also obtained transformed plants using an inducible vector containing RNAi and produced calli in the presence and absence of dexamethasone. The induction of RNAi by dexamethasone reduced the number of calli-producing embryos by 90%. The empty vector control showed no change in the presence or absence of dexamethasone.
Figure 10.
The effect of MtSERF1 knockdown using RNAi on embryo development. Empty vector control (left) and MtSERF1 knockdown (right). Bars = 5 mm. The histogram indicates the reduction in MtSERF1 expression due to RNAi.
Sequence and Phylogenetic Analyses of the Transcription Factor MtSERF1
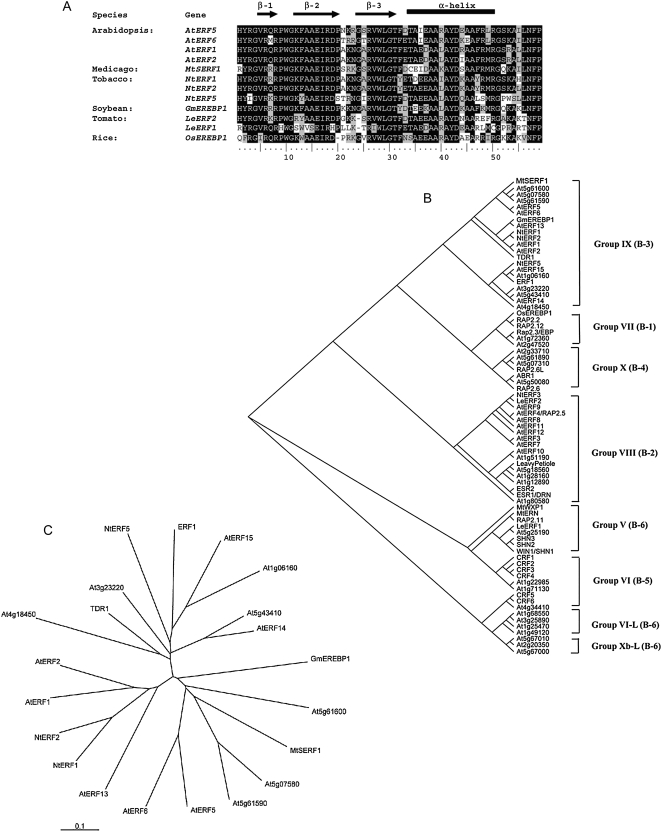
MtSERF1 is a protein of molecular mass 23 kD and contains 201 amino acids. The amino acid sequence of MtSERF1 contains a single AP2/ERF domain as shown by position-specific iterated and pattern hit-initiated BLAST. As indicated by Nakano et al. (2006), this domain is characteristic of the AP2/ERF superfamily and the ERF subfamily contains a single domain. An alignment of this domain with other proteins containing a single AP2/ERF domain shows high similarity (Fig. 11A).
Figure 11.
A, Alignment of the AP2/ERF domain of M. truncatula MtSERF1 with AP2/ERF domains from other plant species. Sequences were aligned with ClustalW implemented in ClustalX (1.8) using default parameters. Black and gray shading of the residues indicates identical and similar amino acid residues, respectively. Gaps required for optimal alignment are indicated by dashes. The black bar and arrows represent predicted α-helix and β-sheet regions, respectively, within the AP2/ERF domain (Allen et al., 1998). B, An unrooted cladogram of MtSERF1 and all known members of the ERF subfamily of Arabidopsis and other well-characterized genes from other species. The names of the genes were given when they are well characterized; otherwise, they are presented as TIGR ID. The tree was generated using the neighbor-joining method (Saitou and Nei, 1987) on ClustalX 1.8 software. Groups were named according to Nakano et al. (2006). Classification by Sakuma et al. (2002) is indicated in parentheses. The analysis is based on the amino acid sequences of the AP2/ERF domain. C, An unrooted phylogenetic tree of MtSERF1 and ERFs clustered in Group IX, using the entire amino acid sequence. Branch lengths are drawn to scale. Bar is estimated amino acid substitutions per site.
To further investigate this uncharacterized ethylene-induced transcription factor, phylogenetic analyses of AP2/ERF domain sequences of all 65 transcription factors of Arabidopsis identified as members of the ERF subfamily (Sakuma et al., 2002; Nakano et al., 2006) and other well-characterized ERFs from other species were conducted. Phylogenetic analyses using the neighbor-joining method (Saitou and Nei, 1987) on ClustalX 1.8 software showed that MtSERF1 belongs to Group IX of the classification of Nakano et al. (2006) or Group B-3 according to the classification of Sakuma et al. (2002), as shown in an unrooted cladogram (Fig. 11B). To find orthologs of MtSERF1 in other species, phylogenetic analyses of the entire amino acid sequences of ERF proteins included in Group IX were performed. As shown in Figure 11C, we found that the MtSERF1 is clustered together with two uncharacterized genes of Arabidopsis, with At5g61590 being the closest ortholog. This gene shares 41% identity with MtSERF1 (data not shown). Both genes also share similar motifs outside the AP2/ERF domain when analyzed using the motif discovery software MEME (http://meme.nbcr.net). These findings suggest that MtSERF1 is distinct from all AP2/ERF domain-containing transcription factors of known function.
DISCUSSION
Transcriptional profiling of the development of somatic embryos from single isolated mesophyll protoplasts of the highly embryogenic M. truncatula genotype 2HA revealed changes in the expression of many transcripts. These data showed increased transcription of ethylene biosynthesis genes and ethylene response genes, which were of interest because of their common involvement in stress and development responses. Subsequent experiments on ethylene response genes identified an ethylene-responsive transcription factor, MtSERF1, which was essential for SE.
Some of the major changes in the microarray data relate to stress, reflected in a range of genes connected to abiotic, biotic, and oxidative stresses. This may have been predicted given that protoplast isolation (Pasternak et al., 2002) and tissue excision (Nolan et al., 2006) associated with the induction of SE is a very stressful, wound-related procedure. Transcriptional profiling in response to mechanical wounding has been carried out in Arabidopsis (Cheong et al., 2002; Delessert et al., 2004) and a diverse group of genes previously related more specifically to wounding, pathogen attack, abiotic stress, and plant hormones are up-regulated. In this study, M. truncatula flavonoid biosynthesis genes were also up-regulated and have also been related to stress protection (Winkel-Shirley, 2002). In M. truncatula embryogenic cultures, there are many stress-related proteins associated with SE (Imin et al., 2004, 2005) as there are in alfalfa (Domoki et al., 2006). In soybean, SE is induced by 2,4-dichlorophenoxyacetic acid in cotyledons and is associated with up-regulation of oxidative stress and defense genes (Thibaud-Nissen et al., 2003). Studies by Che et al. (2006) involving microarray analysis of shoot, root, and callus development in Arabidopsis tissue culture also noted an increased expression of specific stress-related genes.
Among the most highly induced genes in our study was an ethylene biosynthesis gene (Table I). Up-regulation of transcripts of ethylene biosynthesis genes has also been seen in wounding (Cheong et al., 2002; Delessert et al., 2004) and SE in soybean cotyledons (Thibaud-Nissen et al., 2003). ACC synthase was up-regulated on an auxin-rich callus induction medium (Che et al., 2006) in Arabidopsis. We also noted an up-regulation of ethylene response genes and this contributed to ethylene becoming a focus of our studies. In addition to the suite of up-regulated genes related to ethylene, it was of interest to note that it might be expected that, because auxin and cytokinin were present in the medium, auxin and cytokinin response genes would be the only prominently featured hormone-related genes. However, this was not the case. Genes related to ABA, GA, and brassinosteroids were also featured. We have recently discussed the possible roles of these hormones in SE (Rose and Nolan, 2006).
Whereas there is value in focusing on mesophyll protoplasts as a uniform source of starting cells, experimentally leaf explants are commonly used and are experimentally much more straightforward. Leaf explants also produce embryos more quickly, about 40 d earlier than protoplasts. We were able to show that ethylene biosynthesis and ethylene response genes in leaf explants were also up-regulated. The first question that arises is whether the ethylene biosynthesis genes really reflect a requirement for ethylene for SE in M. truncatula. Results with an inhibitor of ethylene biosynthesis (AVG) and ethylene perception (Ag+) strongly support the idea that ethylene is essential for SE in M. truncatula. Consistent with this, the stimulation of ethylene biosynthesis by ACC and MGBG increased SE. This contrasts with the effect of ethylene on auxin-induced in vitro root formation in M. truncatula, where the ethylene-insensitive mutant sickle had enhanced root formation in comparison to wild type (Rose et al., 2006). Reported effects of ethylene on SE are variable and this is not surprising because ethylene concentration and signaling interactions with other hormones (Pierik et al., 2006) are likely to be species, developmentally, and environmentally dependent. However, in a defined experimental system of a developmental process, as with other hormones, there are most likely specific roles to play in the genetic networks (Nemhauser et al., 2006).
To examine the role of ethylene, we examined the expression of ethylene response genes that were up-regulated as cells entered into SE. We specifically focused on TC102138, which, based on our investigations, we designated MtSERF1. This gene is a member of the ERF subfamily based on the classification of Nakano et al. (2006). Further phylogenetic analysis placed MtSERF1 in Group IX of Nakano et al. (2006), which includes the AtERF5 gene induced by wounding in Arabidopsis (Cheong et al., 2002). We have shown that MtSERF1 expression is inhibited by AVG and Ag+, indicating its ethylene dependence. The connection of MtSERF1 to SE is shown by the minimal expression of MtSERF1 in rarely embryogenic Jemalong as opposed to the highly embryogenic 2HA, the localization of MtSERF1 expression to the early embryo and later to a specific shoot pole region of the heart-stage embryo, and the inhibition by of SE by RNAi directed against MtSERF1. It is noteworthy that a number of genes implicated in SE induction are expressed in developing zygotic embryos: SERK1 (Hecht et al., 2001), LEC1 (Lotan et al., 1998), LEC2 (Stone et al., 2001), and WUSCHEL (Zuo et al., 2002). The pattern of MtSERF1 expression in developing ovules of M. truncatula is consistent with expression in the zygotic embryo.
The lack of MtSERF1 expression in Jemalong, rarely embryogenic and near isogenic with respect to 2HA, provides a focus for further analysis. There is a small inhibition of ACS expression and a more strongly reduced ACO expression in Jemalong. This could ultimately lead to reduced signaling and reduced MtSERF1 expression. We also know that Jemalong and 2HA respond to auxin by producing roots, but when cytokinin is added to the auxin only 2HA forms embryos (Nolan et al., 2003) and Jemalong usually forms callus only. The significance of the localization of MtSERF1 expression to the early embryo and to a localized region of the shoot pole of the heart-stage embryo also requires further investigation. We also note that the MtSERF1 promoter contains WUSCHEL-binding sites and WUSCHEL is implicated in the induction of SE, as well as stem cell maintenance in apical meristems (Zuo et al., 2002; Rose and Nolan, 2006).
The finding of a relationship between an ERF subfamily gene and the formation of somatic embryos in vitro is consistent with an emerging picture of the involvement of ERF transcription factors in developmental processes studied in vitro. ENHANCER OF SHOOT REGENERATION1 and 2 (Banno et al., 2001; Ikeda et al., 2006) and RAP2.6L (Che et al., 2006) have a role in shoot regeneration in Arabidopsis. These transcription factors are all members of the AP2/ERF superfamily, as is BABY BOOM, which induces SE when overexpressed in Arabidopsis and Brassica napus (Boutilier et al., 2002). Heterologous expression of BABY BOOM in Nicotiana tabacum enhances regeneration capacity (Srinivasan et al., 2007). BABY BOOM is a member of the AP2 family because it has two repeated AP2/ERF domains (Boutilier et al., 2002; Nakano et al., 2006). Recently, the ERF transcription factor ERN, required for nodulation, has been identified in alfalfa (Middleton et al., 2007). MtSERF1 is in Group IX of the ERF subfamily, whereas ERN is in Group V (Fig. 11B). The AP2/ERF superfamily has a mix of transcription factors that relate to growth and development, abiotic and biotic stressors, and ethylene response (Alonso et al., 2003; Nakano et al., 2006). This may relate to the need to link growth and stress in the evolution of sessile plants.
MATERIALS AND METHODS
Protoplast Isolation and Culture
Protoplasts were isolated from leaves of the highly embryogenic 2HA genotype of Medicago truncatula ‘Jemalong’. A wild-type Jemalong plant frequently produces no embryos. The highest embryo-producing plant we have ever recorded was one embryo per six explants (Nolan et al., 1989). The 2HA genotype was derived from a rare regenerated plant obtained by a single cycle of tissue culture from wild-type Jemalong. This regenerated Jemalong showed enhanced SE, and the seed progeny segregated into types with and without the capacity to produce somatic embryos. Seed from the regenerated Jemalong was used to continue to select for high embryogenicity over four seed generations (Rose et al., 1999). The 2HA genotype can be considered to be a near-isogenic, highly embryogenic mutant of Jemalong. Plants were grown in controlled-environment rooms at low light intensity, as described by Tian and Rose (1999). The flotation procedure utilized for protoplast isolation yields almost exclusively mesophyll protoplasts. Isolated protoplasts were grown in 1% low-melting-point agarose droplets and then transferred to agar plates on P4 10:4 (10 μm naphthylacetic acid [NAA] and 4 μm benzylaminopurine [BAP]) medium for culture as described by Rose and Nolan (1995). For microarray analysis, calli derived from these isolated single protoplasts were taken at the following stages of development (Fig. 1; Supplemental Fig. S1): the cell proliferation stage (40 d of culture), the early globular stage (60 d of culture), and the heart and later stages (80 d of culture).
Cultured Leaf Explants
Cultured M. truncatula leaf explants were obtained from glasshouse-grown 2HA or wild-type Jemalong. Seeds of wild-type Jemalong were originally obtained from the National Medicago Collection, South Australian Research and Development Institute, Adelaide. The standard leaf culture procedure was as described by Nolan et al. (2003). Explants were cultured on P4 10:4 for 3 weeks before transfer to P4 10:4:1 (10 μm NAA, 4 μm BAP, and 1 μm ABA).
16K Oligo Microarray Slides
The Medicago 16K microarray was utilized and has a probe set mapped to Medicago Gene Index Release 8.0 (http//www.tigr.org/docs/tigr/scripts/medicago/ARRAYS/array.TCmapping). The 70-mer oligos were synthesized by Qiagen-Operon and the slides printed at the University of Arizona in the laboratory of Dr. David Galbraith. After printing, the slides were baked for 80 min at 80°C. The oligonucleotide array elements were immobilized by UV cross-linking at 300 mJ, then washed twice with gentle rocking for 2 min each wash, in 2× SSC + 0.2% SDS. The slides were then immersed in boiling hot water for 2 min, blotted briefly, and transferred to ice-cold ethanol for 2 to 5 min. Slides were then dried by centrifugation at 1,500 rpm for 2 to 5 min and finally stored in a light-proof box under cool dry conditions.
RNA Preparation, cDNA Synthesis, and Hybridization of Microarrays
Calli grown from individual protoplasts in an isolation that produced thousands of embryogenic microcalli, consistent with high protoplast quality, were collected at 40, 60, and 80 d after initiation of culture. The calli were frozen in liquid nitrogen and stored at −80°C until RNA was isolated. RNA was isolated as described by Lohar et al. (2006) and stored at −80°C. Total RNA (22 μg/sample) was pooled from three biological replicates giving 66 μg of RNA and 33 μg of RNA was aliquoted into two separate tubes. The Eppendorf tubes containing 33 μg of RNA were thawed on ice, spun dry in a speed vac, and immediately returned to −80°C. The RNA was shipped from Newcastle, Australia, to St. Paul, MN, on dry ice and transferred to −80°C until required. This maintained the quality of the RNA. The 33 μg of RNA were resuspended in 8 μL of nuclease-free double-distilled water and used for cDNA synthesis with a reverse transcriptase primer for labeling with either Cy3 or Cy5 dyes using a 3DNA Array50 kit (Genisphere) as previously described (Lohar et al., 2006).
Experiments were conducted using a regular dye-swap design as described earlier by Lohar et al. (2006). Microarrays for 60 versus 40 d, 80 versus 60 d, and 80 versus 40 d comparisons were hybridized with cDNAs from the two different time points labeled with different dyes. Each hybridization was repeated a total of six times to sample the technical variability, with three repeats of each dye combination to control for dye effects (Lohar et al., 2006).
Microarray Analysis
Methods for array analysis were as described for a 6K microarray (Lohar et al., 2006). Briefly, microarray slides were scanned using an Axon two-laser scanner and image analysis was performed using GenePix (Axon) software. Background-subtracted mean intensities for both tissues were log transformed and normalized before further analysis. Normalization of microarray data was performed using a statistical module developed as a part of Lab Information System, which includes several scripts and modules written in PERL and R languages.
Normalization steps included (1) within-slide normalization using local linear regression (LOWESS function; Yang et al., 2000); and (2) between-slide normalization using four-way ANOVA with replications for multislide dye-swap experiments (Kerr et al., 2000). More detailed description of preprocessing steps, such as log2 transformation of background-subtracted Cy5 and Cy3 intensities, are described by Lohar et al. (2006). Identification of differentially expressed genes was done using SAM software (Stanford), which allows flexible monitoring of the false discovery rate (Tusher et al., 2001). We applied a false discovery rate of <0.1% and the highest q value was <0.06%.
All genes of statistical significance with predicted or known function or that showed significant homology to characterized genes (annotated in the The Institute for Genomic Research database at http://compbio.dfci.harvard.edu/tgi) have been manually divided into 27 classes. Genes that did not fit readily into one of these classes have been classified as “other genes with defined function” and “genes with unknown function.” Supplemental Table S1 lists all the genes incorporated into these classes. To obtain a subset of genes that passed a statistical significance test we have also imposed a fixed ratio threshold of 2.
Real-Time PCR
Total RNA was isolated from calli at different time points and from intact leaves (as a calibrator) using the RNAqueous-4PCR kit (Ambion) according to the manufacturer's instructions. cDNA synthesis was performed using the SuperScript II first-strand synthesis system for RT-PCR (Invitrogen) starting with 2 μg of total RNA with oligo(dT)15 primers. Real-time PCR was performed using the SYBR GreenER qPCR SuperMix Universal kit (Invitrogen) and analyzed in the DNA Engine Opticon 2 continuous fluorescence detection system (Bio-Rad; formerly MJ Research). Primers 5′-TCATACGCCATCATCTCTTAGGT-3′ (forward) and 5′-AGGGGTTGTTTCCTTTGAAGAT-3′ (reverse) were designed to quantify the MtSERF1 expression levels, which were normalized to those of glyceraldehyde-3-P dehydrogenase (GAPDH), primers 5′-TGGTCATCAAACCCTCAACA-3′ (forward) and 5′-CCTCGTTCTTTCCGCTATCA-3′ (reverse), in each sample. To quantify the expression levels of ACS, the primers were 5′-CCCACACAAATTCGCTTCTT-3′ (forward) and 5′-TCACCATGTCCATCACCAGT-3′ (reverse), whereas for ACO the primers were 5′-GGGATTCTTTGAGCTGGTGA-3′ (forward) and 5′-GACGAACATGGAAGGTGCTT-3′ (reverse). PCR cycling conditions included a 94°C heating step for 1 min at the beginning of every run. The tubes were then cycled at 94°C for 30 s, annealed at 60°C for 60 s, and extended at 72°C for 60 s. A melting curve was generated at the end of every run to ensure product uniformity. PCR reactions were performed in triplicate in at least two biological repeats. Transcript abundance was estimated using a modification of the comparative threshold cycle (Ct) method and was calculated as E−ΔΔCt, where ΔΔCt = (Cttarget − CtGAPDH)Time x − (Cttarget − CtGAPDH)Calibrator and E is the estimated amplification efficiency, which was calculated employing the linear regression method on the log(fluorescence) per cycle number data for each amplicon using the LinRegPCR software (Ramakers et al., 2003).
In Situ Hybridization
To generate the RNA probes, a 376-bp fragment specific to MtSERF1 was first amplified by PCR with the primers 5′-CTGTGAAATTGATGCTGCAAA-3′ (forward) and 5′-TGACATAATTGTTGAGCTCACTCC-3′ (reverse). Then, the promoter sequences of T7 and SP6 RNA polymerase were introduced to this fragment by a two-step PCR. The first primers used were 5′-GAGGCCGCGTCTGTGAAATTGATGCTGCAAA-3′ (forward) and 5′-ACCCGGGGCTTGACATAATTGTTGAGCTCACTCC-3′ (reverse). The second set of primers used was 5′-TTATGTAATACGACTCACTATAGGGAGGCCGCGT-3′ (forward) and 5′-CCAATTTAGGTGACACTATAGAAGTACCCGGGGCT-3′ (reverse). This PCR product was subsequently used as a template for in vitro transcription employing T7 and SP6 RNA polymerase to synthesize digoxigenin (DIG)-labeled sense and antisense single-stranded RNA probes, respectively, using a DIG RNA labeling kit (catalog no. 11 093 274 910; Roche Diagnostics GmbH). Two different cytological procedures were used; paraffin embedding and fresh tissue sectioned with a vibratome. For the paraffin procedure, 4- to 5-week-old 2HA calli from leaf explants were fixed in 4% formaldehyde in 0.025 m phosphate buffer at pH 7.2, dehydrated through an ethanol and ethanol:histolene (Fronine Lab Supplies) series, embedded in paraffin, sectioned (8 μm), and hybridized with the DIG-labeled sense and antisense probes according to the manufacturer's instructions. For the fresh tissue procedure, the 2HA embryogenic tissue from leaf explant tissue was embedded in agar and 40-μm sections cut with a vibratome. In both cases, hybridization was detected using a fluorescent antibody enhancer set for DIG detection (catalog no. 176756; Boehringer) and was visualized as a red/purple color after the NBT/BCIP color reaction (Roche Diagnostics). In all cases, no signal over background was observed using control sense-strand probes.
Construction of Constitutive and Inducible RNAi Plasmids
For MtSERF1 RNAi construction, specific sequences in the 3′-end of MtSERF1 mRNA were selected for construction of RNAi fragments. A cDNA fragment of MtSERF1 was amplified by PCR with the primers 5′-CTGTGAAATTGATGCTGCAAA-3′ (forward) and 5′-TGACATAATTGTTGAGCTCACTCC-3′ (reverse). The MtSERF1-specific PCR products were cloned into the vector pCR8/GW/TOPO (Invitrogen). After linearization of the plasmids, the Gateway LR recombination reaction (Invitrogen) was conducted according to the manufacturer's protocol to incorporate the MtSERF1-specific fragment into the binary T-DNA destination vector pH7GW1WG2(II) (Karimi et al., 2002) and pOpOff2(hyg) (Wielopolska et al., 2005) for constitutive and inducible RNAi constructs, respectively. The resulting constructs were introduced into Agrobacterium tumefaciens strain AGL1 by electroporation.
Transformation of M. truncatula
Transformation of M. truncatula 2HA leaf explants was carried out as described by Wang et al. (1996) with some modifications. In brief, leaf pieces were prepared and sterilized according to the method described by Nolan et al. (2003) and dipped into bacterial solution, followed by cocultivation for 2 to 5 d. After cocultivation, the explants were decontaminated by dipping in a solution containing 750 mg/L augmentin (5 parts amoxicillin/L part clavulanic acid; Beecham Laboratories) before plating onto solid medium as described previously in the section on cultured leaf explants. Transformed calli were screened for hygromycin resistance by including hygromycin at 15 mg/L in the medium. Augmentin (500 mg/L) was also added in the medium to eliminate the Agrobacterium. The explants were subcultured on fresh medium every 4 weeks. RNAi constructs were induced by 2.5 μm dexamethasone.
Sequence Analysis and Construction of Phylogenetic Trees
Multiple alignment analyses were performed with ClustalW using a ClustalX 1.8 software package. Phylogenetic trees were constructed using the neighbor-joining method (Saitou and Nei, 1987) included in the ClustalX 1.8 software. Phylogenetic trees were drawn using TreeView (Win32) 1.6.0 software (Page, 1996).
Promoter Sequence Isolation and in Silico Analysis
Isolation of the MtSERF1 promoter was carried out according to the GenomeWalker kit (CLONTECH) with minor modifications. In brief, for the first round of amplification, a biotinylated gene-specific primer and the adaptor primer AP1 were used. Immobilization of the PCR product to streptavidin-coated particles and washing steps were conducted according to the Dynal kilobase BINDER kit (Invitrogen). A one-tenth part of these beads was used for nested PCR as described in the Genome Walker kit and the fragment obtained sequenced. The proximal region of the promoter was analyzed using eukaryotic transcription start site prediction software NNPP, version 2.2 (Reese, 2000, 2001; www.fruitfly.org/seq_tools/promoter.html). Search for core and responsive element motifs was performed in silico by means of the Web Signal Scan Program (Prestridge, 1991; Higo et al., 1999; http://www.dna.affrc.go.jp/Sigscan/signal.html) and MatInspector (Cartharius et al., 2005; http://www.genomatix.de/products/MatInspector).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Histological examination of 40- and 60-d callus used in microarray experiments.
Supplemental Figure S2. The bar graph represents the level of MtSERF1 expression normalized to the GAPDH gene.
Supplemental Table S1. All statistically up-regulated and down-regulated genes at 60 versus 40 d, 80 versus 60 d, and 80 versus 40 d of protoplast culture.
Supplementary Material
Acknowledgments
We wish to thank Yoko Nitanai for assistance with gridding of the microarray signals, Dr. Kim Nolan for assistance with tissue culture, and Dr. Kim Nolan and Dr. Michael Sheahan for helpful discussion.
This work was supported in part by the Australian Research Council Centre of Excellence (grant no. CEO348212) to the University of Newcastle Node of the Centre of Excellence for Integrative Legume Research. Support for microarray analysis was provided by the National Science Foundation Plant Genome project (grant no. 0110206) and the University of Minnesota.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ray J. Rose (ray.rose@newcastle.edu.au).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M (1998) A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J 17 5484–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Banno H, Ikeda Y, Niu QW, Chua NH (2001) Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 13 2609–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MF, Yeung EC (2004) The effects of reduced and oxidized glutathione on white spruce somatic embryogenesis. In Vitro Cell Dev Biol Plant 40 61–66 [Google Scholar]
- Bleecker AB (1999) Ethylene perception and signalling: an evolutionary perspective. Trends Plant Sci 4 269–274 [DOI] [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang LM, Hattori J, Liu CM, van Lammeren AAM, Miki BLA, et al (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14 1737–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21 2933–2942 [DOI] [PubMed] [Google Scholar]
- Chabaud M, de Carvalho-Niebel F, Barker DG (2003) Efficient transformation of Medicago truncatula cv. Jemalong using the hypervirulent Agrobacterium tumefaciens strain AGL1. Plant Cell Rep 22 46–51 [DOI] [PubMed] [Google Scholar]
- Che P, Lall S, Nettleton D, Howell SH (2006) Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol 141 620–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Chang HS, Gupta R, Wang X, Zhu T, Luan S (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol 129 661–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DR (1999) Medicago truncatula—a model in the making! Curr Opin Plant Biol 2 301–304 [DOI] [PubMed] [Google Scholar]
- Delessert C, Wilson IW, Van der Straeten D, Dennis ES, Dolferus R (2004) Spatial and temporal analysis of the local response to wounding in Arabidopsis leaves. Plant Mol Biol 55 165–181 [DOI] [PubMed] [Google Scholar]
- Domoki M, Györgyey J, Biró J, Pasternak TP, Zvara A, Bottka S, Puskás LG, Dudits D, Fehér A (2006) Identification and characterization of genes associated with the induction of embryogenic competence in leaf-protoplast-derived alfalfa cells. Biochim Biophys Acta 1759 543–551 [DOI] [PubMed] [Google Scholar]
- Fehér A, Pasternak TP, Dudits D (2003) Transition of somatic plant cells to an embryogenic state. Plant Cell Tissue Organ Cult 74 201–228 [Google Scholar]
- Harding EW, Tang WN, Nichols KW, Fernandez DE, Perry SE (2003) Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiol 133 653–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass C, Lohrmann J, Albrecht V, Sweere U, Hummel F, Yoo SD, Hwang I, Zhu T, Schäfer E, Kudla J, et al (2004) The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J 23 3290–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Vielle-Calzada JP, Hartog MV, Schmidt EDL, Boutilier K, Grossniklaus U, de Vries SC (2001) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol 127 803–816 [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Banno H, Niu QW, Howell SH, Chua NH (2006) The ENHANCER OF SHOOT REGENERATION 2 gene in Arabidopsis regulates CUP-SHAPED COTYLEDON 1 at the transcriptional level and controls cotyledon development. Plant Cell Physiol 47 1443–1456 [DOI] [PubMed] [Google Scholar]
- Ikeda-Iwai M, Umehara M, Satoh S, Kamada H (2003) Stress-induced somatic embryogenesis in vegetative tissues of Arabidopsis thaliana. Plant J 34 107–114 [DOI] [PubMed] [Google Scholar]
- Imin N, De Jong F, Mathesius U, van Noorden G, Saeed NA, Wang XD, Rose RJ, Rolfe BG (2004) Proteome reference maps of Medicago truncatula embryogenic cell cultures generated from single protoplasts. Proteomics 4 1883–1896 [DOI] [PubMed] [Google Scholar]
- Imin N, Nizamidin M, Daniher D, Nolan KE, Rose RJ, Rolfe BG (2005) Proteomic analysis of somatic embryogenesis in Medicago truncatula. Explant cultures grown under 6-benzylaminopurine and 1-naphthaleneacetic acid treatments. Plant Physiol 137 1250–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7 193–195 [DOI] [PubMed] [Google Scholar]
- Kende H, Zeevaart JAD (1997) The five “classical” plant hormones. Plant Cell 9 1197–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr MK, Martin M, Churchill GA (2000) Analysis of variance for gene expression microarray data. J Comput Biol 7 819–837 [DOI] [PubMed] [Google Scholar]
- Lee MM, Lee SH, Park KY (1997) Effects of spermine on ethylene biosynthesis in cut carnation (Dianthus caryophyllus L.) flowers during senescence. J Plant Physiol 151 68–73 [Google Scholar]
- Lohar DP, Sharopova N, Endre G, Penuela S, Samac D, Town C, Silverstein KAT, VandenBosch KA (2006) Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol 140 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93 1195–1205 [DOI] [PubMed] [Google Scholar]
- Magyar Z, Meszaros T, Miskolczi P, Deak M, Feher A, Brown S, Kondorosi E, Athanasiadis A, Pongor S, Bilgin M, et al (1997) Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. Plant Cell 9 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska-Sawka A, Nothnagel EA (2000) The multiple roles of arabinogalactan proteins in plant development. Plant Physiol 122 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kaló P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al (2007) An ERF transcription factor in Medicago truncatula that is essential for nod factor signal transduction. Plant Cell 19 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 467–475 [DOI] [PubMed] [Google Scholar]
- Nishiwaki M, Fujino K, Koda Y, Masuda K, Kikuta Y (2000) Somatic embryogenesis induced by the simple application of abscisic acid to carrot (Daucus carota L.) seedlings in culture. Planta 211 756–759 [DOI] [PubMed] [Google Scholar]
- Nolan KE, Irwanto RR, Rose RJ (2003) Auxin up-regulates MtSERK1 expression in both Medicago truncatula root-forming and embryogenic cultures. Plant Physiol 133 218–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan KE, Rose RJ, Gorst JE (1989) Regeneration of Medicago truncatula from tissue culture: increased somatic embryogenesis from regenerated plants. Plant Cell Rep 8 278–281 [DOI] [PubMed] [Google Scholar]
- Nolan KE, Saeed NA, Rose RJ (2006) The stress kinase gene MtSK1 in Medicago truncatula with particular reference to somatic embryogenesis. Plant Cell Rep 25 711–722 [DOI] [PubMed] [Google Scholar]
- Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Bioinformatics 12 357–358 [DOI] [PubMed] [Google Scholar]
- Pasternak TP, Prinsen E, Ayaydin F, Miskolczi P, Potters G, Asard H, Van Onckelen HA, Dudits D, Fehér A (2002) The role of auxin, pH, and stress in the activation of embryogenic cell division in leaf protoplast-derived cells of alfalfa. Plant Physiol 129 1807–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, Tholen D, Poorter H, Visser EJW, Voesenek L (2006) The Janus face of ethylene: growth inhibition and stimulation. Trends Plant Sci 11 176–183 [DOI] [PubMed] [Google Scholar]
- Prestridge DS (1991) SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. CABIOS 7 203–206 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339 62–66 [DOI] [PubMed] [Google Scholar]
- Reese MG (2000) Computational prediction of gene structure and regulation in the genome of Drosophila melanogaster. PhD thesis. University of California, Berkeley, CA/University of Hohenheim, Stuttgart, Germany
- Reese MG (2001) Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 26 51–56 [DOI] [PubMed] [Google Scholar]
- Rose RJ (2004) Somatic embryogenesis in plants. In RM Goodman, ed, Encyclopedia of Plant and Crop Science. Marcel Dekker, New York, pp 1165–1168
- Rose RJ, Nolan KE (1995) Regeneration of Medicago truncatula from protoplasts isolated from kanamycin-sensitive and kanamycin-resistant plants. Plant Cell Rep 14 349–354 [DOI] [PubMed] [Google Scholar]
- Rose RJ, Nolan KE (2006) Genetic regulation of somatic embryogenesis with particular reference to Arabidopsis thaliana and Medicago truncatula. In Vitro Cell Dev Biol Plant 42 473–481 [Google Scholar]
- Rose RJ, Nolan KE, Bicego L (1999) The development of the highly regenerable seed line Jemalong 2HA for transformation of Medicago truncatula—implications for regenerability via somatic embryogenesis. J Plant Physiol 155 788–791 [Google Scholar]
- Rose RJ, Wang X-D, Nolan KE, Rolfe BG (2006) Root meristems in Medicago truncatula tissue culture arise from vascular-derived procambial-like cells in a process regulated by ethylene. J Exp Bot 57 2227–2235 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4 406–425 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBS, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290 998–1009 [DOI] [PubMed] [Google Scholar]
- Srinivasan C, Liu ZR, Heidmann I, Supena EDJ, Fukuoka H, Joosen R, Lambalk J, Angenent G, Scorza R, Custers JBM, et al (2007) Heterologous expression of the BABY BOOM AP2/ERF transcription factor enhances the regeneration capacity of tobacco (Nicotiana tabacum L.). Planta 225 341–351 [DOI] [PubMed] [Google Scholar]
- Stasolla C, Belmonte MF, van Zyl L, Craig DL, Liu WB, Yeung EC, Sederoff RR (2004) The effect of reduced glutathione on morphology and gene expression of white spruce (Picea glauca) somatic embryos. J Exp Bot 55 695–709 [DOI] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON 2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Ratet P, Mysore KS (2005) Insertional mutagenesis: a Swiss army knife for functional genomics of Medicago truncatula. Trends Plant Sci 10 229–235 [DOI] [PubMed] [Google Scholar]
- Thibaud-Nissen FO, Shealy RT, Khanna A, Vodkin LO (2003) Clustering of microarray data reveals transcript patterns associated with somatic embryogenesis in soybean. Plant Physiol 132 118–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Rose RJ (1999) Asymmetric somatic hybridisation between the annual legumes Medicago truncatula and Medicago scutellata. Plant Cell Rep 18 989–996 [Google Scholar]
- Touraev A, Vicente O, Heberle-Bors E (1997) Initiation of microspore embryogenesis by stress. Trends Plant Sci 2 297–302 [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBosch KA, Stacey G (2003) Summaries of legume genomics projects from around the globe. Community resources for crops and models. Plant Physiol 131 840–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Rose RJ, Donaldson BI (1996) Agrobacterium-mediated transformation and expression of foreign genes in Medicago truncatula. Aust J Plant Physiol 23 265–270 [Google Scholar]
- Wang KL, Li H, Ecker JR (2002) Ethylene biosynthesis and signaling networks. Plant Cell 14 S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielopolska A, Townley H, Moore I, Waterhouse P, Helliwell C (2005) A high-throughput inducible RNAi vector for plants. Plant Biotechnol J 3 583–590 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5 218–223 [DOI] [PubMed] [Google Scholar]
- Yang YH, Buckley MJ, Dudoit S, Speed TP (2000) Comparison of Methods for Image Analysis on cDNA Microarray Data. Technical Report 2000. Statistics Department, University of California, Berkeley, CA
- Young ND, Shoemaker RC (2006) Genome studies and molecular genetics. Part 1: Model legumes. Exploring the structure, function and evolution of legume genomes. Curr Opin Plant Biol 9 95–98 [DOI] [PubMed] [Google Scholar]
- Yu YB, Yang SF (1979) Auxin-induced ethylene production and its inhibition by aminoethoxyvinylglycine and cobalt ion. Plant Physiol 64 1074–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo JR, Niu QW, Frugis G, Chua NH (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J 30 349–359 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.