Summary
Chromosome alignment and segregation during cell division rely on a highly ordered bipolar microtubule array called the mitotic spindle. The organization of microtubules into bipolar spindles with focused poles during mitosis requires numerous microtubule-associated proteins including both motor and non-motor proteins. Non-motor microtubule-associated proteins display extraordinary diversity in how they contribute to mitotic spindle organization. These mechanisms include regulation of microtubule nucleation and organization, direct and indirect influences on motor function, and control of cell cycle progression. Furthermore, many non-motor spindle proteins display altered expression in cancer cells emphasizing their important roles in cell proliferation.
Introduction
Accurate chromosome segregation during cell division is essential for cell viability. The mitotic spindle, a molecular machine composed primarily of microtubules, is responsible for chromosome segregation. Microtubules within mitotic spindles are highly ordered with their minus ends focused at spindle poles through the combined actions of centrosome-based microtubule nucleation and microtubule motor-based focusing. Dynamic microtubule plus ends extend to the cell cortex to position the spindle and to chromosomes where they mediate chromosome movement by binding to specialized centromeric structures called kinetochores.
The organization of microtubules into the highly ordered bipolar array of the mitotic spindle depends on the activities of numerous motor and non-motor microtubule-associated proteins. Motor proteins have received significant attention because they generate force on microtubules during spindle formation and are potential drug targets for cancer therapy. Indeed, some models for spindle assembly and function are based exclusively on the interactions of different motors with microtubules [1]. However, these models overlook the essential functional roles played by non-motor proteins in the organization and function of mitotic spindles. Non-motor proteins promote the formation and maintenance of mitotic spindles through diverse mechanisms including the nucleation and organization of microtubules, influence on motor function, and regulation of cell cycle control. This diversity may explain why the expression of non-motor spindle proteins is frequently altered in cancer cells. Here we summarize recent data on how the major non-motor spindle proteins contribute to the structural integrity of the mitotic spindle. We also discuss how non-motor spindle proteins participate in cell cycle regulation with implications on cancer.
Cross-linking influence on microtubule stabilization and organization
The salient features of major non-motor spindle proteins are summarized in Table 1. In general, these proteins are relatively large, and many are only expressed during G2/M phase of the cell cycle. Many of these proteins also display substantial phosphorylation during mitosis [2]. These proteins show diverse localization on mitotic spindles including centrosomes, spindle poles, spindle body, central spindle, and kinetochores (Figure 1) where they participate in both microtubule organization and nucleation. Complicating the understanding of these functions, many of these proteins cooperate or interact with each other in some activities.
Table 1.
| Protein Name | Full Name | Ortholog | MW (kDa) | Mitotic Localization | Activity |
|---|---|---|---|---|---|
| Astrin hMAP126 Spag5 DEEPEST | Aster associated protein | M.musculus (Spag5) | ∼135 | spindle poles, kinetochores of bioriented chromosomes | Crosslinks and stabilizes microtubules; Stabilizes cohesin (ref 17, 18) |
| HURP DLG7 | Hepatoma Up-Regulated Protein | C.elegans, D.melanogaster, M.musculus, S.cerevisiae, X.laevis | ∼95 | Kinetochore fibers, adjacent to kinetochores | Stabilizes kinetochore fibers; contributes to chromosome alignment (ref 10) |
| NuMA | NUclear Mitotic Apparatus protein | D.melanogaster (Mud, Asp1*), M.musculus, X.laevis | ∼215 | Spindle Poles | Required for the formation and maintenance of focused spindle poles; part of a complex that inhibits APC/C at spindle poles (ref 4, 40) |
| NuSAP | NUcleolar Spindle-Associated Protein | M.musculus, X.laevis | ∼55 | Central Spindle | Contributes to Nucleation, stabilization and bundling of microtubules near chromosomes; (ref 14, 15, 16) |
| PRC1 | Protein Regulator of Cytokinesis | C.elegans (SPD-1), D.melanogaster (Fascetto/Feo), M.musculus, S.cerevisae (Ase1), S.pombe (Ase1), X.laevis | ∼70 | Central spindle, spindle midbody | Cross-links antiparellel spindle midzone microtubules to stabilize the elongating spindle during anaphase (ref 11, 12) |
| RHAMM HMMR | Receptor of HyAluronon-Mediated Motility | M.musculus, X.laevis (XRHAMM) | ∼70 | Centrosomes, spindle poles, spindle midzone | Induces microtubule nucleation and stability at spindle poles; influences Cyclin B1 activity (ref 26, 28, 39) |
| TACC (1, 2, 3) | Transforming Acidic Coiled-Coil protein | C.elegans (TAC-1), D.melanogaster (D-TACC), M.musculus, X.laevis (Maskin) | ∼150 | Centrosomes, Spindle poles | Promotes microtubule nucleation and stabilization at spindle poles (ref 22, 23, 24, 25, 26) |
| TOGp | Tumor Over-expressed Gene | C.elegans (ZYG-9), D.melanogaster (Minispindles/Msps), M.musculus, S.cerevisae (Stu2), S.pombe (Dis1/Alp14), X.laevis (XMAP215/Dis1) | ∼215 | Centrosomes, Spindle poles | Promotes plus end microtubule dynamics; promotes centrosome and spindle pole stability (ref 19, 20, 21) |
| TPX2 | Targeting Protein for Xklp2 | C.elegans (TPXL-1), D.melanogaster (Asp1*), M.musculus, X.laevis | ∼95 | Spindle Poles, midbody | Cross-links microtubules at poles; promotes microtubule nucleation and maintenance of centrosome integrity; activates AurA (ref 5, 6, 35) |
Asp1 has been shown to localize to spindle poles where it has microtubule nucleation activities similar to that of TPX2 and crosslinking activity involved in spindle pole focusing similar to that of NuMA
Figure 1.
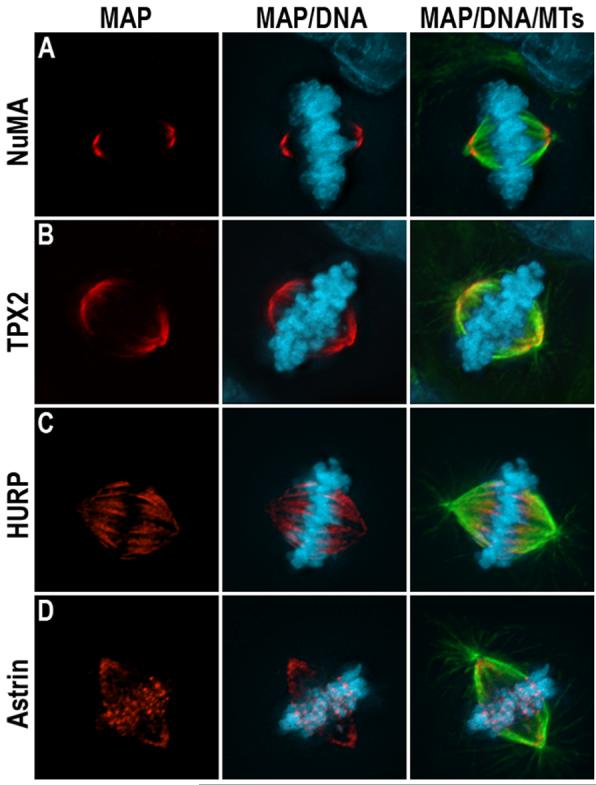
Localization of different non-motor microtubule associated proteins during mitosis in human cells. Microtubules (green), DNA (blue) and either NuMA (A), TPX2 (B), HURP (C), or astrin (D) are shown in red.
Loss of function studies combined with in vitro biochemical experiments demonstrate that many non-motor spindle proteins mechanically cross link microtubules to provide structural support to the mitotic spindle. For example, NuMA binds microtubules directly, and localizes to microtubule minus ends at spindle poles (Figure 1A) [3]. Perturbation of NuMA function leads to splaying of microtubule minus ends at spindle poles indicating that NuMA combines self-association and microtubule binding activities to mechanically cross link microtubules at spindle poles [4]. TPX2 also localizes to the spindle poles (Figure 1B) and binds and bundles microtubules directly. Perturbation of TPX2 function results in poorly focused spindle poles and decreased centrosomal integrity leading to multipolar spindles [5, 6]. The microtubule cross linking activity of NuMA is dominant to that of TPX2, but TPX2 becomes essential for spindle pole organization under conditions where NuMA function is perturbed [7]. HURP also binds microtubules directly, and localizes on the subset of spindle microtubules attached to kinetochores in a unique position adjacent to kinetochores (Figure 1C) [8, 9, 10]. Perturbation of HURP activity decreases the density of microtubules bound to kinetochores suggesting that it may promote kinetochore microtubule stability through microtubule cross linking activity [10]. Finally, PRC1 binds and bundles antiparallel microtubules in the central spindle where it is required for microtubule organization during anaphase/telophase [11, 12]. The strength of microtubule crosslinking formed by each protein and the influence this has on spindle structure appears varied. The turnover of NuMA at spindle poles is relatively slow [3], but that of TPX2 is reported to be relatively fast [13]. Turnover rates for other non-motor spindle proteins have not been reported, but represent an important goal because that will improve understanding of how those cross links contribute to spindle formation and maintenance.
Microtubule nucleation and stabilization
Many non-motor spindle proteins also control the density of microtubules within spindles by regulating microtubule nucleation and/or stability. In some cases, non-motor spindle proteins merely stabilize microtubules against depolymerization akin to conventional microtubule-associated proteins. For example, NuSAP interacts directly with both chromatin and microtubules and most likely promotes spindle formation by selectively stabilizing microtubules associated with chromatin [14, 15, 16]. Similarly, the density of microtubules bound to kinetochores in astrin-deficient cells is reduced [17]. Astrin localizes to kinetochores of bi-oriented chromosomes (Figure 1D) indicating that it may selectively stabilize microtubules bound to kinetochores [18].
Other non-motor spindle proteins contribute to the density of microtubules in spindles directly by regulating microtubule nucleation or polymerization. For example, TOGp (XMAP215) promotes the assembly of microtubules by ushering tubulin subunits into the growing microtubule end and antagonizing the depolymerizing activity of the kinesin-13 protein MCAK [19, 20, 21]. It is unknown if this activity accounts for the disruption of spindle pole and centrosome integrity in TOGp-deficient cells [21]. Furthermore, addition of excess quantities of TPX2 to frog egg extracts (in excess of importin alpha levels) induces microtubule nucleation suggesting that TPX2 can promote microtubule nucleation. This accounts for the reduced microtubule density of spindles in TPX2-deficient cells and/or extracts [5,6]. Whether it does so directly or through activation of other microtubule nucleating complexes is not known. Finally, TACC family members stabilize microtubules and contribute to spindle microtubule density [22, 23, 24, 25, 26]. Some TACC proteins interact with TOGp to promote the targeting of the drosophila TOGp homologue minispindles (Msps) to centrosomes in an Aurora A kinase-dependent manner [23, 27]. A mutant form of D-TACC that cannot be phosphorylated by Aurora A continues to interact with Msps but no longer localizes to centrosomes and microtubule minus ends. Cells expressing this mutant show significant reduction of astral microtubules due to loss of D-TACC/Msps stabilizing activities at centrosomes [23]. Finally, the acidic coiled coil protein RHAMM associates with the microtubule nucleating gamma-tubulin ring complex as well as with TPX2. These interactions permit RHAMM to directly induce microtubule nucleation and contribute to spindle microtubule density [26, 28].
Influences on microtubule motor function
Many non-motor proteins participate in spindle organization by interacting with microtubule motor proteins that exert force on spindle microtubules. In some circumstances, the interaction is direct and the non-motor protein controls the function of the motor protein. This phenomenon is exemplified by the recent demonstration of a heterodimeric complex between the budding yeast Kar3 motor and the non-motor protein Vik1 [29]. Similar direct interactions between motor and non-motor proteins have been documented in mitotic spindles in vertebrate cells. For example, TPX2 binds directly to kinesin XKLP2 and targets that motor to spindle poles where it can perform its mitotic activity [30]. Furthermore, the ability of the minus end-directed motor cytoplasmic dynein to focus microtubule minus ends at spindle poles depends on its association with its activating complex dynactin and the non-motor protein NuMA [4].
Other direct interactions between motor and non-motor proteins have the converse effect with motor activity determining the localization and function of the non-motor spindle protein. For example, whereas PRC1 has been shown to associate with a variety of motors including MKLP, MCAK, and CENP-E in the central spindle, kinesin Kif4 is required to move it to that location to provide the necessary microtubule cross links at anaphase onset [31, 32]. In addition, the minus end-directed kinesin Ncd (HSET) has been shown to drive the non-motor drosophila TOGp homologue minispindles to spindle poles where it participates in regulating spindle microtubule dynamics [27]. Finally, RHAMM is targeted to spindle poles through an interaction with dynein, where, utilizing its role in promoting microtubule nucleation it contributes to both the generation of microtubules and focusing of microtubule minus ends at spindle poles in the presence and absence of centrosomes [28].
In addition to these direct interactions between motor and non-motor spindle proteins, non-motor proteins can influence motor protein function indirectly. This relationship relies upon the ability of non-motor proteins to cross link spindle microtubules. Microtubule cross links generated by non-motor proteins create a load (static friction) that can oppose sliding between adjacent microtubules. If the cross links are sufficiently stable and numerous, they will create a load that exceeds the force exerted by the motor, thereby influencing the activity of the motor without directly contacting the motor. This idea has been borne out experimentally in two systems. First, in fission yeast, microtubule cross links generated by the non-motor Ase1 (PRC1) in the central spindle oppose the ability of the kinesin Klp2p to slide microtubules apart to support spindle bipolarity [33]. Second, in mammalian mitotic extracts, exaggerated motor activity can be overcome with additional microtubule cross linking activity contributed by NuMA [34]. The breadth of this indirect mode of influence of non-motor proteins on motors is currently unknown because of the dearth of studies examining both motors and non-motor contributions simultaneously. This indirect mode of regulation may be quite widespread or fairly rare if it requires motors and non-motor proteins with unique biophysical properties (motor: dwell time, force-velocity relationship, and processivity; non-motor: microtubule on/off kinetics and cross link stiffness).
Cell Cycle Regulation
Recent data are uncovering important mechanisms through which non-motor spindle proteins regulate cell cycle progression. The Aurora kinase family has important roles in regulating mitotic spindle assembly and accurate chromosome segregation. Aurora A is a key regulator of centrosome maturation and spindle assembly, and its activity and localization are regulated by non-motor spindle proteins. For example, TPX2 is a potent activator of Aurora A kinase activity [35] and may serve to locally activate the kinase within the spindle [35, 36, 37]. TACC proteins also interact with Aurora A, and that interaction helps Aurora A regulate microtubule nucleation and stabilization at centrosomes [25]. Similarly, PRC1 is required to localize Aurora A to the central spindle [11]. Cyclin B1 localization and, presumably, cdk1 activity are also associated with non-motor spindle proteins. For example, TOGp interacts directly with Cyclin B1 [38] and RHAMM induces cell cycle delay in G2/M phase by suppressing Cyclin B1 activity when it is either over-expressed or depleted [39].
Timely mitotic exit also requires the action of the non-motor spindle proteins astrin and NuMA. Cells deficient in astrin are delayed in progression through mitosis and frequently display multipolar spindles. However, after extended mitotic delay, sister chromatids begin to separate without overtly transitioning to anaphase. Separase appears to become activated in these cells indicating that astrin may participate in the regulation of Separase activation [17]. Other recent data shows that NuMA associates with the anaphase promoting complex/cyclosome (APC/C) inhibitor Emi1 in addition to dynactin and cytoplasmic dynein [40]. This complex appears to inhibit APC/C at spindle poles, thereby preventing spindle-associated Cyclin B degradation until all chromosomes are properly attached to spindle microtubules. NuMA’s primary function is to maintain spindle pole organization in response to strong forces generated by kinetochores [7] raising the possibility that NuMA may react to changes in force at the spindle pole in response to chromosome biorientation.
Cancer
Most non-motor spindle proteins have been reported as highly expressed in human tumors. Indeed, many non-motor spindle proteins were identified and originally named based on their expression profiles in cancer (HURP, Hepatoma upregulated protein; TACC, tumor-associated coiled-coil protein; ch-TOGp, colonic and hepatic tumor overexpressed gene). HURP, NuMA, PRC1, TACC, RHAMM and TPX2 are all highly expressed in some cancers [41, 42, 43, 44, 45, 46, 47]. In fact, TPX2 and PRC1 are proteins whose over-expression is most highly correlated with the cancer phenotype of chromosomal instability [47]. Also, the NuMA1 gene maps to one of the most frequently amplified chromosomal segments in cancer cells [42] and a unique allele shows strong genetic linkage to heritable breast cancer [48]. Further, both up-regulation and down-regulation of TACC family members has been linked to breast cancer, while TACC gene rearrangements are seen in multiple myelomas [43]. The correlation between expression of non-motor spindle proteins and cancer is beginning to be confirmed through direct experimentation. For example, over-expression of HURP in 393T cells causes enhanced cell growth under low serum conditions [41], and depletion of Prc1 has been shown to slow the growth of several breast cancer cell lines [49]. In addition, depletion of TPX2 reduces the survival of tumor cells expressing the activated K-Ras oncogene, but not cells expressing wild-type K-Ras [50].
Summary
Non-motor spindle proteins fulfill diverse functions during mitosis, and emerging evidence indicates that each protein plays multiple roles in mitosis, often combining both structural and regulatory functions. Such multi-tasking represents an experimental challenge to unequivocally deciphering the functional contribution that each non-motor spindle protein makes to spindle organization and function. However, these combined activities most likely hold the key toward understanding why non-motor spindle proteins are commonly over expressed in human tumors. Interestingly, motor proteins are not highly over expressed in tumors indicating that motors are present in concentrations greater than necessary for their cellular roles, and that non-motor spindle proteins are limiting for proper spindle formation. Questions remaining open relate to the determination of the kinetics of microtubule binding for each non-motor protein and how that binding is regulated, the identification of binding and or catalytic sites of action of each protein, and understanding the interrelationship between microtubule binding and cell cycle regulatory properties. Answering these questions will generate significant insight into spindle assembly mechanisms and will pave the way toward targeting these proteins for chemotherapy.
Acknowledgements
We apologize to those researchers whose primary work is not cited here as a consequence of editorial constraints on length. Work in the authors’ laboratory is funded by the National Institutes of Health (GM51542).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000;407(6800):41–7. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- 2•.Nousiainen M, Sillje HHW, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. PNAS. 2006;103(14):5391–96. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The authors using mass spectrometry to map phosphorylation sites on all major spindle-associated proteins.
- 3.Kisurina-Evgenieva O, Mack G, Du Q, Macara I, Khodjakov A, Compton DA. Multiple mechanisms regulate NuMA dynamics at spindle poles. J Cell Sci. 2004;117(26):6391–400. doi: 10.1242/jcs.01568. [DOI] [PubMed] [Google Scholar]
- 4.Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW. Formation of Spindle Poles by Dynein/Dynactin-dependent Transport of NuMA. J Cell Biol. 2000;149(4):851–62. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wittman T, Matthias W, Karsenti E, Vernos I. TPX2, A Novel Xenopus MAP involved in Spindle Pole Organization. J Cell Biol. 2000;149(7):1405–18. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrett S, Auer K, Compton DA, Kapoor TM. hTPX2 Is Required for Normal Spindle Morphology and Centrosome Integrity during Vertebrate Cell Division. Curr Biol. 2002;12:2055–59. doi: 10.1016/s0960-9822(02)01277-0. [DOI] [PubMed] [Google Scholar]
- 7••.Manning AL, Compton DA. Mechanisms of spindle-pole organization are influenced by kinetochore activity in mammalian cells. Curr Biol. 2007;17(3):260–5. doi: 10.1016/j.cub.2006.11.071. [DOI] [PubMed] [Google Scholar]
- The authors demonstrate that different microtubule cross linking proteins organize mitotic spindles, and the use of each depends on how much force is being exerted on spindle microtubules.
- 8.Sillje HH, Nagel S, Korner R, Nigg EA. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol. 2006;16(8):731–42. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 9.Koffa MD, Casanova CM, Santarella R, Kocher T, Wilm M, Mattaj IW. HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol. 2006;16(8):743–54. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 10.Wong J, Fang G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J Cell Biol. 2006;173(6):879–91. doi: 10.1083/jcb.200511132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamashita A, Sato M, Fujita A, Yamamoto M, Toda T. The roles of fission yeast ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol Biol Cell. 2005;16(3):1378–95. doi: 10.1091/mbc.E04-10-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu C, Lau E, Schwarzenbacher R, Bossy-Wetzel E, Jiang W. Spatiotemporal control of midzone formation by PRC1 in human cells. PNAS. 2006;103(16):6196–201. doi: 10.1073/pnas.0506926103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang P, Jacobson MK, Mitchison TJ. Poly (ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 14.Raemakers T, Ribbeck K, Beaudouin J, Annaert W, Van Camp M, Stockmans I, Smets N, Bouillon R, Ellenberg J, Carmeliet G. NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J Cell Biol. 2003;162(6):1017–29. doi: 10.1083/jcb.200302129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ribbeck K, Groen AC, Santarella R, Bohnsack MT, Raemakers T, Kocher T, Gentzel M, Gorlich D, Wilm M, Carmeliet G, et al. NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol Biol Cell. 2006;17(6):2646–60. doi: 10.1091/mbc.E05-12-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ribbeck K, Raemakers T, Carmeliet G, Mattaj I. A Role for NuSAP in Linking Microtubules to Mitotic Chromosomes. Curr Biol. 2007;17:230–6. doi: 10.1016/j.cub.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 17••.Thein KH, Kleylein-Sohn J, Nigg EA, Gruneberg U. Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. J Cell Biol. 2007;178:345–354. doi: 10.1083/jcb.200701163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The authors demonstrate that the non-motor spindle protein astrin participates in regulating cell cycle exit by controlling separase activity.
- 18.Mack GJ, Compton DA. Analysis of mitotic microtubule-associated proteins using mass spectrometry identifies astrin, a spindle-associated protein. PNAS. 2001;98(25):14434–39. doi: 10.1073/pnas.261371298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charrasse S, Schoeder M, Gauthier-Rouviere C, Ango F, Cassimeris L, Gard DL, Larroque C. The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J Cell Sci. 1998;111(1):1371–83. doi: 10.1242/jcs.111.10.1371. [DOI] [PubMed] [Google Scholar]
- 20.Kinoshita K, Arnal I, Desai A, Drechsel DN, Hyman AA. Reconstitution of physiological microtubule dynamics using purified components. Science. 2001;294(5545):1340–3. doi: 10.1126/science.1064629. [DOI] [PubMed] [Google Scholar]
- 21.Cassimeris L, Morabito J. TOGp, the Human Homolog of XMAP215/Dis1, Is Required for Centrosome Integrity, Spindle Pole Organization, and Bipolar Spindle Assembly. Mol Biol Cell. 2004;15:1580–90. doi: 10.1091/mbc.E03-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gergely F, Karlsson C, Still I, Cowell J, Kilmartin J, Raff JW. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. PNAS. 2000;97(26):14352–7. doi: 10.1073/pnas.97.26.14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barros TP, Kinoshita K, Hyman AA, Raff JW. Aurora A activates D-TACC-Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J Cell Biol. 2005;170(7):1039–46. doi: 10.1083/jcb.200504097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee MJ, Gergely F, Jeffers K, Peak-Chew SY, Raff JW. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behavior. Nat Cell Biol. 2001;3:643–649. doi: 10.1038/35083033. [DOI] [PubMed] [Google Scholar]
- 25.Peset I, Seiler J, Sardon T, Bejarano LA, Rybina S, Vernos I. Function and regulation of Maskin, a TACC family protein, in microtubule growth during mitosis. J Cell Biol. 2005;170(7):1057–66. doi: 10.1083/jcb.200504037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maxwell CA, Keats JJ, Crainie M, Sun X, Yen T, Shiyuba E, Hendzel M, Chan G, Pilarski LM. RHAMM is a Centrosomal Protein That Interacts with Dynein and Maintains Spindle Pole Stability. Mol Biol Cell. 2003;14:2262–76. doi: 10.1091/mbc.E02-07-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cullen CF, Ohkura H. Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat Cell Biol. 2001;3(7):637–42. doi: 10.1038/35083025. [DOI] [PubMed] [Google Scholar]
- 28.Groen AC, Cameron LA, Coughlin M, Miyamoto DT, Mitchison TJ, Ohi R. XRHAMM Functions in Ran-Dependent Microtubule Nucleation and Pole Formation during Anastral Spindle Assembly. Curr Biol. 2004;14:1801–11. doi: 10.1016/j.cub.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 29•••.Allingham JS, Sproul LR, Rayment I, Gilbert SP. Vik1 modulates microtubule-Kar3 interactions through a motor domain that lacks an active site. Cell. 2007;128(6):1161–72. doi: 10.1016/j.cell.2006.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The authors show that the motor activity of Kar3 is regulated by the direct physical association with the non-motor protein Vik1.
- 30.Wittmann T, Boleti H, Antony C, Karsenti E, Vernos I. Localization of the kinesin-like protein Xklp2 to spindle poles requires a leucine zipper, a microtubule-associated protein and dynein. J Cell Biol. 1998;143(3):673–85. doi: 10.1083/jcb.143.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurasawa Y, Earnshaw Y, Mochizuki Y, Dohmae N, Todokoro K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 2004;23(16):3237–48. doi: 10.1038/sj.emboj.7600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu C, Jiang W. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. PNAS. 2005;102(2):343–8. doi: 10.1073/pnas.0408438102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•••.Janson ME, Loughlin R, Loiodice I, Fu C, Brunner D, Nedelec FJ, Tran PT. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 2007;128(2):357–68. doi: 10.1016/j.cell.2006.12.030. [DOI] [PubMed] [Google Scholar]
- The authors show that microtubule cross links generated by Ase1 regulate the ability of the motor Klp2p to slide microtubules in spindles. This demonstrates how non-motor proteins can regulate motor function without directly binding to the motor.
- 34.Chakravarty A, Howard L, Compton DA. A mechanistic model for the organization of microtubule asters by motor and non-motor proteins in a mammalian mitotic extract. Mol Biol Cell. 2004;15(5):2116–32. doi: 10.1091/mbc.E03-08-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eyers PA, Maller JL. Regulation of Xenopus Aurora A activation by TPX2. J Biol Chem. 2004;279(10):9008–15. doi: 10.1074/jbc.M312424200. [DOI] [PubMed] [Google Scholar]
- 36.Kufer TA, Sillje HH, Korner R, Gruss OJ, Meraldi P, Nigg EA. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol. 2002;158(4):617–23. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLuca M, Lavia P, Guarguaglini G. A Functional Interplay Between Aurora-A, Plk1, and TPX2 at Spindle Poles. Cell Cycle. 2006;5(3):296–303. doi: 10.4161/cc.5.3.2392. [DOI] [PubMed] [Google Scholar]
- 38.Charrasse S, Lorca T, Doree M, Larroque C. The Xenopus XMAP215 human homologue TOG proteins interact with cyclin B1 to target p34cdc2 to microtubules during mitosis. Exp Cell Res. 2000;254(2):249–56. doi: 10.1006/excr.1999.4740. [DOI] [PubMed] [Google Scholar]
- 39.Mohaptra S, Yang X, Wright JA, Turley EA, Greenberg AH. Soluble hyaluronan receptor RHAMM induces mitotic arrest by suppressing Cdc2 and cyclin B1 expression. J Exp Med. 1996;183(4):113–8. doi: 10.1084/jem.183.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Ban KH, Torres JZ, Miller JJ, Mikhailov A, Machury MV, Tung JJ, Rieder CL, Jackson PK. The END network couples spindle pole assembly to inhibition of the anaphase-promoting complex/cyclosome in early mitosis. Dev Cell. 2007;13(1):29–42. doi: 10.1016/j.devcel.2007.04.017. [DOI] [PubMed] [Google Scholar]
- This article shows that the non-motor protein NuMA participates in regulating cell cycle exiting as part of a complex containing Emi1 that controls APC/C activity.
- 41.Tsou AP, Yang CW, Huang CY, Yu RC, Lee YC, Chang CW, Chen BR, Chung YF, Fann MJ, Chi CW, et al. Identification of a novel cell cycle regulated gene, HURP, overexpressed in human hepatocellular carcinoma. Oncogene. 2003;22(2):298–307. doi: 10.1038/sj.onc.1206129. [DOI] [PubMed] [Google Scholar]
- 42.Sun QY, Schatten H. Role of NuMA in vertebrate cells: review of an intriguing multifunctional protein. Front Biosci. 2006;11:1137–46. doi: 10.2741/1868. [DOI] [PubMed] [Google Scholar]
- 43.Gergely F. Centrosomal TACCtics. BioEssays. 2002;24:915–25. doi: 10.1002/bies.10162. [DOI] [PubMed] [Google Scholar]
- 44.Still IH, Vince P, Cowell JK. The third member of the transforming acidic coiled coil-containing gene family, TACC3, maps in 4p16, close to translocation breakpoints in multiple myeloma, and is upregulated in various cancer cell lines. Genomics. 1999;58(2):165–70. doi: 10.1006/geno.1999.5829. [DOI] [PubMed] [Google Scholar]
- 45.Maxwell CA, Rasmussen E, Zhan F, Keats JJ, Adamia S, Strachan E, Crainie M, Walker R, Belch A, Pilarski LM, et al. RHAMM expression and isoform balance predict aggressive disease and poor survival in multiple myeloma. Blood. 2004;104(4):1151–58. doi: 10.1182/blood-2003-11-4079. [DOI] [PubMed] [Google Scholar]
- 46.Maxwell CA, Keats JJ, Belch AR, Pilarski LM, Reiman T. Receptor for Hyaluronan-Mediated Moyility Correlates with Centrosome Abnormalities in Multiple Myeloma and Maintains Mitotic Integrity. Cancer Res. 2005;65(3):850–60. [PubMed] [Google Scholar]
- 47.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi A. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–48. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 48.Kammerer S, Roth RB, Hoyal CR, Reneland R, Marnellos G, Kiechle M, Schwarz-Boeger U, Griffiths LR, Ebner F, Rehbock J, et al. Association of the NuMA region on chromosome 11q13 with breast cancer susceptibility. PNAS. 2005;102(6):2004–9. doi: 10.1073/pnas.0409806102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimo A, Nishidate T, Ohta T, Fukada M, Makamura Y, Katagiri T. Elevated expression of protein regulator of cytokinesis 1, involved in the growth of breast cancer cells. Cancer Sci. 2007;98(2):174–81. doi: 10.1111/j.1349-7006.2006.00381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morgan-Lappe SE, Tucker LA, Huang X, Zhang Q, Sarthy AV, Zakula D, Vernetti L, Schurdak M, Wang J, Fesik SW. Identification of Ras-Related Nuclear Protein, Targeting Protein for Xenopus Kinesin-like Protein 2, and Stearoyl-CoA Desaturase 1 as Promising Cancer Targets from an RNAi-Based Screen. Cancer Res. 2007;67(9):4390–98. doi: 10.1158/0008-5472.CAN-06-4132. [DOI] [PubMed] [Google Scholar]


