Abstract
Developing tissues require cells to undergo intricate processes to shift into appropriate niches. This requires a functional connection between adhesion-mediating events at the cell surface and a cytoskeletal reorganization to permit directed movement. A small number of proteins are proposed to link these processes. Here, we identify one candidate, Cindr, the sole Drosophila melanogaster member of the CD2AP/CIN85 family (this family has been previously implicated in a variety of processes). Using D. melanogaster retina, we demonstrate that Cindr links cell surface junctions (E-cadherin) and adhesion (Roughest) with multiple components of the actin cytoskeleton. Reducing cindr activity leads to defects in local cell movement and, consequently, tissue patterning and cell death. Cindr activity is required for normal localization of Drosophila E-cadherin and Roughest, and we show additional physical and functional links to multiple components of the actin cytoskeleton, including the actin-capping proteins capping protein alpha and capping protein beta. Together, these data demonstrate that Cindr is involved in dynamic cell rearrangement in an emerging epithelium.
Introduction
By recruiting proteins into complexes, adaptor proteins create nodes of regulation and activity. The founding member of the CD2AP/CIN85 family of adaptor proteins was initially isolated in a yeast interaction screen as a binding partner of the T cell receptor CD2 (Dustin et al., 1998), independently from the kidney (MET-1; Lehtonen et al., 2000) and as a ligand for p130Cas (Kirsch et al., 1999). The homologue CIN85 was identified as a partner of the E3 ubiquitin ligase Cbl (Take et al., 2000) and separately as SETA and Ruk (Bogler et al., 2000; Gout et al., 2000). Many roles have been ascribed to the CD2AP/CIN85 family but its function in situ remains poorly understood. Here, we examine the sole Drosophila melanogaster CD2AP/CIN85 orthologue, cindr.
The phenotype of CD2AP knockout mice is chiefly one of tissue degeneration: cardiac hypertrophy, splenic and thymic atrophy, glomerular sclerosis, and a loss of podocyte foot processes (Shih et al., 1999). The CD2AP/CIN85 family is primarily proposed to function in endocytosis to down-regulate receptor tyrosine kinase activity (Dikic, 2003). This model arises from coimmunoprecipitation and interaction experiments that have identified a wealth of CD2AP/CIN85 interactors, colocalization studies performed in culture or tissue, and in vitro assays. CIN85 constitutively associates with endophilin and, on growth factor stimulation, complexes with Cbl to mediate receptor down-regulation. Furthermore, interactions between CD2AP/CIN85 and other trafficking proteins have been described including AP-2, Dab2, Rab4, PAK2, ALIX, and ESCRT-1 (Chen et al., 2000; Brett et al., 2002; Cormont et al., 2003; Kowanetz et al., 2003; Kurakin et al., 2003; Schmidt et al., 2003; Usami et al., 2007).
Recent work has also suggested a relationship between CD2AP/CIN85 proteins and actin. They have been detected in actin-rich regions of podocytes and cultured cells and have been found to bind actin in vitro to promote actin bundling (Kirsch et al., 2001; Welsch et al., 2001, 2005; Lehtonen et al., 2002; Cormont et al., 2003; Schmidt et al., 2003; Schiwek et al., 2004; Gaidos et al., 2007; Gauthier et al., 2007). CD2AP has also been reported to bind the actin-capping protein CPα/β dimer and inhibit its function in vitro (Hutchings et al., 2003; Bruck et al., 2006) and anillin at the actin-rich cleavage furrow (Monzo et al., 2005). CD2AP activity is required for migration of rat gastric mucosal cells (Mustonen et al., 2005) and polarization of the cytoskeleton during T cell receptor activation (Dustin et al., 1998). The role and mechanism by which CD2AP/CIN85 regulates cytoskeletal dynamics within an epithelium in situ remains unclear.
Furthermore, the CD2AP/CIN85 family has also been reported to bind the adhesion molecules E-cadherin and nephrin (Shih et al., 1999, 2001; Palmen et al., 2002; Lehtonen et al., 2004; Mustonen et al., 2005). Nephrin and NEPH-1 form the backbone of the slit diaphragm, a specialized junction that traverses podocyte foot processes in the mammalian kidney (Huber and Benzing, 2005; Patari-Sampo et al., 2006). Direct interactions between CD2AP, nephrin, and podocin and between CD2AP and the podocyte-specific actin-bundling protein synaptopodin are essential for slit diaphragm integrity. In addition, a protein complex containing nephrin, cadherin, p120-catenin, ZO-1, and CD2AP has been isolated from Madin-Darby canine kidney cells and mouse glomerular lysates (Lehtonen et al., 2004). Collectively, these data suggest that CD2AP may have a role in anchoring junctions to the cytoskeleton or in regulating actin dynamics at this important intersection.
The challenge remains to understand how different roles of CD2AP/CIN85 are integrated in the organism, which interactors are recruited into CD2AP/CIN85 complexes, and how these are regulated. Here, we show that targeted reduction of cindr in the pupal fly eye resulted in defects in overall patterning due to aberrant local cell movements. We link these defects to misregulation of actin dynamics and mislocalization of Drosophila E-Cadherin (DE-Cad) and the fly NEPH-1 orthologue Roughest (Rst) which, with its binding partner Hibris (Hbs; a D. melanogaster Nephrin orthologue), is a central mediator of cell–cell adhesion in the pupal retina. We link Cindr functionally and physically to orthologues of capping protein alpha (Cpa) and capping protein beta (Cpb) and further explore the role of Cindr in modulating the actin cytoskeleton during eye maturation. Our data support a primary role for cindr in linking junction and actin regulation and help account for many of the phenotypes ascribed to mutations in mammalian CD2AP/CIN85.
Results
Expression and localization of cindr
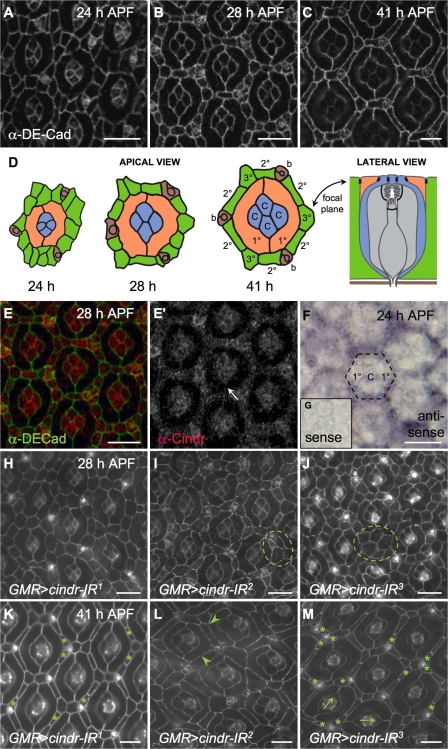
The developing D. melanogaster pupal eye is one of the most thoroughly characterized developing epitheliums, and it provides a useful model to explore the role of CD2AP/CIN85 proteins. Emergence of the pupal eye is shown in Fig. 1 (A–D). Ommatidia emerge in the larval eye as photoreceptor neurons and (glial-like) “cone cells” form the ommatidial core. Between the ommatidia lies an initially undifferentiated pool of “interommatidial precursor cells” (IPCs). From the IPCs emerge 1°, 2°, and 3° (primary, secondary, and tertiary) pigment cells, which are support cells that will eventually optically insulate each ommatidium. These pigment cells find their niche through cell rearrangements, and we focus on their movements in this work. During early pupariation, each ommatidial core is enwrapped by two 1° pigment cells. Shortly afterward, 2° and 3° pigment cells, along with sensory bristles, rearrange into an interweaving hexagonal lattice that organizes the ommatidia into a precise honeycomb (Fig. 1). These patterning events occur 18–42 h after puparium formation (APF).
Figure 1.
Patterning the D. melanogaster pupal retina requires Cindr. (A–C and E–G) Wild-type Canton S retinae dissected at the indicated time points. DE-Cad marks AJs. (D) Illustration of ommatidia from A-C showing 1° cells (orange), 2° and 3° cells (green), cone cells (blue), photoreceptors (gray), and bristles (brown). AJs are illustrated in black. Lateral view was inspired by Tepass and Harris (2007). (E) 28-h-APF Canton S retina with Cindr (red) and DE-Cad (green) shown. (E′) Cindr fills the cytoplasm of lattice and cone cells and localizes to AJs; the arrow indicates membrane between two 1° cells. (F) In situ analysis at 24 h APF to detect cindr transcripts; broken line indicates a single ommatidium. (G) Control sense probe. Retinae dissected at 28 (H–J) and 41 h (K–M) APF expressing cindr-IR transgenes as indicated. DE-Cad marks AJs. Mutant phenotypes include failure of IPCs to resolve into single file (circled), excess IPCs (asterisks), discontinuous DE-Cad (arrowheads), and ommatidial misrotation (arrows). Bars 10 μm.
These precise movements require the junction-associated protein DE-Cad and the adhesion protein Rst, raising the question as to how these surface proteins can influence cell movements. In our search for new factors that regulate this patterning process, we identified CG31012 in silico as the single D. melanogaster orthologue of vertebrate CD2AP and CIN85 and christened the locus cindr (CIN85 and CD2AP orthologue). The annotation of this locus predicts four isoforms (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200706108/DC1). We identified at least two isoforms using a rabbit polyclonal antibody generated against the common central region. Antibody specificity was confirmed by Western analysis of a deficiency that overlaps cindr (Fig. S1 B) and by detection of ectopic or reduced Cindr levels in the third larval instar wing disc and pupal eye (Figs. S2 and S3).
In tissue, Cindr transcript and protein was expressed ubiquitously at all stages examined (Fig. 1, E and F). The protein itself was found primarily at the apical surface and in vesicles near the surface. For example, undifferentiated embryonic and larval tissue contained puncta of Cindr in close association with the adherens junctions (AJs; Fig. S2 and not depicted). As cells emerged from the IPC pool, the concentration of puncta increased in the emerging 2° and 3° cells 18–41 h APF but remained low in emerging 1° cells (Figs. 1 E, S3 C, and S5, B′–H′, available at http://www.jcb.org/cgi/content/full/jcb.200706108/DC1). Cindr levels were high in the photoreceptor neurons and cone cells as well as the sensory bristles (Figs. 1 and S3). Low levels were also observed near the fenestrated basement membrane at the base of the retina (Fig. S3 E).
Reducing Cindr activity leads to junctional instability
The available D. melanogaster cindr P insertion–mediated alleles did not sufficiently perturb the locus to generate significant phenotypes. We therefore targeted the locus using RNAi. We generated multiple transgenic fly lines with inverted repeat (IR) transgenes (Fig. S2 A) that specifically targeted the long (PC, cindr-IR1), long plus mediate (PC and PD, cindr-IR2), or all three transcripts (cindr-IR3) when expressed using the GAL4/UAS system (Brand and Perrimon, 1993). Based on quantitative real-time PCR (qrt-PCR; Table I), immunocytochemistry (Fig. S2), and phenotype (see Fig. 1, I–M), cindr-IR2 was the most effective at reducing transcript and protein levels. Ubiquitous expression of cindr-IR2 and cindr-IR3 led to embryonic and early larval lethality, respectively; cindr-IR1 generated no observable adult phenotype.
Table I. qrt-PCR analysis of relative reduction of cindr transcripts using RNAi.
| Mean relative expressiona (SDb) primer set |
|||
|---|---|---|---|
| Genotypec | B | D | K |
| da>cindr-IR1 | d | 0.45 (0.05) | 0.71 (0.04) |
| da>cindr-IR2 | 0.31 (0.15) | Not doned | Not done |
| da>cindr-IR3 | 0.76 (0.25) | Not done | 0.68 (0.31) |
Compared to control cDNA isolated from da>GFP tissue.
n = 3 per two independent cDNA preparations.
da, daughterless-GAL4.
Primer set recognizes cindr-IR transgene.
When both cindr-IR2 and cindr-IR3 transgenes (glass multimer reporter [GMR]>cindr-IR2+3) were driven together in the eye for at least 24 h, virtually no Cindr was detected as the animal progressed to pupal stages (Fig. S3 B′). GMR>cindr-IR2+3 directed less decrease in Cindr protein during larval development and no developmental defects were observed, which suggests that the protein can show significance perdurance (unpublished data). Therefore, we cannot rule out a role for Cindr during the larval stages of eye development.
In wild-type pupal eyes, the process of patterning IPCs into a precise arrangement of 1°, 2°, and 3° pigment cells is mostly complete by 28 h APF (Fig. 1 B). This process was delayed or lost when cindr activity was reduced in GMR>cindr-IR2 and GMR>cindr-IR3 eyes; GMR>cindr-IR1 eyes exhibited a lower level of defects (Fig. 1, H–M). Cells were misplaced, pattern elements within the ommatidial field were misoriented, and the number of pigment cells was variable.
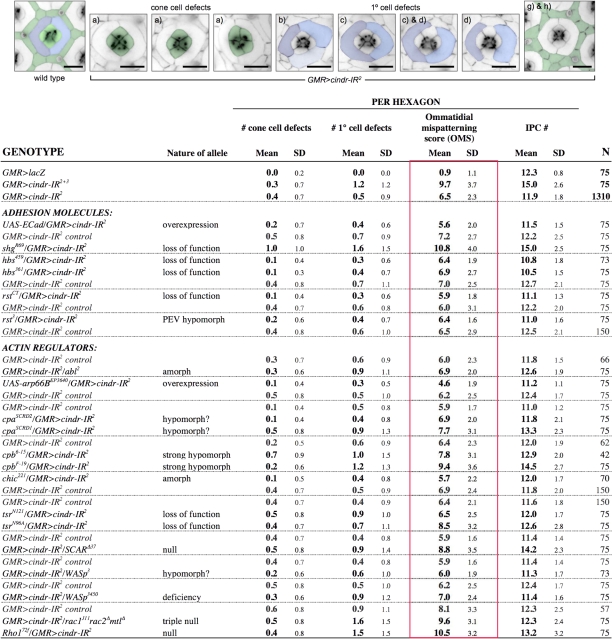
Quantitation of these defects at 41 h APF, when pigment cell patterning is completed, emphasized the broad nature of the patterning defects. For example, an additional one to two pigment cells surrounded 23% of GMR>cindr-IR1 ommatidia (n = 159) and 46% of GMR>cindr-IR2 ommatidia; 16% of GMR>cindr-IR3 ommatidia were surrounded by an additional three to five cells (n = 204). The most posterior fifth of GMR>cindr-IR2 retinae showed still more severe defects: 81% of ommatidia had a shortfall of one to five surrounding IPCs (n = 86; compare Figs. 1 L and 5 A). To further quantitate these phenotypes, we developed a mean “ommatidial mispatterning score” (OMS) system that accounted for errors in patterning, cell number, and rotation (Fig. 2). Focusing on ommatidia at the center of the eye for consistency, we found that reducing cindr activity (GMR>cindr-IR2+3) led to defects in each of these aspects (Fig. 2). Control GMR>lacZ eyes were scored with an OMS of 0.9 errors per ommatidial cluster (n = 75); the OMS for cindr-IR2+3 ommatidia was 9.7 errors per ommatidium (n = 75) and the OMS for cindr-IR2 was 6.4 (n = 1,310). This analysis highlights the requirement by retinal cells for normal cindr activity during their maturation into a functional neuroepithelium.
Figure 2.
Analysis of patterning defects observed in retinae dissected at 41 h APF. The OMS (boxed in red) is the mean of the total number of defects observed per ommatidium. These defects include (a) errors in the arrangement of cone cells (number of cone cell defects; cone cells are pseudo-colored green in examples); aberrant (b) number, (c) size, and (d) junctional integrity of 1° cells (number of 1° cell defects; 1° cells are pseudo-colored blue in examples); (e) errors in ommatidial rotation (see examples in Fig. 1 M); (f) incorrect number or position of bristles (note bristle misplacement in Fig. 1 L); (g) errors in the number or specification of 3° cells; and number of (h) additional IPCs (pseudo-colored green) or (i) missing IPCs (see Fig. 1 L). The total number of IPCs per ommatidium is also given separately for each genotype analyzed. Since the number of rotation errors (0.1 per hexagon; n = 1,310), bristle errors (∼1.5 per hexagon), and 3° cell errors (∼2.4 per hexagon) observed in GMR>cindr-IR2 retinae were unmodified in all genetic interactions, these values are not shown separately. Bars, 10 μm.
Reducing cindr activity led to instability of cells within the pupal eye
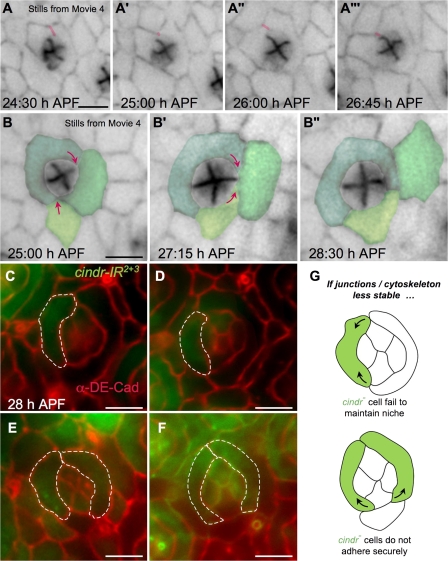
Our analysis with fixed tissue indicated a requirement for Cindr as pigment cells moved into their proper niches. However, the patterning process in the pupal eye is rapid, progressive, and highly dynamic. Unlike cell culture, exploring the role of factors such as Cindr during epithelial maturation is difficult to achieve in situ, especially dynamically and with subcellular resolution. For example, the cindr patterning defects we observed could be accounted for by cell movements that are reduced, increased, unstable, or misdirected. An important advantage to using the fly eye as a model for exploring Cindr function is the recent establishment of live imaging to observe events in situ (Larson et al., 2008), and we took advantage of this technology to answer several questions about the role of Cindr in the patterning process. We used fluorescently labeled α-catenin–GFP, a protein associated with DE-Cad, to visualize junctions in situ.
As expected, in live imaging movies of control eyes, we observed two emerging 1° cells enwrapping each ommatidium as each 1° cell formed a stable junction with its 1° partner. The result was a stereotypical “collar” of two 1° cells surrounding each ommatidium's central cone cell quartet (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200706108/DC1). Initial recruitment of 1° cells in GMR>cindr-IR22+3 eyes commenced normally and many, though not all, fledgling 1° cells successfully encircled a central cone cell quartet (Video 2). However, these emerging 1° cells commonly failed to maintain their niches: 1° cells often lost contact with their partners, a loss typically coupled with dissolution of their shared junction. As a result, one proto-1° cell was often replaced by another cell to generate a new 1° cell. Remarkably, this instability of the 1°–1° interface continued throughout the stages of IPC patterning, hours after 1° cells have normally completed their movements (Fig. 3, A and B; and Video 3). Similar junctional instability and 1° cell replacements were rarely observed in control eyes. These results are consistent with a model in which loss of cindr activity permits cell movement but prevents cells from maintaining their niche through stable junctions. We note that cells that are not undergoing cell movements, such as cone cells, do not show similarly destabilized junctions.
Figure 3.
Cindr is required to stabilize cells during patterning. (A and B) Progressive views of single ommatidia from Video 3 (available at http://www.jcb.org/cgi/content/full/jcb.200706108/DC1); α-catenin–GFP marks AJs. (A–A‴) A fluctuating 1°–1° interface is highlighted in pink. (B–B″) Selected cells are pseudo-colored; arrows indicate the direction of membrane protrusion as a pre-1°cell is displaced. (C–F) Small clones of cells expressing cindr-IR2+3 (labeled with GFP). DE-Cad is shown in red. Broken lines outline mutant 1° cells. (G) Cartoons illustrating the expected phenotypic outcomes if one or both 1° cells are mutant for cindr. Bars, 5 μm.
A failure to maintain proper position and stable junctions was also observed in the emerging arrays of 2° and 3° pigment cells. The movement of the IPCs in these older cindr-IR22+3–expressing retinae was similarly stochastic: excessive switching of cell–cell contacts, severe alterations in apical cell profiles, and a failure of IPCs to stably rearrange stepwise into single file (Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200706108/DC1). The result was a failure to achieve coherent patterning. In contrast, in control eyes, IPCs moved smoothly and with small local movements to gradually establish the hexagonal array of 2° and 3° cells (Video 4). These abnormal cell movements and contacts were again suggestive of unstable and transient junctions in cindr-IR22+3 retinae and also indicate a failure of proper morphogenesis.
We observed further evidence of unstable junctions by examining small clonal patches of cells expressing cindr-IR22+3. Isolated cindr-IR22+3 1° cells were consistently smaller than their wild-type neighbors (Fig. 3, C, D, and G). Paired cindr-IR22+3 1° cells frequently retracted, permitting a neighboring wild-type IPC direct contact with the central cone cells (Fig. 3, E–G) and often leading to an ectopic third 1° cell.
Cindr is functionally linked to DE-Cad
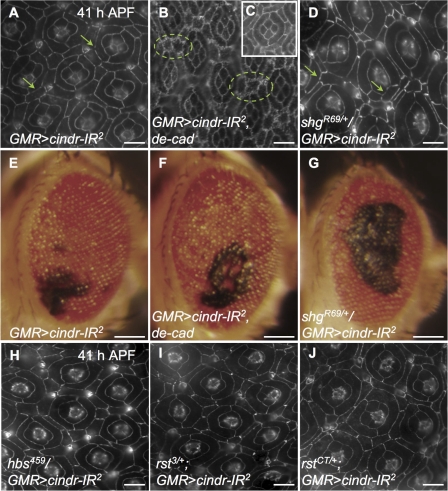
Our results indicate a role for Cindr in mediating surface events. A subset of Cindr protein was found associated with DE-Cad (Figs. 1 E, S3, and S5), a central component of the AJ, and loss of cindr activity led to unstable junctions and abnormal cell movements. What is the relationship between Cindr and DE-Cad? Modifying a phenotype by removing one genomic copy of a gene is a standard and powerful technique to demonstrate that two loci function together in a common process. DE-Cad is encoded by the shotgun (shg) locus: the GMR>cindr-IR2 mispatterning phenotype (OMS = 7.2) was enhanced in a heterozygous shgR69/+ background (OMS = 10.8; Figs. 4 D and 2). In addition, large patches of black tissue were observed in the eyes of 68.3% (n = 97) of these adult flies (Fig. 4 G); this was a significant enhancement of the penetrance and extent of degenerative tissue that was observed in GMR>cindr-IR2 alone (12.6%, n = 254; Fig. 4 E). This blackened eye phenotype is suggestive of late epithelial cell death and indicates that the relationship between Cindr and DE-Cad extends to later stages of development.
Figure 4.
cindr interacts with adhesion loci. (A) GMR>cindr-IR2 retina; DE-Cad was discontinuous (arrows). (B) GMR>cindr-IR2, de-cad. Mispatterning was rescued by expression of de-cad; cell membranes are elaborated and “frilly” (circled). (C) GMR>de-cad. Ectopic DE-Cad alone did not affect patterning. (D) shgR69/+/GMR>cindr-IR2. Enhanced mispatterning and discontinuous DE-Cad (arrows). (E–G) Adult eyes with black regions indicating tissue degeneration; genotypes are indicated. (H–J) Mild enhancement of cindr-IR2 in hbs or rst heterozygotes. (H) hbs459/GMR>cindr-IR2. (I) rst3/+;GMR>cindr-IR2. (J) rstCT/+;GMR>cindr-IR2. All tissue was dissected at 41 h APF; DE-Cad detection is shown. Adults were photographed 1 d after eclosion. Bars: (A–D and H–J) 10 μm; (E–G) 100 μm.
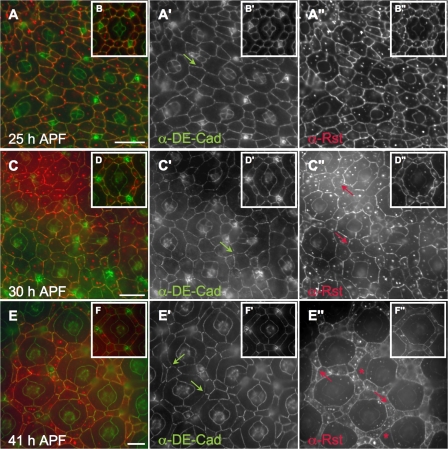
In wild-type pupal eye tissue, DE-Cad protein is found at high continuous levels between all apical cell profiles. An exception is between emerging 2° and 3° cells when the level of DE-Cad present at AJs decreases during their rearrangement into a hexagonal lattice (Fig. 5, B′ and D′; Bao and Cagan, 2005). This decrease may represent a necessary step to allow for local cell rearrangements; higher levels of DE-Cad are normally restored after cell patterning is completed and cell movements end (Fig. 5 F′). This progressive loss of DE-Cad between 2° and 3° cells was disrupted when cindr activity was decreased (GMR>cindr-IR2 and GMR>cindr-IR2+3): abnormally weakly stained and discontinuous DE-Cad was observed in all cell types and all time points examined (Figs. 1 I and 5, A, C, and E). Later, at 41 h APF, mildly discontinuous DE-Cad was still observed in over ∼25% of the retina, most often in posterior regions corresponding with the most severe patterning defects (Figs. 1 L and 5 E). This discontinuous localization of DE-Cad was enhanced in a heterozygous shg background (Fig. 4 D and not depicted), most markedly at the “tricellular junctions” where three cells interface.
Figure 5.
Deregulation of DE-Cad and Rst distribution when Cindr is reduced. (A, C, and E) GMR>cindr-IR2+3 retina processed to detect DE-Cad (green in overlay panel) and Rst (red) dissected at 25, 30, and 41 h APF. DE-Cad was irregularly distributed (green arrows). Rst remains strongly localized to IPC–IPC membranes (red arrows) and, by 41 h APF, was cytoplasmic in IPCs except for a subset that have large apical profiles resembling those of 1° cells (asterisks). These cells may have temporarily contacted cone cells earlier and received 1°-inductive signals (Miller and Cagan, 1998). (B, D, and F) Control GMR>lacZ ommatidia showing wild-type distribution of DE-Cad and Rst. Bars 10 μm.
Our localization studies indicate that Cindr acts to both permit dynamic regulation of DE-Cad during cell movement and to stabilize its presence at the cell surface. If this view is correct, expressing ectopic DE-Cad should rescue cindr-IR patterning defects. Importantly, ectopic DE-Cad strongly rescued the GMR>cindr2 mispatterning phenotype and reduced the cindr-IR2 OMS from 7.2 to 5.6 (Figs. 2 and 4 B). This provides further evidence that a central role for Cindr in situ is to mediate events at the AJs.
Previous work reported that CD2AP can interact in vitro with E-cadherin via its SH3 domains (Lehtonen et al., 2004; Mustonen et al., 2005). We found that ubiquitous expression of a Cindr-PB construct, which retained only the C-terminal half of the full-length protein but lacked SH3 domains, was distributed in a striking perimembrane pattern but was apparently excluded from the AJ; full-length Cindr-PC or Cindr-PD isoforms were localized normally when similarly expressed (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200706108/DC1, and not depicted). This suggests that Cindr's SH3 domains are required specifically for membrane localization. Further consistent with this view, a complimentary N-terminal construct (Cindr-ΔPB) that includes the three SH3 domains was normally localized including to the AJs (Fig. S4 and not depicted). Although this data suggests that Cindr may interact directly with DE-Cad through its SH3 domains, we failed to detect a Cindr–DE-Cad interaction in conditions that permitted binding to other proteins (see Materials and methods).
Two additional observations are notable regarding the relationship between Cindr and DE-Cad. First, in addition to rescuing the GMR>cindr-IR2 phenotype, ectopic DE-Cad accumulated at the apical membrane, which became elaborated (Fig. 4 B). The mammalian orthologue CIN85 has been implicated in regulating endocytosis of receptors such as those for EGF and PDGF (Dikic and Giordano, 2003). The membrane elaboration of ectopic DE-Cad may reflect a role for Cindr in its endocytosis; however, this model would not account for the loss of endogenous DE-Cad at the surface when cindr activity was reduced (Fig. 5). Second, Western analysis did not detect any changes in overall levels of DE-Cad (or Rst, see following section) in GMR>cindr-IR2+3 pupal eyes (unpublished data), again supporting the view that Cindr acts primarily on DE-Cad's localization and not its stability. Together, our data indicate that Cindr acts in situ in part by regulating dynamic junctional remodeling and DE-Cad function during a period of dynamic cell movement.
Cindr is functionally linked to the adhesion protein Rst
In addition to regulating AJs we found a functional link between Cindr and Rst, a transmembrane protein and NEPH-1 orthologue that mediates cell–cell adhesion during IPC patterning (Schneider et al., 1995). Rst acts with its binding partner Hbs to play a central role in directing the cell movements required for 2°/3° patterning (Bao and Cagan, 2005). Initially expressed at all IPC–IPC boundaries (Fig. 5 B″) by 22 h APF, Rst protein becomes expressed primarily in 2° and 3° cells as the 2°/3° hexagonal lattice emerges (Reiter et al., 1996). Rst protein is localized predominantly to AJs and, to a greater extent than DE-Cad, becomes progressively capped to the interface between 2°/3° cells and 1° cells (Fig. 5, D″ and F″; and Fig. S3 C) due to its physical interaction with Hbs, which is expressed in 1° cells (Bao and Cagan, 2005).
Cindr protein colocalized with Rst at the AJ (Fig. S3 C). Reducing Cindr blocked the dynamic redistribution of Rst protein: In GMR>cindr-IR2+3 pupal eyes, Rst remained at steady diffuse levels at the junctions of most IPC–IPC contacts through the stages of hexagonal lattice emergence (Fig. 5, C″ and E″). We also observed abnormally high levels of diffuse cytoplasmic Rst. Similar to shg, Western analysis indicated that Rst protein was present at normal levels and that reducing the gene dosage of rst or hbs did not restore proper Rst protein localization (unpublished data). This indicates that levels of Rst protein are not responsible for the observed cindr-IR defects in Rst localization.
Our genetic modifier data also supported a functional interaction between Cindr and Rst/Hbs. Pupae heterozygous for hbs361, hbs459, rst3, or rstCT exhibited mild but consistent suppression of the GMR>cindr-IR2 phenotype (Fig. 4, H, I, and J; and not depicted). The prevalence of cone and 1° cell defects was decreased (Fig. 2) and the number of 2° and 3° cells was reduced to slightly below normal.
Together, these data provide a functional link between Cindr and the surface proteins DE-Cad and Rst during the stages of dynamic AJ and adhesion remodeling and cell movement.
Cindr acts with regulators of the actin cytoskeleton
In the course of searching for binding partners for Cindr that are active in the eye, we identified components of the actin cytoskeleton. Of note, the patches of dead tissue we observed in eyes of GMR>cindr-IR2+3 adults (Fig. 4 E) phenocopied those reported for alleles of the actin-related loci cpa and cpb (Delalle et al., 2005). We also note a similarity between the twisted, thickened, short, and bent thoracic macrochaetea observed when cindr activity was reduced (see Fig. 8 I) and similar defects found for actin regulatory loci such as cpb (Hopmann et al., 1996; Hopmann and Miller, 2003; Frank et al., 2006).
Figure 8.
Cindr localizes to and regulates F-actin. (A, B, D, and E) Canton S retinae dissected at times as indicated. Tissue was incubated in phalloidin to detect F-actin (white). Insets show detection of Cindr (green), DE-Cad (blue), and F-actin (red) in selected fields. (C and F) GMR>cindr-IR2+3 retina; phalloidin is shown. (G) High-magnification image of a single protruding ommatidial bristle at 41 h APF. The retina was torn before imaging to remove tissue below the bristle (top right). Cindr (green) is concentrated at the tip of the protruding bristle. Phalloidin (red) and DE-Cad (blue) are also shown. (H) Dorsal thorax of adult Canton S fly. (I) apterous>cindr IR2+3 adult thorax. Macrochaetea are missing, hooked, thickened, or stunted (arrows). (J) 28-h-APF pupal eye mosaic for cpascrd1. The lack of nuclear GFP labels mispatterned mutant cells. DE-Cad (blue, J′) was discontinuous (arrows) and phalloidin (red, J″) detects extremely elevated levels of polymerized actin. Bars: (A–G and J) 5 μm; (H and I) 100 μm.
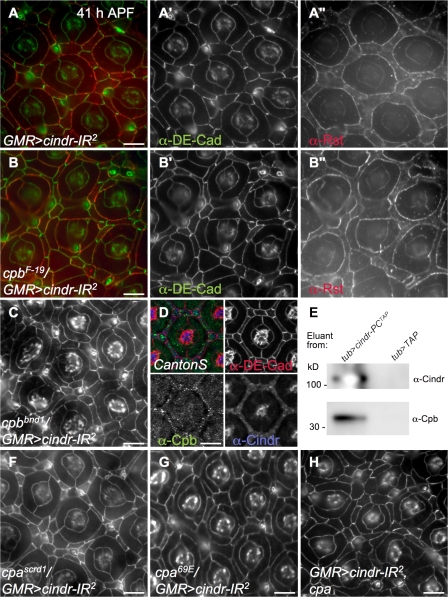
CPα/CPβ heterodimers reversibly bind barbed, fast-growing ends of actin filaments to prevent further elongation or depolymerization (Isenberg et al., 1980; Hopmann et al., 1996). Demonstrating a functional link between cindr and the D. melanogaster orthologues cpa/cpb, we found that multiple alleles of either cpa or cpb strongly and dominantly enhanced the patterning defects of GMR>cindr-IR2 (Figs. 2 and 5, B, C, F, and G). Similarly, we observed an enhancement of the cindr-IR phenotype when ectopic cpa or cpb was concurrently expressed (Fig. 6 H and not depicted); neither protein gave rise to a detectable phenotype when misexpressed alone (not depicted). The ability of both increased and decreased cpa/cpb activity to enhance cindr-dependent phenotypes indicates that the balance of Cindr and Cpa/Cpb is important. Cpb protein was expressed in all cell types within the pupal eye (Fig. 6 D) and the distribution and levels of Cpb were not detectably altered in a cindr-IR context (not depicted).
Figure 6.
cindr interacts with cpb and cpa. (A) GMR>cindr-IR2 retina probed for DE-Cad (green, A′) and Rst (red, A″). (B) cpbF-19/GMR>cindr-IR2. Mispatterning was strongly enhanced. (C) cpbbnd1/GMR>cindr-IR2. (D) In wild-type retinae, Cpb (green) and Cindr (blue) colocalized. DE-Cad is shown in red. (E) Western analysis of final eluants of coimmunopreciptations from embryos expressing cindr-PCTAP or TAP only probed with α-Cindr (top) or α-Cpb (bottom). (F–H) cindr-IR2 was strongly enhanced in trans to cpa heterozygotes or when cpa was overexpressed. (F) cpascrd1/GMR>cindr-IR2. (G) cpa69E/GMR>cindr-IR2. (H) GMR>cindr-IR2, cpa. All tissue was dissected at 41 h APF; DE-Cad is only shown in C, F, G, and H. Bars, 10 μm.
Further, mass spectrometry analysis of embryonic protein precipitates identified Cpa and Cpb as physically interacting with a tandem affinity purification (TAP)-tagged isoform of Cindr (Fig. 6 E). This is consistent with recent work indicating that mammalian CPα and CPβ binds CD2AP (Hutchings et al., 2003; Bruck et al., 2006) and, indeed, a potential CPα/CPβ binding site has been partly conserved in Cindr (Fig. S1 C).
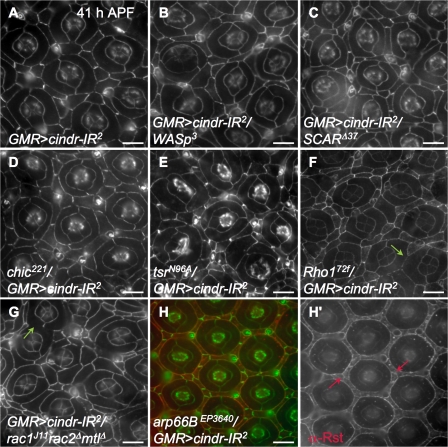
These results strongly link Cindr function to actin remodeling, and genetic tests with other components of the actin remodeling machinery support this view. We examined Wiskott-Aldrich syndrome protein (WASp), suppressor of cAR (SCAR), profilin (Chickadee [Chic], required for the addition of actin monomers to filaments), cofilin (Twinstar [Tsr], promotes F-actin depolymerization), and the tyrosine kinase Abelson (Siripala and Welch, 2007a,b). The cindr-IR2 phenotype was enhanced in retinae heterozygous for mutations in each of these loci (Figs. 2 and Figs.7, and not depicted). The small GTPases Rho and Rac are central regulators of actin-based cytoskeletal dynamics (Hall, 2005) and we also observed an enhancement of GMR>cindr-IR2 in individuals heterozygous for Rho172 or a Rac1J11 Rac2Δ MtlΔ triple mutation (Figs. 2 and 7, F and G). Mutations in each of these seven loci increased the mean IPC number and enhanced the number of cone and/or 1° cell errors in the presence of GMR>cindr-IR22 and mildly but detectably increased the OMS value.
Figure 7.
cindr interacts with many actin regulatory loci. DE-Cad shown in A–G and is shown in green in H. (A) GMR>cindr-IR2 retina. Mispatterning was modified by removing a single copy of loci for (B) WASp, (C) SCAR, (D) chic, (E) tsr, (F) Rho, or (G) rac. DE-Cad was often spotty or absent where 1° cells from neighboring ommatidia made contact (green arrows). (H) Ectopic expression of arp66B rescued aberrant patterning but Rst (H and H′, red) remained at IPC–IPC boundaries (red arrows). All tissues were dissected at 41 h APF. Bars, 10 μm.
Conversely, ectopic expression of the actin-related protein (Arp) 2/3 component arp66B led to strong rescue of GMR>cindr-IR22; interestingly, errant localization of Rst to IPC–IPC membrane interfaces was not corrected in this genetic context despite its phenotypic rescue (Figs. 2 and 7, H and H′). Expression of arp66B alone did not disrupt patterning (unpublished data). This result further emphasizes that reducing cindr activity leads to reduction of actin regulation. In mammalian systems, the interaction between CD2AP and the Arp2/3 complex is indirect and mediated by association with Cortactin (Cort; Lynch et al., 2003) but the cindr-IR retinal phenotype was unmodified in a cortM7 or cortD4 heterozygous background (unpublished data).
Cindr regulates actin dynamics
In addition to regulating events at the surface, our genetic and biochemical data strongly indicated that Cindr is used by retinal cells to regulate actin cytoskeleton remodeling during the patterning process. The dynamics of actin remodeling have not been previously described in the pupal eye. We therefore used phalloidin to explore the role of actin dynamics in patterning and its regulation by Cindr (Figs. 8 and S5). During early development stages (18–22 h APF), filamentous F-actin was found predominantly at cell membranes in pre-1° cells and IPCs (Fig. S5 A″); as development progressed, the levels of membrane- and AJ-associated F-actin increased (Fig. S5, B″–G″), reaching a peak 27–30 h APF (Fig. 8 B). Importantly, Cindr foci were observed to colocalize with enriched phalloidin staining at the AJ (Fig. S5). Concurrently, numerous bundles of F-actin formed within the IPCs and eventually the 1° cells, radiating from the AJs to densely fill the apical cytoplasm (Figs. 8 E and S5, B″–H″). Membrane-associated actin decreased after 30 h APF, and by 41 h APF, little or no membrane-associated F-actin remained between IPCs and 1° cells, though it remained at AJs of other cell types (Fig. 8 E).
In summary, actin showed stereotyped and dynamic polymerization and AJ localization during the period of maximal IPC movement, and the dynamics of Cindr localization roughly mirrored F-actin throughout this period. This coevolving of F-actin and Cindr localization was also observed in other cell types including the cone cells and sensory bristles (Figs. 8 and S5). The emerging bristles were especially instructive: as the tip of each bristle shaft emerged, F-actin bundles are generated from successive short linear actin modules that form at the growing tip of each bristle and elongate to graft together (Guild et al., 2003). Cindr puncta colocalized along the length of these elongating F-actin shafts but particularly at newer distal ends, which suggests a role for Cindr in active actin polymerization (Fig. 8 G) and was reflected in malformed thoracic macrochaetae when Cindr was reduced during their development (Fig. 8 I).
Strongly reducing cindr activity (GMR>cindr-IR2+3) had a marked effect on F-actin during IPC patterning: at 28 h APF, the density of F-actin fibrils and spikes filling the cytoplasm of cindr-IR IPCs was markedly increased (Fig. 8 C). At later stages of patterning, a more dense and disorganized cytoskeletal network was observed in IPCs and the radiating arrangement of actin fibrils in 1° cells was absent at 41 h APF (Fig. 8 F). Within cone cells, F-actin levels were mildly elevated at 28 h APF, and distinct F-actin spikes failed to radiate from cell membranes (Fig. 8 C). These data directly support a role for Cindr in regulating dynamic actin organization rather than regulating actin polymerization per se.
This failure of proper dynamic changes to the actin cytoskeleton presumably accounts for the abnormal cell movements, unstable cell junctions, and severe mispatterning observed when Cindr activity was reduced. To further assess this model, we compared IPCs' requirement for Cindr with their requirement for capping proteins, which have been previously demonstrated to regulate F-actin dynamics. We examined discrete clones of tissue that were homozygous mutants for null alleles of cpa or cpb. Abnormally high levels of F-actin were observed to radiate from the membranes of mutant cells, extending with apparent continuity into neighboring wild-type cells (Fig. 8 J, and not depicted). When generated early, clones showed errors in photoreceptor and cone cell organization, reduced cell viability, and pools of undifferentiated cells (unpublished data). When generated later to circumvent differentiation defects, DE-Cad was localized discontinuously around the cell circumference (Fig. 8 J and unpublished data). These phenotypes were similar to, though more severe than, those observed in cells with a partial reduction of cindr activity (e.g., Figs. 5 and 8 C) and confirm that a disorganized cytoskeleton can disrupt both the AJ and IPC patterning.
Discussion
Our evidence indicates that Cindr provides a functional link between dynamically regulated surface adhesion and the cytoskeletal changes required for normal pupal eye patterning. Loss of cindr activity led to misplacement of retinal support cells, which adopted shapes uncharacteristic for their niche in the retinal field. The reasons for this became apparent when we examined cell behavior in live tissue. Reducing cindr prevented 1° cells from maintaining enwrapment of the cone cells; instead, cindr 1° cells were unable to firmly establish this niche and frequently retracted, allowing neighboring IPCs to have direct contact with the cone cells. Similar instability was observed in the remaining IPCs fated to establish the 2°/3° hexagonal lattice. Histology demonstrated further changes both to AJ components and the actin cytoskeleton. Although we cannot rule out additional roles for Cindr such as regulation of endocytosis, we did not observe evidence for such a role during cells' rearrangements.
In wild-type tissue, Hbs and Rst are localized exclusively to 1°–IPC interfaces during IPC patterning; heterophilic interactions between these molecules are thought to direct the rearrangement of IPCs into single rows around each ommatidium (Reiter et al., 1996; Bao and Cagan, 2005; Grzeschik and Knust, 2005). In cindr-IR tissue, we observed a mislocalization of Rst to the entire IPC circumference. Such mislocalization would impede the generation of a preferential adhesive force, disrupting the direction or flow of cell movement and subsequent patterning. We also detected irregularities in the localization of DE-Cad around the circumference of retinal cells when Cindr was reduced. This presumably resulted in uneven or unreliable junctional stability that further destabilized the dynamic switching of cell positions. Genetic interactions with the loci for rst, hbs, and shg confirmed that these loci cooperate with cindr during patterning.
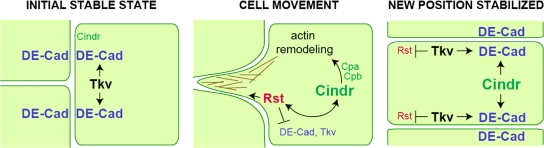
We also demonstrate that the actin cytoskeleton is dynamically remodeled during pupal eye patterning and that reducing cindr activity led to a change in the details of cytoskeletal dynamics. In wild-type tissue, polymerized actin was initially detected almost exclusively at AJs of pre-1° cells and IPCs. These actin rings intensified as cells became rearranged and then, remarkably, once patterning was established, membrane-associated F-actin strongly diminished. The functional significance of these changes is likely linked to concurrent modification of Rst- and AJ-mediated adhesion. For example, the levels of both DE-Cad and Armadillo (β-catenin) decrease between IPCs as they are rearranged, which would serve to facilitate Rst-mediated IPC movements toward 1° cells (Fig. 9, middle; Bao and Cagan, 2005). We show data indicating that the actin cytoskeleton is coordinately reinforced at AJs as adhesion is weakened. This may serve to maintain the surface integrity of the retinal cells while junctional strength decreases and the tissue is remodeled. Once patterning is achieved, the BMP receptor Thickvein then acts to restrengthen AJs (Cordero et al., 2007); the reduction of F-actin may reflect the reduced need for a dense actin ring. Throughout this process, Cindr acts as a pivotal regulator to coordinate AJ modification and actin polymerization.
Figure 9.
Model of proposed Cindr regulatory network. Cindr shifts from the membrane to the cytoplasm to connect evolving events at the surface to regulation of the actin cytoskeleton. The result is coordinated cell movement. The ligands for Rst (Hbs) and Thickvein (Dpp) are not depicted.
Several data presented in this paper suggest that Cindr acts directly to regulate actin. First, the intensity and distribution of cytoplasmic Cindr puncta in retinal cells tracks that of F-actin in IPCs during development. Second, the striking dynamics of F-actin polymerization are lost, presumably helping account for their abnormal cell movements. Third, strong genetic interactions were observed between cindr-IR and multiple components of the actin regulatory machinery. Fourth, we coimmunoprecipitated the two capping protein subunits Cpa and Cpb together with Cindr from D. melanogaster embryos. And, finally, several phenotypes are shared between tissue mutant for cindr-IR and tissue mutant for cpa or cpb: pupal eye mispatterning, gaps in the distribution of DE-Cad around the circumference of retinal cells, bristle malformation, and tissue degeneration.
Our results also emphasize the important role of the actin cytoskeleton in regulating or maintaining AJ integrity (Mege et al., 2006). However, our data also argue that Cindr regulates the localization of transmembrane adhesion proteins at least in part independently of the cytoskeleton (Fig. 7): reducing the genetic component of actin regulators enhanced the patterning defects of cindr-IR but not the disruption to DE-Cad, and ectopic Arp66B rescued cindr-IR mispatterning but did not rescue aberrant localization of Rst. In the absence of Cindr, miscoordination of the actin machinery together with aberrant localization of junctional complexes is likely the underlying cause of tissue mispatterning during development. Similarly, deregulation of actin and junction instability is apparently cell lethal in more mature tissue, eventually leading to degeneration of mutant tissue. This may be analogous to the degeneration of mammalian podocytes that has been associated with mutations in CD2AP (Schiffer et al., 2004; Peters et al., 2006; Woroniecki et al., 2006). How Cindr itself is regulated during development remains an open question.
Materials and methods
D. melanogaster stocks
The following stocks were used (described in Flybase, http://flybase.bio.indiana.edu/): abelson2; arp66BEP3640; UAS–α-catenin–GFP; UAS–de-cadherin–GFP, and shgR69 (gifts from R. Carthew, Northwestern University, Evanston, IL); chic221; cortM7, and cortD4 (gifts from P. Rorth, Temasek Life Sciences Laboratory Limited, National University of Singapore, Kent Ridge, Singapore); cpa69E, cpa1074E, and UAS-cpa (gifts from F. Janody, Instituto Gulbenkian, Oeiras, Portugal); cpascrd1, cpascrd2, and cpbbnd1 (gifts from I. Hariharan, University of California, Berkeley, Berkeley, CA); cpbM14 and UAS-cpb (gifts from P. Garrity, Brandeis University, Waltham, MA); cpb6-15; cpbF19; Df(3R)8194; hbs459; hbs361; UAS-hbs-IR, UAS-rst-IR, and UAS-rst (gifts from S. Bao, Mount Sinai School of Medicine, New York, NY); rac1J11 rac2Δ mtlΔ; Rho172; rst3; rstCT; SCARΔ37; SCARK03107; tsrN96A; tsrN121A; WASp3 and the WASp deficiency Df(3R)3450 (gifts from E. Schejter, Weizmann Institute of Science, Rehovot, Israel); UAS-GFP and UAS-lacZ; apterous-GAL4 (gift from S. Cohen, Temasek Life Sciences Laboratory Limited, National University of Singapore); daughterless-GAL4 (gift from J. Zallen, Sloan Kettering Institute, New York, NY); GMR-GAL4; patched-GAL4; and tubulin-GAL4. The progeny of hsFLP; cindr-IR2+3 (see following section) crossed to Act5C>y+>GAL4, UAS-GFPS65T, and hsFLP; FRT42D, cpascrd1/CyO crossed to FRT42D, Ubi-GFPS65T were heat-shocked to generate FLP/FRT-mediated mitotic clones (Xu and Rubin, 1993).
Transgenic lines
For cindr-PC expression constructs, we amplified a 2.7-kb fragment from cDNA clone RH08284 (Drosophila Genomics Resource Center) using GAATCGCGGCCGCCATGGAAAAC (5′/631) and GATTCATCTAGACTCAAACTTGC (3′/3,306). These introduced Not1–Xba1 sites for insertion into pUAST N-T TAP (Wu et al., 2005) or pUAST-EGFP (Parker et al., 2001). For GFP-tagged cindr-PD and cindr-PB, CGGCAGCGGCCGCCATGTTGCCAC or GCACCGCGGCCGCCATGACCACAAC were paired with 3′/3,306; for cindr-ΔPB, we used 5′/631 and GAGTTCGTTCTAGACTAGTGGCTGG.
Fragments of cindr were amplified from genomic Canton-S cDNA, to generate UAS-cindr-IR constructs as previously described (Bao and Cagan, 2006) targeting 472–1,022 bp (cindr-IR1), 1,016–1,518 bp (cindr-IR2), and 1,664–2,246 bp (cindr-IR3). Transgenic lines were generated by standard P element–mediated transformation methods. In this paper, we used lines carrying multiple transgenes: cindr-IR1.18+18, cindr-IR2.21+23, cindr-IR3.73+81, and cindr-IR 2.21+23, 3.63+76, which we refer to as cindr-IR1, cindr-IR2, cindr-IR3, and cindr-IR2+3, respectively.
qrt-PCR
Efficacy of the cindr-IR transgenes was assayed by qrt-PCR of two independent preparations of cDNA from embryos or L1 larvae in which RNAi transgenes were driven using da-GAL4. Primers sets were: B, GAGCTGGATCTCCAGAAAGG and CGTTGCCATTGCTACTGACT; D, GCCACAGCCACAGTACCA and CTCCCGATCCCGAAACTAT; and K, GATCACCAGTAGCTCCCACA and AGGCGAAACACTTCGTTGAT. Each qrt-PCR experiment was repeated three times. Melt curve analysis showed a single peak for all samples and PCR products were verified using restriction analysis. Two primer sets recognizing rp49 were used as internal controls.
Cindr antibody
A fragment of Cindr common to all isoforms was amplified from RH08284 using GACCGGGAGCAGTAGCAATAGC and GTCGTTGAGAATTCCACAAACAT and inserted into pGEX-2T. The GST-Cindr384–740 fusion protein was raised in BL-21 cells for generation of a rabbit polyclonal antibody (Proteintech Group, Inc).
Western analysis of embryos
50 embryos homozygous for Df(3R)Exel8194 (a deletion encompassing cindr) and 50 heterozygous embryos were gathered 6–8 h after egg laying, homogenized in sample buffer, and separated on a NuPage 4–12% gradient SDS-polyacrylamide gel (Invitrogen). Rabbit anti-Cindr was used at 1:1,000 and HRP-conjugated secondary antibodies were used at 1:5,000 (Cell Signaling Technology).
Coimmunoprecipitation and mass spectrometry
cindr-PCTAP (or for controls, TAP only) was expressed using tubulin-GAL4 and isolated from embryos 2–24 h after egg laying. Dechorionated embryos were washed, lysed in a modified Tris buffer (Veraksa et al., 2005), and incubated for 20 min on ice. The lysate was then cleared by a 5-min centrifugation at 5,500 rpm and processed as described previously (Rigaut et al., 1999). Protein isolated from 2.5 ml of embryos was sufficient for LC-MS mass spectrometry (Siteman Cancer Center Proteomics Core Facility, Washington University). Coimmunoprecipitation of Cpb/Cpa with Cindr was confirmed by Western analysis of the final eluant; rat anti-Cpb was used at 1:50 (Hopmann et al., 1996).
In situ hybridization, immunofluorescence, and live imaging
Pupae were collected at 0 h APF and maintained at 25°C until dissection as described previously (Bao and Cagan, 2005). For in situ hybridization, digoxin-labeled RNA probes were synthesized from the entire cindr-coding region from RH08284. For immunofluorescence, primary antibodies were rabbit anti-Cindr (1:100), rat anti–DE-Cad2 (1:20, DSHB), mouse anti-Armadillo N2 7A1 (1:10; Developmental Studies Hybridoma Bank), mouse anti-Rst Mab24A5.1 (1:50; Schneider et al., 1995), and rat anti-Cpb (1:50). To visualize polymerized actin, rhodamine phalloidin was included in primary antibody incubations (1:100; Invitrogen). Secondary antibodies were conjugated Alexa 488 and 568 (Invitrogen) and Cy5 (Jackson ImmunoResearch Laboratories). The mounting media was Vectashield (Vector Laboratories). Samples were analyzed at RT with either an objective microscope (Axioplan2; Carl Zeiss, Inc.) using a Plan-Apochromat 63× 1.40 NA oil objective (Carl Zeiss, Inc.), camera (Quantix; Photometrics), and Image-Pro Plus 5.1 software (Media Cybernetics, Inc) or using a confocal system (TCS SP2; Leica) with constituent software and Plan-Apochromat 63× 1.32 NA or Plan-Apochromat 100× 1.40 NA oil objective lenses (Leica). Results were processed for publication using Photoshop (Adobe) with minimal adjustment of brightness or contrast applied to the whole image and analyzed for mispatterning errors as described in Fig. 2. Live imaging was performed as described previously (Larson et al., 2008) for GMR-GAL4/cindr-IR3; UAS-α-catenin-GFP/cindr-IR2 (experimental) and GMR-GAL4/+; UAS-α-catenin-GFP/UAS-lacZ (control) pupae. Images were gathered every 15 min at multiple focal planes using an Axiophot microscope (Carl Zeiss Inc.) and composite images for each time point were assembled in Photoshop, from which QuickTime (Apple) movies were generated.
Online supplemental material
Fig. S1 shows predicted Cindr isoforms, sequence, and Western analysis of embryos. Fig. S2 illustrates the cindr-IR transgenes and qrt-PCR primer sets and shows verification of the Cindr antibody and IR transgenes in the wing disc. Fig. S3 shows detection of Cindr in GMR>cindr-IR2+3 retinae and at the apical, basal, and rhabdomere planes in a 41-h-APF Canton-S ommatidium. Fig. S4 shows localization of GFP-tagged Cindr isoforms. Fig. S5 shows F-actin over a time course of development. Videos 1 and 4 are time lapse studies of control retina from 18–22 and 23–27 h APF, respectively. Videos 2 and 3 are of experimental cindr-IR retinae from 18–22 and 24–28 h APF, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200706108/DC1.
Supplemental Material
Acknowledgments
We thank all members of the Cagan Laboratory for lively discussion. We give thanks to David Larson for perfecting the live imaging protocol and Sujin Bao for advice on RNAi. We thank Andrey Shaw and Serawit Bruck for discussion, Jim Skeath for the use of his confocal microscope, Reid Townsend and Petra Gilmore for MS-SPEC analysis, and numerous fellow Drosophilists for generously sending reagents, often across international borders. We apologize to authors whose work could not be cited due to space constraints.
This work was funded by National Institutes of Health grant R01 EY1149.
Abbreviations used in this paper: AJ, adherens junction; APF, after puparium formation; Arp, actin-related protein; Chic, Chickadee; Cort, Cortactin; Cpa, capping protein alpha; Cpb, capping protein beta; DE-Cad, Drosophila E-cadherin; GMR, glass multimer reporter; Hbs, Hibris; IPC, interommatidial precursor cell; IR, inverted repeat; OMS, ommatidial mispatterning score; qrt-PCR, quantitative real-time PCR; Rst, Roughest; SCAR, suppressor of cAR; shg, shotgun; TAP, tandem affinity purification; Tsr, Twinstar; WASp, Wiskott-Aldrich syndrome protein.
References
- Bao, S., and R. Cagan. 2005. Preferential adhesion mediated by Hibris and Roughest regulates morphogenesis and patterning in the Drosophila eye. Dev. Cell. 8:925–935. [DOI] [PubMed] [Google Scholar]
- Bao, S., and R. Cagan. 2006. Fast cloning inverted repeats for RNA interference. RNA. 12:2020–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogler, O., F.B. Furnari, A. Kindler-Roehrborn, V.W. Sykes, R. Yung, H.J. Huang, and W.K. Cavenee. 2000. SETA: a novel SH3 domain-containing adapter molecule associated with malignancy in astrocytes. Neuro. Oncol. 2:6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A.H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118:401–415. [DOI] [PubMed] [Google Scholar]
- Brett, T.J., L.M. Traub, and D.H. Fremont. 2002. Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure. 10:797–809. [DOI] [PubMed] [Google Scholar]
- Bruck, S., T.B. Huber, R.J. Ingham, K. Kim, H. Niederstrasser, P.M. Allen, T. Pawson, J.A. Cooper, and A.S. Shaw. 2006. Identification of a novel inhibitory actin-capping protein binding motif in CD2-associated protein. J. Biol. Chem. 281:19196–19203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B., S.C. Borinstein, J. Gillis, V.W. Sykes, and O. Bogler. 2000. The glioma-associated protein SETA interacts with AIP1/Alix and ALG-2 and modulates apoptosis in astrocytes. J. Biol. Chem. 275:19275–19281. [DOI] [PubMed] [Google Scholar]
- Cordero, J.B., D.E. Larson, C.R. Craig, R. Hays, and R. Cagan. 2007. Dynamic decapentaplegic signaling regulates patterning and adhesion in the Drosophila pupal retina. Development. 134:1861–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormont, M., I. Meton, M. Mari, P. Monzo, F. Keslair, C. Gaskin, T.E. McGraw, and Y. Le Marchand-Brustel. 2003. CD2AP/CMS regulates endosome morphology and traffic to the degradative pathway through its interaction with Rab4 and c-Cbl. Traffic. 4:97–112. [DOI] [PubMed] [Google Scholar]
- Delalle, I., C.M. Pfleger, E. Buff, P. Lueras, and I.K. Hariharan. 2005. Mutations in the Drosophila orthologs of the F-actin capping protein alpha- and beta-subunits cause actin accumulation and subsequent retinal degeneration. Genetics. 171:1757–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic, I. 2003. Mechanisms controlling EGF receptor endocytosis and degradation. Biochem. Soc. Trans. 31:1178–1181. [DOI] [PubMed] [Google Scholar]
- Dikic, I., and S. Giordano. 2003. Negative receptor signalling. Curr. Opin. Cell Biol. 15:128–135. [DOI] [PubMed] [Google Scholar]
- Dustin, M.L., M.W. Olszowy, A.D. Holdorf, J. Li, S. Bromley, N. Desai, P. Widder, F. Rosenberger, P.A. van der Merwe, P.M. Allen, and A.S. Shaw. 1998. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell. 94:667–677. [DOI] [PubMed] [Google Scholar]
- Frank, D.J., R. Hopmann, M. Lenartowska, and K.G. Miller. 2006. Capping protein and the Arp2/3 complex regulate nonbundle actin filament assembly to indirectly control actin bundle positioning during Drosophila melanogaster bristle development. Mol. Biol. Cell. 17:3930–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidos, G., S. Soni, D.J. Oswald, P.A. Toselli, and K.H. Kirsch. 2007. Structure and function analysis of the CMS/CIN85 protein family identifies actin-bundling properties and heterotypic-complex formation. J. Cell Sci. 120:2366–2377. [DOI] [PubMed] [Google Scholar]
- Gauthier, N.C., P. Monzo, T. Gonzalez, A. Doye, A. Oldani, P. Gounon, V. Ricci, M. Cormont, and P. Boquet. 2007. Early endosomes associated with dynamic F-actin structures are required for late trafficking of H. pylori VacA toxin. J. Cell Biol. 177:343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout, I., G. Middleton, J. Adu, N.N. Ninkina, L.B. Drobot, V. Filonenko, G. Matsuka, A.M. Davies, M. Waterfield, and V.L. Buchman. 2000. Negative regulation of PI 3-kinase by Ruk, a novel adaptor protein. EMBO J. 19:4015–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik, N.A., and E. Knust. 2005. IrreC/rst-mediated cell sorting during Drosophila pupal eye development depends on proper localisation of DE-cadherin. Development. 132:2035–2045. [DOI] [PubMed] [Google Scholar]
- Guild, G.M., P.S. Connelly, L. Ruggiero, K.A. Vranich, and L.G. Tilney. 2003. Long continuous actin bundles in Drosophila bristles are constructed by overlapping short filaments. J. Cell Biol. 162:1069–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A. 2005. Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 33:891–895. [DOI] [PubMed] [Google Scholar]
- Hopmann, R., and K.G. Miller. 2003. A balance of capping protein and profilin functions is required to regulate actin polymerization in Drosophila bristle. Mol. Biol. Cell. 14:118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopmann, R., J.A. Cooper, and K.G. Miller. 1996. Actin organization, bristle morphology, and viability are affected by actin capping protein mutations in Drosophila. J. Cell Biol. 133:1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, T.B., and T. Benzing. 2005. The slit diaphragm: a signaling platform to regulate podocyte function. Curr. Opin. Nephrol. Hypertens. 14:211–216. [DOI] [PubMed] [Google Scholar]
- Hutchings, N.J., N. Clarkson, R. Chalkley, A.N. Barclay, and M.H. Brown. 2003. Linking the T cell surface protein CD2 to the actin-capping protein CAPZ via CMS and CIN85. J. Biol. Chem. 278:22396–22403. [DOI] [PubMed] [Google Scholar]
- Isenberg, G., U. Aebi, and T.D. Pollard. 1980. An actin-binding protein from Acanthamoeba regulates actin filament polymerization and interactions. Nature. 288:455–459. [DOI] [PubMed] [Google Scholar]
- Kirsch, K.H., M.M. Georgescu, T. Shishido, W.Y. Langdon, R.B. Birge, and H. Hanafusa. 2001. The adapter type protein CMS/CD2AP binds to the proto-oncogenic protein c-Cbl through a tyrosine phosphorylation-regulated Src homology 3 domain interaction. J. Biol. Chem. 276:4957–4963. [DOI] [PubMed] [Google Scholar]
- Kowanetz, K., J. Terzic, and I. Dikic. 2003. Dab2 links CIN85 with clathrin-mediated receptor internalization. FEBS Lett. 554:81–87. [DOI] [PubMed] [Google Scholar]
- Kurakin, A.V., S. Wu, and D.E. Bredesen. 2003. Atypical recognition consensus of CIN85/SETA/Ruk SH3 domains revealed by target-assisted iterative screening. J. Biol. Chem. 278:34102–34109. [DOI] [PubMed] [Google Scholar]
- Larson, D.E., Z. Liberman, and R.L. Cagan. 2008. Cellular behavior in the developing Drosophila pupal retina. Mech. Dev. 125:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtonen, S., A. Ora, V.M. Olkkonen, L. Geng, M. Zerial, S. Somlo, and E. Lehtonen. 2000. In vivo interaction of the adapter protein CD2-associated protein with the type 2 polycystic kidney disease protein, polycystin-2. J. Biol. Chem. 275:32888–32893. [DOI] [PubMed] [Google Scholar]
- Lehtonen, S., F. Zhao, and E. Lehtonen. 2002. CD2-associated protein directly interacts with the actin cytoskeleton. Am. J. Physiol. Renal Physiol. 283:F734–F743. [DOI] [PubMed] [Google Scholar]
- Lehtonen, S., E. Lehtonen, K. Kudlicka, H. Holthofer, and M.G. Farquhar. 2004. Nephrin forms a complex with adherens junction proteins and CASK in podocytes and in Madin-Darby canine kidney cells expressing nephrin. Am. J. Pathol. 165:923–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, D.K., S.C. Winata, R.J. Lyons, W.E. Hughes, G.M. Lehrbach, V. Wasinger, G. Corthals, S. Cordwell, and R.J. Daly. 2003. A Cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J. Biol. Chem. 278:21805–21813. [DOI] [PubMed] [Google Scholar]
- Mege, R.M., J. Gavard, and M. Lambert. 2006. Regulation of cell-cell junctions by the cytoskeleton. Curr. Opin. Cell Biol. 18:541–548. [DOI] [PubMed] [Google Scholar]
- Miller, D.T., and R.L. Cagan. 1998. Local induction of patterning and programmed cell death in the developing Drosophila retina. Development. 125:2327–2335. [DOI] [PubMed] [Google Scholar]
- Monzo, P., N.C. Gauthier, F. Keslair, A. Loubat, C.M. Field, Y. Le Marchand-Brustel, and M. Cormont. 2005. Clues to CD2-associated protein involvement in cytokinesis. Mol. Biol. Cell. 16:2891–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustonen, H., A. Lepisto, S. Lehtonen, E. Lehtonen, P. Puolakkainen, and E. Kivilaakso. 2005. CD2AP contributes to cell migration and adhesion in cultured gastric epithelium. Biochem. Biophys. Res. Commun. 332:426–432. [DOI] [PubMed] [Google Scholar]
- Palmen, T., S. Lehtonen, A. Ora, D. Kerjaschki, C. Antignac, E. Lehtonen, and H. Holthofer. 2002. Interaction of endogenous nephrin and CD2-associated protein in mouse epithelial M-1 cell line. J. Am. Soc. Nephrol. 13:1766–1772. [DOI] [PubMed] [Google Scholar]
- Parker, L., S. Gross, and L. Alphey. 2001. Vectors for the expression of tagged proteins in Drosophila. Biotechniques. 31:1280–1282, 1284, 1286. [DOI] [PubMed] [Google Scholar]
- Patari-Sampo, A., P. Ihalmo, and H. Holthofer. 2006. Molecular basis of the glomerular filtration: nephrin and the emerging protein complex at the podocyte slit diaphragm. Ann. Med. 38:483–492. [DOI] [PubMed] [Google Scholar]
- Peters, I., I. Tossidou, J. Achenbach, R. Woroniecki, M. Mengel, J.K. Park, M. Paschy, K. de Groot, H. Haller, and M. Schiffer. 2006. IGF-binding protein-3 modulates TGF-beta/BMP-signaling in glomerular podocytes. J. Am. Soc. Nephrol. 17:1644–1656. [DOI] [PubMed] [Google Scholar]
- Reiter, C., T. Schimansky, Z. Nie, and K.F. Fischbach. 1996. Reorganization of membrane contacts prior to apoptosis in the Drosophila retina: the role of the IrreC-rst protein. Development. 122:1931–1940. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032. [DOI] [PubMed] [Google Scholar]
- Schiffer, M., P. Mundel, A.S. Shaw, and E.P. Bottinger. 2004. A novel role for the adaptor molecule CD2-associated protein in transforming growth factor-beta-induced apoptosis. J. Biol. Chem. 279:37004–37012. [DOI] [PubMed] [Google Scholar]
- Schiwek, D., N. Endlich, L. Holzman, H. Holthofer, W. Kriz, and K. Endlich. 2004. Stable expression of nephrin and localization to cell-cell contacts in novel murine podocyte cell lines. Kidney Int. 66:91–101. [DOI] [PubMed] [Google Scholar]
- Schmidt, M.H., B. Chen, L.M. Randazzo, and O. Bogler. 2003. SETA/CIN85/Ruk and its binding partner AIP1 associate with diverse cytoskeletal elements, including FAKs, and modulate cell adhesion. J. Cell Sci. 116:2845–2855. [DOI] [PubMed] [Google Scholar]
- Schneider, T., C. Reiter, E. Eule, B. Bader, B. Lichte, Z. Nie, T. Schimansky, R.G. Ramos, and K.F. Fischbach. 1995. Restricted expression of the irreC-rst protein is required for normal axonal projections of columnar visual neurons. Neuron. 15:259–271. [DOI] [PubMed] [Google Scholar]
- Shih, N.Y., J. Li, V. Karpitskii, A. Nguyen, M.L. Dustin, O. Kanagawa, J.H. Miner, and A.S. Shaw. 1999. Congenital nephrotic syndrome in mice lacking CD2-associated protein. Science. 286:312–315. [DOI] [PubMed] [Google Scholar]
- Shih, N.Y., J. Li, R. Cotran, P. Mundel, J.H. Miner, and A.S. Shaw. 2001. CD2AP localizes to the slit diaphragm and binds to nephrin via a novel C-terminal domain. Am. J. Pathol. 159:2303–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siripala, A.D., and M.D. Welch. 2007. a. SnapShot: actin regulators I. Cell. 128:626. [DOI] [PubMed] [Google Scholar]
- Siripala, A.D., and M.D. Welch. 2007. b. SnapShot: actin regulators II. Cell. 128:1014. [DOI] [PubMed] [Google Scholar]
- Take, H., S. Watanabe, K. Takeda, Z.X. Yu, N. Iwata, and S. Kajigaya. 2000. Cloning and characterization of a novel adaptor protein, CIN85, that interacts with c-Cbl. Biochem. Biophys. Res. Commun. 268:321–328. [DOI] [PubMed] [Google Scholar]
- Tepass, U., and K.P. Harris. 2007. Adherens junctions in Drosophila retinal morphogenesis. Trends Cell Biol. 17:26–35. [DOI] [PubMed] [Google Scholar]
- Usami, Y., S. Popov, and H.G. Gottlinger. 2007. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J. Virol. 81:6614–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraksa, A., A. Bauer, and S. Artavanis-Tsakonas. 2005. Analyzing protein complexes in Drosophila with tandem affinity purification-mass spectrometry. Dev. Dyn. 232:827–834. [DOI] [PubMed] [Google Scholar]
- Welsch, T., N. Endlich, W. Kriz, and K. Endlich. 2001. CD2AP and p130Cas localize to different F-actin structures in podocytes. Am. J. Physiol. Renal Physiol. 281:F769–F777. [DOI] [PubMed] [Google Scholar]
- Welsch, T., N. Endlich, G. Gokce, E. Doroshenko, J.C. Simpson, W. Kriz, A.S. Shaw, and K. Endlich. 2005. Association of CD2AP with dynamic actin on vesicles in podocytes. Am. J. Physiol. Renal Physiol. 289:F1134–F1143. [DOI] [PubMed] [Google Scholar]
- Woroniecki, R.P., M. Schiffer, A.S. Shaw, F.J. Kaskel, and E.P. Bottinger. 2006. Glomerular expression of transforming growth factor-beta (TGF-beta) isoforms in mice lacking CD2-associated protein. Pediatr. Nephrol. 21:333–338. [DOI] [PubMed] [Google Scholar]
- Wu, C., Y.P. Wairkar, C.A. Collins, and A. DiAntonio. 2005. Highwire function at the Drosophila neuromuscular junction: spatial, structural, and temporal requirements. J. Neurosci. 25:9557–9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, T., and G.M. Rubin. 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 117:1223–1237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.