Abstract
Histone modification in chromatin is one of the key control points in gene regulation in eukaryotic cells. Protein complexes composed of histone acetyltransferase or deacetylase, WD40 repeat protein, and many other components have been implicated in this process. Here, we report the identification and functional characterization of HOS15, a WD40-repeat protein crucial for repression of genes associated with abiotic stress tolerance through histone deacetylation in Arabidopsis. HOS15 shares high sequence similarity with human transducin-beta like protein (TBL), a component of a repressor protein complex involved in histone deacetylation. Mutation of the HOS15 gene renders mutant plants hypersensitive to freezing temperatures. HOS15 is localized in the nucleus and specifically interacts with histone H4. The level of acetylated histone H4 is higher in the hos15 mutant than in WT plants. Moreover, the stress inducible RD29A promoter is hyperinduced and associated with a substantially higher level of acetylated histone H4 in the hos15 mutant under cold stress conditions. Our results suggest a critical role for gene activation/repression by histone acetylation/deacetylation in plant acclimation and tolerance to cold stress.
Keywords: cold responsive gene expression, freezing tolerance, histone H4 deacetylation, WD 40 protein
Plants respond to environmental stresses by altering the expression of many genes. The altered status of gene expression is closely associated with an acclimated condition that renders the plant more stress tolerant. Among the genetic mechanisms leading to these gene expression changes, chromatin remodeling has until recently received little attention. In eukaryotes, transcriptional control of gene expression occurs within the context of chromatin that is composed of nucleosomal subunits. Each nucleosome consists of ≈146 bp of DNA wrapped twice around an octamer of core histones (H2A, H2B, H3, and H4). Individual nucleosomes and more compacted structures in chromatin are refractory to transcription. Chromatin remodeling, which alters accessibility of genes to transcriptional regulatory proteins, is now recognized as a central component of gene regulation associated with a metastable (epigenetic) genetic status.
Two general types of chromatin-modifying complexes have been described. One type covalently modifies histone N-terminal tails protruding from the nucleosome core (e.g., by acetylation, methylation, or phosphorylation) (1, 2). The second class controls the ATP-dependent nucleosome-remodeling complexes that noncovalently modify and reposition nucleosomes in the chromatin structure (3, 4). Acetylation and deacetylation of lysine residues in the N termini of histones represent extensively studied types of chromatin modifications that have been shown to play fundamental roles in diverse chromatin-based processes. Hyperacetylation of histones H2B, H3, and H4 has been generally associated with transcriptionally active chromatin (5), whereas the chromatin of inactive regions is enriched in deacetylated histones (6). Additional support for the existence of a direct molecular link between histone acetylation status and transcription regulation (7–9) is provided by the finding that many transcriptional coactivators, including GCN5, PCAF, CBP/p300, and SRC-1/ACTR, possess intrinsic histone acetyltransferase activity. Moreover, transcriptional repressors such as NuRD, SIN3, Groucho/Tup1, and SMRT/N-CoR associate with histone deacetylases (7–9). In plants, histone modification has been shown to be involved in metastable changes required to maintain altered cellular and tissue properties after several rounds of mitosis (10). For example, repression of FLC gene expression during vernalization in Arabidopsis was reported to be associated with histone deacetylation and methylation (11, 12).
Because plants must produce new cells and continue growth during and after adaptation (acclimation) to new environments, it could be expected that chromatin-mediated metastable genetic changes are involved in the process. However, evidence for the involvement of chromatin remodeling in gene expression and adaptation responses to the environment remains elusive. Here, we report that HOS15, a WD40-repeat protein, functions to control gene expression through histone deacetylation in chromatin. HOS15 was identified in a forward genetic screen for mutations that alter abiotic stress signaling (13, 14). The hos15 mutant plants accumulate higher levels of transcripts of many stress-regulated genes and are hypersensitive to freezing temperature. HOS15 interacts specifically with and promotes deacetylation of histone H4, indicating that chromatin remodeling plays an important role in gene regulation in plant responses and tolerance to abiotic stresses.
Results
Isolation of the hos15 Mutant.
A large population of Arabidopsis plants expressing a RD29A::LUC fusion gene (13) were mutagenized with T-DNA, and the T2 progeny were screened for altered expression of RD29A::LUC in response to low temperature, ABA, or osmotic treatment. The hos15 mutant showed a substantially higher level of RD29A::LUC expression in response to all treatments (Fig. 1). All of 67 F1 plants from a hos15 x WT cross exhibited a WT phenotype. The selfed F2 progeny segregated ≈3:1 (439:144, WT to mutant), indicating that the hos15 mutation is recessive and in a single nuclear gene.
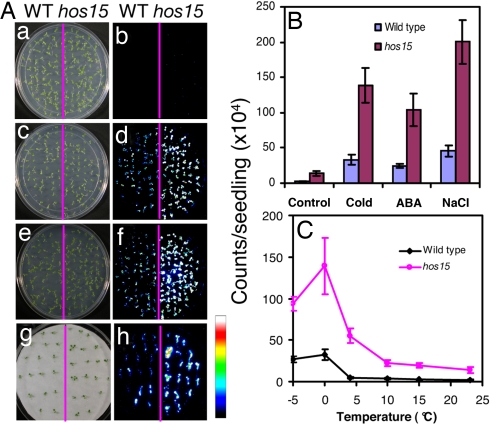
Fig. 1.
The hos15 mutation enhances RD29A::LUC expression. (A) RD29A::LUC expression was quantitatively measured as luminescence intensity (counts per plant). One-week-old WT and hos15 seedlings and their luminescence images taken without treatment (Control) or after treatment at 0°C for 24 h (Cold), 100 μM ABA for 3 h (ABA), and 300 mM NaCl for 5 h (NaCl). The scale bar at Right shows the luminescence intensity from dark blue (lowest) to white (highest). (B) Quantification of RD29A::LUC expression of plants shown in A. (C) RD29A::LUC expression in WT and hos15 plants under different temperatures. Error bars are standard deviation (n = 20).
HOS15 Is a Negative Regulator of Stress-Regulated Gene Expression.
Consistent with the RD29A::LUC expression patterns, induction of the luciferase transcript and the endogenous RD29A gene transcript was substantially higher in the hos15 mutant than WT after stress treatments (Fig. 2A). The expression of two other stress-responsive genes, COR15A and ADH1, was also more induced in hos15 plants in response to stress treatments (Fig. 2A). We also examined whether the expression levels of the CBF transcription factors are altered in the hos15 mutant plants. There was only a slight reduction in transcript levels of CBF1, CBF2, and CBF3 in the hos15 mutant plants 2 h after cold stress (Fig. 2B). This suggests that HOS15 affects the expression of cold regulon genes by a mechanism other than by controlling CBF transcription.
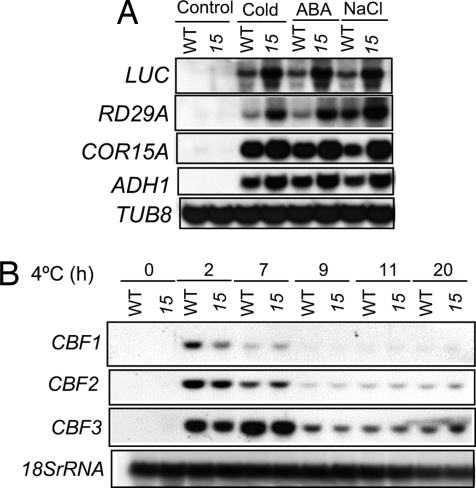
Fig. 2.
Regulation of gene expression in hos15 plants. (A) The plants were stress-treated as follow: Control, room temperature; cold, 0°C for 24 h; ABA, 100 μM ABA for 3 h; NaCl, 300 mM NaCl for 5 h. β-tubulin 8 (TUB8) was used as the loading control. (B) Gene expression under cold treatment (4°C) with light (16 h of light; 8 h of darkness) for indicated time points. 18S rRNA gene was used as the loading control.
To identify other targets of HOS15 gene regulation, comprehensive analysis of gene expression profiles in the hos15 plants under cold stress was achieved through the use of Affymetric microarrays. The microarray data were analyzed with affylmGUI software included in the R package (15–17). A total of 136 genes showed higher expression levels by at least 2-fold in hos15 plants compared with WT when exposed to cold [supporting information (SI) Dataset S1]. These genes did not appear to be restricted to specific functional categories because they are annotated to encode proteins with diverse cellular functions. The expression of five genes (At5g64000, At5g61900, At2g18660, At1g35230, and At4g36110) randomly selected from the 136 was validated by real-time RT-PCR analyses (Fig. S1).
By comparing our results with the published microarray data, we found that, among the 136 genes that are more expressed in hos15 compared with WT plants after cold treatment, 39 were identified as cold-induced in WT plants (Dataset S2 and refs. 18–22). The proportion of cold-induced genes that are hyperinduced in hos15 (28.7%) is much higher than that for genes in general, which range from 2.3% to 10.8% (18–22). Interestingly, database comparisons also revealed that among the 136 genes, 8 whose transcripts are more abundant in cold-treated hos15 plants compared with WT plants are actually down-regulated in WT plants by cold (Dataset S3 and refs. 18–22). Real-time RT-PCR analysis of four of these genes confirmed that their transcripts were more abundant in hos15 plants with or without cold treatment but were reduced in WT after cold stress (Fig. S2). All of these except PR5 have unknown functions (23, 24). Statistical analysis and database comparisons also revealed that expression of 27 genes with various functions was reduced in hos15 plants after cold treatment compared with WT (Dataset S4). None of these are cold-inducible and only four of them (At5g24780, At2g3770, At3g28270, and At1g62510) are down-regulated by cold in WT (18–22). Real-time RT-PCR analysis of four of these genes confirmed that their transcripts were reduced in hos15 plants under cold treatment (Fig. S3). Real-time RT-PCR analysis confirmed that expression of At5g67320/HOS15 is disrupted in hos15 (see Fig. 4D), as first indicated by microarray analyses (see Fig. 4). Our microarray results suggest that relief from gene repression caused by the hos15 mutation may allow the continued expression of putative negative effectors of cold acclimation that should be repressed during cold exposure. Also, only 1 of the 39 cold-induced and none of the suppressed genes in hos15 plants appears in the CBF regulon (18–22). This is consistent with the interpretation that HOS15 does not solely function through the well established ICE/CBF cold induction pathway (25).
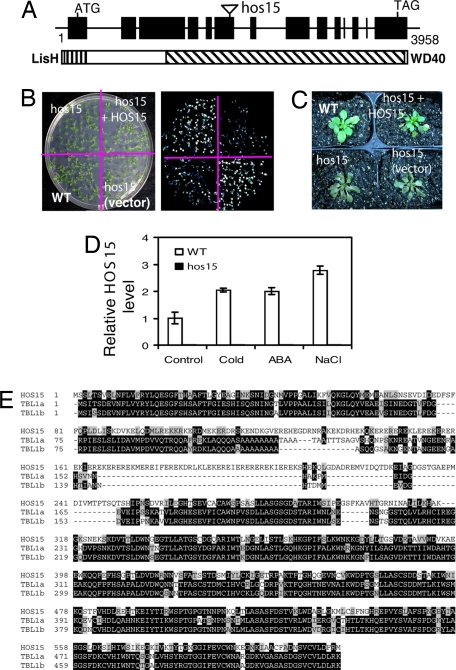
Fig. 4.
HOS15 encodes a WD40 protein. (A) Structure of HOS15 gene and its gene product. Positions are relative to the translation start site. The hos15 mutation is also indicated. The HOS15 protein contains two conserved domains: LisH (for Lissencephaly type-1-like homology motif, located between amino acids 5–37) and WD40 (the eight WD40-repeats are located as follow: 254–293, 309–352, 354–393, 396–434, 437–476, 479–527, 530–569, and 572–610). (B) Luminescence image of cold-treated (0°C for 24 h) hos15, WT, a representative line of hos15 transformed with an empty vector pCAMBIA1200 [hos15 (vector)], and a representative homozygous line of hos15 transformed with WT HOS15 gene (hos15 + HOS15). (C) Freezing tolerance of the plants shown in B at −5°C for 4 h. The plants were cold-acclimated (4°C for 7 d) before stress treatment. Photographs were taken 8 d after the treatment. (D) Comparison of HOS15 (accession no. AAL15328) with transducin-beta-like 1 (TBL1) proteins from human [TBL1a (O60907) and TBL1b (NP_078941.2)]. (E) HOS15 is slightly induced by stress treatments. Control, room temperature; cold, 0°C for 24 h; ABA, 100 μM ABA for 3 h; NaCl, 300 mM NaCl for 4 h.
hos15 Plants Are Specifically Hypersensitive to Freezing.
The hos15 plants were unchanged in their tolerance to NaCl and to exogenous application of ABA (Fig. S4). hos15 plants also displayed similar tolerance to heat stress (Fig. S5) and to oxidative stress conferred by H2O2 (Fig. S6). However, by both visual assessment and by an electrolyte leakage assay, the basal freezing tolerance and tolerance after cold acclimation of hos15 plants is reduced (Fig. 3).
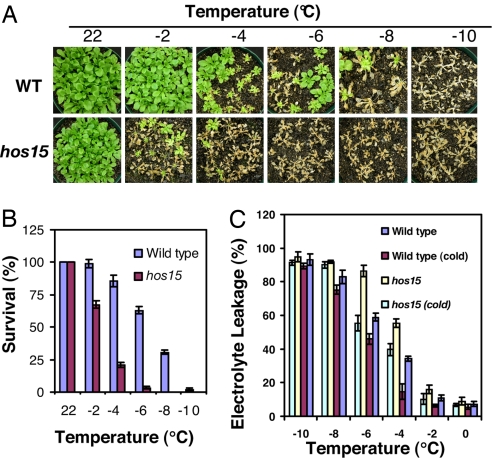
Fig. 3.
hos15 plants are more sensitive to freezing temperatures. (A) Plants are shown after freezing treatment at the indicated temperatures. The photos were taken 1 week after the freezing treatments. (B) Quantification of survival of plants shown in A. Error bars are standard deviation (n = 80–100). (C) Leakage of electrolytes in hos15 and WT plants when treated at indicated temperatures below freezing. WT (cold), hos15 (cold): cold-acclimated (4°C for 7 d) WT and hos15 plants. Error bars are standard deviation (n = 6).
HOS15 Encodes a WD40 Protein.
Using TAIL-PCR, a T-DNA insertion was located in the sixth exon of a predicted gene K8K14.4 (At5g67320) (Fig. 4A). A genomic fragment containing the WT K8K14.4 gene was used to complement the hos15 gene. Forty-five out of 52 homozygous hos15 plants transformed with the WT K8K14.4 gene in the T2 generation displayed WT phenotypes (Fig. 4 B and C). Thus, disruption of At5g67320 in hos15 plants appears to be responsible for the observed altered phenotypes.
Analyses of two HOS15 full-length cDNA sequences of G4G5T7and M66M07STM (GenBank accession no. BE526642) indicated that the HOS15 gene contains 14 exons and 13 introns (Fig. 4A). Real-time RT-PCR analysis revealed that the HOS15 gene is slightly up-regulated by cold, NaCl and ABA treatments (Fig. 4D). Amino acid sequence analysis suggested that HOS15 encodes a protein containing a LisH motif and eight WD40-repeats (Fig. 4A). HOS15 shares high similarity with human transducin beta-like 1 (TBL1) proteins (26–28) (Fig. 4E).
HOS15 Is Important for Histone Deacetylation.
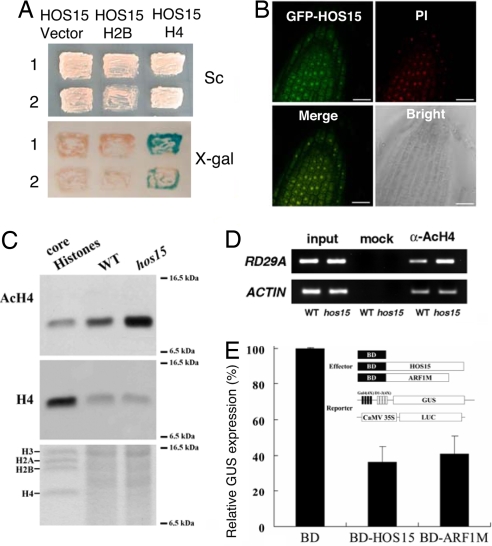
In humans, TBL1 interacts with histones and functions in a repressor protein complex to regulate gene activity. Fig. 5A shows that HOS15 can interact with histone H4 but not histone H2B, which appears to be different from human TBL1 that can interact with both histone H4 and H2B (27). Differences in N-terminal sequences between TBL1 and HOS15 may explain the different histone interaction preferences of these two proteins (29). However, an interaction of HOS15 with H2B may not easily be observed in the yeast nucleus, as used in our assay. Confocal microscopy revealed that a HOS15-GFP fusion protein is predominantly localized in the nucleus as expected for a repressor complex protein (Fig. 5B).
Fig. 5.
HOS15 is important for histone deacetylation. (A) (Upper) two independent yeast cotransformant strains carrying both of HOS15 and vector control (Left), HOS15 and H2B (Center), and HOS15 and H4 (Right) were streaked onto selective media. (Lower) activation of the lacZ reporter gene is indicated by the formation of blue colonies on plates containing X-Gal. (B) Confocal microscope images of root tip cells of transgenic plants transformed with GFP-HOS15 construct. Propidium iodide (PI) staining indicates the positions of nuclei. (C) Western blot analysis with anti-acetylated H4 antibody (Top) revealed that hos15 plants accumulate elevated levels of acetylated H4 compared with WT. Core histones were used as immunoblot control. Western blot, using anti-H4 antibody (Middle) and Coomassie Brilliant Blue staining (Lower) indicate equal amount of proteins were loaded. (D) Chromatin immunoprecipitation (ChIP) analysis, using antibody recognizing actylated histone H4 (α-AcH4). RD29A promoter DNA associated with immunoprecipitated histone was amplified by PCR. Actin served as an internal control. Input: rescued genomic DNA before adding anti-AcH4 antibody (α-AcH4). Mock: ChIP without anti-α-AcH4 antibody. (E) HOS15 functions as a transcriptional repressor. Arabidopsis leaf protoplasts were cotransfected with a reporter gene and two effector genes. Effector genes containing yeast Gal4 DBD (BD) fused in-frame with either HOS15 or ARF1M (control).
hos15 mutant plants accumulated a higher level of nuclear tetra-acetylated histone H4 compared with WT plants (Fig. 5C) after cold treatment. Thus, HOS15 not only interacts with histone H4 but also regulates the acetylation status of histone H4. Moreover, using a chromatin immunoprecipitation (ChIP) assay, we analyzed acetylation of histone H4 associated with the RD29A promoter, one of the hyperinduced genes in the hos15 mutant. Chromatin of WT and hos15 plants was immunoprecipitated by using antibody against tetra-acetylated H4. As shown in Fig. 5D, RD29A promoter DNA was more abundant in precipitated chromatin from the hos15 mutant than from WT plants. Thus, HOS15 appears to function in the repression of RD29A expression by facilitating deacetylation of histone H4 associated with the RD29A promoter.
HOS15 has Repressor Activity.
Because HOS15 appears to function as a part of a transcriptional corepressor complex, we used a transient expression assay to test the potential gene repression function of HOS15. HOS15 fused to the yeast Gal4 BD effector and a constitutively expressed reporter gene containing four upstream Gal4 DNA binding sites (Gal4 (4X)-D1–3(4X)-GUS) were cotransfected into Arabidopsis protoplasts (Fig. 5E) (30). As expected, HOS15 caused repression of the reporter gene expression by a similar amount (≈60%) conferred by a known repressor protein, ARF1M (Fig. 5E). This result provides direct evidence that HOS15 has a repression function in vivo.
Discussion
Our results suggest that HOS15 functions as a repressor to control gene expression important to cold tolerance through chromatin modification. Sequence analysis indicates that HOS15 protein shares highest similarity with the human protein TBL1 (Fig. 4E), a component of the human SMRT/N-CoR gene repressor complex that is involved in modification of chromatin structure by its associated histone deacetylases (HDACs). TBL1 acts as a bridge between the corepressor protein and histones by interacting with both of them within the complex. It is then able to facilitate histone deacetylation that is catalyzed by HDAC3 at specific DNA locations. The N terminus of TBL1 interacts with the N termini of both H2B and H4 histones, and the first three C-terminal WD40-repeats of TBL1 interact with the corepressor complex (27, 29). Our results suggest that HOS15 may have a similar function. HOS15 is localized in the nucleus as revealed by HOS15-GFP localization results (Fig. 5B). HOS15 can interact with histone H4 (Fig. 5A). The hos15 mutant accumulates more acetylated histone H4 than WT plants (Fig. 5C). Moreover, in hos15 mutant plants the promoter of the RD29A gene is more active and is associated with histone H4 that is more acetylated (Figs. 2A and 5D). Therefore, it appears that RD29A and likely other genes involved in stress tolerance are repressed through a function of HOS15 in histone deacetylation.
HOS15 is one of 237 predicted WD40-repeat proteins in Arabidopsis (31). WD40-repeat proteins have been found in many protein complexes, such as histone modification complexes like the N-CoR/SMRT, NuRD, and SIN3 repressors (32) and the Grouncho repressosome (7). Recent biochemical purification of N-CoR and SMRT has demonstrated that both SMRT and N-CoR exist as large protein complexes with an estimated size of 1.5–2.0 MDa and are primarily associated with TBL and HDAC (26, 27, 29, 33). Even though HOS15 resembles TBL1 and appears to function as a component in a protein repressor complex, BLAST searches against Arabidopsis databases failed to identify any significant homologs of proteins that interact with TBL1, i.e., NCoR/SMART corepressor, suggesting that these components in a repressor complex involving HOS15 do not share a high level of sequence identity with those in animal cells.
The involvement of the chromatin acetylation/deacetylation process in cold tolerance (34) suggests that before and during cold acclimation, histone acetylation (reduced corepressor activity) of positive effector genes of cold tolerance, and deacetylation (increased corepressor activity) of negative effector genes of cold tolerance help determine the cold tolerance status of the plant. Genes that fail to become repressed in the hos15 mutant background (Fig. S2) are candidates for such negative effectors that could contribute to the cold sensitivity of the hos15 mutant, even though it exhibits hyperinduction of many cold regulon genes. There were a number of genes with lower levels of expression in hos15, and some of these could be key positive effectors, so their reduced expression may contribute to the freezing sensitivity of the mutant. The precise negative and positive effectors influenced by the hos15 mutation that may explain the observed cold sensitivity of the mutant are unknown. However, it is clear that modification of these targets impact cold tolerance more than the up-regulated COR genes, because the hos15 mutation suppresses any influence of their overexpression and causes loss of cold tolerance. As we have indicated for other cold sensitive mutants (14), hyperexpression of COR genes also may result from an increased signal resulting from hypersensitivity to cold.
Chromatin remodeling is associated with the control of gene expression to establish different epigenetic states of the cell. Epigenetic changes are normally considered to be the main basis of cell type identity and organization (e.g., production of petal cells versus leaf cells during the change from vegetative to floral status). In fact, histone modification has been shown to control the cold-induced (vernalization) flowering response (35). From our results, it is now apparent that altered epigenetic status also is involved in the establishment of a condition of tolerance to different environments as suggested (36).
It is hard to conceive of very specific gene targets for hos15, considering the known general function of the repressor complex, unless a large number of different complexes form from various combinations of different family members of the components, including the HOS15/TBL1-like protein family. Alternatively, partial loss of function of the repressor complex could simply shift its activity to a target discrimination status, influenced by environmental factors such as cold. Several other gene products with traditionally understood “housekeeping” functions also present specific phenotypes upon mutation (37). Identification of other components of the repressor protein complex involving HOS15 and its direct target genes and their functions will certainly reveal more about the role of chromatin structure dynamics in plant cold tolerance.
Materials and Methods
Isolation of the hos15 Mutant.
A T-DNA insertional population of Arabidopsis thaliana plants (ecotype C24) expressing the homozygous transgene RD29A::LUC (13; referred to as WT) was generated by using Agrobacterium tumefaciens-mediated transformation. T2 seeds of transgenic plants were used to screen for mutants exhibiting altered expression of RD29A::LUC in response to cold, ABA, and/or osmotic stress by using a luminescence imaging system as described in ref. 13. The hos15 mutant was identified as showing higher expression of RD29A::LUC gene in response to cold, salt, and ABA treatments.
Freezing Tolerance.
Cold-acclimated and nonacclimated hos15 and WT plants grown in soil at the rosette stage were used for a whole plant freezing tolerance tests and an electrolyte leakage assay as described in refs. 14, 38, and 39.
Northern Blot Analysis.
For gene regulation studies, 2-week-old WT and hos15 seedlings grown on germination medium [1× Murashige and Skoog salts, 2% sucrose (pH 5.7)] were untreated or treated with either low temperature, ABA, or NaCl. Total RNA extraction and subsequent RNA gel analysis were performed as described in ref. 40. The gene specific probes were amplified by PCR with primers as listed in Dataset S5.
Cloning of HOS15 and Construction of Plasmids.
DNA flanking the left border of the inserted T-DNA in hos15 plants was isolated by thermal asymmetric interlaced (TAIL) PCR (40). The HOS15 gene was amplified by PCR, using primers 15A and 15B. The resulting PCR fragment was cloned into the binary vector pCAMBIA1200 between the KpnI and XbaI sites and the identity of the insert was confirmed by sequencing. The resulting construct was then introduced into hos15 mutant plants through Agrobacterium tumefaciens-mediated (strain GV3101) T-DNA transformation. Primary transformants were isolated on MS medium containing 50 mg/liter hygromycin (Invitrogen). Progenies of these transformants were evaluated for RD29A::LUC expression and freezing tolerance.
The coding region of HOS15 was amplified from the full-length cDNA clone G5G4T7 (accession no. N96838) by PCR with the primers 15C and 15D. The PCR fragment was fused in-frame at the c-terminus of GFP in the pEGAD vector. The resulting construct was introduced into Arabidopsis WT plants through Agrobacterium tumefaciens-mediated (strain GV3101) T-DNA transformation. The analysis of GFP expression of the transgenic plants that were resistant to 5 mg/liter bialaphos was performed with a confocal laser-scanning microscope as described in ref. 41. To visualize the positions of nuclei, the seedlings were incubated in the dark at 4°C for 30 min in the propidium iodide (PI) staining solution [0.2 M Tris·HCl (pH 7.5), 4 mM MgCl2, 0.5% (vol/vol) Triton X-100, 0.05 mg/ml propidium iodide, 0.05 mg/ml RNase, and 28.8 mM β-mercaptoethanol].
Real-Time PCR Analysis.
Real-time RT-PCR analysis was performed as described in ref. 42. Primers used for real-time RT-PCR analysis are listed in Dataset S5.
Yeast Two-Hybrid Assays.
The HOS15 coding region was amplified by PCR and cloned in-frame between the NcoI and SmaI sites of pAS2, resulting in the bait plasmid pAS2-HOS15. HTB1 [histone H2B (At1g07790)] and HFO1 [histone H4 (At3g46320)] coding regions were amplified by PCR and in-frame cloned into pACT2 between the BamHI and XhoI sites to generate prey constructs, pACT2-HTB1 and pACT2-HFO1. Plasmid DNA of bait and prey constructs was transformed into the Saccharomyces cerevisiae strain Y190. Individual colonies of transformants were streaked on agar plates containing synthetic complete (SC) media lacking tryptophan and leucine and grown for 24 h. Yeast cells were transferred onto a nitrocellulose transfer membrane (NT BioTrace, 0.45 mm), and β-galactosidase (β-gal) filter assays were performed as described in ref. 43.
Determination of Histone Acetylation.
Nuclei were isolated from 500 mg of Arabidopsis seedling tissues by using Honda's buffer as described in ref. 44. Purified nuclei were resuspended and briefly sonicated in PBS buffer. Twenty micrograms of nuclear protein and 1 μg of purified core histones from chicken (Upstate Biotechnology) were separated by SDS/PAGE and blotted onto a PVDF membrane (Millipore). Anti-tetra-acetylated-histone H4 (1:5,000) or anti-histone H4 (1:2,000) (Upstate Biotechnology) primary antibodies, anti-rabbit horseradish peroxidase coupled antibody, and the ECL system (Amersham) were used to detect acetylated and unacetylated histone H4.
ChIP Assay.
The ChIP assay was performed following the protocol established in ref. 45 with 300 mg of 2-week-old plants, and the same anti-tetra-acetylated-histone H4 antibody that was used for Western blot analysis. Precipitated DNA was dissolved in 75 μl of TE and 2 μl of was used for PCR. Quantitative PCR was used to determine the amounts of genomic DNA immunoprecipitated in the ChIP experiments. The primers used in ChIP assays are listed in Dataset S5.
Gene Repression Assay.
The reporter construct containing the GUS reporter gene used in this study was described in ref. 30. Effector genes were under the control of the CaMV 35S double enhancer promoter followed by a translational enhancer from the Tobacco mosaic virus 5′ leader sequence (46) and a 3′ nopaline synthase untranslated region. The fusion constructs were introduced into Arabidopsis leaf protoplasts from 2-week-old seedlings by PEG–mediated transformation as described in ref. 47. The fluorometric GUS activity assay was performed as described in ref. 48. The control plasmid carrying the luciferase gene under the control of the 35S promoter was used as an internal control to normalize the data for eliminating variations in the experimental conditions (49).
Acknowledgments.
This work was supported in part by National Institutes of Health Grants R01GM070795 and R01GM059138 (to J.-K. Z.), Plant Diversity Research Center of the 21st Century Frontier Research Program of MOST Grant PF0330401–00, Environmental Biotechnology National Core Research Center Project of KOSEF Grant R15–2003-012-01002-00, Biogreen 21 project of the Rural Development Administration Grant 20070301034030 from Korea, National Science Foundation Plant Genome Award DBI0223905/9813360, and United States Department of Agriculture Southwest Consortium Award 2006-34186-16976. This is Purdue University Agricultural Program Paper 2008-18293. J.C. was supported by scholarships from the Brain Korea 21 Program, Ministry of Education and Human Resources Development, Korea.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801029105/DCSupplemental.
References
- 1.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Wu J, Grunstein M. 25 years after the nucleosome model: Chromatin modifications. Trends Biochem Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- 3.Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 4.Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 6.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 7.Courey AJ, Jia S. Transcriptional repression: The long and the short of it. Genes Dev. 2001;15:2786–2796. doi: 10.1101/gad.939601. [DOI] [PubMed] [Google Scholar]
- 8.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 9.Jepsen K, Rosenfeld MG. Biological roles and mechanistic actions of corepressor complexes. J Cell Sci. 2002;115:689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- 10.Reyes JC. Chromatin modifiers that control plant development. Curr Opin Plant Biol. 2006;9:21–27. doi: 10.1016/j.pbi.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 11.Bastow R, et al. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- 12.Sung S, Amasino RM. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature. 2004;427:159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- 13.Ishitani M, Xiong L, Stevenson B, Zhu J-K. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: Interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1997;9:1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu J, et al. An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc Natl Acad Sci USA. 2004;101:9873–9878. doi: 10.1073/pnas.0403166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 17.Wu Z, Irizarry RA, Gentleman R, Martinez Murillo F, Spencer F. A model based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
- 18.Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreps JA, et al. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Provart NJ, et al. Gene expression phenotypes of Arabidopsis associated with sensitivity to low temperatures. Plant Physiol. 2003;132:893–906. doi: 10.1104/pp.103.021261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee BH, Henderson DA, Zhu J-K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell. 2005;17:3155–3175. doi: 10.1105/tpc.105.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]
- 23.Midoro-Horiuti T, Brooks EG, Goldblum RM. Pathogenesis-related proteins of plants as allergens. Ann Allergy Asthma Immunol. 2001;87:261–271. doi: 10.1016/S1081-1206(10)62238-7. [DOI] [PubMed] [Google Scholar]
- 24.van Loon LC, Rep M, Pieterse CM. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- 25.Chinnusamy V, Zhu J, Zhu J-K. Gene regulation during cold acclimation in plants. Physiol Plant. 2006;126:52–61. [Google Scholar]
- 26.Guenther MG, et al. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- 28.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 29.Yoon HG, et al. Purification and functional characterization of the human N-CoR complex: The roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell. 2001;13:2809–2822. doi: 10.1105/tpc.010289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Nocker S, Ludwig P. The WD-repeat protein superfamily in Arabidopsis: Conservation and divergence in structure and function. BMC Genomics. 2003;4:50–60. doi: 10.1186/1471-2164-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–356. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- 33.Li J, et al. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vlachonasios KE, Thomashow MF, Triezenberg SJ. Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell. 2003;15:626–638. doi: 10.1105/tpc.007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood CC, et al. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA. 2006;103:14631–14636. doi: 10.1073/pnas.0606385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molinier J, Ries G, Zipfel C, Hohn B. Transgeneration memory of stress in plants. Nature. 2006;442:1046–1049. doi: 10.1038/nature05022. [DOI] [PubMed] [Google Scholar]
- 37.Xiong L, et al. Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell. 2001;1:771–781. doi: 10.1016/s1534-5807(01)00087-9. [DOI] [PubMed] [Google Scholar]
- 38.Ristic Z, Ashworth EN. Changes in leaf ultrastructure and carbohydrates in Arabidopsis thaliana (Heynh) cv. Columbia during rapid cold acclimation. Protoplasma. 1993;172:111–123. [Google Scholar]
- 39.Ishitani M, Xiong L, Lee H, Stevenson B, Zhu J-K. HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell. 1998;9:1151–1161. doi: 10.1105/tpc.10.7.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu J, et al. An enhancer mutant of Arabidopsis salt overly sensitive 3 mediates both ion homeostasis and the oxidative stress response. Mol Cel Biol. 2007;27:5214–5224. doi: 10.1128/MCB.01989-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 43.Halfter U, Ishitani M, Zhu J-K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weigel D, Glazebrook J. Arabidopsis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- 45.Johnson L, Cao X, Jacobsen S. Interplay between Two Epigenetic Marks: DNA Methylation and Histone H3 Lysine 9 Methylation. Curr Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- 46.Skuzeski JM, Nichols LM, Gesteland RF. Analysis of leaky viral translation termination codons in vivo by transient expression of improved beta-glucuronidase vectors. Plant Mol Biol. 1990;15:65–79. doi: 10.1007/BF00017725. [DOI] [PubMed] [Google Scholar]
- 47.Baek D, et al. Bax-induced cell death of Arabidopsis is meditated through reactive oxygen-dependent and -independent processes. Plant Mol Biol. 2004;56:15–27. doi: 10.1007/s11103-004-3096-4. [DOI] [PubMed] [Google Scholar]
- 48.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoo JH, et al. Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. J Biol Chem. 2005;280:3697–3706. doi: 10.1074/jbc.M408237200. [DOI] [PubMed] [Google Scholar]