Abstract
The transcriptional coactivator PGC-1α is a potent regulator of several metabolic pathways, including, in particular, the activation of oxidative phosphorylation and mitochondrial biogenesis. Recent evidence suggests that increasing PGC-1α activity may have beneficial effects in various conditions, including muscular dystrophy, diabetes, and neurodegenerative diseases. We describe here a high-throughput screen to identify small molecules that induce PGC-1α expression in skeletal muscle cells. A number of drug classes are identified, including glucocorticoids, microtubule inhibitors, and protein synthesis inhibitors. These drugs induce PGC-1α mRNA, and the expression of a number of genes known to be regulated by PGC-1α. No induction of these target genes is seen in PGC-1α −/− cells, demonstrating that the drugs act through PGC-1α. These data demonstrate the feasibility of high-throughput screening for inducers of PGC-1α. Moreover, the data identify microtubule inhibitors and protein synthesis inhibitors as modulators of PGC-1α and oxidative phosphorylation.
Keywords: colchicine, high throughput
Coactivators are proteins that dock on transcription factors and alter chromatin structure and the transcription machinery to stimulate gene expression (reviewed in ref. 1). It is likely that all transcription factors must dock one or more coactivators to initiate transcription. Most studies of gene regulation have focused on the modulation of transcription factors; more recently, however, coactivators have emerged as potent regulatory targets of physiological stimuli and hormones (2).
PGC-1α is the best-studied example of such a regulated coactivator. PGC-1α was first identified as a cold-inducible PPAR-γ-binding protein in brown fat (3). Since then, it has become apparent that PGC-1α can bind to, and coactivate, most members of the nuclear receptor family, and many other transcription factors. PGC-1α activates transcription by recruiting several proteins that have histone acetyltransferase activity, including CBP, p300, and SRC-1 (4), and the Mediator protein complex, which is thought to recruit RNA polymerase II (5).
PGC-1α has a variety of biological activities in different tissues, and most of these activities are linked to various aspects of oxidative metabolism. The expression of PGC-1α in white fat cells gives them many of the properties of brown fat cells, including mitochondrial biogenesis and expression of UCP-1. PGC-1α is induced by exercise in skeletal muscle (e.g., refs. 6–9), where it induces mitochondrial biogenesis, angiogenesis (41), and a switch in fibers toward more oxidative types I and IIa (10). PGC-1α is induced in the fasted liver, where it induces gluconeogenesis and β-oxidation of fatty acids (11, 12). In all of these cases, PGC-1α increases a core program of mitochondrial biogenesis and respiration, and ancillary programs that go along with increased respiration in each tissue.
Elevating PGC-1α activity may be beneficial in a number of diseases. PGC-1α −/− mice are prone to several neurodegenerative diseases (13), suggesting a neuroprotective role for PGC-1α. The same mice also have a strong predisposition to cardiac failure (14, 15), and PGC-1α levels decline in a number of rodent models of heart dysfunction (16–20), suggesting a cardioprotective role for PGC-1α as well. Transgenic expression of PGC-1α in skeletal muscle protects from the atrophy associated with denervation, a model for ALS (21). The same mice also have a markedly improved recovery of blood flow after ligation of the femoral artery, a model of chronic ischemia (41). Finally, transgenic expression of PGC-1α in skeletal muscle also ameliorates the muscle damage and reduced locomotive function evident in mdx mice, a model of Duchenne's muscular dystrophy (22).
These observations provide a very strong impetus to search for drugs that can induce PGC-1α. PGC-1α is highly regulated in a number of tissues; this indicates that pathways exist that impinge on PGC-1α expression, and suggests that they might be targets for small molecules. We describe here a high-throughput screening method for the identification of small molecules that can regulate gene expression. When applied to PGC-1α, the method identified a number of compounds able to induce PGC-1α, and a genetic and cell biological program regulated by PGC-1α.
Results
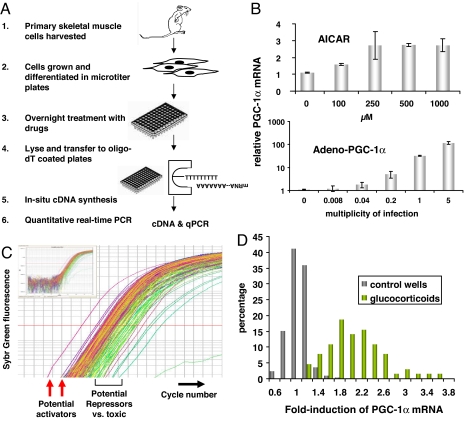
An outline of the screen for chemical activators of PGC-1α gene expression is shown in Fig. 1A. In brief, primary satellite skeletal muscle cells are harvested from mouse hindlimb, grown in culture, plated into 96-well plates, and stimulated to differentiate for 3 days. The wells are then treated with compounds for 24 h. After this, the cells are washed and lysed, and the total lysate is transferred to 384-well plates coated covalently with oligo(dT), to which poly(A)-tailed mRNA hybridizes. After extensive washing, the bound mRNA is reverse transcribed in situ to yield cDNA, and this is used as template for quantification of gene expression by real-time PCR. The measured expression of PGC-1α mRNA is then normalized to the measured expression of TATA box-binding protein (TBP), which acts as an internal control. In this way, the response of any chosen gene to treatment with each compound in a given compound library can be determined.
Fig. 1.
Screen for small molecules that induce PGC-1α. (A) Schema of the screen. (B Upper) Myotubes were treated with the indicated doses of AICAR for 24 h, and levels of PGC-1α mRNA were measured. (B Lower) PGC-1α mRNA levels in myotubes 48 h after infection with adenovirus encoding for PGC-1α. (C) Representative graph of qPCR amplification curves detecting PGC-1α mRNA levels from one 384-well qPCR plate of the screen. (Inset) Shown are the complete curves. (D) Bin diagram of fold induction of PGC-1α in DMSO-control wells and in wells containing a synthetic glucocorticoid.
Primary skeletal muscle cells were chosen for the screen for a number of reasons. First, these cells are not embryonic and not immortalized. Second, PGC-1α can be induced in these cells in response to certain stimuli, such as AMP-activated protein kinase (AMPK) activators (23). And third, levels of PGC-1α expression in these cells are 10 times higher than in clonal myoblastic cell lines (e.g., C2C12s) and much closer to those found in intact skeletal muscle (data not shown). The measurement of endogenous gene expression was also chosen for a number of reasons. First, the common alternative of using promoter-luciferase fusion constructs is severely limited when the precise makeup of the promoter in question is not known. Second, gene expression on plasmids is artificial and fails to take into account chromatin modifications. Third, only by looking at endogenous gene expression can effects by distant elements, such as enhancers, be detected. And fourth, assays based on promoter-luciferase constructs cannot detect effects on gene expression due to mRNA stability, splicing, or other posttranscriptional events.
The feasibility of evaluating PGC-1α gene expression in 96-well plates of skeletal muscle cells was tested by treating cells with either 5-aminoimidazole-4-carboxamide-1-β-d-ribofuranoside (AICAR), an AMPK activator known to induce PGC-1α in these cells (23), or by infecting the cells with varying doses of adenovirus encoding for PGC-1α. As shown in Fig. 1B, doses of AICAR as low as 100 μM, and infection with virus with a multiplicity of infection as low as 0.04, could reliably detect increased PGC-1α in this format. Next, the assay variability was determined by measuring PGC-1α expression in 640 wells that were “mock”-treated with DMSO vehicle. The standard deviation of the measured Ct value was 0.45 (Table 1). The standard deviation was further reduced to 0.31 when the values were normalized to the internal control, TBP (Table 1). This variability is quite small, yielding a Z factor of 0.6 for detecting increases in PGC-1α expression of as little as 3-fold. Indeed, in the 640 control samples, not one had a PGC-1α expression that differed >2-fold from the mean.
Table 1.
Results of the primary screen
| n | Standard deviation, cycles |
Number of positives |
||||
|---|---|---|---|---|---|---|
| TBP | PGC-1α | TBP minus α | >2× induced | >3× induced | ||
| Controls | 640 | 0.36 | 0.45 | 0.31 | 0 (0%) | 0 (0%) |
| All wells | 3,840 | 0.41 | 0.53 | 0.42 | 82 (2.5%) | 23 (0.7%) |
*Levels of PGC-1α and TBP mRNAs were evaluated. The standard deviations of the cycle number at which SYBR green fluorescence reached an arbitrary threshold are indicated for the control wells and the test wells.
†Number of hits, as defined by either 2-fold or 3-fold induction over the average signal in the 640 control wells.
We used this method to screen a library of 3,120 compounds, made up of the Spectrum Collection (2,000 cmpds) and the Prestwick Chemical Library (1,120 cmpds). Together these collections include a total of 2,490 unique compounds, including ≈40% of all Food and Drug Administration (FDA)-approved drugs, with most of the remaining compounds being known bioactives. Samples from most known drug classes are represented in the group. Ten 384-well plates were treated as described, and the expression of PGC-1α (and TBP as control) was evaluated. A number of the wells in each plate were treated only with vehicle (DMSO), to be used as plate-specific negative controls. The remainder of the wells were treated with compounds in the micromolar range. Fig. 1C shows representative qPCR amplification curves for PGC-1α for one of these plates; potential activators are identified as wells where PGC-1α cDNA is amplified earlier than the in the other wells, whereas wells where PGC-1α is amplified later reflect either inhibition of PGC-1α mRNA expression or toxicity to the cells. The latter can be separated based on whether expression of TBP is also affected. Seventy-eight distinct compounds that induce PGC-1α expression 2-fold or greater were identified from the library of compounds (Table 2). When a compound identified as a “hit” was represented more than once in the library, it was identified as positive in each representation, strongly supporting the reproducibility of the screen. A plate containing candidate compounds was generated and used to retest the findings from the primary screen. Only 24 of the original 78 compounds failed to induce PGC-1α >2-fold in this repeat of the assay (Table 2), underscoring the fidelity of the method.
Table 2.
Small molecules identified
| n* | Fold induction† | Confirmed positives‡ | |
|---|---|---|---|
| Glucocorticoids | 37 | 2.00–3.65 | 3 of 3 |
| Tubulin inhibitors | 9 | 2.12–5.34 | 3 of 3 |
| Protein synthesis inhibitors | 5 | 3.61–73.64 | 3 of 3 |
| Others | 31 | 2.02–3.22 | 7 of 31 |
*n, number of molecules identified in that class.
†Fold induction, over the average signal of the 640 control wells.
‡Confirmed positives indicates molecules that induced PGC-1α >2-fold in a repeat of the screen.
It is interesting to note what the false-negative rate of the screen would be under various settings. If we use the strongest hit in the screen as a positive control (emetine, 74-fold) and 2-fold induction of PGC-1α as a threshold for defining a hit, then the false-negative rate is essentially zero (data not shown). On the other hand, a weaker inducer, like a glucocorticoid, is more likely to fall below a predetermined threshold. 37 of the original hits were synthetic or naturally occurring glucocorticoids. There were, in fact, 64 glucocorticoids in the library used (data not shown). Of these, 37 (58%) induced PGC-1α >2-fold, 59 (92%) induced PGC-1α >1.5-fold, and all of them induced PGC-1α >1.3-fold (Fig. 1D). Thus, even for very mild inducers of PGC-1α expression, the false-negative rate is very low (e.g., 8% for a cutoff of >1.5-fold induction, or 0% for a cutoff of >1.3-fold). The PGC-1α promoter contains canonical binding sites for the glucocorticoid receptor (GR), and the GR has been shown to induce PGC-1α expression in liver (12, 24); the data here demonstrate this to be true in muscle cells as well.
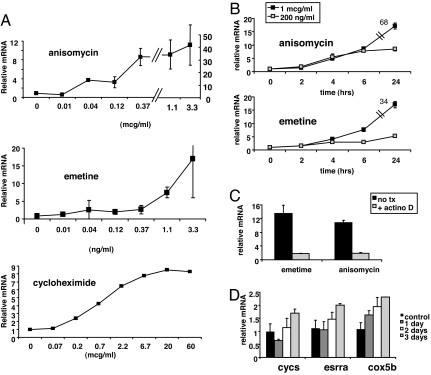
The most potent inducers of PGC-1α were emetine, anisomycin, and cephaline, all three of which are protein synthesis inhibitors. Emetine and cephaline are the two main ingredients of sodium of ipecac, whereas anisomycin is an antifungal agent often used for academic purposes as an activator of MAPK. Treatment of primary skeletal muscle myotubes with varying doses of anisomycin for 16 h demonstrated that as little as 40 ng/ml of drug significantly induced PGC-1α expression, whereas higher doses induced PGC-1α as much as 40-fold (Fig. 2A). Similarly, 40 ng/ml of emetine induced PGC-1α 4-fold, whereas 3.3 μg/ml induced PGC-1α 18-fold. Cycloheximide is a well known inhibitor of protein synthesis that did not score in the screen here. Nevertheless, it too induced PGC-1α up to 10-fold (Fig. 2A), confirming that inhibition of protein synthesis likely mediates the induction of PGC-1α. It is not clear why cycloheximide did not score in the primary screen. Both anisomycin and emetine induced PGC-1α within 4 h but had no effect at 2 h (Fig. 2B), indicating that the mechanism of action is not an immediate-early response sometimes seen with inhibition of protein synthesis. Treatment with actinomycin-D, an inhibitor of transcription, completely blocked the induction of PGC-1α (Fig. 2C), demonstrating that induction occurred at the transcriptional level (or, less likely, by a profound effect on mRNA half-life). To test the effects of anisomycin on genes known to be regulated by PGC-1α, cells were treated with anisomycin for up to 3 days. Only low doses of anisomycin were used, because higher doses would completely block all synthesis of new protein. As shown in Fig. 2D, this led to significant induction at 3 days of several well known PGC-1α target genes, including esrra (ERRα), cycs (cytochrome c), and cox5b (a subunit of cytochrome oxidase). That mild inhibition of protein synthesis induces PGC-1α mRNA without totally blocking its translation suggests that the mechanism involved is more sensitive than simply a general reduction in protein translation.
Fig. 2.
Protein synthesis inhibitors induce PGC-1α in primary skeletal muscle cells. (A) Myotubes were treated with the indicated doses of anisomycin, emetine, or cycloheximide, and levels of PGC-1α mRNA were evaluated after 16 h. (B) Myotubes were treated with 200 ng/ml or 1 μg/ml of anisomycin or emetine for the indicated times, and PGC-1α mRNA was measured. (C) PGC-1α mRNA levels in myotubes treated with anisomycin or emetine after pretreatment for 30 min with actinomycin D. (D) Myotubes were treated with 30 ng/ml of anisomycin for 1–3 days, as indicated, and the mRNA levels of cycs, esrra, and cox5b were measured by qPCR.
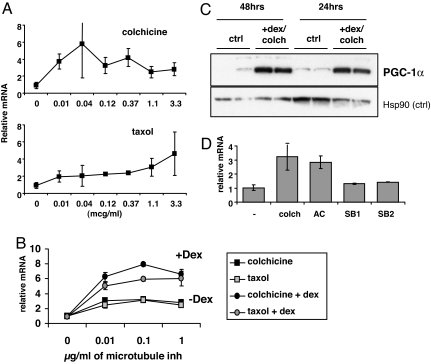
The most frequently identified inducers of PGC-1α in this screen, outside the glucocorticoids, were microtubule inhibitors. These included colchicine, taxol, podophylotoxin, and nocodazole and a number of similar “-azoles,” all of which induced PGC-1α 2- to 5-fold (Table 1). Interestingly, both microtubule destabilizers (e.g., colchicine) and stabilizers (e.g., taxol) scored positively. Treatment of primary skeletal muscle myotubes with varying doses of colchicine for 16 h demonstrated that as little as 10 ng/ml of drug induced PGC-1α mRNA 3-fold (Fig. 3A). Similarly, 10 ng/ml of taxol induced PGC-1α expression 2-fold; higher doses did not increase PGC-1α expression much more. However, treating cells with a combination of a microtubule inhibitor and a glucocorticoid had a synergistic effect, inducing PGC-1α mRNA as much as 8-fold (Fig. 3B). Dexamethasone alone induces PGC-1α only ≈1.3- to 2-fold (Fig. 3B and data not shown). The amount of PGC-1α protein was similarly induced, as determined by Western blotting (Fig. 3C). Microtubule inhibitors have been reported to affect MAPK signaling (25–27), and p38 MAPK is known to phosphorylate and stabilize PGC-1α (28). Treating cells with p38 MAPK inhibitors before exposure to colchicine abrogated the induction of PGC-1α by colchicine (Fig. 3D). Inhibition of adenylate cyclase, a known modulator of PGC-1α expression, had no impact (Fig. 3D). Hence, microtubule inhibitors induce PGC-1α expression and protein levels, at least, in part, by activation of p38 MAPK.
Fig. 3.
Tubulin inhibitors induce PGC-1α in primary skeletal muscle cells and synergize with glucocorticoids. (A) Myotubes were treated with the indicated doses of colchicine or taxol, and levels of PGC-1α mRNA were evaluated after 16 h. (B) PGC-1α mRNA levels after treatment for 16 h with the indicated doses of taxol or colchicine with and without 1 μM dexamethasone. (C) Myotubes were treated with 30 ng/ml colchicine + 1 μM dexamethasone for 24 or 48 h, and PGC-1α protein levels were measured by Western blotting. (D) PGC-1α mRNA levels in myotubes treated with colchicine + 1 μM dexamethasone after pretreatment for 30 min with the indicated inhibitors. AC, adenylase cyclase inhibitor SQ 22536, p38 MAPK inhibitors SB1, sb203580, and SB2, sb202190.
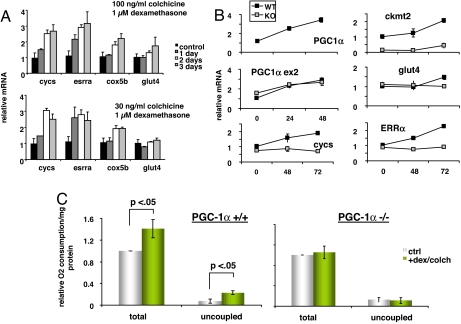
To evaluate the effects of microtubule inhibitors on genes known to be regulated by PGC-1α, differentiated primary skeletal muscle cells were treated with colchicine and dexamethasone for up to 3 days. This led to significant induction of a number of PGC-1α target genes, including esrra, cycs, and cox5b (Fig. 4A). The requirement for PGC-1α in the induction of these genes was tested by comparing the response of primary skeletal muscle cells from wild-type and PGC-1α −/− mice. Three days of treatment induced PGC-1α target genes in wild-type cells, but not in PGC-1α −/− cells (Fig. 4B). Taken together, these data indicate that treating cells with a combination of a microtubule inhibitor and a glucocorticoid induces a program of genes involved in oxidative phosphorylation by induction of PGC-1α.
Fig. 4.
Tubulin inhibitors + glucocorticoids induce oxidative phosphorylation genes and activity in a PGC-1α-dependent fashion. (A) Myotubes were treated with the indicated drugs for 1–3 days, as indicated, and the mRNA levels of cycs, esrra, cox5b, and glut4 were measured by qPCR. (B) Primary skeletal muscle cells were prepared from PGC-1α +/+ and −/− animals, differentiated into myotubes, and treated with 30 ng/ml colchicine + 1 μM dexamethasone. After 48 or 72 h, PGC-1α mRNA levels were measured. (C) Total and uncoupled respiration in PGC-1α +/+ and −/− myotubes treated for 72 h with 30 ng/ml colchicine + 1 μM dexamethasone.
Finally, to examine whether the activation of this PGC-1α-dependent genetic program led to physiological changes in oxidative phosphorylation, cellular respiration was measured by using an oxygen-sensing Clarke electrode. Treatment of cells with colchicine and dexamethasone for 3 days led to a significant 30% increase in total respiration (Fig. 4C). Uncoupled respiration, as measured after the addition of the complex V poison oligomycin, more than doubled (Fig. 4C). These changes mirror those observed after the introduction of ectopic PGC-1α in cells (29). Importantly, colchicine and dexamethasone had no effect on respiration in PGC-1α −/− cells, strongly suggesting that PGC-1α is required for the effect (Fig. 4C).
Discussion
We have described here a high-throughput method for identifying small molecules that induce PGC-1α gene expression. By using this method, we identify microtubule inhibitors, glucocorticoids, and protein synthesis inhibitors, as potent inducers of PGC-1α expression. High-throughput evaluation of gene expression is generally done by using plasmid-based luciferase assays (30). These approaches suffer from significant shortcomings, including the artificial nature of unchromatinized plasmids and the assumptions that must be made when constructing reporter plasmids. Methods to assay endogenous gene expression, including array-based and bead-based approaches, often have limited sensitivity/specificity profiles, significant time and cost demands that prohibit true high throughput, and/or the need for highly specialized equipment (e.g., refs. 30–35). The screen described here is simple and relatively inexpensive, allowing for scaling up to larger libraries of compounds. No machinery other than standard liquid transfer robotics is needed. Induction of gene expression by as little as 2-fold can reliably be detected with a very low false-positive rate. Hence, the screen boasts both a high sensitivity and specificity. The method is also adaptable to measuring any gene of interest and, in principle, to cells other than the primary skeletal muscle cells used here.
Transcription factors have not typically been successfully targeted in small-molecule screens. In part this is because of their nuclear location, but in large part it reflects the fact that transcription factors are generally not highly regulated and most do not bind ligands (a number of nuclear receptors are a notable exception). PGC-1α, on the other hand, is highly modulated in a number of physiological contexts, such as in the liver in response to fasting and in muscle in response to exercise. A number of intracellular pathways are known to impinge on PGC-1α, including signaling by cAMP, AMPK, Ca2+, and p38 MAPK (23, 24, 28, 36–38). It therefore seemed plausible that small molecules that regulate PGC-1α expression could be identified. The results shown here demonstrate that this is indeed the case. It will be of great interest to screen larger libraries, containing a diversity of compounds, to search for modulators of PGC-1α.
Microtubule inhibitors have wide clinical use, most often as antiproliferatives for purposes ranging from antifungal to antineoplastic. Colchicine is also used clinically for the treatment of gout. Its anti-inflammatory action is thought to be mediated mostly on inflammatory cells in affected joints. The mechanism of its action remains incompletely understood. Evidence indicates that PGC-1α may have anti-inflammatory activities (39); the data presented here suggest the notion that colchicine may inhibit inflammation at least in part by induction of PGC-1α.
Glucocorticoids are also widely used as anti-inflammatory agents. Interestingly, the combination of a glucocorticoid and a microtubule inhibitor had a synergistic effect on the induction of PGC-1α. Combination therapy is burgeoning as an approach to harness the usefulness of established drugs. Full activation of pathways may require multiple hits. Moreover, in principle, different drugs that synergize on a particular pathway could be used in smaller doses, thereby minimizing side effects.
Being able to induce PGC-1α pharmaceutically holds great clinical promise. The induction of PGC-1α in skeletal muscle by genetic means improves angiogenesis and recovery from ischemia (41), blocks the atrophy triggered by denervation (21), and improves indices of muscle damage in a model of Duchenne's muscular dystrophy (22). PGC-1α may also have important cardioprotective and neuroprotective effects (13–15). Simultaneous induction of PGC-1α in all tissues in the body may harbor some side effects, such as induction of gluconeogenesis in the liver (11, 12). Ideally, drugs that target only specific tissues will be identified. At the same time, certain side effects would certainly be tolerated in the treatment of lethal and incurable diseases such as DMD.
Methods
Cells and Reagents.
Primary skeletal muscle cells were harvested from 1-wk-old C57BL/6 mice and grown as described in ref. 40. Cells were amplified through four to five passages and frozen in batches. C2C12 and 10T1/2 cells were acquired from American Type Culture Collection. The drug library was from the Broad Institute. All other drugs were from Sigma. Oligo(dT)-coated plates were from Qiagen. RT and qPCR reagents were from Applied Biosystems. Adenovirus encoding for PGC-1α has been described. Oligonucleotides were purchased from Integrated DNA Technologies. Respiration assays were performed as described in ref. 29.
Primary Screen and qPCR.
Cells were thawed in batches immediately into 96-well plates and allowed to grow for 24 h. One hundred nanoliters of compound was then pin-transferred into each well (final concentration, in general, 10 μM) by using a CyBi-Well (CyBio) equipped with a 96-pin array. Twenty-four hours later, cells were washed once in PBS and then lysed in 80 μl of lysis buffer (Qiagen) by using a Multidrop Combi (Thermo Electron). After 5 min, 30 μl of cell lysate was transferred to 384-well oligo(dT)-coated plates (using CyBi-Well) and incubated at room temperature for 90 min. Plates were washed three times with washing buffer (Qiagen) and reverse transcription was performed in the same well, according to the manufacturer's instructions (Applied Biosystems) in a 5-μl volume, after which the cDNA-containing mixture was diluted with 20 μl of water. Two microliters of this solution were then transferred to 384-well qPCR plates (using CyBi-Well), and 3 μl of master buffer containing PCR oligos and Cyber-containing mix (Applied Biosystems) were added (using Multidrop Combi). Quantitative real-time PCR was performed by using an ABI 7900HT instrument. qPCR data were analyzed by using the ΔΔCt method, with TBP as internal control and pooled DMSO-treated samples as external control. For more details on this protocol, see Wagner and Arany (42). All error bars shown are +/− SEM.
Acknowledgments.
This work is supported by National Institutes of Health Grants HL079172 and NS059440 (to Z.A.) and DK54477 and DK61562 (to B.M.S.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Hermanson O, Glass CK, Rosenfeld MG. Nuclear receptor coregulators: Multiple modes of modification. Trends Endocrinol Metab. 2002;13:55–60. doi: 10.1016/s1043-2760(01)00527-6. [DOI] [PubMed] [Google Scholar]
- 2.Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 3.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 4.Puigserver P, et al. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 5.Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 6.Koves TR, et al. PPARgamma coactivator-1alpha -mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem. 2005;280(39):33588–33598. doi: 10.1074/jbc.M507621200. [DOI] [PubMed] [Google Scholar]
- 7.Cluberton LJ, McGee SL, Murphy RM, Hargreaves M. Effect of carbohydrate ingestion on exercise-induced alterations in metabolic gene expression. J Appl Physiol. 2005;99:1359–1363. doi: 10.1152/japplphysiol.00197.2005. [DOI] [PubMed] [Google Scholar]
- 8.Baar K, et al. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- 9.Norrbom J, et al. PGC-1alpha mRNA expression is influenced by metabolic perturbation in exercising human skeletal muscle. J Appl Physiol. 2004;96:189–194. doi: 10.1152/japplphysiol.00765.2003. [DOI] [PubMed] [Google Scholar]
- 10.Lin J, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 11.Herzig S, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 12.Yoon JC, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 13.Lin J, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Arany Z, et al. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1:259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Arany Z, et al. Transverse aortic constriction leads to accelerated heart failure in mice lacking PPAR-γ coactivator 1α. Proc Natl Acad Sci USA. 2006;103:10056–10091. doi: 10.1073/pnas.0603615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehman JJ, Kelly DP. Gene regulatory mechanisms governing energy metabolism during cardiac hypertrophic growth. Heart Fail Rev. 2002;7:175–185. doi: 10.1023/a:1015332726303. [DOI] [PubMed] [Google Scholar]
- 17.Garnier A, et al. Depressed mitochondrial transcription factors and oxidative capacity in rat failing cardiac and skeletal muscles. J Physiol. 2003;551:491–501. doi: 10.1113/jphysiol.2003.045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finck BN, Lehman JJ, Barger PM, Kelly DP. Regulatory networks controlling mitochondrial energy production in the developing, hypertrophied, and diabetic heart. Cold Spring Harbor Symp Quant Biol. 2002;67:371–382. doi: 10.1101/sqb.2002.67.371. [DOI] [PubMed] [Google Scholar]
- 19.Sano M, et al. Activation of cardiac Cdk9 represses PGC-1 and confers a predisposition to heart failure. EMBO J. 2004;23:3559–3569. doi: 10.1038/sj.emboj.7600351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Czubryt MP, McAnally J, Fishman GI, Olson EN. Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1 alpha) and mitochondrial function by MEF2 and HDAC5. Proc Natl Acad Sci USA. 2003;100:1711–1716. doi: 10.1073/pnas.0337639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandri M, et al. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci USA. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Handschin C, et al. PGC-1alpha regulates the neuromuscular junction program and ameliorates Duchenne muscular dystrophy. Genes Dev. 2007;21:770–783. doi: 10.1101/gad.1525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci USA. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci USA. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subbaramaiah K, Hart JC, Norton L, Dannenberg AJ. Microtubule-interfering agents stimulate the transcription of cyclooxygenase-2. Evidence for involvement of ERK1/2 AND p38 mitogen-activated protein kinase pathways. J Biol Chem. 2000;275:14838–14845. doi: 10.1074/jbc.275.20.14838. [DOI] [PubMed] [Google Scholar]
- 26.Lee LF, Li G, Templeton DJ, Ting JP. Paclitaxel (Taxol)-induced gene expression and cell death are both mediated by the activation of c-Jun NH2-terminal kinase (JNK/SAPK) J Biol Chem. 1998;273:28253–28260. doi: 10.1074/jbc.273.43.28253. [DOI] [PubMed] [Google Scholar]
- 27.Wang TH, et al. Microtubule-interfering agents activate c-Jun N-terminal kinase/stress-activated protein kinase through both Ras and apoptosis signal-regulating kinase pathways. J Biol Chem. 1998;273:4928–4936. doi: 10.1074/jbc.273.9.4928. [DOI] [PubMed] [Google Scholar]
- 28.Puigserver P, et al. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Mol Cell. 2001;8:971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 29.St-Pierre J, et al. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells. J Biol Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 30.Inglese J, et al. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 31.Stegmaier K, et al. Gene expression-based high-throughput screening (GE-HTS) and application to leukemia differentiation. Nat Genet. 2004;36:257–263. doi: 10.1038/ng1305. [DOI] [PubMed] [Google Scholar]
- 32.Wei G, et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell. 2006;10:331–342. doi: 10.1016/j.ccr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Lamb J, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 34.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 35.Kris RM, et al. High-throughput, high-sensitivity analysis of gene expression in Arabidopsis. Plant Physiol. 2007;144:1256–1266. doi: 10.1104/pp.107.098681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohas LM, et al. A fundamental system of cellular energy homeostasis regulated by PGC-1alpha. Proc Natl Acad Sci USA. 2007;104:7933–7938. doi: 10.1073/pnas.0702683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barger PM, Browning AC, Garner AN, Kelly DP. p38 mitogen-activated protein kinase activates peroxisome proliferator-activated receptor alpha: A potential role in the cardiac metabolic stress response. J Biol Chem. 2001;276:44495–44501. doi: 10.1074/jbc.M105945200. [DOI] [PubMed] [Google Scholar]
- 38.Knutti D, Kressler D, Kralli A. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proc Natl Acad Sci USA. 2001;98:9713–9718. doi: 10.1073/pnas.171184698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Handschin C, et al. Abnormal glucose homeostasis in skeletal muscle-specific PGC-1alpha knockout mice reveals skeletal muscle-pancreatic beta cell crosstalk. J Clin Invest. 2007;117:3463–3474. doi: 10.1172/JCI31785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 41.Arany Z, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature. 2008;451(7181):1008–1012. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- 42.Wagner BK, Arany Z. High-throughput real-time PCR for detection of gene expression levels. In: Clemons P, Tollida N, Wagner B, editors. Cell-Based Assays in High Throughput Screening. Totowa, NJ: Humana Press; 2008. in press. [Google Scholar]