Abstract
Major differences exist between plants and animals both in the extent of microRNA (miRNA)-based gene regulation and the sequence complementarity requirements for miRNA-messenger RNA pairing. Whether these differences affect how these sites evolve at the molecular level is unknown. To determine the extent of sequence variation at miRNAs and their targets in a plant species, we resequenced 16 miRNA families (66 miRNAs in total) and all 52 of the characterized binding sites for these miRNAs in the plant model Arabidopsis (Arabidopsis thaliana), accounting for around 50% of the known miRNAs and binding sites in this species. As has been shown previously in humans, we find that both miRNAs and their target binding sites have very low nucleotide variation and divergence compared to their flanking sequences in Arabidopsis, indicating strong purifying selection on these sites in this species. Sequence data flanking the mature miRNAs, however, exhibit normal levels of polymorphism for the accessions in this study and, in some cases, nonneutral evolution or subtle effects on predicted pre-miRNA secondary structure, suggesting that there is raw material for the differential function of miRNA alleles. Overall, our results show that despite differences in the architecture of miRNA-based regulation, miRNAs and their targets are similarly constrained in both plants and animals.
Changes in gene regulation have long been thought to be important to evolutionary diversification (King and Wilson, 1975). Extensive variation in gene expression has been documented both within and across many species (e.g. in primates; Enard et al., 2002; Whitney et al., 2003; Morley et al., 2004), Fundulus (Oleksiak et al., 2002, 2005), and Drosophila (Jin et al., 2001; Rifkin et al., 2003), though in most cases the regulatory mechanisms and phenotypic consequences of this diversity are unknown. Although changes in transcriptional regulation are likely a major source of gene expression diversity, variability in posttranscriptional regulation may also contribute (Chen and Rajewsky, 2007).
MicroRNAs (miRNAs) are small RNAs approximately 21 nucleotides long with complementarity to specific regions in messenger RNAs (mRNAs) and are important posttranscriptional regulators of gene expression in eukaryotes (Carrington and Ambros, 2003). Binding of miRNAs to target mRNAs triggers the cleavage, translational repression, or deadenylation of these targets (Zhang et al., 2007). Once transcribed, miRNAs are processed into small, stem-loop precursors (pre-miRNAs) that are further processed by RNaseIII-type endonucleases (Drosha and Dicer in animals, and DICER-LIKE1 [DCL1] in plants) and methylated to form mature miRNAs (Jones-Rhoades et al., 2006). Mature miRNAs join with ARGONAUTE to form RNA-induced silencing complexes that can subsequently target specific mRNAs (Jones-Rhoades et al., 2006).
The number of miRNAs per eukaryotic genome varies by species. For instance, miRBase presently lists 114, 117, and 326 miRNA genes in Arabidopsis (Arabidopsis thaliana), Caenorhabditis elegans, and humans, respectively (Griffiths-Jones, 2004; Griffiths-Jones et al., 2006). Whereas animals have many unique mature miRNA sequences and small miRNA families, the situation is reversed in plants, which typically have a small number of unique miRNA sequences and large miRNA families (Li and Mao, 2007). Though many miRNAs have been identified across eukaryotes, a recent high-throughput sequencing experiment suggests that additional, lowly expressed miRNAs may exist that have escaped previous molecular and bioinformatics approaches (Fahlgren et al., 2007).
The total number of genes targeted by miRNAs is also highly variable and genome specific. Only 1% of all protein-coding genes appear to be miRNA targets in Arabidopsis (Rhoades et al., 2002; Jones-Rhoades and Bartel, 2004), whereas at least 20% of all protein-coding genes are likely miRNA targets in animals (Lewis et al., 2003, 2005; Grun et al., 2005; Krek et al., 2005; Lall et al., 2006). Furthermore, differences exist between plants and animals in the number of genes targeted by each miRNA. For example, whereas each Drosophila miRNA has on average over 50 predicted targets (Grun et al., 2005), most Arabidopsis miRNAs have six targets or fewer (Jones-Rhoades et al., 2006).
Differences in miRNA function also exist between plants and animals. In animals, the complementarity between the first six to eight bases of a target to a miRNA are most important to binding (Rajewsky, 2006). In contrast, plants require nearly complete complementarity across the entire miRNA and binding target (Schwab et al., 2005, 2006). In addition, although in animals miRNA binding sites are almost exclusively within 3′ untranslated regions (UTRs), most plant miRNAs have binding sites within coding exons (Chen and Rajewsky, 2007).
The extent to which miRNAs contribute to phenotypic evolution is unclear, but evidence suggests they could play an important role. Although essential miRNA-target site interactions have been conserved for >400 million years in plants and animals, e.g. miR165/166 and Class III HD-ZIP (homeodomain Leu zipper) genes in land plants (Floyd and Bowman, 2004) and the let-7 miRNA and the lin-41 mRNA in metazoans (Pasquinelli et al., 2000), others appear to be species or clade specific and may function in evolutionarily derived traits (Bonnet et al., 2004; Axtell and Bartel, 2005).
Little is known about the microevolution of miRNA-target site interactions, though a few studies have documented functional polymorphisms at these sites. A single nucleotide polymorphism (SNP) that results in a de novo miRNA binding site has been shown to underlie a quantitative trait locus for muscularity in sheep (Clop et al., 2006), whereas an SNP in an existing miRNA binding site may cause Tourette's syndrome in humans (Abelson et al., 2005). Additionally, an SNP has been identified in Human herpesvirus that affects Drosha processing of an miRNA precursor (Gottwein et al., 2006).
Genome-wide surveys of miRNA and miRNA binding site polymorphism have only been conducted in humans. These studies have shown levels of polymorphism at miRNAs and their targets are lower than at coding or neutral regions; the mutations at these sites exhibit a general signature of purifying selection (Chen and Rajewsky, 2006; Saunders et al., 2007). One of these studies has also shown evidence for positive selection on some miRNA binding sites based on long-range haplotype signatures (Saunders et al., 2007), implying that beneficial miRNA-target site polymorphisms may exist.
Predictions can be made about the expected level of miRNA and miRNA binding site sequence variation in plants relative to humans based on the functional differences between plant and animal miRNAs that are described above. Plant miRNAs typically have fewer mRNA targets and more miRNA family members than animal miRNAs, which may lead to reduced constraint on and higher sequence diversity in miRNA sequences in plants. However, as most plant miRNAs perform important functions in development and physiology, and spatiotemporal functional differences may exist among plant miRNA family members making each independently essential, constraint on plant miRNAs may parallel that observed in humans. As for miRNA binding sites, constraint is likely to be strong across the entire miRNA binding site in plants due to the importance of the entire binding site in miRNA-mRNA pairing. Additionally, plant miRNA binding sites may experience additional constraint due to the presence of these sites largely in coding exons in plants.
We assess the levels and patterns of nucleotide polymorphism in miRNAs and their binding sites in the model plant Arabidopsis by resequencing more than half of the characterized miRNAs and binding targets in this species from 24 diverse accessions. We find significantly reduced genetic variation at these sites relative to flanking sequence, with only four SNPs and an insertion/deletion (indel) present in our sample. However, we do find substantial variation flanking miRNAs both within Arabidopsis and between it, and the closely related out-group Arabidopsis lyrata. Interestingly, four miRNAs exhibit nonneutral patterns of molecular variation, and numerous SNPs are predicted to have subtle effects on pre-miRNA secondary structure. Our results suggest that mutations within mature miRNAs and their binding sites do not contribute substantially to gene expression and phenotypic variation in this model plant species, but that ample variation flanks mature miRNAs that could contribute to the evolutionary diversification of these key regulatory genes.
RESULTS
SNPs and Nucleotide Divergence in Arabidopsis miRNAs
We investigated the sequence variation of 66 miRNAs belonging to 16 miRNA families, as well as 52 mRNA binding site targets that represented all the validated targets for these miRNAs (Table I). On average, we resequenced four miRNAs per family and three target sites per miRNA family in a set of 24 accessions. These miRNAs were selected because: (1) their interactions with mRNA targets have been functionally characterized, and/or (2) they target transcripts of genes with known roles in development. Altogether, these comprise over 55% of the presently described miRNAs and 40% of the validated binding sites in Arabidopsis, based on data in Jones-Rhoades et al. (2006).
Table I.
miRNAs and targets included in this study
| miRNA Familya | No. of Copies | Target Family | No. of Targets | Verified Targets |
|---|---|---|---|---|
| miR156/157 | 12 | SBP | 11 | SPL2, SPL3, SPL4, SPL10 |
| miR159/319 | 6 | MYB and TCP | 13 | MYB33, MYB65, TCP2, TCP3, TCP4, TCP10, TCP24 |
| miR160 | 3 | ARF | 3 | ARF10, ARF16, ARF17 |
| miR162 | 2 | DICER | 1 | DCL1 |
| miR164 | 3 | NAC | 6 | CUC1, CUC2, NAC1, At5g07680, At5g61430 |
| miR165/166 | 9 | Class III HD-ZIP | 5 | PHB, PHV, REV, ATHB-8, ATHB-15 |
| miR167 | 4 | ARF | 2 | ARF6, ARF8 |
| miR168 | 2 | ARGONAUTE | 1 | AGO1 |
| miR170/171 | 4 | SCL | 3 | SCL6-III, SCL6-IV |
| miR172 | 5 | AP2 | 6 | AP2, TOE1, TOE2, TOE3 |
| miR393 | 2 | F-box | 5 | TIR1, AFB1, AFB2, AFB3, At3g23690 |
| miR394 | 2 | F-box | 2 | At1g27340 |
| miR395 | 6 | APS | 3 | APS1, APS4 |
| miR396 | 2 | GRF | 7 | GRL1, GRL2, GRL3, GRL7, GRL8, GRL9 |
| miR398 | 3 | CSD | 3 | CSD1, CSD2, At3g15640 |
| miR403 | 1 | ARGONAUTE | 1 | AGO2 |
Table adapted from Jones-Rhoades et al. (2006).
For each miRNA, we sequenced on average 489 bp, with approximately 133 bp of pre-miRNA (based on pre-miRNA predictions in miRBase), as well as about 180 and 176 bp of upstream and downstream flanking sequence, respectively. The average level of SNP per site (θ; Watterson, 1975) at these miRNAs is 0.0004 ± 0.0003 (mean ± se of the mean), or one SNP every 2.5 kb of miRNA sequence. Underscoring this low sequence diversity, only two miRNAs, miR156d and miR395f, were actually found to be polymorphic. The microRNA miR156d has a single SNP segregating at 10% frequency (found in both the Ei-2 and Ll-0 accessions), whereas there is a single SNP in miR395f found in the Cvi-0 accession (4% frequency; Fig. 1). The miR156d SNP is at miRNA-mRNA mismatch position and is unlikely to affect binding, whereas the miR395f minor allele disrupts a complementary position relative to the major allele (Fig. 1). Sequence comparisons with the closely related out-group A. lyrata show that both of these polymorphisms are derived within Arabidopsis. No fixed differences exist between Arabidopsis and A. lyrata at the examined miRNAs (i.e. K = 0). Levels of both nucleotide polymorphism and divergence at these sites are significantly below background levels of θ = 0.0055 ± 0.0002 and K = 0.085 ± 0.005 as assessed using 1,213 previously resequenced genome-wide loci (Wilcoxon rank sum test; P < 0.0001 for both polymorphism and divergence).
Figure 1.
A to D, Polymorphisms occurring in Arabidopsis miR156d (A), miR395f (B), the AFB1 binding site for miR393 (C), and the TOE3 binding site for miR172 (D). For the miRNA polymorphisms, an example binding site is included for reference. The targeting mature miRNA sequence is included for reference purposes with the binding site polymorphisms. Binding sites are portrayed 5′ → 3′, whereas miRNAs are presented 3′ → 5′. Polymorphic positions are colored gray and marked by a box. Major and minor describe the high and low-frequency alleles, respectively. Complementary positions (i.e. nonmismatches or gaps) are depicted with black lines. G:U mismatch positions, which may contribute to miRNA-target binding, are marked with a black circle. Note pairing symbols are based on major allele states.
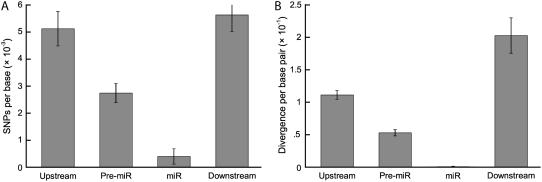
We also estimated levels of nucleotide diversity for the sequences flanking the miRNAs, including the pre-miRNAs and the upstream and downstream flanking sequence. Nucleotide polymorphism levels at these sites are substantially higher than those observed in the miRNAs themselves, with a mean θ = 0.0025 ± 0.0003 for the pre-miRNA, and mean θ = 0.0051 ± 0.0006 and 0.0054 ± 0.0006 for the upstream and downstream flanking sequences, respectively (Fig. 2). Although no indel polymorphisms were observed in the mature miRNA sequences, numerous indels were detected in the pre-miRNAs and flanking sites (0.7 per kilobase pair at pre-miRNAs, 1.7 per kilobase pair for upstream sequence, and 2 per kilobase pair for downstream flanking sequence). Levels of nucleotide divergence are also higher at these sites, with mean K = 0.052 ± 0.007 for pre-miRNA, 0.11 ± 0.014 for upstream sequence, and 0.2 ± 0.026 at downstream sites (Fig. 2). The dramatically reduced intraspecific polymorphism and interspecific divergence at mature miRNAs, and to a lesser extent pre-miRNAs, suggests that purifying selection is the predominant evolutionary force that acts on miRNAs in Arabidopsis.
Figure 2.
A and B, Mean levels of polymorphism (A) and divergence (B) at miRNAs and flanking regions. se bars are included. Nonterminated error bars indicate that the sum of the mean and the error occurs above the maximum of the y axis.
Levels and Patterns of Nucleotide Polymorphism and Divergence in miRNA Target Binding Sites
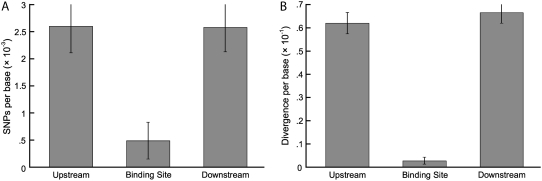
We also estimated polymorphism and divergence at the target binding sites of the miRNAs we examined (Table I). In all cases, these sites had been previously validated as the target sites of specific miRNAs (Jones-Rhoades et al., 2006, and refs. therein). Of these binding sites, 47 are in exons, two are in 5′ UTRs, and three are in 3′ UTRs. Six of the exonic binding sites and one of the 5′ UTR binding sites are interrupted by introns and, consequently, require splicing to bind with their complementary miRNAs. For each binding site, we sequenced on average 476 bp, with the miRNA binding site at the center of the sequenced region.
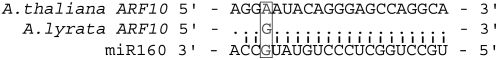
Like their cognate miRNAs, we also observe significantly low polymorphism levels at the miRNA binding sites relative to background polymorphism, with mean nucleotide diversity equal to 0.0005 ± 0.0003 (Wilcoxon rank sum test; P < 0.0001). Only two binding sites of the 52 we studied—in the AUXIN SIGNALING F-BOX1 (AFB1) and TARGET OF EAT3 (TOE3) genes—are polymorphic (Fig. 1). The AFB1 binding site, which is targeted by miR393, has a single SNP segregating at 12% frequency (in the Edi-0, Ga-0, and Ll-0 accessions). The AFB1 minor allele converts a miRNA-mRNA match position to a mismatch position relative to the major allele (Fig. 1). TOE3 is targeted by miR172 and this binding site has a 7-bp deletion and an SNP that cosegregate at 4% frequency in our sample (found in the Gy-0 accession). The TOE3 binding site deletion, however, is partially recovered in the mRNA due to upstream sequence similarity, resulting in only a single base-pair deletion and a SNP in the mature transcript (Fig. 1). Although the low-frequency TOE3 polymorphisms are derived mutations, the derived mutation at AFB1 is the common SNP allele. Nucleotide divergence at target binding sites is K = 0.003 ± 0.002, which is significantly lower than the genome-wide average (Wilcoxon rank sum test; P < 0.0001). Only one binding site—in AUXIN RESPONSE FACTOR10 (ARF10), which is targeted by miR160—exhibits a fixed sequence difference between species. This substitution occurs at a mismatch position in the miRNA-mRNA pairing sequence (Fig. 3).
Figure 3.
The nucleotide substitution that occurs between Arabidopsis and A. lyrata in ARF10. The ARF10 binding site is portrayed 5′ → 3′, whereas the sequence for miR160, which targets ARF10, is presented 3′ → 5′. The variable position is colored gray and marked by a box. Complementary positions (i.e. nonmismatches or gaps) are depicted with black lines. G:U mismatch positions, which may contribute to miRNA-target binding, are marked with a black circle. Note pairing symbols are based on major allele states.
Levels of nucleotide variation were also reduced at miRNA binding sites relative to their flanking sequences (mean θ = 0.0028 ± 0.0005 and 0.0033 ± 0.0005 for upstream and downstream flanking sequences, respectively; see Fig. 4). These flanking nucleotide diversity values are low in comparison to data surrounding mature miRNAs, and are likely due to the location of many of these binding sites in coding exons. To correct for this, we also calculated silent site nucleotide diversity (θsilent) for miRNA binding sites and their flanking sequences. These estimates (θsilent = 0.0015 ± 0.0007, 0.0069 ± 0.0026, and 0.0053 ± 0.0009 for miRNA binding sites, upstream sequence, and downstream sequence, respectively) are higher than those for uncorrected nucleotide diversity estimates. The relative levels of variation across the site classes remain similar to uncorrected values, however, because nucleotide diversity at the binding sites is still much lower than at flanking sites. Additionally, divergence was much lower at binding sites than at upstream (K = 0.062 ± 0.005) or downstream (K = 0.067 ± 0.001) sites (Fig. 4).
Figure 4.
A and B, Mean levels of polymorphism (A) and divergence (B) at miRNA binding sites and flanking regions. se bars are included. Nonterminated error bars indicate that the sum of the mean and the error occurs above the maximum of the y axis.
Summary Statistics of the Nucleotide Site-Frequency Spectrum
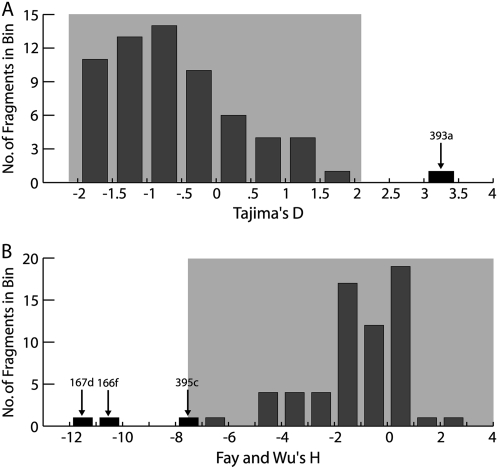
Although miRNAs and their target binding sites have little variation, we observe normal levels of sequence variation in regions flanking miRNAs. Selection on these polymorphisms or those linked to them could generate nonneutral patterns of linked sequence variation. To examine this possibility, we calculated Tajima's D (Tajima, 1989) and Fay and Wu's H (Fay and Wu, 2000) for the entire sequence fragment containing each miRNA, as well as for the Nordborg et al. (2005) genome-wide fragments. Using the Nordborg data as an empirical distribution, these tests identify four fragments that possess extreme values for either Tajima's D or Fay and Wu's H. MiR393a has a high Tajima's D value (D = 3.39; empirical P < 0.001; Fig. 5). Significant Tajima's D values can be indicative of balancing selection or extreme population stratification at a locus. MiR166f, miR167d, and miR395c have low Fay and Wu's H values (H = −10.03, −11.44, and −7.22 for miR166f, miR167d, and miR395c, respectively; empirical P < 0.05 for each miRNA; Fig. 5) when compared to the empirical distribution, which can be indicative of positive selection. These results suggest that polymorphisms at or linked to miRNA genes may be targets of selection, though these findings must be regarded cautiously given the complex demography and pattern of linkage disequilibrium of this species (Nordborg et al., 2005; Schmid et al., 2005).
Figure 5.
A and B, Distribution of Tajima's D (A) and Fay and Wu's H (B) values across all miRNA resequencing fragments. Significant fragments are labeled and colored black. Light gray boxes represent the range of the middle 95% or the top 95% of the empirical distribution for Tajima's D and Fay and Wu's H, respectively, determined using genome-wide background fragments from Nordborg et al. (2005).
Secondary Structure Predictions of Pre-miRNA Haplotypes Using Biologically Relevant Temperatures
To evaluate the possible impacts of SNPs on pre-miRNA secondary structure, we computationally predicted the secondary structure and Gibbs free energy (ΔG) of the pre-miRNA from each observed haplotype using the mfold program (Walter et al., 1994; Zuker, 2003). Due to both seasonal variation in temperature and geographic differences in climate, Arabidopsis experiences a broad range of temperatures in the wild that may be important to consider when predicting secondary structures for organic macromolecules. We selected for analysis two temperatures—5°C and 20°C—that are tolerable extremes of the range of temperatures that Arabidopsis experiences naturally.
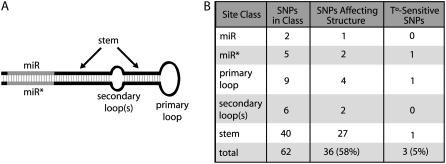
Of the 66 miRNAs we looked at overall, only 35 had SNPs segregating in their predicted pre-miRNAs; 62 total SNPs were identified across these pre-miRNAs. Using the predicted pre-miRNA secondary structure for the haplotype of the Columbia (Col-0) accession of Arabidopsis for each miRNA, we determined the structural context of each SNP within its respective pre-miRNA. The vast majority of the SNPs were located in double-stranded stem regions—40 in the general stem and seven in the miRNA or miRNA* (Fig. 6). Of the remaining 15 SNPs, nine were located in the primary loop at the top of the pre-miRNA stem-loop molecule and the remaining six were in secondary loops occurring along the stem of the molecule (Fig. 6).
Figure 6.
A and B, Locations of SNPs within pre-miRNA molecules. Structural context classifications are displayed in A. In B, SNP counts per site class are presented. The number of SNPs affecting secondary structure in either 5°C or 20°C is listed, as well as the number of SNPs that displayed effects only at 5°C (labeled T°-sensitive SNPs in the image). Note that none of the SNPs had dramatic effects on pre-miRNA secondary structure and that in all cases the general integrity of the molecule was maintained. SNP effects on secondary structure were subtle and included the addition or subtraction of a secondary loop or the enlargement or shrinking of a primary or secondary loop.
We predicted pre-miRNA secondary structure at both 5°C and 20°C, which represents a sampling of the temperature extremes Arabidopsis might be expected to experience during its lifecycle. Of the pre-miRNA SNPs, 33 (53%) were predicted to alter pre-miRNA secondary structure at both temperatures relative to the Col-0 pre-miRNA allele (Fig. 6). All predicted secondary structure changes were subtle (i.e. addition or subtraction of small loops along the stem; two nucleotide enlargement or shrinking of primary or secondary stem loops) and appeared to maintain the general integrity of the pre-miRNA stem-loop molecule. SNPs disrupting secondary structure occurred in all structural contexts of pre-miRNAs (Fig. 6). For 26 SNPs (42% of all pre-miRNA SNPs), pre-miRNA secondary structure was entirely maintained across pre-miRNA alleles. Ten of these SNPs had no structural effect because they occurred within loops. The other 16 SNPs that did not affect secondary structure were all located along the pre-miRNA stem and fell into one of five classes (SNP counts in parentheses): (1) occurring within mismatch positions (two SNPs), (2) creating a nondisruptive mismatch from a match (seven SNPs), (3) creating a match from a nondisruptive mismatch (one SNP), (4) a purine transition (A ↔ G) with the pairing base a U (three SNPs), and (5) a pyrimidine transition (C ↔ U) with the pairing base a G (three SNPs). Three SNPs (5% of all SNPs) had predicted structural effects at 5°C, but not at 20°C. These SNPs occur in a loop in miR156d, at an A ↔ G with U pairing site (class 4) in miR157c, and at a C ↔ U with G pairing site (class 5) in miR164a.
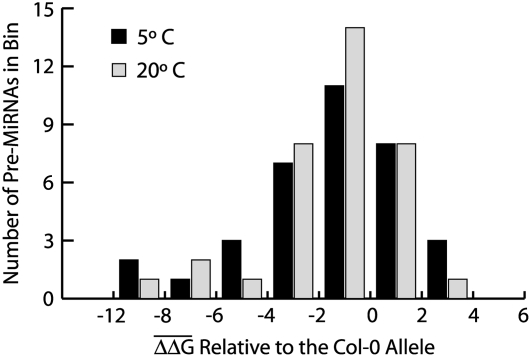
To more quantitatively assess the effects of SNPs on pre-miRNA stability, we next measured ΔG for each pre-miRNA haplotype and calculated the difference in ΔG (ΔΔG) between the Col-0 allele and the non-Col-0 alleles. For loci with more than two alleles, the mean ΔΔG was calculated across all values for the locus. Ninety-one percent of the loci (32 of 35) had a mean ΔΔG that fell within the range of −6 to 4 kcal/mol at both temperatures (Fig. 7). On average, mean ΔΔG was −1.64 kcal/mol and −1.53 kcal/mol at 5°C and 20°C, respectively, suggesting that most SNPs destabilize pre-miRNAs in relation to the Col-0 allele. Interestingly, temperature has a clear effect on ΔΔG as values are more dispersed around the mean at 5°C relative to 20°C, suggesting that the destabilizing effects of polymorphisms on RNA secondary structure are enhanced by cold temperature.
Figure 7.
Distribution of mean ΔΔG values. The 35 resequenced loci with SNPs detected in the pre-miRNA were included.
DISCUSSION
Posttranscriptional regulation of gene expression is a common phenomenon across eukaryotes, but the extent to which variability in this process contributes to diversity in gene expression and phenotype is unclear (Chen and Rajewsky, 2007). Examples in humans (Abelson et al., 2005), sheep (Clop et al., 2006), and Human herpesvirus (Gottwein et al., 2006) suggest that functional polymorphisms at miRNAs and miRNA binding sites do exist. Genomic surveys of polymorphism at miRNAs and their targets can determine the prevalence of such variants across a species. To date, genome-wide levels of nucleotide variation at miRNAs and miRNA binding sites have only been assessed in humans (Chen and Rajewsky, 2006; Saunders et al., 2007), and the low levels of polymorphism at these sites indicates strong purifying selection on these regulatory RNAs and their targets.
MiRNAs in the model plant Arabidopsis have been implicated in several developmental processes, including flowering time (Aukerman and Sakai, 2003; Achard et al., 2004), juvenile/adult transition (Wu and Poethig, 2006), leaf shape (Nikovics et al., 2006), and adaxial/abaxial polarity (Emery et al., 2003). Our results indicate that these Arabidopsis miRNAs and their binding sites evolve under strong sequence constraint. Indeed, in all these genes, only two miRNA SNPs, two binding site SNPs, and a binding site indel were detected in our sample, with most of these polymorphisms being low-frequency derived polymorphisms. Additionally, only one substitutional difference exists between Arabidopsis and A. lyrata at these sites.
Overall, our results support that, like in humans, the predominant force acting on Arabidopsis miRNAs and their targets is purifying selection. Despite the presence of more copies of each mature miRNA sequence in the Arabidopsis genome than in the human genome and the smaller number of mRNA targets per miRNA in plants, plant miRNAs exhibit very strong purifying selection comparable to that observed in humans (Saunders et al., 2007). The prediction that miRNA sequences should be conserved across their entirety in plants, which is contrary to patterns of sequence variation in human miRNAs, does hold true. This corroborates the findings of molecular biology experiments that document plants' requirements for sequence complementarity across the miRNA-mRNA pairing sequence (Schwab et al., 2005, 2006). Unlike in humans (Saunders et al., 2007), no moderate frequency binding site polymorphisms segregate in our sample, suggesting that miRNA binding site variation is unlikely to contribute to phenotypic diversity in Arabidopsis.
The degree of purifying selection on miRNAs and their target binding sites can be assessed by comparing levels of variation at miRNAs and their targets to levels of amino-acid-changing variation in protein-coding genes. The mean level of nonsynonymous polymorphism (θnsyn) for the miRNA target gene exons resequenced in this study is 0.002 ± 0.0003, which is over 4-fold higher than nucleotide diversity values at miRNAs and their binding sites. This is also observed at the interspecific level; the mean rate of nonsynonymous substitution (Ka) between Arabidopsis and A. lyrata is 0.026 ± 0.003, which is nearly 30-fold higher than mean K for miRNAs and approximately 10-fold higher than K values at miRNA binding sites. These comparisons indicate that purifying selection on miRNAs and their binding sites is stronger than it is for amino acid changes in protein-coding genes.
The strong sequence constraint of miRNAs and their binding sites suggest that evolutionary changes in these sequences are unlikely to be major contributors to natural variation in Arabidopsis. We have, however, identified a small number of rare miRNA and target site polymorphisms that may have functional effects, and have shown that substantial flanking variation exists both within Arabidopsis and between it and A. lyrata. Overall, our results imply that the roles of miRNA-target interactions in plant function are essential and are subject to strong purifying selection, but that variation flanking these sites could contribute to regulatory diversity at these genes and their downstream targets.
Empirical and computational approaches have shown that pre-miRNA secondary structure is important to the processing and maturation of miRNAs (Zeng et al., 2005; Ritchie et al., 2007). Our analyses of the effects of SNPs on predicted pre-miRNA secondary structure suggests that another element contributing to sequence constraint at and near miRNAs may be selection for the maintenance of the pre-miRNA stem loop; 42% of all detected SNPs were predicted to have no effect on pre-miRNA secondary structure. Of the numerous SNPs that were identified with structural effects, all had subtle effects, maintaining the general integrity of the stem-loop molecule. These results together imply that purifying selection culls mutations with strong effects on pre-miRNA secondary structure. This finding is similar to results from diverse organisms, such as bacteria (Katz and Burge, 2003), flies (Kirby et al., 1995), and mammals (Chamary and Hurst, 2005), showing constraint on other types of RNA molecules.
Of note is that most detected pre-miRNA polymorphisms appear to have a destabilizing effect on RNA secondary structure because the majority of the studied loci have nonzero ΔΔG values. These destabilizing effects appear to be partially mediated by temperature, a point supported both by the increased dispersion of ΔΔG at 5°C relative to 20°C and the identification of three polymorphisms with structural effects at 5°C and not 20°C. These findings suggest that the use of biologically relevant temperature, which in Arabidopsis represents the range of environmental temperatures it might experience during a growing season, may be an important consideration when predicting RNA or protein secondary structure. Indeed, temperature could mediate gene regulation in nature through its effects on secondary structure.
MiRNAs comprise a key class of regulatory loci in eukaryotic systems, and we are beginning to understand the evolutionary forces that govern the diversification of these genes. Our work suggests that despite fundamental differences in miRNA-based regulation, miRNAs and their targets are similarly constrained in both plants and animals. The possibility of variation in cis-regulation or processing of miRNAs in Arabidopsis and other species merits further attention, though documenting such functional variation, if it exists, will be technically challenging due to the presence of multiple copies of many mature miRNAs. We have shown, however, that the raw material for such variation does exist in Arabidopsis and that it may be responsive to temperature, laying the groundwork for future experiments focused on potential molecular functional variation at miRNAs in Arabidopsis.
MATERIALS AND METHODS
PCR and DNA Sequencing
miRNAs and binding targets were chosen as a subset of those listed in Jones-Rhoades et al. (2006). miRNA precursors were determined based on references provided at the miRBase Web site (http://microrna.sanger.ac.uk/). The Arabidopsis (Arabidopsis thaliana) accessions used in this study were chosen to span the geographic range of the species and are included in Supplemental Data S1. All primers were designed from the Col-0 genome sequence using Primer3 (Rozen and Skaletsky, 2000). Primer pairs were designed to amplify products between 400 and 600 bp in length and are provided in Supplemental Data S1. The amplified regions were centered on the miRNA precursor listed in miRBase or the miRNA binding site. All PCR primers were blasted against the Col-0 genome sequence on the The Arabidopsis Information Resource Web site (www.arabidopsis.org) to ensure that only the targeted genomic region would be amplified. PCR and sequencing was done as previously described (Olsen et al., 2006) by Cogenics. On average, 23 individuals were successfully sequenced per miRNA and miRNA binding site.
Sequence Analysis
Sequences were initially aligned and edited using the Phred and Phrap programs (Codon Code) and BioLign version 2.09.1 (Tom Hall, Ibis Therapeutics). Additional manual alignment and polymorphism identification were conducted in BioEdit version 7.0.5 (Tom Hall, Ibis Therapeutics). Reported summary statistics were calculated in either Microsoft Excel, DnaSP version 4.1.0 (Rozas et al., 2003), or Variscan (Vilella et al., 2005) and are included in Supplemental Data S1. Site classifications (i.e. as miRNA, pre-miRNA, upstream, or downstream) were made based on information in miRBase. Nucleotide diversity was calculated as θ, the population mutation rate per locus based on the number of segregating sites (Watterson, 1975). Nucleotide substitution rates (K) were calculated in Variscan based on the Jukes-Cantor model. Silent site variation was calculated based on all mutations that did not affect the amino acid sequence of the protein encoded by a target mRNA, including intronic and UTR sequence. The empirical distribution for Tajima's D (Tajima, 1989), as well as the background level of polymorphism for the accessions in this study, was generated using 1,213 previously published, genome-wide resequencing fragments (Nordborg et al., 2005). Tajima's D was calculated because it can be useful in detecting both positive selection and balancing selection. We also calculated Fay and Wu's H (Fay and Wu, 2000) because it previously was shown to be less biased by demography in Arabidopsis than other tests evaluating the site frequency spectrum (Schmid et al., 2005). A. lyrata sequence data was obtained for all fragments in this study, as well as for 100 randomly selected fragments from the Nordborg et al. (2005) study to generate the background distribution of K and Fay and Wu's H. The A. lyrata data were acquired by using the Trace Archive database Mega BLAST search function at National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast/mmtrace.shtml). The top one or two hits per fragment were assembled and aligned to the Arabidopsis multiple alignments. No significant hits were found corresponding to miR395a, although there was a region of sequence similarity found for the upstream region of this fragment. Any sites initially found to be diverged between Arabidopsis and A. lyrata at the miRNA binding sites were reexamined by additional BLAST searching. A. lyrata traces used in this study are included in Supplemental Data S1. It should be noted that the study of Nordborg et al. (2005) used 96 accessions, 24 of which are included in this study. To account for this, we only used sequence data corresponding to the accessions in this study to generate empirical distributions. Wilcoxon rank sum tests, ANOVAs, and regressions were conducted in JMP version 5 (SAS).
Secondary Structure Prediction
The program mfold v2.3 was used to predict the pre-miRNA secondary structure and the ΔG for each naturally occurring pre-miRNA haplotype (excluding those differentiated from the Col-0 allele by indels). Comparison of SNP locations to the predicted Col-0 structure at 5°C was used to identify the structural context of each SNP. In cases where multiple structures were predicted for a particular pre-miRNA haplotype, ΔG was calculated as the average of all these predictions. ΔΔG was then calculated for each locus by subtracting the non-Col-0 allele's ΔG from the Col-0 allele's ΔG. Because the number of haplotypes per pre-miRNA was variable, we calculated a mean ΔΔG per pre-miRNA, which was simply the average of all ΔΔG values for that locus.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EU549868 to EU551085 (miRNAs) and EU548273 to EU549692 (binding sites).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Data S1. Information on the accessions, primers, A. lyrata genome sequence trace files, background resequencing fragments, and miRNA and miRNA binding site molecular population genetics results used in this study.
Supplementary Material
Acknowledgments
We thank Daisuke Saisho and members of the Purugganan laboratory for assistance with this project and manuscript. We also thank Kevin Chen for reading a draft of this manuscript.
This work was supported by a U.S. Department of Education Graduate Assistance in Areas of National Need Fellowship and a National Science Foundation Graduate Research Fellowship (to I.M.E.), and by grants from the National Science Foundation Frontiers in Integrated Biological Research and Plant Genome Research Programs and the U.S. Department of Defense (to M.D.P.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ian M. Ehrenreich (ehrenreich@ncsu.edu).
The online version of this article contains Web-only data.
References
- Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al (2005) Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science 310 317–320 [DOI] [PubMed] [Google Scholar]
- Achard P, Herr A, Baulcombe DC, Harberd NP (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131 3357–3365 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Bartel DP (2005) Antiquity of microRNAs and their targets in land plants. Plant Cell 17 1658–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E, Wuyts J, Rouze P, Van de Peer Y (2004) Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc Natl Acad Sci USA 101 11511–11516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301 336–338 [DOI] [PubMed] [Google Scholar]
- Chamary JV, Hurst LD (2005) Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals. Genome Biol 6 R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N (2006) Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet 38 1452–1456 [DOI] [PubMed] [Google Scholar]
- Chen K, Rajewsky N (2007) The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet 8 93–103 [DOI] [PubMed] [Google Scholar]
- Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, et al (2006) A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet 38 813–818 [DOI] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13 1768–1774 [DOI] [PubMed] [Google Scholar]
- Enard W, Khaitovich P, Klose J, Zollner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R, et al (2002) Intra- and interspecific variation in primate gene expression patterns. Science 296 340–343 [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2 e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu CI (2000) Hitchhiking under positive Darwinian selection. Genetics 155 1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd SK, Bowman JL (2004) Gene regulation: ancient microRNA target sequences in plants. Nature 428 485–486 [DOI] [PubMed] [Google Scholar]
- Gottwein E, Cai X, Cullen BR (2006) A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J Virol 80 5321–5326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S (2004) The microRNA Registry. Nucleic Acids Res 32 D109–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34 D140–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N (2005) microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol 1 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Riley RM, Wolfinger RD, White KP, Passador-Gurgel G, Gibson G (2001) The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat Genet 29 389–395 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14 787–799 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57 19–53 [DOI] [PubMed] [Google Scholar]
- Katz L, Burge CB (2003) Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res 13 2042–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC (1975) Evolution at two levels in humans and chimpanzees. Science 188 107–116 [DOI] [PubMed] [Google Scholar]
- Kirby DA, Muse SV, Stephan W (1995) Maintenance of pre-mRNA secondary structure by epistatic selection. Proc Natl Acad Sci USA 92 9047–9051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al (2005) Combinatorial microRNA target predictions. Nat Genet 37 495–500 [DOI] [PubMed] [Google Scholar]
- Lall S, Grun D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, et al (2006) A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol 16 460–471 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120 15–20 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115 787–798 [DOI] [PubMed] [Google Scholar]
- Li A, Mao L (2007) Evolution of plant microRNA gene families. Cell Res 17 212–218 [DOI] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG (2004) Genetic analysis of genome-wide variation in human gene expression. Nature 430 743–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P (2006) The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18 2929–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3 e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleksiak MF, Churchill GA, Crawford DL (2002) Variation in gene expression within and among natural populations. Nat Genet 32 261–266 [DOI] [PubMed] [Google Scholar]
- Oleksiak MF, Roach JL, Crawford DL (2005) Natural variation in cardiac metabolism and gene expression in Fundulus heteroclitus. Nat Genet 37 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Caicedo AL, Polato N, McClung A, McCouch S, Purugganan MD (2006) Selection under domestication: evidence for a sweep in the rice waxy genomic region. Genetics 173 975–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al (2000) Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408 86–89 [DOI] [PubMed] [Google Scholar]
- Rajewsky N (2006) microRNA target predictions in animals. Nat Genet (Suppl) 38 S8–S13 [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110 513–520 [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Kim J, White KP (2003) Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet 33 138–144 [DOI] [PubMed] [Google Scholar]
- Ritchie W, Legendre M, Gautheret D (2007) RNA stem-loops: to be or not to be cleaved by RNAse III. RNA 13 457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19 2496–2497 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the www for general users and for biologist programmers. In S Krawetz, S Misener, eds, Bioinformatics Methods and Protocols: Methods for Molecular Biology. Humana Press, Totowa, NJ, pp 365–386 [DOI] [PubMed]
- Saunders MA, Liang H, Li WH (2007) Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA 104 3300–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid KJ, Ramos-Onsins S, Ringys-Beckstein H, Weisshaar B, Mitchell-Olds T (2005) A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics 169 1601–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8 517–527 [DOI] [PubMed] [Google Scholar]
- Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella AJ, Blanco-Garcia A, Hutter S, Rozas J (2005) VariScan: analysis of evolutionary patterns from large-scale DNA sequence polymorphism data. Bioinformatics 21 2791–2793 [DOI] [PubMed] [Google Scholar]
- Walter AE, Turner DH, Kim J, Lyttle MH, Muller P, Mathews DH, Zuker M (1994) Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci USA 91 9218–9222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7 256–276 [DOI] [PubMed] [Google Scholar]
- Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, Brown PO (2003) Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci USA 100 1896–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR (2005) Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J 24 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang Q, Pan X (2007) MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol 210 279–289 [DOI] [PubMed] [Google Scholar]
- Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.