Abstract
Protein C inhibitor (PCI) is a serpin with many roles in biology, including a dual role as pro- and anticoagulant in blood. The protease specificity and local function of PCI depend on its interaction with cofactors such as heparin-like glycosaminoglycans (GAGs) and thrombomodulin (TM). Both cofactors significantly increase the rate of thrombin inhibition, but GAGs serve to promote the anticoagulant activity of PCI, and TM promotes its procoagulant function. To gain insight into how PCI recognition of thrombin is aided by these cofactors, we determined a crystallographic structure of the Michaelis complex of PCI, thrombin, and heparin to 1.6 Å resolution. Thrombin interacts with PCI in an unusual fashion that depends on the length of PCI's reactive center loop (RCL) to align the heparin-binding sites of the two proteins. The principal exosite contact is engendered by movement of thrombin's 60-loop in response to the unique P2 Phe of PCI. This mechanism of communication between the active site of thrombin and its recognition exosite is previously uncharacterized and may relate to other thrombin substrate–cofactor interactions. The cofactor activity of heparin thus depends on the formation of a heparin-bridged Michaelis complex and substrate-induced exosite contacts. We also investigated the cofactor effect of TM, establishing that TM bridges PCI to thrombin through additional direct interactions. A model of the PCI–thrombin–TM complex was built and evaluated by mutagenesis and suggests distinct binding sites for heparin and TM on PCI. These data significantly improve our understanding of the cofactor-dependent roles of PCI in hemostasis.
Keywords: hemostasis, serpin, crystallography, thrombomodulin, exosite
Protein C inhibitor (PCI) is a member of the serpin family of protease inhibitors (for recent reviews see refs. 1 and 2), first described for its ability to inhibit the anticoagulant-activated protein C (APC) (3, 4). The procoagulant function of plasma PCI was further supported by the finding that the thrombin–thrombomodulin (TM) complex, responsible for activation of protein C, is also selectively inhibited by PCI (5). However, PCI is also capable of inhibiting several of the procoagulant proteases generated as part of the hemostatic response, raising the possibility of a dual role for PCI in regulating blood coagulation (for reviews see refs. 6 and 7). Elevated PCI levels have been associated with venous thrombosis (8) and myocardial infarction (9), and the PCI–APC complex level is a sensitive marker for thrombotic events (10). The function of PCI in hemostasis depends on cofactors (for review see ref. 11). Heparin and similar glycosaminoglycans (GAGs) accelerate the inhibition of several proteases, including APC and thrombin, and TM accelerates the inhibition of thrombin. Determining how these cofactors influence the rate of protease inhibition by PCI is critical to understanding the various roles of PCI in regulating blood coagulation.
PCI utilizes the well characterized serpin mechanism of protease inhibition (12). It is best described as a two-step reaction where the rate of inhibition depends on the formation of the initial recognition (Michaelis) complex, and in the second step both serpin and protease undergo significant conformational rearrangement leading to a covalently linked final complex. The serpin mechanism is especially well suited for inhibiting coagulation factors because the conformational change induced in the protease can result in a loss of cofactor binding. An important example of this is the induced disorder of exosite I on thrombin upon complexation with serpins (13, 14). Thrombin can thus be removed from its high-affinity interaction with TM through inhibition by PCI. Regulation of serpin activity by cofactors is generally achieved by altering the rate of formation and stability of the Michaelis complex (15). Heparin-like GAGs accelerate the rate of APC and thrombin inhibition by PCI, and TM selectively accelerates the inhibition of thrombin. Heparin is thought to bridge PCI to thrombin in a manner similar to that recently seen in the crystal structure of the antithrombin (AT)-thrombin-heparin ternary complex (16, 17), but the APC-PCI bridged complex may involve a more intimate shared site on heparin (18, 19). In either case, cofactor activity depends on both serpin and protease binding to the same heparin chain, with acceleration of inhibition conferred by both improved rate of diffusion and stabilization of the encounter complex by the heparin bridge. The cofactor activity of TM relies on its tight interaction with thrombin and possible exosite interactions between TM and PCI. Thrombin is known to bind to the EGF domains 5 and 6 of TM, and PCI is thought to interact with EGF domain 4 (20), although a direct interaction between TM and PCI has not been demonstrated.
The structures of native (18) and reactive center loop (RCL)-cleaved (19) PCI revealed the general serpin fold and some important additional features relevant to cofactor and protease binding. The region recognized by proteases as a substrate loop, the RCL, is unusually long and flexible in PCI. This can influence protease binding in two important ways: First, the rate of Michaelis complex formation will be reduced because of the inherent mobility of the RCL. In addition, there will be a large entropic cost for RCL binding by a protease, predictably reducing the stability of the Michaelis complex. These factors combine to increase the Km for proteases, a fact borne out by the generally low rates of protease inhibition by PCI, and necessitate cofactors and/or exosite interactions to improve the Km. The second function of a long, flexible RCL is to increase the reach of a tethered protease to engage exosites on a larger surface of PCI. Another structural difference between PCI and other serpins is the location of the heparin-binding site along helix H (18, 21–23). All other heparin-binding serpins (e.g., antithrombin, heparin cofactor II, plasminogen activator inhibitor I, protease nexin 1) use a basic helix D for heparin binding (11). The heparin-binding site of PCI on helix H is near to the RCL and thus in close proximity to an attacking protease. This can have additional effects on protease recognition, such as abrogating (24) or accelerating complex formation by altering the surface of PCI encountered by proteases.
To understand how PCI recognizes thrombin and the accelerating effect of cofactors, we determined a 1.6-Å crystal structure of the Michaelis complex between thrombin and PCI in the presence of a bridging heparin chain. Thrombin is docked on PCI in an unusual position that aligns its heparin binding site (exosite II) with that of PCI (helix H). The position of thrombin is allowed by the extra RCL residues C-terminal to the P1–P1′ bond [substrate residues are numbered Pn N-terminally and Pn′ C-terminally of the P1–P1′ scissile bond, and the corresponding cavities in the protease are numbered Sn and Sn′ (25)] and engenders significant exosite interactions that help stabilize the complex. A model of the PCI–thrombin–TM Michaelis complex was built on the structure and suggested favorable direct interactions between TM and PCI that were supported by mutagenesis and biochemical studies. This work reveals how PCI recognition of thrombin is facilitated by cofactors and supports a dual role for PCI in striking the proper hemostatic balance.
Results and Discussion
Overall Structure.
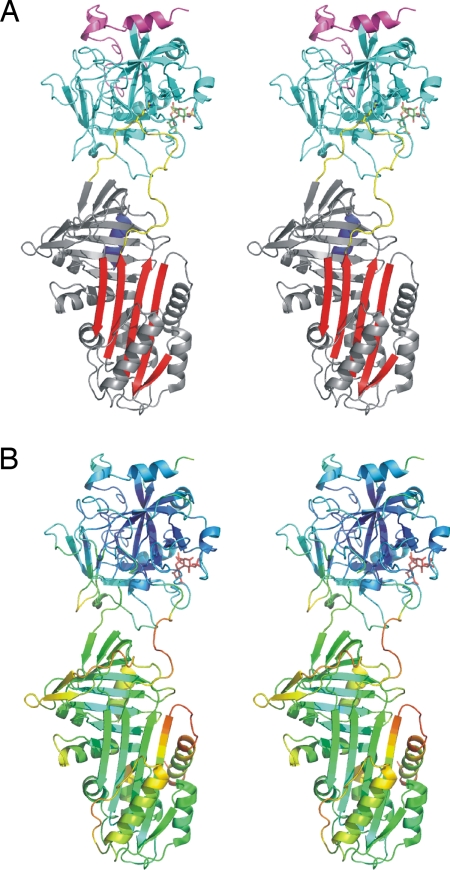
We obtained high-quality diffraction data from a single crystal of the PCI, S195A thrombin, heparin 14-mer complex to 1.6 Å resolution. Molecular replacement with our recently published structure of native PCI (18) and S195A thrombin from 1JOU (26) placed one complex in the asymmetric unit, and iterative rounds of refinement and rebuilding resulted in the final model (Table 1 and Fig. 1). The gross conformational features of thrombin were not altered by its complexation by PCI and heparin, but significant movement of the 35- and 60-loops was observed [supporting information (SI) Fig. S1]. The structure of PCI in the complex was nearly identical to that of native PCI (Cα RMSD of ≈0.6 Å), with no evidence of preinsertion of the hinge region into β-sheet A. Thus, as with other crystal structures of serpin–protease Michaelis complexes, the serpin component did not change conformation in response to formation of the Michaelis complex. However, we did observe a high degree of conformational mobility in PCI in the complex, reflected by poor electron density in several regions and unusually high B-factors. The relative flexibility of PCI in the complex is nicely illustrated by coloring the ribbon diagram according to main-chain temperature factors (B-factors, Fig. 1B). The figure demonstrates a low overall B-factor for thrombin (harmonically averaged value of ≈17.5 Å2) and a twofold higher average B-factor for PCI (≈35.0 Å2). The highest levels of conformational mobility were in regions known to be involved in β-sheet A expansion (27): the hinge region of the RCL, the top of s2A, helix D, and helix F. PCI thus seems to be a molecule on the “edge,” poised for rapid RCL insertion. The implication is that the formation of the Michaelis complex somehow induces instability in PCI while it awaits the triggering event of formation of the acyl enzyme intermediate.
Table 1.
Data processing, refinement, and model (3B9F)
| Crystal | |
| Space group | P1 |
| Cell dimensions, Å | a = 44.05; b = 48.82; c = 97.87 |
| Angles, ° | α = 78.73; β = 81.49; γ = 77.68 |
| Solvent content, % | 43.9 |
| Data processing statistics | |
| Wavelength, Å | 1.06 (Diamond, beam line I03) |
| Resolution, Å | 37.50–1.60; 1.69–1.60 |
| Total reflections | 305,107; 25,620 |
| Unique reflections | 95,914; 13,603 |
| Multiplicity | 3.2; 1.9 |
| 〈I/σ (I)〉 | 12.9; 1.8 |
| Completeness, % | 93.6; 90.3 |
| Rmerge* | 0.052; 0.445 |
| Model | |
| Number of atoms modeled | |
| Protein | 5,493 |
| Heparin | 31 |
| Water | 523 |
| Carbohydrate | 24 |
| Glycerol molecules | 5 |
| Sulphate Ions | 3 |
| Average B-factor, Å2 | 30.2 |
| Refinement statistics | 30.95–1.60 Å; 1.70–1.60 Å |
| Reflections in working/free set | 92,977/2,928; 15,020/460 |
| R-factor2/R-free, % | 20.8/23.4; 32.5/33.7 |
| R.m.s. deviation of bonds, Å/angles, ° from ideality | 0.005/1.3 |
| Ramachandran plot; residues | |
| Most-favored region, % | 87.4 |
| Additionally allowed region, % | 11.8 |
| Generously allowed region, % | 0.8 |
| Disallowed region, % | 0 |
*Rmerge = Σhkl Σi|Ihkl − 〈Ihkl〉|/Σhkl Σi 〈Ihkl〉.
†R-factor = Σhkl||Fobs- − -Fcalc||/Σhkl|Fobs|.
Fig. 1.
Stereo ribbon diagrams of the PCI–thrombin–heparin Michaelis complex. (A) The complex is shown with PCI in the standard orientation with the yellow RCL on top and the red β-sheet A facing. The P1 Arg residue is shown as yellow rods, and the heparin binding helix H is at the back of the PCI in blue. Thrombin is on top, with its light chain colored magenta and the heavy chain colored cyan. The heparin disaccharide built into electron density is shown as green rods. (B) The same view as in A but colored according to temperature factor (B-factor from blue to red) to illustrate the mobile regions.
PCI–Thrombin Contacts.
As shown in Fig. 1, thrombin docks onto the top of PCI (when the serpin is oriented in the standard fashion) and engages the RCL within its active site. The distance between thrombin and the body of PCI is greater that that seen when thrombin is complexed with either heparin cofactor II (HCII) (28) or antithrombin (AT) (17) (Fig. 2). PCI has a unique four residue P′ insert that extends to effectively keep thrombin at “arms length.” Based on the structure of native PCI, we predicted that its extra long RCL would allow exosite contacts not available to other serpins, and this is indeed what we observe. The orientation of thrombin in complex with PCI differs significantly from that observed with either HCII or AT (Fig. 2). In complexes with HCII and AT, thrombin made exosite contacts that locked thrombin into the observed position. In a similar manner, the thrombin–PCI Michaelis complex buries a combined surface of 2,090 Å2, 865 Å2 of which is due to exosite contacts (defined as contacts outside the principal substrate binding pocket S4–S3′). The principal exosite involves the 60-loop of thrombin and strands 2 and 3 of β-sheet C on PCI, with Asp60E interpolating between the positively charged side chains of Lys-277, Lys-278, and Arg-279 to allow several direct and water-mediated hydrogen bonds (Fig. 2 B and C). The importance of this exosite is supported by mutagenesis of both the 60-loop of thrombin (29) and residues 276 and 277 of PCI (22). In contrast to the other structures of thrombin–serpin Michaelis complexes, thrombin's γ-loop makes no contact with PCI and cannot be modeled into electron density.
Fig. 2.
The position of thrombin and its interactions with PCI. (A) The position of thrombin in complex with PCI is compared with that found in previous complexes with HCII and AT. The stereoview is colored as in Fig. 1A, with thrombin from the HCII complex colored orange and that from the AT complex in pink (semitransparent, heavy chains only). The heparin-binding C-terminal helix is colored green for all thrombins to help illustrate the difference in orientation. In the PCI–thrombin complex, the heparin-binding helices of thrombin and PCI are aligned at the back. (B) The surface of thrombin is shown in the standard orientation and colored green to illustrate the contact surface with PCI (yellow rods). The RCL of PCI binds in the active-site cleft from P6–P5′, and 2F,o − F,c electron density contoured at 1σ is shown for residues P4–P3′. The main exosite contact between the 60-loop of thrombin and PCI is also shown. (C) A close-up of the exosite contact between the 60-loop of thrombin and β-sheet C residues of PCI (colored as in B). Direct and water-mediated hydrogen bonds are shown as broken red lines.
The contacts between the RCL of PCI and the active-site cleft of thrombin are extensive and in many ways typical. Of principal importance for thrombin recognition is the burying of the P1 Arg side chain into the acidic S1 pocket and the interaction of the P4 Phe side chain in the aryl binding S4 site. The P2 Phe residue of PCI, however, is unique among known substrates and inhibitors of thrombin, and its occupancy of the S2 pocket cannot occur without significant movement of the 60-loop (Fig. S1 and Fig. S2). It has been proposed that part of the TM-mediated effect is an allosteric enlargement of the S2 pocket (5). Indeed, the structure of the thrombin-TM456 complex (inhibited with EGRCK) (30) reveals an S2 pocket of intermediate size between that observed in our PCI complex and that of PPACK-inhibited thrombin (31) (Fig. S2), consistent with the hypothesis of a partially allosteric mechanism. Because the 60-loop is the principal exosite for PCI recognition, it is possible that in the absence of TM the role of the bulky P2 Phe in PCI is to induce the formation of this exosite contact.
Heparin Bridge.
Although the crystals were grown in the presence of a bridging 14-mer heparin chain, only one heparin disaccharide could be placed in electron density (Fig. 1 and Fig. S3). The disaccharide was found docked to exosite II of thrombin in an orientation and position consistent with a previous structure of thrombin bound to natural heparin (32). No density was observed in the putative heparin-binding region of PCI and the side chains of residues known to interact with heparin could not be fully modeled into electron density, suggesting an equilibrium between multiple heparin-binding modes within the crystal. Such an explanation is supported by the weak affinity of PCI for heparin coupled with an environment within the crystal conducive to multiple heparin conformations. Indeed, when crystallographically related complexes were generated, we observed a large highly basic tunnel formed by the alignment of exosite I and II of thrombin and helix H and β-sheet C of PCI. As shown in Fig. S4, the 14-mer heparin chain fits comfortably into this tunnel and can assume multiple distinct positions that would preclude its being resolved in the crystal structure, despite the high resolution. The juxtaposition of these basic regions in the crystal suggests that heparin is serving to alleviate electrostatic repulsion, thereby allowing formation of crystal contacts. Therefore, we conclude that the crystals were formed from heparin-bridged complexes and that heparin adopted a new multistate equilibrium within the crystal lattice. The alignment of the heparin-binding sites of thrombin and PCI supports this conclusion (Fig. 3). When a 14-mer heparin chain is superimposed onto the disaccharide modeled into electron density, it fully occupies the heparin-binding sites of thrombin and PCI with only minor adjustments to glycosidic bond torsion angles. Fourteen saccharide units would appear to be the minimal heparin length capable of bridging and fully occupying the two binding sites, consistent with previous biochemical studies (33).
Fig. 3.
Stereo representations of the PCI–thrombin complex bridged by a 14-mer heparin chain. (A) The ribbon diagram is rotated 150° relative to the view shown in Fig. 1A to illustrate the alignment of the heparin-binding regions. A 14-mer heparin chain (green rods) is placed onto the modeled disaccharide to yield a bridged complex with fully occupied heparin-binding sites. (B) The electrostatic surface of the complex in the same orientation as in A reveals the continuous basic (blue) heparin-binding sites of thrombin (Upper) and PCI (Lower).
Model of the Thrombomodulin Complex.
It has been suggested that the basis of TM acceleration of thrombin inhibition by PCI is a direct interaction between the fourth EGF domain of TM and a positively charged surface on PCI, possibly the heparin-binding site on helix H (20). However, these studies were conducted by transplanting the H-helix of PCI onto AT and studying the properties of the chimera. To determine whether direct PCI–TM interactions occur, we examined the dependence of thrombin inhibition on TM concentration (Fig. 4A). We found that the observed rate constant for thrombin inhibition depended on TM concentration in a bell-shaped fashion, supporting a direct interaction between TM and PCI. The positive part of the curve nicely verifies the known Kd of thrombin for TM456 (14 ± 2 nM) and the negative slope fits a Kd of 24 ± 8 μM, corresponding to the dissociation constant of PCI for TM. Because thrombin binds tightly to EGF domains 5 and 6 of TM, it is likely that a direct interaction between EGF domain 4 and PCI is responsible, to some degree, for the accelerating effect of TM on thrombin inhibition.
Fig. 4.
Binding of TM to PCI assessed by biochemical and mutagenesis studies. (A) The effect of TM concentration on rate of thrombin inhibition by PCI reveals a bell-shaped dependence. The initial step reflects the high-affinity binding of TM to thrombin and peaks when the complex is 1:1. The down slope reflects the competitive binding of PCI to TM. The solid line is the nonlinear fit to a two-site-specific binding equation. (B) The site on PCI responsible for TM binding was determined by assessing the TM-accelerated rates of thrombin inhibition for several PCI species: rPCI is the recombinant background on which all point mutations were made, and pPCI is plasma-derived PCI that is glycosylated at three sites including Asn-230. Open bars indicate rate constants obtained in the absence of cofactor; horizontal striped bars indicate the presence of TM456; and the cross-hatched bar is for TM and 8-mer heparin (as described in Materials and Methods).
To gain insight into how TM interacts with PCI, we created a model of the complex by superimposing the thrombin–TM structure (30) onto thrombin in our structure. The fit was reasonable, with EGF domain 5 making favorable contacts and the negatively charged face of EGF4 docking onto a positively charged surface of PCI. There were minor clashes between residues 355 and 357 in EGF4 with Glu-253 and Gly-254 of PCI, which were easily relieved by taking advantage of the flexibility between EGF domains 4 and 5. The model suggested several favorable interactions. Of particular importance was the predicted salt-bridge and hydrogen bonds between the rigid EGF5 domain (residues Asp-400 and Asn-402) with Arg-362 in strand 1C of PCI. In addition, Glu-253 and Lys-255 of PCI were in position to make contacts with EGF4. We tested this model by creating charge-reversal mutations at these and other positions on PCI and determining their effect on TM acceleration of thrombin inhibition (Fig. 4B). We can also predict from the model that heparin binding or glycosylation at Asn-230 would not adversely affect the TM-enhanced rate of thrombin inhibition, because EGF4 is positioned ≈20 Å and 12 Å away from these sites, respectively. However, as shown in Fig. 4B, glycosylation of Asn-230 (in plasma-derived PCI) and 8-mer heparin binding both adversely affect the ability of TM to enhance thrombin inhibition. This suggests that EGF4 docks closer to Asn-230 and the heparin binding site than can be accounted for in the model. This conclusion was also supported by the modest effect of mutations at Glu-253 and Lys-255 and the dramatic effect of the Arg229Glu mutation (≈20-fold reduction in rate constant). The lack of effect of the Arg362Glu variant was most informative, because Arg-362 should be making intimate favorable contacts with the rigid EGF5 domain of TM. To account for this observation, it is necessary to rotate the entire thrombin-TM complex by 25° (Fig. 5). This nicely removes Arg-362 from the interaction site with TM while maintaining the exosite contacts described above and also satisfying the observations summarized in Fig. 4B. Interestingly, the glycosylation at Asn-230 of PCI appears to act as a barrier between the TM-binding site on sheet C and the heparin-binding site along helix H. Although the presence of heparin can affect the formation of the complex with TM, there is no predicted direct contact between the two molecules in our model, and simultaneous occupancy by the two cofactors is thus feasible. This is consistent with biochemical data showing that the presence of the chondroitin sulfate moiety on TM marginally improves the rate of thrombin inhibition by PCI (5).
Fig. 5.
Stereoview of the models of the PCI–thrombin–TM456 complex, oriented to highlight the proposed interaction surface. Residues from Fig. 4B are colored according to their effect on TM activation of thrombin inhibition, with red indicating no effect, yellow a modest effect, and green a large effect. The initial model of the complex was generated by superimposing the thrombin–TM456 structure on thrombin in complex with PCI. The position of TM456 in this complex (semitransparent orange) suggested contacts between EGF5 and Arg-362 (red balls on right) and significant overlap with residues Glu-253 and Lys-255 (yellow balls on right). The biochemical data did not support this model. Another model was created by rotating the whole thrombin–TM456 complex by 25° to the left (thrombin colored as before and TM456 in orange). This model places EGF4 in intimate contact with Arg-229 (green balls) and near residues colored in yellow. The only glycosylation site in this region is Asn-230, and its N-linked carbohydrate is shown as green rods (positioned by superimposing the structure of RCL-cleaved plasma PCI).
Conclusions
PCI is a promiscuous serpin that depends on cofactors to confer its tissue-specific activities. The apparent pro- and anticoagulant roles of PCI in blood coagulation are both mediated through the inhibition of thrombin. The anticoagulant action of PCI occurs on cell surfaces dense in GAGs that accelerate thrombin inhibition by supporting the formation of the bridged Michaelis complex revealed by our crystal structure. This complex is unique and can be formed only by the alignment of the heparin-binding sites of the two proteins through the exosite contacts describe above and by exploiting the extra long P′ region of PCI. On cell surfaces rich in TM, PCI functions as a procoagulant by specifically recognizing and inhibiting the thrombin–TM complex, thus shutting down APC formation. Using biochemical techniques, we demonstrate a direct PCI–TM interaction through EGF4 and show with modeling and mutagenesis that EGF4 binds an exosite adjacent to, but not overlapping, the heparin-binding site of PCI. These studies provide significant insights into the mechanisms regulating the important but enigmatic serpin, PCI.
Materials and Methods
Materials.
Size-fractionated heparin was purchased from Iduron. Plasma-derived human thrombin was purchased from Haematologic Technologies, and plasma PCI was purified as before (19). Crystallization reagents were purchased from Hampton Research, and chromogenic substrate S2238 was purchased from Chromogenix.
Protein Production and Purification.
Human PCI, N-terminally truncated by 16 residues, was produced in Escherichia coli and purified as described (18). This material is referred to as recombinant PCI in the text and is the background for all described PCI mutants. Mutations were made by using the Stratagene site-directed mutagenesis protocol and were verified by DNA sequencing at Lark Technologies. Recombinant human S195A thrombin was produced from baby hamster kidney cells as described (34). The TM fragment containing EGF domains 4–6 (corresponding to residues 345–465 of human thrombomodulin) was prepared from E. coli as described in SI Materials and Methods.
Crystallization, Data Collection, and Refinement.
The ternary complex of S195A thrombin, recombinant PCI, and 14-mer heparin was formed in a molar ratio of 1:1:1.2 and concentrated to 3.9 mg/ml in 20 mM Tris, 20 mM NaCl (pH 7.4). Diffraction-quality crystals were obtained from 1:1 ratio drops containing complex and 0.12 M magnesium sulfate, 12% PEG3350. Crystals were cryoprotected in 20% PEG3350, 0.05 M magnesium sulfate, and 25% glycerol and flashed cooled to 100 K in the vapor nitrogen stream. Data were collected from a single crystal at Diamond Light Source beam line I03 and processed by using Mosflm, Scala, and Truncate (35). The structure was solved by molecular replacement with Phaser (36) by using S195A thrombin (1JOU monomer AB), and native PCI (2OL2) as search models. Initial maps were calculated after running ARP/wARP (version 7.0) (37) to minimize model bias. Refinement was carried out by using CNS (version 1.0) (38), and XtalView (39) was used for model building. Data processing and refinement statistics are given in Table 1. Figures were made by using Pymol (40), and the interaction area was calculated by the protein–protein interaction server (www.biochem.ucl.ac.uk/bsm/PP/server). Thrombin numbering is based on chymotrypsin, with insertion loops indicated by sequential letters. Coordinates and structure factors are deposited in the Protein Data Bank under PDB ID code 3B9F.
Thrombin-Inhibition Assay.
Thrombin inhibition was assayed at room temperature in 20 mM Tris·HCl (pH 7.4), 100 mM NaCl, 0.2% BSA, and 0.1% PEG8000. Plasma-derived human thrombin (5 nM) and recombinant PCI and its variants (50–100 nM) were used in the assay. For TM acceleration, 5 nM recombinant TM456 was preincubated with thrombin before PCI was added. In the assay of pPCI/8-mer, pPCI was preincubated with 5 μM size-fractionated 8-mer heparin before being added to the inhibition assay. All assays were carried out in 96-well plates. Briefly, thrombin, with or without TM456, was inhibited with PCI for 15 seconds to 10 min, and the reaction was stopped by adding a large excess of substrate S2238. Residual thrombin activity was measured by S2238 hydrolysis, and the inhibition rate was calculated from the linear plot of the natural log of residual thrombin activity versus time. The second-order rate constant was calculated by dividing the observed rate constant by the total PCI concentration (conditions were pseudo first-order in PCI). Each rate constant was measured at least twice, and the average with standard error is reported. In the TM inhibition bell-shaped curve assay, thrombin was preincubated with 0–61.5 μM TM456 before PCI was added into the inhibition assay. The observed rate constants from four measurements were averaged and plotted against TM456 concentration. The up and down portions of the bell curve were fitted simultaneously to a two-site-specific binding equation by using GraphPad Prism.
Acknowledgments.
This work was supported by the Medical Research Council (U.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates and structure factors are deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3B9F).
This article contains supporting information online at www.pnas.org/cgi/content/full/0711055105/DCSupplemental.
References
- 1.Huntington JA. Shape-shifting serpins—advantages of a mobile mechanism. Trends Biochem Sci. 2006;31:427–435. doi: 10.1016/j.tibs.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 3.Marlar RA, Griffin JH. Deficiency of protein C inhibitor in combined factor V/VIII deficiency disease. J Clin Invest. 1980;66:1186–1189. doi: 10.1172/JCI109952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki K, Nishioka J, Hashimoto S. Protein C inhibitor: Purification from human plasma and characterization. J Biol Chem. 1983;258:163–168. [PubMed] [Google Scholar]
- 5.Rezaie AR, Cooper ST, Church FC, Esmon CT. Protein C inhibitor is a potent inhibitor of the thrombin–thrombomodulin complex. J Biol Chem. 1995;270:25336–25339. doi: 10.1074/jbc.270.43.25336. [DOI] [PubMed] [Google Scholar]
- 6.Geiger M. Protein C inhibitor, a serpin with functions in- and outside vascular biology. Thromb Haemostasis. 2007;97:343–347. [PubMed] [Google Scholar]
- 7.Rau JC, Beaulieu LM, Huntington JA, Church FC. Serpins in thrombosis, hemostasis and fibrinolysis. J Thromb Haemostasis. 2007;5(Suppl 1):102–115. doi: 10.1111/j.1538-7836.2007.02516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meijers JC, Marquart JA, Bertina RM, Bouma BN, Rosendaal FR. Protein C inhibitor (plasminogen activator inhibitor-3) and the risk of venous thrombosis. Br J Haematol. 2002;118:604–609. doi: 10.1046/j.1365-2141.2002.03652.x. [DOI] [PubMed] [Google Scholar]
- 9.Carroll VA, et al. Plasma protein C inhibitor is elevated in survivors of myocardial infarction. Arterioscler Thromb Vasc Biol. 1997;17:114–118. doi: 10.1161/01.atv.17.1.114. [DOI] [PubMed] [Google Scholar]
- 10.Kolbel T, Strandberg K, Mattiasson I, Stenflo J, Lindblad B. Activated Protein C–Protein C inhibitor complex: A new biological marker for aortic aneurysms. J Vasc Surg. 2006;43:935–939. doi: 10.1016/j.jvs.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 11.Huntington JA. Heparin activation of serpins. In: Garg HG, Linhardt RJ, Hales CA, editors. Chemistry and Biology of Heparin and Heparan Sulfate. Oxford: Oxford Univ Press; 2005. pp. 367–398. [Google Scholar]
- 12.Huntington JA, Read RJ, Carrell RW. Structure of a serpin–protease complex shows inhibition by deformation. Nature. 2000;407:923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 13.Bock PE, Olson ST, Bjork I. Inactivation of thrombin by antithrombin is accompanied by inactivation of regulatory exosite I. J Biol Chem. 1997;272:19837–19845. doi: 10.1074/jbc.272.32.19837. [DOI] [PubMed] [Google Scholar]
- 14.Fredenburgh JC, Stafford AR, Weitz JI. Conformational changes in thrombin when complexed by serpins. J Biol Chem. 2001;276:44828–44834. doi: 10.1074/jbc.M108710200. [DOI] [PubMed] [Google Scholar]
- 15.Izaguirre G, Swanson R, Raja SM, Rezaie AR, Olson ST. Mechanism by which exosites promote the inhibition of blood coagulation proteases by heparin-activated antithrombin. J Biol Chem. 2007;282:33609–33622. doi: 10.1074/jbc.M702462200. [DOI] [PubMed] [Google Scholar]
- 16.Huntington JA. Mechanisms of glycosaminoglycan activation of the serpins in hemostasis. J Thromb Haemost. 2003;1:1535–1549. doi: 10.1046/j.1538-7836.2003.00305.x. [DOI] [PubMed] [Google Scholar]
- 17.Li W, Johnson DJ, Esmon CT, Huntington JA. Structure of the antithrombin–thrombin–heparin ternary complex reveals the antithrombotic mechanism of heparin. Nat Struct Mol Biol. 2004;11:857–862. doi: 10.1038/nsmb811. [DOI] [PubMed] [Google Scholar]
- 18.Li W, Adams TE, Kjellberg M, Stenflo J, Huntington JA. Structure of native protein C inhibitor provides insight into its multiple functions. J Biol Chem. 2007;282:13759–13768. doi: 10.1074/jbc.M701074200. [DOI] [PubMed] [Google Scholar]
- 19.Huntington JA, Kjellberg M, Stenflo J. Crystal structure of protein C inhibitor provides insights into hormone binding and heparin activation. Structure (London) 2003;11:205–215. doi: 10.1016/s0969-2126(02)00944-9. [DOI] [PubMed] [Google Scholar]
- 20.Yang L, Manithody C, Walston TD, Cooper ST, Rezaie AR. Thrombomodulin enhances the reactivity of thrombin with protein C inhibitor by providing both a binding site for the serpin and allosterically modulating the activity of thrombin. J Biol Chem. 2003;278:37465–37470. doi: 10.1074/jbc.M307243200. [DOI] [PubMed] [Google Scholar]
- 21.Kuhn LA, et al. Elucidating the structural chemistry of glycosaminoglycan recognition by protein c inhibitor. Proc Natl Acad Sci USA. 1990;87:8506–8510. doi: 10.1073/pnas.87.21.8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirk RA, Elisen MG, Meijers JC, Church FC. Role of the h helix in heparin binding to protein c inhibitor. J Biol Chem. 1994;269:28690–28695. [PubMed] [Google Scholar]
- 23.Neese LL, Wolfe CA, Church FC. Contribution of basic residues of the d and h helices in heparin binding to protein C inhibitor. Arch Biochem Biophys. 1998;355:101–108. doi: 10.1006/abbi.1998.0716. [DOI] [PubMed] [Google Scholar]
- 24.Ecke S, Geiger M, Binder BR. Heparin binding of protein-C inhibitor—analysis of the effect of heparin on the interaction of protein-C inhibitor with tissue kallikrein. Eur J Biochem. 1997;248:475–480. doi: 10.1111/j.1432-1033.1997.00475.x. [DOI] [PubMed] [Google Scholar]
- 25.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 26.Huntington JA, Esmon CT. The molecular basis of thrombin allostery revealed by a 1.8 Å structure of the “slow” form. Structure (London) 2003;11:469–479. doi: 10.1016/s0969-2126(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 27.Whisstock JC, Skinner R, Carrell RW, Lesk AM. Conformational changes in serpins: I. The native and cleaved conformations of alpha(1)-antitrypsin. J Mol Biol. 2000;296:685–699. doi: 10.1006/jmbi.1999.3520. [DOI] [PubMed] [Google Scholar]
- 28.Baglin TP, Carrell RW, Church FC, Esmon CT, Huntington JA. Crystal structures of native and thrombin-complexed heparin cofactor II reveal a multistep allosteric mechanism. Proc Natl Acad Sci USA. 2002;99:11079–11084. doi: 10.1073/pnas.162232399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fortenberry YM, et al. Essential thrombin residues for inhibition by protein C inhibitor with the cofactors heparin and thrombomodulin. J Thromb Haemost. 2007;5:1486–1492. doi: 10.1111/j.1538-7836.2007.02574.x. [DOI] [PubMed] [Google Scholar]
- 30.Fuentes-Prior P, et al. Structural basis for the anticoagulant activity of the thrombin–thrombomodulin complex. Nature. 2000;404:518–525. doi: 10.1038/35006683. [DOI] [PubMed] [Google Scholar]
- 31.Bode W, et al. The refined 1.9 Å crystal structure of human alpha-thrombin: Interaction with d-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment. EMBO J. 1989;8:3467–3475. doi: 10.1002/j.1460-2075.1989.tb08511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carter WJ, Cama E, Huntington JA. Crystal structure of thrombin bound to heparin. J Biol Chem. 2004;280:2745–2749. doi: 10.1074/jbc.M411606200. [DOI] [PubMed] [Google Scholar]
- 33.Pratt CW, Whinna HC, Church FC. A comparison of three heparin-binding serine proteinase inhibitors. J Biol Chem. 1992;267:8795–8801. [PubMed] [Google Scholar]
- 34.Ye J, Rezaie AR, Esmon CT. Glycosaminoglycan contributions to both protein C activation and thrombin inhibition involve a common arginine-rich site in thrombin that includes residues arginine 93, 97, and 101. J Biol Chem. 1994;269:17965–17970. [PubMed] [Google Scholar]
- 35.Leslie AWG. Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography. 1992:26. [Google Scholar]
- 36.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 37.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 38.Brunger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 39.McRee DE. A visual protein crystallographic software system for x11/xview. J Mol Graphics. 1992;10:44–46. [Google Scholar]
- 40.DeLano W. The PyMOL Molecular Graphics System. San Diego: DeLano Scientific; 2002. [Google Scholar]