Abstract
We propose a novel theory for the evolution of polyandry driven by genetic benefits to females whose offspring interbreed. In species with an ecology characterized by frequent colonization of new habitat patches, consanguineous matings may be common during the early stages of colonization, but genetic diversity may grow as new colonizers arrive. We show that with levels of inbreeding depression similar to those found in predominantly inbreeding populations, a polyandrous female can benefit her descendants since matings among her brood are mainly between half siblings rather than full siblings. We examine the invasion by a polyandrous phenotype using explicit genetic models in which costs of inbreeding are themselves subject to selection. In common with other models of inbreeding, we find that underlying high levels of inbreeding tend to purge deleterious recessive alleles, and hence these are unlikely to maintain sufficient inbreeding depression to favour polyandry. However, if costs of inbreeding are due to overdominance, biologically realistic levels of inbreeding depression result in genetic benefits large enough to favour polyandry provided it is not too costly. The potential significance of polyandry as a mechanism to reduce inbreeding in grandchildren will depend upon the genetic basis of inbreeding depression in natural, inbreeding populations.
Keywords: deleterious recessive, inbreeding, overdominance, polyandry, modelling, multiple mating
1. Introduction
Across species, it is much more common to find females mating with more than one male than to find strict monogamy (taxonomic reviews in Birkhead & Møller 1998). This pattern demands an explanation because polyandry has such broad implications, affecting mate choice, mate competition, sexual conflict, speciation and a host of other issues (Bretman & Tregenza 2005). The puzzle is that females ought to be able to get all the sperm they need for a lifetime's reproduction from a single mating, so why do they incur the inevitable costs associated with mating with multiple males? A number of explanations have been proposed based on the idea that costs of mating are more than balanced by direct benefits to the female or genetic benefits to their offspring. Direct benefits include nuptial gifts provided at mating (Vahed 1998), future paternal care (Kempenaers 1993; Ihara 2002) and reduced male harassment (Thornhill & Alcock 1983). Genetic benefits can be divided into three classes: (i) polyandrous females are able to bias paternity towards males with genes that confer higher fitness or genes that are more compatible with the female's genome (e.g. Thornhill & Alcock 1983; Madsen et al. 1992; Zeh & Zeh 1996); (ii) polyandry is a means of bet-hedging against variation in sire quality or compatibility (e.g. Yasui 1998); and (iii) offspring live together and benefit from increased genetic diversity due to niche separation or disease resistance (e.g. Tooby 1982; Robinson 1992).
Here we propose a fourth class of genetic benefit that may explain polyandry in species that are subject to periodic episodes of high inbreeding risk. The basis of our proposal is that polyandrous females have offspring that are half siblings rather than full siblings. Hence, if a female's offspring are likely to mate with one another then polyandry may be advantageous because it reduces the degree of inbreeding in a female's grandchildren. This does not require any paternity biasing by females who are assumed to simply mate and fertilize their eggs at random.
This new class of explanation differs fundamentally from previous hypotheses regarding a role for inbreeding in explaining polyandry (Stockley et al. 1993), which fall into two classes. The paternity biasing hypothesis is based on the potential for polyandrous females to mate with both related and unrelated males, but to either invest more in their outbred offspring (Stockley et al. 1993) or preferentially fertilize their eggs with sperm from the unrelated males (Stockley 1997; Tregenza & Wedell 2002; Bretman et al. 2004). If females are capable of either of these behaviours then polyandry could be selected because it provides an opportunity for females to bias investment. The reduced variance hypothesis is based on the fact that if there is a risk of mating with a relative, polyandrous females will have the same arithmetic mean level of inbreeding in their offspring as do monandrous females but will experience lower between-brood variance. Yasui (1998) has shown that there are benefits of this type of genetic bet-hedging but that they are likely to be vanishingly small. Hosken & Blanckenhorn (1999) have pointed out that if the relationship between genetic diversity within a brood and mean fitness is exponential then it could also benefit females to mate polyandrously, but such relationships seem unlikely and have never been documented. Our proposal is distinct from previous inbreeding–polyandry hypotheses because it does not require paternity or investment bias, or accelerating benefits of brood diversity, but stems instead from reductions in the level of inbreeding in a female's grand-offspring, not her offspring.
Situations where females are exposed to periodic inbreeding are likely to occur in many multivoltine invertebrates with populations that are structured through the existence of discrete resource patches such as food plants or parasite hosts. In these situations, females may leave crowded resource patches having mated to several males and go in search of an empty patch. Their offspring will then be restricted to siblings as potential mates until a second female finds the patch and produces offspring. A specific example of this scenario might be an insect such as a flour beetle (Tribolium sp.) or other stored product pest whose ecology consists of adult females leaving a large established population to search for new habitat patches (such as an uninfested sack of flour).
Invertebrates living in seasonal environments may also experience episodic variation in inbreeding risk. For instance, many herbivorous insects and their predators have high host specificity and population sizes which increase over a few generations from spring to late summer. In some species males die at the onset of winter and mated females overwinter as adults. Examples include predatory bugs Orius sauteri and Orius minutus (Ito & Nakata 1998), several species of aphidophagous syrphids (hoverflies; Schneider 1969) and the mosquitoes Anopheles maculipennis and Culex pipiens (Danilevsky 1966). In species where females mate during periods of high population density and then overwinter and lay eggs the next spring, it seems likely that inbreeding risk may be high initially and decline as the populations expand.
There are also a range of social and eusocial species with ecologies that create the potential for cyclical inbreeding and outbreeding; for instance, in some termite species including Mastotermes darwiniensis, and several species of the genus Reticulitermes, females found a colony with a single male but presumably have the opportunity to mate with more than one unrelated male. As the colony grows it expands and can bud off smaller foraging colonies which may become isolated from the main nest. When this happens, previously non-reproductive workers develop into secondary sexuals but their only options as mates are their full siblings (Thorne et al. 1999). These new colonies then develop to produce sexuals which leave the nest and have the opportunity to outbreed once more. In social spiders, a single female frequently founds a new colony which may then persist for several generations. Molecular population genetic data indicate that within colonies females mate with their brothers (Roeloffs & Riechert 1988; Smith & Hagen 1996; Evans & Goodisman 2002), but evidence for polyandry in some female nest founders and in laboratory studies indicates that new colonies may be founded by polyandrous females who have had the opportunity to mate with males that are not their immediate relatives (Evans & Goodisman 2002).
Here we use mathematical models to investigate genetic benefits of polyandry in a species where there is a significant level of breeding between half and full siblings. We begin by developing a heuristic model to estimate the genetic benefits of polyandry in a population subject to inbreeding depression. We then consider two explicit genetic mechanisms leading to costs of inbreeding: deleterious recessive traits and overdominance. We track the prevalence of different genotypes at one or more loci, and use this to calculate the level of inbreeding depression that evolves, and the rate at which a polyandrous phenotype might invade.
2. The model
We consider a diploid species with non-overlapping generations. There are L unlinked loci at each of which there can be two alleles A and B. We assume that the probability that an individual reaches sexual maturity is multiplied by a factor x, 1 or y for each locus whose genotype is respectively AA, AB or BB. We are interested in two specific cases: (i) deleterious recessive y=1, x<1 and (ii) overdominance y=x. We consider all cases from lethal (x=0) to slightly deleterious (x close to 1). We assume that A alleles mutate into B alleles, and vice versa, at a rate μ per generation.
Each female mates with a fixed number of males and produces a brood; the phenotype determining the number m of mates is inherited by all female offspring (a more complex inheritance mechanism would not affect our conclusions regarding stable strategies). We assume that there are alternating generations of inbreeding and outbreeding. At even generations, females choose their mates at random from the population as a whole; at odd generations, females may only mate with (randomly chosen) males from the same brood. This model captures qualitatively the phenomenon of increased breeding between siblings, while the outbreeding generation allows for a mutant phenotype to invade the wild-type population.
We assume that the fecundity is high so that broods contain large numbers of individuals. We assume, however, that the population size is kept constant by a limiting process which occurs just before the outbreeding generation. The case of a large population can therefore be simplified by considering only the average numbers of individuals of each genotype. The details of the mathematical analysis for infinite population are to be found in appendix 1 in the electronic supplementary material. The results for finite population size, where broods vary stochastically in size and composition, have been obtained by individual-based simulation.
3. Heuristic argument
Let us first develop a simple argument to estimate the potential genetic benefits of polyandry in a population subject to inbreeding depression. Inbreeding depression is usually defined as δ=(Wo−Ws)/Wo, Ws being the relative fitness of inbred and Wo the fitness of outbred individuals (Keller & Waller 2002). We are specifically interested in within-population inbreeding depression, i.e. the outbreeding individuals pick mates at random from the same population.
Assume deleterious, recessive traits at a number of loci, and that reproductive success is a product of factors from each locus. Most deleterious alleles will be at heterozygous loci and inbreeding depression will primarily be caused by loci where both alleles are identical by descent. The number of such loci will be proportional to the relatedness r of the parents so that the number of offspring is proportional to (Ws/Wo)r=(1−δ)r. If we take Ws to refer to matings between genetically identical individuals, then full-sib matings reduce the fitness by (1−δ)1/2, and half-sib matings by (1−δ)1/4, relative to outbreeding individuals.
In our model, at even generations, a female breeds with unrelated males, hence the size of her brood does not depend on the number of mates she took. However, at odd generations, all mates are chosen from the same brood. Hence, if she takes one mate then these matings are between full sibs, whereas if she takes two then half are between half sibs. Inbreeding therefore reduces the number of grandchildren by in the first case and by in the second. Since these benefits are accrued over two generations, the relative fitness of ‘biandry’ over monandry is the square root of this ratio. The relative fitness benefits are therefore . When δ is small, this gives approximately
| (3.1) |
i.e. the fitness benefits are only one-sixteenth of the inbreeding depression in the population. For other types of genetic costs to inbreeding, the full formula for (W2−W1)/W1 might be different, but we expect the result (3.1) to be reasonably robust because benefits are due to one-half of the breeding events being between individuals of relatedness one-quarter rather than one-half, and these benefits are realized over two generations .
More generally, if a female mates with m males, then her progeny have fitness . The relative fitness benefit of taking m mates rather than m−1 mates would then be
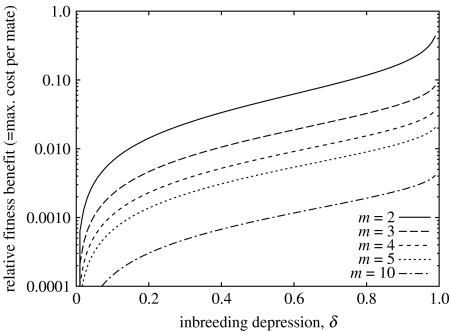
| (3.2) |
Equation (3.2) is plotted in figure 1 for different values of m. Inbreeding depression is often found to be in the region of δ≈0.2, even for populations with a significant amount of inbreeding (Byers & Waller 1999; Keller & Waller 2002; Boakes et al. 2007). This would imply a fitness benefit of 1.4% to the biandry. This reduces as m increases, being 0.46% at m=3, 0.23% at m=4 and 0.021% at m=10.
Figure 1.
Expected fitness benefits of choosing m rather than m−1 mates, for different levels of inbreeding depression δ.
Suppose now that there is a fitness cost κ for each mate the female takes, i.e. the fitness of a female taking m+1 mates is reduced by a factor (1−κ) relative to one taking m. In order for m mates to be favoured over m−1, the cost κ must be less than the relative fitness benefit in equation (3.2). Polyandry is therefore only favoured when costs per mating are of the order of a few per cent or below.
4. Deleterious recessive traits
Here we consider the case where costs of inbreeding are due to deleterious recessive alleles, i.e. y=1 and 0≤x<1. Deleterious alleles are lost from the population through selection, so if the mutation rate μ is small these alleles will be rare. We use this as the basis for an approximation method to calculate the equilibrium properties of this system, the details of which are given in appendix 1.3 in the electronic supplementary material. We find that, in an infinite monandrous population, the frequency of the deleterious allele at odd generations is
| (4.1) |
where the notation ‘O(μ2)’ means ‘terms of order μ2 or smaller’. The number of loci with genotype AA, AB and BB is multinomially distributed with means
For a random mating population, we would expect p≈μ1/2. Therefore, inbreeding purges deleterious alleles so that their prevalence is lower by a factor approximately μ1/2 relative to the random mating case. Our approximation breaks down in the limit where the per-generation cost 1−x of the deleterious allele is comparable to the mutation rate μ, where equation (4.1) predicts that p approaches unity. This limit is explored in more detail in appendix 1.3 in the electronic supplementary material.
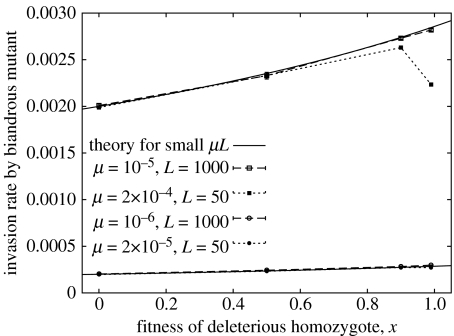
Now consider the case of a rare biandrous mutant in an infinite wild-type monandrous population. The invasion rate r is the relative difference in population growth per generation R of the mutant and wild-type, r=(Rmut/Rwild)−1. Results from simulations of an effectively infinite wild-type population are plotted in figure 2 for four combinations of μ and L, and 0<x<1. We show in appendix 1.3 in the electronic supplementary material that, for infinite N and small μL, the invasion rate is
| (4.2) |
We can see in figure 2 that this analytical prediction (displayed as a solid line) is a good approximation to the simulation results when μ is small, but breaks down for larger μ when x is close to 1.
Figure 2.
Invasion rates for a biandrous mutant into an infinite monandrous population. The solid lines show the theoretical prediction , valid for small μL. The upper data have μL=10−2 and the lower data have μL=10−3; open symbols and dashed lines have L=1000 recessive loci, filled symbols and dotted lines have L=50 recessive loci.
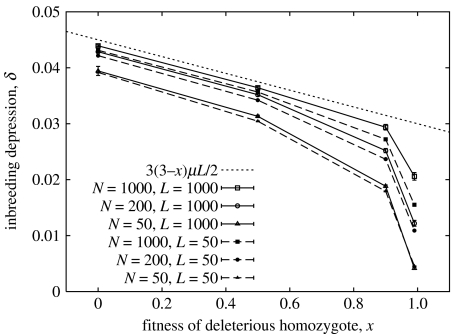
Since the invasion rate is the relative fitness advantage of the biandrous phenotype, we need r to be greater than the fitness cost κ per mate in order for polyandry to be favoured. The values of μ and L in figure 2 are chosen to be at the high end of a biologically plausible range, but we find that, since the invasion rates are very small, polyandry can only evolve if the cost per mate is correspondingly small. This can be understood in terms of the level of inbreeding depression in the population, which is illustrated in figure 3 for the largest value of μL that was used in figure 2. If an individual breeds with another that is genetically identical at a given locus, then its fitness is multiplied by x, (3+x)/4 or 1 depending on whether its genotype is AA, AB or BB. On the other hand, an outbreeding individual has probability p of receiving an A allele at that locus from each parent, so has fitness 1+O(p2L). We show in appendix 1.3 in the electronic supplementary material that the inbreeding depression in an infinite population is therefore
| (4.3) |
which is shown as a dotted line in figure 3. Note that the heuristic argument in §3 does not calculate the invasion rate exactly, as shown by the fact that the curves in figures 2 and 3 do not have the same shape. The reason for this is that the genotype frequencies in the invading population differ from those in the wild-type population, as discussed in appendix 1.3 in the electronic supplementary material. The true invasion rate in equation (4.2) differs from the approximation using equations (3.1) and (4.3) by a factor 64/(3(10−3x)(3−x)), which varies between 0.71 and 1.52 for 0<x<1.
Figure 3.
Inbreeding depression for μL=10−2, as a function of fitness x of the deleterious homozygote, for finite population size N. The dotted line shows the analytical prediction δ=(3(3−x)/2)μL valid for N=∞ and small μL. Filled symbols and dashed lines have L=50 recessive loci, mutation rate μ=2×10−4; open symbols and solid lines have L=1000 loci and μ=10−5.
Figure 3 also shows simulation results for the inbreeding depression in finite populations, to show how this deviates from the infinite-N result in equation (4.3). We see that the analytical approximation works well for small x and large N, but breaks down when x is close to 1, especially for smaller values of N. This is partly explained by the breakdown of equation (4.1) for finite μ and small 1−x, but a further reason is that deleterious alleles can become fixed in the population when their effect (1−x) is small and N is finite (appendix 3.1 in the electronic supplementary material). Once fixed, these alleles no longer contribute to within-population inbreeding depression and the effective number of loci L is reduced. Consequently, irrespective of the population size, inbreeding depression due to deleterious recessive alleles depends on sufficiently large values of μL.
5. Overdominant traits
Another possible cause of inbreeding depression is overdominance—higher fitness of heterozygotes. Here, we consider the case of L symmetrically overdominant loci, where either homozygote AA or BB has fitness x<1 relative to the heterozygote AB. Since selection favours polymorphism, both the alleles tend to be maintained at a high abundance even in the absence of mutation. In this section, we assume a low mutation rate that nevertheless prevents fixation of either allele at any locus (see appendix 3.2 in the electronic supplementary material). For an infinite monandrous population, loci are statistically independent (appendix 2.1 in the electronic supplementary material), and the frequency of each genotype is readily obtained by numerical solution of the recurrence equations (appendix 1.4 in the electronic supplementary material). Inbreeding depression in an infinite population is shown in appendix 1.4 in the electronic supplementary material to be
| (5.1) |
Biologically realistic levels of inbreeding depression of the order δ≈0.2 can be achieved in this model with x≈0.91, 0.95 and 0.98 for L=10, 20 and 50, respectively. We therefore expect that polyandry can evolve for parameters in this range, provided each mating incurs a fitness cost κ of less than 1 or 2%. A more detailed discussion of this model, including the existence of parameter regimes where mixed strategies can evolve, can be found in appendix 1.4 in the electronic supplementary material.
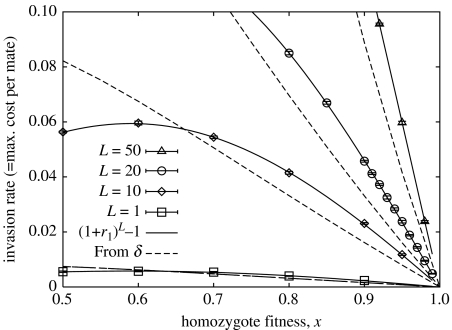
The invasion rate of a rare biandrous mutant, measured from stochastic simulations, is illustrated in figure 4. The invasion rate r1 for the case L=1 can be straightforwardly computed by iteration of the recurrence equations, as shown in appendix 1.4 in the electronic supplementary material. It turns out that the loci are not strictly independent in the biandrous case (appendix 2.2 in the electronic supplementary material), but nevertheless the invasion rate for general L is apparently given to a good accuracy by r(L)=(1+r1)L−1, as shown by the solid lines in figure 4. Dashed lines show the predicted invasion rate from the heuristic argument, based on equations (3.1) and (5.1). Again, we see that this argument is not exact, though its predictions are comparable to the true value.
Figure 4.
The invasion rate of a rare biandrous mutant in a monandrous wild-type population given L overdominant loci. This should equate to the largest fitness cost per mating event for which biandry is favoured over monandry. Points, with 1 s.d. error bars: based on simulations of a wild-type population of size 10 000, with a small population of mutant females. Solid line: [(1+r1)L−1], where r1 is the relative growth rate for the single locus case. Dashed line: the approximate prediction [(1/2){(1−δ)−1/4+1}]1/2−1 calculated from the inbreeding depression δ.
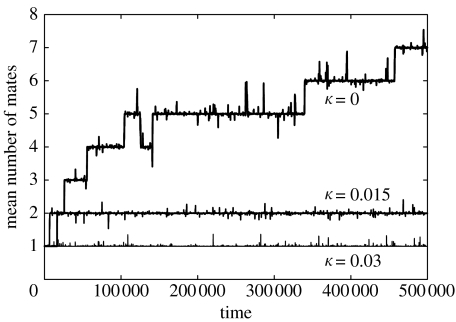
If the female incurs a fitness cost κ for each mate she takes, we expect that polyandry will still be the favoured strategy provided κ is smaller than the invasion rate r that would occur if mating were not costly. In figure 5, we illustrate the evolution of polyandry in a finite population of N=1000 individuals, for 20 loci with x=0.95. Each female has a maternally inherited polyandry phenotype which determines the number of mates she takes, and mutates by ±1 with a probability 10−5 per generation. The y-axis plots the population average of the polyandry phenotype. If there are no costs to mating (κ=0) then the population becomes increasingly polyandrous, and biandry is found to be the stable strategy when costs are finite (κ=0.015) but not too large (κ=0.03).
Figure 5.
Evolution of polyandry for different costs per mating κ. The population has N=1000 individuals with L=20 overdominant loci, with homozygote fitness x=0.95, and polymorphism at these loci is maintained by mutations between alleles at a rate 10−6 per allele per generation. The polyandry phenotype is maternally inherited and mutates by ±1 at a rate 10−5 per generation.
6. Discussion
Our aim has been to propose a novel potential adaptive benefit of polyandry and to examine the magnitude of the benefit conferred by a ‘polyandry gene’ with explicit assumptions about inbreeding risk and the genetics of inbreeding depression. We have shown that polyandry can indeed give genetic fitness benefits in populations with some level of inbreeding depression δ. However, these benefits are of the order of approximately δ/16; hence, if δ=0.2, polyandry will not be favoured if the cost per mate exceeds 1 or 2%. Empirical evidence (Thornhill 1993; Charlesworth & Charlesworth 1999) suggests that costs of inbreeding are primarily due to the expression of deleterious recessive alleles. As would be expected, in our model, inbreeding tends to purge deleterious recessive alleles and hence inbreeding depression and the corresponding genetic benefits due to polyandry are low. We find that polyandry is only favoured if fitness costs per mating are less than 2μL/(10−3x). For high values such as μ=10−5 and L=1000, we would need κ<0.0029 for polyandry to be favoured by this mechanism. The result is relatively insensitive to whether the trait is lethal (x=0) or mildly deleterious (x close to 1) because the less efficient purging of alleles in the latter case is counterbalanced by their smaller effect. If the trait were only partially rather than completely recessive, we expect purging to be even more efficient, and inbreeding depression to be still lower.
A major unresolved question in the study of inbreeding and the maintenance of genetic variation is how significant inbreeding depression persists within natural inbreeding populations (Lande & Schemske 1985; Charlesworth & Charlesworth 1987; Husband & Schemske 1996). In addition to purging, slightly deleterious mutations are subject to random fixation in finite populations, and the effective population size is reduced by inbreeding and linkage between loci (Keller & Waller 2002), all of which will tend to reduce the costs of inbreeding relative to random mating. One potential explanation is that a significant proportion of inbreeding depression is caused by overdominant loci, which are not subject to purging. Overdominance has been much debated as an explanation for inbreeding depression (Charlesworth & Charlesworth 1987) and theory suggests that the assumption made in our model, that the relative fitness of both homozygotes are very similar, may allow maintenance of polymorphisms that will otherwise tend to be lost in inbreeding populations (Nagylaki 1976). Moreover, in our model, we find that significant inbreeding depression is maintained even when overdominance is not symmetric. Ferreira & Amos (2006) have recently found evidence for 12–25 overdominant loci in Drosophila melanogaster. We find an inbreeding depression of for 10–20 overdominant loci with 5–9% heterozygote advantage and a range of values for x and y where the time to fixation in population of 1000 individuals is more than 106 generations (appendix 3.2 in the electronic supplementary material), suggesting that a mutation rate of 10−6 per generation would be sufficient to maintain polymorphism. Under these circumstances, polyandry can evolve when the costs of taking an additional mate are less than 1–2% of total fitness, a level likely to be greatly in excess of the cost of a single mating for most species.
Our model is primarily heuristic since it assumes alternating generations of inbreeding and outbreeding. This is likely to be rare in nature, although as we point out above there are a number of invertebrate groups that are likely to experience cyclical changes in inbreeding risk across generations. Our argument can be extended to a situation where there is some inbreeding at all generations. If a fraction h of all matings is between members of the same brood, the fitness benefit of biandry is predicted to be (the alternating-generations model corresponds to h=1/2 as there is full inbreeding every two generations). This suggests that higher levels of inbreeding may be more likely to favour polyandry, though this may be offset by greater purging of deleterious alleles and hence lower inbreeding depression. A final analysis of the importance of polyandry in reducing inbreeding in grandchildren will depend upon progress in understanding the more fundamental issues of the purging of deleterious recessives and the maintenance of inbreeding depression in inbred populations.
Acknowledgments
We thank T. Evans and T. Bilde for their advice on natural history, Y. Wong and S. Goodman for several useful discussions, and D. Hosken, J. Hodgson and two anonymous referees for their constructive comments on an earlier version of this manuscript. T.T. is funded by a Royal Society Fellowship. This work was partly supported by the Leverhulme Trust and the European Social Fund.
Supplementary Material
Detailed mathematical analysis of the model used in the paper
References
- Birkhead T.R, Møller A.P. Academic Press; London, UK: 1998. Sperm competition and sexual selection. [Google Scholar]
- Boakes E.H, Wang J, Amos W. An investigation of inbreeding depression and purging in captive pedigreed populations. Heredity. 2007;98:172–182. doi: 10.1038/sj.hdy.6800923. doi:10.1038/sj.hdy.6800923 [DOI] [PubMed] [Google Scholar]
- Bretman A, Tregenza T. Measuring polyandry in wild populations: a case study using promiscuous crickets. Mol. Ecol. 2005;14:2169–2179. doi: 10.1111/j.1365-294X.2005.02556.x. doi:10.1111/j.1365-294X.2005.02556.x [DOI] [PubMed] [Google Scholar]
- Bretman A, Wedell N, Tregenza T. Molecular evidence of post-copulatory inbreeding avoidance in the field cricket Gryllus bimaculatus. Proc. R. Soc. B. 2004;271:159–164. doi: 10.1098/rspb.2003.2563. doi:10.1098/rspb.2003.2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers D.L, Waller D.M. Do plant populations purge their genetic load? Effects of population size and mating history on inbreeding depression. Annu. Rev. Ecol. Syst. 1999;30:479–513. doi:10.1146/annurev.ecolsys.30.1.479 [Google Scholar]
- Charlesworth B, Charlesworth D. The genetic basis of inbreeding depression. Genet. Res. 1999;74:329–340. doi: 10.1017/s0016672399004152. doi:10.1017/S0016672399004152 [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Syst. 1987;18:237–268. doi:10.1146/annurev.es.18.110187.001321 [Google Scholar]
- Danilevsky A.S. Tokyo University Press; Tokyo, Japan: 1966. Photoperiodism and seasonal development of insects. [Google Scholar]
- Evans T.A, Goodisman M.A.D. Nestmate relatedness and population genetic structure of the Australian social crab spider Diaea ergandros (Araneae: Thomisidae) Mol. Ecol. 2002;11:2307–2316. doi: 10.1046/j.1365-294x.2002.01623.x. doi:10.1046/j.1365-294X.2002.01623.x [DOI] [PubMed] [Google Scholar]
- Ferreira A´.G.A, Amos W. Inbreeding depression and multiple regions showing heterozygote advantage in Drosophila melanogaster exposed to stress. Mol. Ecol. 2006;15:3885–3893. doi: 10.1111/j.1365-294X.2006.03093.x. doi:10.1111/j.1365-294X.2006.03093.x [DOI] [PubMed] [Google Scholar]
- Hosken D.J, Blanckenhorn W.U. Female multiple mating, inbreeding avoidance, and fitness: it is not only the magnitude of costs and benefits that counts. Behav. Ecol. 1999;10:462–464. doi:10.1093/beheco/10.4.462 [Google Scholar]
- Husband B.C, Schemske D.W. Evolution of the magnitude and timing of inbreeding depression in plants. Evolution. 1996;50:54–70. doi: 10.1111/j.1558-5646.1996.tb04472.x. doi:10.2307/2410780 [DOI] [PubMed] [Google Scholar]
- Ihara Y. A model for evolution of male parental care and female multiple mating. Am. Nat. 2002;160:235–244. doi: 10.1086/341019. doi:10.1086/341019 [DOI] [PubMed] [Google Scholar]
- Ito K, Nakata T. Diapause and survival in winter in two species of predatory bugs. Orius sauteri and O. minutus. Entomol. Exp. Appl. 1998;89:271–276. doi:10.1023/A:1003507708633 [Google Scholar]
- Keller L.F, Waller D.M. Inbreeding effects in wild populations. Trends Ecol. Evol. 2002;17:230–241. doi:10.1016/S0169-5347(02)02489-8 [Google Scholar]
- Kempenaers B. A case of polyandry in the blue-tit—female extra-pair behavior results in extra male help. Ornis Scand. 1993;24:246–249. doi:10.2307/3676741 [Google Scholar]
- Lande R, Schemske D.W. The evolution of self-fertilization and inbreeding depression in plants 1. Genetic models. Evolution. 1985;39:24–40. doi: 10.1111/j.1558-5646.1985.tb04077.x. doi:10.2307/2408514 [DOI] [PubMed] [Google Scholar]
- Madsen T, Shine R, Loman J, Hakansson T. Why do female adders copulate so frequently? Nature. 1992;355:440–441. doi:10.1038/355440a0 [Google Scholar]
- Nagylaki T. A model for the evolution of self-fertilization and vegetative reproduction. J. Theor. Biol. 1976;58:55–58. doi: 10.1016/0022-5193(76)90138-7. doi:10.1016/0022-5193(76)90138-7 [DOI] [PubMed] [Google Scholar]
- Robinson G.E. Regulation of division-of-labor in insect societies. Annu. Rev. Entomol. 1992;37:637–665. doi: 10.1146/annurev.en.37.010192.003225. doi:10.1146/annurev.en.37.010192.003225 [DOI] [PubMed] [Google Scholar]
- Roeloffs R, Riechert S.E. Dispersal and population genetic structure of the cooperative spider, Agelena consociata, in West African rainforest. Evolution. 1988;42:173–183. doi: 10.1111/j.1558-5646.1988.tb04117.x. doi:10.2307/2409125 [DOI] [PubMed] [Google Scholar]
- Schneider F. Bionomics and physiology of aphidophagous Syrphidae. Annu. Rev. Entomol. 1969;14:103–124. doi:10.1146/annurev.en.14.010169.000535 [Google Scholar]
- Smith D.R, Hagen R.H. Population structure and interdemic selection in the cooperative spider Anelosimus eximius (Araneae: Theridiidae) J. Evol. Biol. 1996;9:589–608. doi:10.1046/j.1420-9101.1996.9050589.x [Google Scholar]
- Stockley P. No evidence of sperm selection by female common shrews. Proc. R. Soc. B. 1997;264:1497–1500. doi: 10.1098/rspb.1997.0207. doi:10.1098/rspb.1997.0207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockley P, Searle J.B, Macdonald D.W, Jones C.S. Female multiple mating behaviour in the common shrew as a strategy to reduce inbreeding. Proc. R. Soc. B. 1993;254:173–179. doi: 10.1098/rspb.1993.0143. doi:10.1098/rspb.1993.0143 [DOI] [PubMed] [Google Scholar]
- Thorne B.L, Traniello J.F.A, Adams E.S, Bulmer M. Reproductive dynamics and colony structure of subterranean termites of the genus Reticulitermes (Isoptera Rhinotermitidae): a review of the evidence from behavioral, ecological, and genetic studies. Ethol. Ecol. Evol. 1999;11:149–169. [Google Scholar]
- Thornhill N.W. University of Chicago Press; Chicago, IL: 1993. The natural history of inbreeding and outbreeding: theoretical and empirical perspectives. [Google Scholar]
- Thornhill R, Alcock J. Harvard University Press; Cambridge, MA: 1983. The evolution of insect mating systems. [Google Scholar]
- Tooby J. Pathogens, polymorphism, and the evolution of sex. J. Theor. Biol. 1982;97:557–576. doi: 10.1016/0022-5193(82)90358-7. doi:10.1016/0022-5193(82)90358-7 [DOI] [PubMed] [Google Scholar]
- Tregenza T, Wedell N. Polyandrous females avoid costs of inbreeding. Nature. 2002;415:71–73. doi: 10.1038/415071a. doi:10.1038/415071a [DOI] [PubMed] [Google Scholar]
- Vahed K. The function of nuptial feeding in insects: a review of empirical studies. Biol. Rev. Camb. Philos. Soc. 1998;73:43–78. doi:10.1017/S0006323197005112 [Google Scholar]
- Yasui Y. The ‘genetic benefits’ of female multiple mating reconsidered. Trends Ecol. Evol. 1998;13:246–250. doi: 10.1016/s0169-5347(98)01383-4. doi:10.1016/S0169-5347(98)01383-4 [DOI] [PubMed] [Google Scholar]
- Zeh J.A, Zeh D.W. The evolution of polyandry I: intragenomic conflict and genetic incompatibility. Proc. R. Soc. B. 1996;263:1711–1717. doi:10.1098/rspb.1996.0250 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed mathematical analysis of the model used in the paper