Abstract
Gene expression, like many biological processes, is subject to noise. This noise has been measured on a global scale, but its general importance to the fitness of an organism is unclear. Here, I show that noise in gene expression in yeast has evolved to prevent harmful stochastic variation in the levels of genes that reduce fitness when their expression levels change. Therefore, there has probably been widespread selection to minimise noise in gene expression. Selection to minimise noise, because it results in gene expression that is stable to stochastic variation in cellular components, may also constrain the ability of gene expression to respond to non-stochastic variation. I present evidence that this has indeed been the case in yeast. I therefore conclude that gene expression noise is an important biological trait, and one that probably limits the evolvability of complex living systems.
Keywords: evolution, gene expression, noise, robustness
Introduction
Noise in gene expression is the stochastic variation in the expression level of a gene under a constant environmental condition (Raser and O'Shea, 2005). Recently, it has become possible to quantify the levels of noise in the expression of a gene, and rapid progress has been made in understanding how this noise might be regulated (for reviews, see Raser and O'Shea, 2005; Kaufmann and van Oudenaarden, 2007). In particular, in yeast, gene expression noise has been measured on a global scale and shown to vary widely between different genes, functional classes of genes and genes regulated by different regulatory mechanisms (Bar-Even et al, 2006; Newman et al, 2006). However, despite these global experiments, the overall importance of noise in living systems is still unclear.
Whereas the expression levels of some genes can be altered without any apparent phenotypic effect, decreasing (Giaever et al, 2002; Deutschbauer et al, 2005) or increasing (Sopko et al, 2006) the expression of other genes can be very harmful. Therefore, it has been predicted that if noise in gene expression is a physiologically relevant trait, it might be minimised to prevent harmful stochastic variation in the levels of these ‘dosage-sensitive' genes (Cook et al, 1998; Fraser et al, 2004). The availability of global measurements of both gene expression noise (Newman et al, 2006) and dosage-sensitive genes (Giaever et al, 2002; Deutschbauer et al, 2005; Sopko et al, 2006) mean that this prediction can now be systematically tested.
In their global analysis of gene expression noise in yeast, Newman and co-workers found that essential genes tend to have lower noise than nonessential genes (Newman et al, 2006). Batada and Hurst highlighted that this finding was consistent with selection to minimise noise for dosage-sensitive genes (Batada and Hurst, 2007a). In further support of this hypothesis, they showed that noise tends to be lower for haploinsufficient genes (i.e., genes that reduce growth when their dosage is decreased by half in heterozygotes) than for haplosufficient essential genes, and that genes that produce a strong growth defect when deleted tend to have lower noise than those producing a weak growth defect (Batada and Hurst, 2007a).
Although the lower mean noise reported for essential and haploinsufficient genes is consistent with the noise being minimised to avoid harmful stochastic variation in the expression of dosage-sensitive genes, the interpretation of these results is complicated by the existence of many variables that are known to correlate with both noise (Bar-Even et al, 2006; Newman et al, 2006) and gene essentiality (Jeong et al, 2001; Pal et al, 2003; Papp et al, 2004; Chen and Xu, 2005; Gustafson et al, 2006), including gene expression levels, regulatory mechanisms, protein interactions, and protein functions. The relationship between dosage-sensitivity and noise may therefore not be a direct one.
In this report, I first perform a more detailed analysis of the relationship between gene expression noise and gene dosage-sensitivity. Most importantly, I show that genes with high noise are depleted of both genes that reduce fitness when their expression is increased as well as of those that reduce fitness when their expression is reduced. These two classes of genes are largely independent, which together with the previous evidence (Newman et al, 2006; Batada and Hurst, 2007a) makes it very likely that the relationship between dosage-sensitivity and gene expression noise is a direct one. It therefore seems that noise in gene expression has indeed been widely minimised by natural selection to prevent stochastic variation in the levels of dosage-sensitive genes.
Having established this, I then investigate whether selection to minimise noise has had any long-term consequences for the evolution of gene expression in yeast. I present evidence that the requirement to minimise noise may have limited the ability of genes to change expression in response to genetic perturbations. Moreover, the need to limit noise may also have restricted the extent to which gene expression can change between species. I conclude that noise in gene expression is an important biological trait, and one that may also limit the long-term evolvability of an organism.
Results and discussion
Genes sensitive to either a decrease or to an increase in expression have low noise
Stochastic variation in the expression of genes has been predicted to be more harmful for genes that reduce fitness when their expression levels are altered (Cook et al, 1998; Fraser et al, 2004). In their global analysis of gene expression noise, Newman and co-workers indeed found that genes that are harmful when they are deleted tend to have lower noise than other genes (Newman et al, 2006; Batada and Hurst, 2007a). However, they also found many other features that had similar or stronger correlations with noise (Newman et al, 2006). Given that there are also many similar features (including expression levels, regulatory mechanisms, protein interactions, and protein functions) that have been associated with gene essentiality (Jeong et al, 2001; Pal et al, 2003; Papp et al, 2004; Chen and Xu, 2005; Gustafson et al, 2006), it is therefore not clear whether the relationship between gene expression noise and essentiality is a direct or an indirect effect.
To resolve this ambiguity, I turned to a second set of genes that would be expected to have low noise if stochastic variation in gene expression is harmful—genes which are harmful when their expression is increased. If noise has indeed been minimised to prevent harmful stochastic variation in gene expression, then these genes would also be expected to have low noise. The set of genes that are toxic when they are overexpressed in yeast do not significantly overlap those that are harmful when they are deleted (Sopko et al, 2006). They also encode proteins with very different properties and cellular functions (Sopko et al, 2006; Semple et al, 2008) and so represent a good independent test of a direct relationship between dosage-sensitivity and gene expression noise.
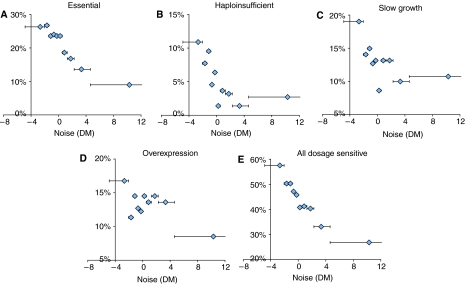
As shown in Figure 1, just as is seen for genes that reduce fitness when they are deleted (essential genes, haploinsufficient genes, genes required for normal growth, Figure 1A–C), the proportion of genes that inhibit growth when they are overexpressed is much lower for genes with high noise (DM) than for genes with low noise (Figure 1D). For example, among the 236 genes that have DM<−2, 26% are essential, 10% are haploinsufficient, 19% are required for normal growth, and 16% are harmful when overexpressed. In contrast of the 359 genes that have DM>3, only 3% are essential, 0.3% are haploinsufficient, 3% are required for normal growth, and 4% are toxic when over-expressed (P<10−5 for all phenotypes, Fisher's exact test). That is, both genes that are sensitive to a decrease or to an increase in expression very rarely have high noise. These two independent sets of genes have very different properties, and so this result strongly suggests that the relationship between noise and dosage-sensitivity is a direct one. This conclusion is also supported by comparing the levels of noise between genes with different severities of fitness defect (Batada and Hurst, 2007a, Supplementary Figure 1). Noise in gene expression therefore appears to be tuned to minimise stochastic variation in the expression levels of dosage-sensitive genes. Either there must exist mechanisms to prevent stochastic variation in the expression levels of these genes, or there has been selection to prevent these genes from evolving regulatory mechanisms that would result in high noise.
Figure 1.
Noise is minimised for dosage-sensitive genes. The relationship between gene expression and the proportion of genes that are (A) essential, (B) haploinsufficient, (C) inhibit growth when deleted or (D) inhibit growth when overexpressed. The result when considering all dosage-sensitive genes is shown in (E). The percentage of genes with each phenotype is shown for each of 10 equally sized bins of genes ranked according to an expression-level-adjusted measure of noise (DM). The range of each bin is shown, except for the maximum of the top bin, which extends to 61.0.
Essential genes may be highly expressed to limit noise
Noise in gene expression has been found to correlate inversely with gene expression levels (Bar-Even et al, 2006; Newman et al, 2006). The measure of noise used here (DM) (Newman et al, 2006) is designed to compensate for this effect, and indeed the trends seen here are not due to variations in gene expression levels (Supplementary Figure 2). However, increasing gene expression levels does represent a simple mechanism to reduce noise, and this may partially explain why essential genes tend to have high expression levels (Pal et al, 2003). In contrast, this seems unlikely to represent a valid strategy to reduce noise levels for genes that are harmful when they are overexpressed, and indeed these genes have low expression levels (our unpublished data), and so must use other mechanisms to reduce noise.
Proteins with more protein interactions have lower noise
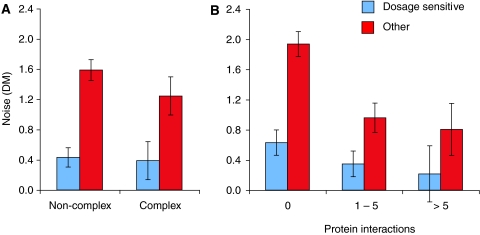
Gene essentiality and gene expression levels both also correlate with the number of protein interactions made by a gene product (Jeong et al, 2001; Pal et al, 2003). The relationship between noise and dosage-sensitivity is still very strong even after accounting for the number of protein interactions made by a gene product or for its membership of a protein complex (Figure 2), which again suggests that this is a direct effect. However, it can also be seen in Figure 2 that genes with more protein interactions do tend to have lower noise even after accounting for dosage sensitivity. Such an effect has previously been predicted on theoretical grounds (Fraser et al, 2004), and I suggest here three possible explanations for this effect. First, the number of protein interactions may be a variable capturing fitness defects that have not yet been measured in the laboratory. Second, the reduced noise of protein complex subunits may reflect selection to reduce harmful ‘imbalance' in the stoichiometry of protein complexes (Papp et al, 2003; Fraser et al, 2004). Third, the reduced noise of protein complex subunits may be a consequence of active mechanisms that rapidly degrade protein complex subunits that have not been stably associated into complexes. That is, the stability of assembled complexes themselves, combined with active degradation methods, may be responsible for the observed low noise.
Figure 2.
Proteins with more interactions have lower noise. The relationship between noise and dosage sensitivity is seen both for genes that are part of MIPS protein complexes and for other genes (A). It is also seen when controlling for the number of protein interactions (B). P-values for differences between dosage-sensitive and other genes: 2 × 10−4 (complex subunits), 9 × 10−9 (non-complex subunits), 4 × 10−7 (zero protein interactions), 0.05 (1–5 protein interactions), and 1 × 10−3 (>5 protein interactions) (Wilcoxon rank sum test). In addition, genes without protein interactions have a higher mean noise than genes with protein interactions for both dosage-sensitive and other genes (P=1 × 10−3 and P=5 × 10−8, respectively). The error bars represent measure±s.e.
Noise may limit the evolvability of gene expression
I have shown above that noise in gene expression has probably been minimised to prevent harmful stochastic variation in the expression of dosage-sensitive genes. A gene with low gene expression noise must, by definition, be insensitive to stochastic variation in the levels of cellular components. This may either reflect the expression of a gene being ‘insulated' from cellular networks or the existence of mechanisms or network motifs that function to reduce stochastic variation. These same genes are therefore also likely to be insensitive to non-stochastic alterations in the levels of cellular components, including those resulting from genetic mutations. That is, genes with low levels of noise in gene expression may also have expression levels that change little in response to random mutagenesis, and that are restrained in their ability to vary throughout evolution. This in turn would be reflected in these genes having expression patterns that only evolve slowly between species. In this way selection to minimise noise may constrain the long-term ‘evolvability' of living systems (Wagner, 2005). This intuitive prediction is supported by theoretical work using artificial gene networks, which shows that selection to minimise noise can result in gene expression that is stable to genetic perturbations (Ciliberti et al, 2007).
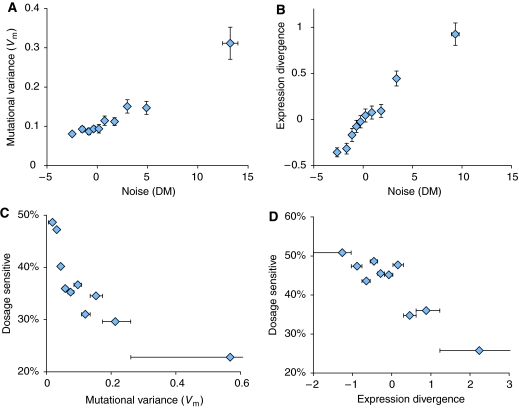
To address this prediction that selection to minimise noise may constrain the evolvability of gene expression, I used genome-wide data measuring both the global response of gene expression to random mutagenesis in mutation accumulation experiments (‘mutational variance') (Landry et al, 2007) and the divergence of gene expression between closely related yeast species (‘expression divergence') (Tirosh et al, 2006). If selection to control noise results in gene expression that is stable to genetic perturbations, then both of these variables should correlate well with noise. Moreover, if noise is minimised for dosage-sensitive genes, then dosage-sensitive genes should have expression patterns that are stable to random mutagenesis, and that evolve slowly between species. All of these predictions are upheld by the available data. Both mutational variance (Figure 3A, Spearman's Rank correlation coefficient ρ=0.27, P=1.08 × 10−14, n=776) and expression divergence (Figure 3B, ρ=0.30, P=2.2 × 10−16, n=1750) correlate very well with gene expression noise in yeast. Moreover, also consistent with the predictions, the proportion of dosage-sensitive genes is much higher for both genes with low mutational variance (Figure 3C) and for genes with low expression divergence between species (Figure 3D). That the same trends are seen with these two very different measures of expression divergence, measured both within and between species, adds confidence to these conclusions.
Figure 3.
Mutational variance (Vm) and expression divergence between species are restricted for dosage-sensitive genes with low noise. Both mutational variance, a measure of the change in a gene's expression in response to random mutagenesis (Landry et al, 2007) (A), and the divergence in expression between yeast species (Tirosh et al, 2006) (B), negatively correlate with noise (Spearman's Rank correlation coefficient, ρ=0.27, P=1.08 × 10−14, n=776, and ρ=0.30, P=2.2 × 10−16, n=1750, respectively). Error bars represent measure±s.e. (C) The relationship between mutational variance and the percentage of dosage-sensitive genes. The percentage of dosage-sensitive genes is shown for ten equally sized bins of genes arranged according to their mutational variance. (D) The same plot but comparing dosage-sensitivity to gene expression divergence between yeast species, again for ten equally sized bins of yeast genes. The ranges of each bin are shown, except for the maximum of the top bins, which extend to 4.05 and 9.02, respectively.
Direct selection for robustness to mutation is not expected under most conditions, because the single mutations being considered do not reduce fitness and so cannot be selected in most realistic conditions (Nowak et al, 1997; Wagner, 2000, 2005). In contrast, if noise reduces fitness, then there can be direct selection to minimise noise in gene expression. Therefore, as predicted by simulations (Ciliberti et al, 2007), I propose that selection to minimise noise in the expression of dosage-sensitive genes has resulted in these genes having expression mechanisms that are also stable to genetic perturbations and that therefore evolve slowly between species.
Conclusions
In summary, the data presented here demonstrate that noise in gene expression is tuned to minimise harmful stochastic variation in the expression levels of dosage-sensitive genes. Noise is thus an important biological trait, and one that has probably been subject to direct natural selection. Moreover, in agreement with theoretical predictions, the available data sets in yeast suggest that selection to minimise noise may also have constrained the long-term evolvability of gene expression in this species.
Materials and methods
The following phenotype data sets were used: essential genes (Mewes et al, 2006), haploinsufficient genes (Deutschbauer et al, 2005), genes with a slow growth phenotypes in rich media (Giaever et al, 2002), and genes with overexpression phenotypes (Sopko et al, 2006). Noise measurements are from Newman et al (2006), who used GFP reporter constructs to measure the levels of noise in the expression of >2500 yeast genes. Noise correlates with expression levels (Bar-Even et al, 2006; Newman et al, 2006), so an expression level-adjusted measure of noise (DM) (Newman et al, 2006)) is used throughout this work. Mutational variance (Vm) measurements, a measure of the divergence in the expression level of a gene in mutation accumulation experiments, were taken from Landry et al (2007). Measurements of expression divergence between closely related yeast species were taken from Tirosh et al (2006). Protein interaction data used are the high-confidence (i.e., supported by more than one piece of evidence; Bertin et al, 2007; Batada et al, 2007b) subset of the literature-curated yeast protein interactome (Reguly et al, 2006). Literature-curated protein complexes were downloaded from MIPS (Mewes et al, 2006). Statistical tests were performed using the R package (http://www.R-project.org).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure Legends
Acknowledgments
This work was funded by the Spanish Ministry of Education and Science (MEC) through the EMBL-CRG Systems Biology Unit and by the Institució Catalana de Recerca i Estudis Avançats (ICREA).
References
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O'Shea E, Pilpel Y, Barkai N (2006) Noise in protein expression scales with natural protein abundance. Nat Genet 38: 636–643 [DOI] [PubMed] [Google Scholar]
- Batada NN, Hurst LD (2007a) Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat Genet 39: 945–949 [DOI] [PubMed] [Google Scholar]
- Batada NN, Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hurst LD, Tyers M (2007b) Still stratus not altocumulus: further evidence against the date/party hub distinction. PLoS Biol 5: e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin N, Simonis N, Dupuy D, Cusick ME, Han JD, Fraser HB, Roth FP, Vidal M (2007) Confirmation of organized modularity in the yeast interactome. PLoS Biol 5: e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Xu D (2005) Understanding protein dispensability through machine-learning analysis of high-throughput data. Bioinformatics 21: 575–581 [DOI] [PubMed] [Google Scholar]
- Ciliberti S, Martin OC, Wagner A (2007) Robustness can evolve gradually in complex regulatory gene networks with varying topology. PLoS Comput Biol 3: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DL, Gerber AN, Tapscott SJ (1998) Modeling stochastic gene expression: implications for haploinsufficiency. Proc Natl Acad Sci USA 95: 15641–15646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer AM, Jaramillo DF, Proctor M, Kumm J, Hillenmeyer ME, Davis RW, Nislow C, Giaever G (2005) Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 169: 1915–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB (2004) Noise minimization in eukaryotic gene expression. PLoS Biol 2: e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Gustafson AM, Snitkin ES, Parker SC, DeLisi C, Kasif S (2006) Towards the identification of essential genes using targeted genome sequencing and comparative analysis. BMC Genomics 7: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabasi AL, Oltvai ZN (2001) Lethality and centrality in protein networks. Nature 411: 41–42 [DOI] [PubMed] [Google Scholar]
- Kaufmann BB, van Oudenaarden A (2007) Stochastic gene expression: from single molecules to the proteome. Curr Opin Genet Dev 17: 107–112 [DOI] [PubMed] [Google Scholar]
- Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL (2007) Genetic properties influencing the evolvability of gene expression. Science 317: 118–121 [DOI] [PubMed] [Google Scholar]
- Mewes HW, Frishman D, Mayer KF, Munsterkotter M, Noubibou O, Pagel P, Rattei T, Oesterheld M, Ruepp A, Stumpflen V (2006) MIPS: analysis and annotation of proteins from whole genomes in 2005. Nucleic Acids Res 34: D169–D172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS (2006) Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846 [DOI] [PubMed] [Google Scholar]
- Nowak MA, Boerlijst MC, Cooke J, Smith JM (1997) Evolution of genetic redundancy. Nature 388: 167–171 [DOI] [PubMed] [Google Scholar]
- Pal C, Papp B, Hurst LD (2003) Genomic function: rate of evolution and gene dispensability. Nature 421: 496–497; Discussion 497–498 [DOI] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD (2003) Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197 [DOI] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD (2004) Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature 429: 661–664 [DOI] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK (2005) Noise in gene expression: origins, consequences, and control. Science 309: 2010–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hon GC, Myers CL, Parsons A, Friesen H, Oughtred R, Tong A, Stark C, Ho Y, Botstein D, Andrews B, Boone C, Troyanskya OG, Ideker T, Dolinski K, Batada NN, Tyers M (2006) Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J Biol 5: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple JI, Vavouri T, Lehner B (2008) A simple principle concerning the robustness of protein complex activity to changes in gene expression. BMC Syst Biol 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, Boone C, Andrews B (2006) Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell 21: 319–330 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Weinberger A, Carmi M, Barkai N (2006) A genetic signature of interspecies variations in gene expression. Nat Genet 38: 830–834 [DOI] [PubMed] [Google Scholar]
- Wagner A (2000) The role of population size, pleiotropy and fitness effects of mutations in the evolution of overlapping gene functions. Genetics 154: 1389–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A (2005) Robustness and Evolvability in Living Systems. Princeton, NJ: Princeton University Press [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure Legends