Abstract
Systems analyses have facilitated the characterization of metabolic networks of several organisms. We have reconstructed the metabolic network of Leishmania major, a poorly characterized organism that causes cutaneous leishmaniasis in mammalian hosts. This network reconstruction accounts for 560 genes, 1112 reactions, 1101 metabolites and 8 unique subcellular localizations. Using a systems-based approach, we hypothesized a comprehensive set of lethal single and double gene deletions, some of which were validated using published data with approximately 70% accuracy. Additionally, we generated hypothetical annotations to dozens of previously uncharacterized genes in the L. major genome and proposed a minimal medium for growth. We further demonstrated the utility of a network reconstruction with two proof-of-concept examples that yielded insight into robustness of the network in the presence of enzymatic inhibitors and delineation of promastigote/amastigote stage-specific metabolism. This reconstruction and the associated network analyses of L. major is the first of its kind for a protozoan. It can serve as a tool for clarifying discrepancies between data sources, generating hypotheses that can be experimentally validated and identifying ideal therapeutic targets.
Keywords: flux balance analysis, infectious disease, Leishmania major, metabolic network, systems biology
Introduction
Computational modeling and systems analysis techniques can facilitate the interrogation of biological systems on the genome scale. Using available genomic, proteomic and metabolomic data, models of biochemical networks with predictive capabilities can be constructed (Forst, 2006). Computational analysis of these reconstructed networks can give much insight into the biological system and yield a set of hypotheses that can be experimentally validated. For example, simulating gene knockouts and assessing robustness of particular enzyme-catalyzed reactions enable evaluation of the efficacy of potential drug targets (Reed et al, 2006a; Jamshidi and Palsson, 2007).
The metabolic networks of several organisms, including Escherichia coli, Helicobacter pylori, Saccharomyces cerevisiae and Homo sapiens have been reconstructed, and the reconstructions of many others are currently underway (Schilling et al, 2002; Forster et al, 2003; Reed et al, 2003, 2006a; Borodina and Nielsen, 2005; Duarte et al, 2007; Feist et al, 2007; Oberhardt et al, 2008). These network models have facilitated the reconciliation of discrepancies between heterogeneous data sets (Reed et al, 2006a). Such systematic compilation has enabled precise characterization of cellular networks and aided in efforts to refine genome annotation (Feist et al, 2006; Reed et al, 2006a, 2006b). The resultant in silico models have predicted growth rates and gene essentiality under different medium conditions and have characterized the use of alternative carbon sources by particular organisms (Schilling et al, 2002; Feist et al, 2006, 2007).
Here, we report the reconstruction of the metabolic network of the parasite Leishmania major Friedlin, the causative agent of cutaneous leishmaniasis. Closely related Leishmania spp. cause diffuse cutaneous, mucocutaneous and visceral forms of the disease. Overall, leishmaniasis has an annual incidence rate of two million cases and causes approximately 59 000 deaths worldwide each year (Davies et al, 2003). The vector for this disease is a female phlebotomine sandfly (Lipoldova and Demant, 2006). The morphological stages of Leishmania have been previously described in detail (Molyneux and Killick-Kendrick, 1987). Leishmania exist in flagellated form known as the promastigote within the sandfly gut. Upon contact with the mammalian host, promastigotes enter macrophages and transform into non-flagellated amastigotes (see Figure S1 in Supplementary Information I) (Lipoldova and Demant, 2006).
This reconstruction and the associated network analyses represent the first constraint-based model for a protozoan. The metabolic network reconstruction presented here accounts for 560 genes, 1112 reactions and 1101 metabolites. The reconstruction is highly compartmentalized with eight unique subcellular localizations accounted for in the model. We have proposed novel single and double gene deletion predictions resulting in a lethal phenotype. Additionally, we have validated predictions of gene essentiality in the network with published knockout studies from highly related Leishmania and Trypanosoma species, and we demonstrated the utility of the reconstruction in hypothesized refinements of the L. major genome annotation. Other results include a novel prediction of an in silico minimal medium to support growth, characterization of network robustness and efficacy of drug targets in the presence of enzyme inhibitors, evaluation of morphological stage-specific metabolism and examination of the iterative design phase in model building. This type of systems analysis not only provides a platform for data integration and hypothesis generation to further scientific research in infectious disease but also facilitates the identification of therapeutic drug targets against devastating tropical diseases such as leishmaniasis.
Results
Properties of the metabolic network
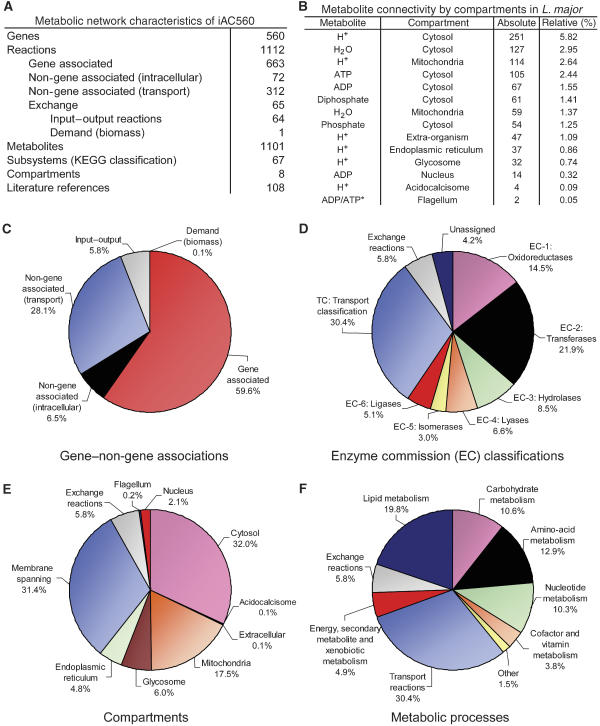
The iAC560 metabolic network reconstruction (see section on naming convention in Materials and methods) of L. major accounts for 560 genes spanning the 36 chromosomes of the genome, approximately 6.7% of all genes present in the organism (see Figure 1A). The model included 1112 reactions of which 1047 were metabolic and 65 were exchange. Of the exchange reactions, 64 were input–output exchanges that allowed extracellular metabolites in the medium to enter the system or end products of metabolism to be excreted out of the system. The remaining exchange reaction was the biomass demand used as a drain for essential metabolites to characterize growth of the in silico system. iAC560 also accounted for 1101 metabolites, a high number compared with other metabolic reconstructions (Borodina and Nielsen, 2005; Reed et al, 2006a). This was mostly due to the highly compartmentalized nature of the model; a total of eight unique compartments were included in the model, namely the cytosol, acidocalcisome, extracellular space, mitochondria, glycosome, endoplasmic reticulum, flagellum and nucleus. Additionally, 108 literature references providing biochemical, physiological and other validating information were used in reconstructing the metabolic network of L. major.
Figure 1.
Network characteristics of iAC560. Fundamental characteristics of the iAC560 metabolic network are summarized. In (A) the total number of genes, reactions, metabolites, subsystems (via the KEGG classification scheme), compartments and literature references used in the model are listed. In (B), the most highly connected metabolites in every compartment of the model are presented. The absolute value indicates the total number of times a particular metabolite is participating in reactions within the model. The relative connectivity is equal to the absolute connectivity divided by the sum of the absolute connections of all metabolites in the network. (C–F) The iAC560 reactions grouped by gene–non-gene associations, enzyme commission (EC) classifications, compartments or subcellular localizations, and metabolic processes, respectively. *ADP and ATP have equal connectivity in the flagellum.
The metabolites with the highest connectivity across different compartments were identified (see Figure 1B). The absolute value indicates the number of times a particular metabolite occurred in any reaction of the stoichiometric matrix (S-matrix). The relative value indicates percent connectivity of the metabolite with respect to the entire metabolic network. The hydrogen ion (or proton) was the most connected metabolite in six out of eight compartments. This result emphasizes the role of proton-dependent reactions within the network. In the metabolic network reconstruction of S. cerevisiae, another eukaryotic multi-compartment organism, the hydrogen ion was also the most highly connected metabolite (Forster et al, 2003). Other highly connected metabolites in L. major included ATP, ADP, H2O, phosphate and diphosphate, which are known to participate in numerous metabolic reactions.
The distribution of reactions across gene–non-gene associations, enzyme commission (EC) classifications, compartments and metabolic processes was calculated (see Figures 1C–F). The majority of metabolic reactions included in the reconstruction were associated with genes (see Figure 1C). The remainder comprised intracellular non-gene-associated reactions, inter-compartment transport reactions and transporters to the environment. Importantly, the intracellular non-gene-associated reactions comprised only 6.5% of all the reactions present in the model. This grouping contained reactions that were spontaneous (those that proceeded without the presence of enzymes) and reactions added for proper functioning of the computational model. There were insufficient gene annotations or literature evidence for the latter. Also, many transport reactions, which were added to provide access to enzymatic reactions segregated in multiple locations within the cell, did not have known gene associations. As shown in Figure 1D, transferases, or enzymes that aid in transferring a functional moiety from one metabolite to another, constituted the greatest portion of enzymatic reactions in the model. For example, hexokinase (EC: 2.7.1.1) transfers a phosphate from ATP to glucose to yield glucose 6-phosphate and ADP. Interestingly, the order of abundance of reactions grouped by EC classifications in L. major was identical to that of S. cerevisiae, emphasizing the high degree of conservation in metabolic processes (Forster et al, 2003).
The cytosol and membrane-spanning groups accounted for 63.4% of all reactions (see Figure 1E). The membrane-spanning group included inter-compartment transporters and transporters to the environment in addition to certain oxidative phosphorylation reactions that spanned the mitochondrial and acidocalcisomal (a vacuole-like compartment first discovered in trypanosomatids; Docampo et al, 2005) membranes. The glycosome included approximately 6.0% of reactions. The first part of glycolysis is compartmentalized in the glycosome, which also includes enzymatic reactions from nucleotide and lipid metabolism among others (Hart and Opperdoes, 1984; Opperdoes and Michels, 1993; Opperdoes and Szikora, 2006). Not considering transport reactions, the percentage of intracellular reactions participating in lipid metabolism was the greatest when compared to reactions across other metabolic subsystems (see Figure 1F). This was partly due to the presence of two fatty acid synthesis mechanisms present in trypanosomes: an endoplasmic reticulum-based elongase pathway with fatty acyl groups attached to coenzyme-A (CoA) and a mitochondrial type II pathway with fatty acyl groups attached to acyl carrier protein (Lee et al, 2007).
Single and double gene deletion predictions
Genes in iAC560 were ‘knocked-out' and classified as lethal, growth-reducing or having no effect. Figure 2 describes the results of single and double gene deletion predictions, both performed under defined medium conditions (see Merlen et al, 1999; Schuster and Sullivan, 2002). Specifically, Figure 2A illustrates that 12% of all single gene knockouts are lethal and 10% are growth reducing. For 2% of all gene knockouts, growth is between 0 and 90%, and in 8% of the cases, growth is between 90 and 100%. Figure 2B classifies all lethal single gene knockouts according to the metabolic process that they are involved in. Approximately 83% of all lethal genes belonged to three metabolic processes: lipid, carbohydrate or amino-acid metabolism, highlighting how critical these are to general function.
Figure 2.
Single and double gene deletion predictions. The results of in silico gene deletion predictions are summarized. (A) Single gene deletion predictions are classified into four categories with respect to wild type: lethal (0%), growth reducing (>0% and <90%), growth reducing (>90% and <100%) and no effect (100%). For each case, the percentage of growth is indicated in the parentheses. (B) The lethal single gene deletions grouped according to metabolic processes. (C) The total number of lethal single and double gene deletions. Lethal double gene deletions can be classified as trivial lethal or non-trivial lethal. Trivial lethal implies that at least one of the genes involved is lethal in a single gene deletion. In non-trivial lethal double deletions, both genes involved are not lethal individually as single gene deletions but are lethal in combination.
Of particular interest are genes that are lethal to L. major but are not present in human. For example, LmjF05.0350, which codes for trypanothione reductase, was lethal in silico. This enzyme uses a cofactor N1,N8-bis (L-γ-glutamyl-L-hemicystinyl-glycyl) spermidine, otherwise known as trypanothione (Fairlamb et al, 1985). This cofactor is only present in Kinetoplastida (Krauth-Siegel et al, 2003), the class of organisms to which Leishmania belongs. Additionally, trypanothione reductase is an essential enzyme for the survival of Leishmania spp. (Tovar et al, 1998; Krauth-Siegel et al, 2003). Therefore, trypanothione metabolism is being studied as a potential for therapeutic design and targeting (Benson et al, 1992; Dumas et al, 1997; Chan et al, 2002; Fairlamb, 2003; Krauth-Siegel et al, 2003). Model predictions for trypanothione reductase were consistent with published literature. In addition, tryparedoxin peroxidase (Alphey et al, 2000; Fairlamb, 2003) and fumarate reductase (Chen et al, 2001) encoded by LmjF15.1040 and LmjF35.1180, respectively, both lethal genes in silico, are also likely targets for antileishmanial drugs as these enzymes are not present in mammalian cells.
Furthermore, the set of lethal gene knockouts include LmjF31.2940 and LmjF21.0845, coding for squalene synthase and hypoxanthine-guanine phosphoribosyltransferase, respectively. There are inhibitors for both enzymes: 3-(biphenyl-4-yl)-3-hydroxyquinuclidine (BPQ-OH) and ER27856 for squalene synthase and 4-hydroxy-3,4-pyrazolopyrimidine (allopurinol) for hypoxanthine-guanine phosphoribosyltransferase. These drugs have been experimentally tested against trypanosomatids (Urbina et al, 2002; Urbina and Docampo, 2003; Braga et al, 2004). The squalene synthase inhibitors affect the sterol biosynthesis pathway, taking advantage of the trypanosomatid requirement for specific endogenous sterols (e.g. ergosterol) (Urbina et al, 2002; Braga et al, 2004). Allopurinol takes advantage of the inability of the trypanosomatid species to synthesize purines de novo (Urbina and Docampo, 2003; Berriman et al, 2005). Each of the remaining in silico lethal genes represents an individual hypothesis that can be experimentally tested and validated.
In addition to single gene deletion predictions, all possible double gene deletions were also simulated (see Figure 2C). Out of 156 520 double deletions, 19 341 were lethal. Another 19 285 double deletions were trivial lethal, i.e. at least one of the genes involved was lethal in a single deletion. There were 56 non-trivial lethal double deletions, i.e. both genes involved were not lethal individually as single deletions, but were lethal only in combination. An analysis of double gene deletion effects on the metabolic network revealed that 57.6% of the reactions affected participate in either lipid or carbohydrate metabolism. The remaining 42.4% of the reactions are involved in nucleotide, energy, amino acid, vitamin and cofactor, secondary metabolite and xenobiotic metabolism. These 56 double deletions are additional hypotheses that can be experimentally validated and represent predictions of genetic interactions and synthetic lethality that have proven highly useful in published reports (Boone et al, 2007). The large fraction of double deletions that were not lethal is possibly indicative of the high degree of redundancy in the network. See Supplementary Information II for a complete listing of all non-trivial lethal double deletions.
Validation of reaction knockout predictions
The viability of selected simulations of gene knockouts was compared with published data. Table I lists the complete set of lethal and non-lethal knockouts obtained from literature and the corresponding in silico predictions in four different medium conditions, namely: minimal medium; minimal medium and glucose; minimal medium, glucose and other amino acids; and defined medium (see Merlen et al, 1999; Schuster and Sullivan, 2002). The rows colored gray, yellow and white indicate agreement with literature, partial agreement with literature and disagreement between iAC560 predictions and experimental data, respectively. The minimal medium predictions had the greatest overall compliance with 72.4% agreement. Predictions in defined medium had the lowest overall compliance with 65.5% agreement. The other two medium conditions had overall compliance of 69.0%. Most often the experimental knockouts were done in varying medium conditions. It is difficult to characterize and replicate in silico the exact environmental conditions that were used in the experimental systems. This is a likely cause for the discrepancy seen between the in silico and experimental systems. Additionally, another cause for discrepancy can be attributed to the fact that experimental data were obtained from related Leishmania and Trypanosoma organisms, and not entirely from L. major. In future iterations of the model, discrepancies that exist in knockout data may be corrected by addressing gene–protein-reaction (GPR) relationships, kinetic constraints or gaps in the network.
Table 1.
Validation of reaction knockout predictions
 |
Improving genome annotation through reconstruction
The manual process of compiling and integrating data from heterogeneous sources such as public databases and published literature facilitates in clarifying discrepancies and providing functional annotation to previously uncharacterized genes (Feist et al, 2006). Table II indicates all of the annotation refinements proposed in the iAC560 metabolic network. Out of 25 new annotations, 17 were previously characterized as ‘hypothetical proteins' in public databases. The rest had either an incorrect annotation, localization or EC classification.
Table 2.
Table of annotation refinements
| Gene | Annotation compiled from KEGG/GeneDB | Proposed iAC560 annotation | Evidence | ||||
|---|---|---|---|---|---|---|---|
| Putative identification | Localization | EC classification | Identification | Localization | EC classification | ||
| LmjF02.0500 | Hypothetical protein, conserved | Unknown | Unknown | Glycerate kinase | Cytosol | EC: 2.7.1.31 | BLAST search shows amino-acid sequence similarity with genes from Yersinia pseudotuberculosis and Streptococcus agalactiae. Localization hypothesized. EC number obtained from ExPASy ENZYME database |
| LmjF04.1160 | Fructose-1,6-bisphosphatase, cytosolic, putative | Cytosol | EC: 3.1.3.11 | Fructose-1,6-bisphosphatase | Glycosome | EC: 3.1.3.11 | See Naderer et al (2006); Opperdoes and Szikora (2006). PTS detected and isoelectric point is greater than 8.5 |
| LmjF05.0090 | Hypothetical protein, conserved | Unknown | Unknown | NUDIX hydrolase dihydroneopterin triphosphate pyrophosphohydrolase/hydrolase | Nucleus | EC: 3.6.1.- | BLAST search shows amino-acid sequence similarity with two annotated genes each from Trypanosoma cruzi and T. brucei. Localization hypothesized |
| LmjF16.0340 | Hypothetical protein, conserved | Endoplasmic reticulum, membrane | Unknown | 3-Beta-hydroxysteroid-delta(8), delta(7) -isomerase | Endoplasmic reticulum | EC: 5.3.3.5 | BLAST search snows amino-acid sequence similarity with genes from Bos taurus, Homo sapiens and Cavia porcellus. EC number obtained from ExPASy ENZYME database |
| LmjF16.1290 | Hypothetical protein, conserved | Unknown | Unknown | Diacylglycerol kinase | Mitochondria, cytosol | EC: 2.7.1.107 | BLAST search shows amino-acid sequence similarity with two annotated genes from T. cruzi. Localization hypothesized. EC number obtained from ExPASy ENZYME database |
| LmjF20.0970 | Hypothetical protein, conserved | Unknown | Unknown | 1,2-Dihydroxy-3-keto- 5-methylthiopentene dioxygenase | Cytosol | EC: 1.13.11.54 | BLAST search shows amino-acid sequence similarity with genes from Gallus gallus, Mus musculus, Rattus norvegicus, Schizosaccharomyces pombe, Aspergillus terreus, B. taurus, Arabidopsis thaliana, H. sapiens, Xenopus laevis, Danio rerio and X. tropicalis. Localization hypothesized. EC number obtained from ExPASy ENZYME database |
| LmjF23.0370 | Hypothetical protein, conserved | Unknown | Unknown | Cytochrome c oxidase subunit 10 | Mitochondria | EC: 1.9.3.1 | BLAST search shows amino-acid sequence similarity with one T. brucei cytochrome c oxidase subunit 10 genes and 3 T. cruzi genes. Localization hypothesized. EC number obtained from ExPASy ENZYME database |
| LmjF23.1480 | Hypothetical protein, conserved | Unknown | Unknown | Alanine racemase | Cytosol | EC: 5.1.1.1 | BLAST search shows amino-acid sequence similarity with genes from Ostreococcus tauri, Aspergillus clavatus, A. fumigatus, Neosartorya fischeri (see Panizzutti et al, 2006—Leishmania amazonensis). EC number obtained from ExPASy ENZYME database |
| LmjF25.2010 | Hypothetical protein, conserved 2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase | Unknown | Unknown | 2,4-Dihydroxyhept-2-ene-1,7-dioic acid aldolase; 4-hydroxy-2-ketopimelate aldolase | Mitochondria | EC: 4.1.2.- | BLAST search shows amino-acid sequence similarity with genes from Escherichia coli, Shigella sonnei, S. boydii and S. flexneri. Localization hypothesized |
| LmjF28.1840 | Hypothetical protein, conserved | Unknown | Unknown | Methylthioribulose-1-phosphate dehydratase | Cytosol | EC: 4.2.1.109 | BLAST search shows amino-acid sequence similarity with genes from Bacillus subtilis, B. licheniformis and Stigmatella aurantiaca. Localization hypothesized. EC number obtained from ExPASy ENZYME database |
| LmjF29.1830 | Dihydrolipoamide dehydrogenase, putative | Cytoplasm | EC: 1.8.1.4 | Dihydrolipoamide dehydrogenase | Mitochondria | EC: 1.8.1.4 | See Hannaert et al (2003b) (part of the pyruvate dehydrogenase complex) |
| LmjF31.1780 | Hypothetical protein, conserved | Unknown | Unknown | Sphingosine N-acyltransferase | Endoplasmic reticulum | EC: 2.3.1.24 | BLAST search shows amino-acid sequence similarity with gene from Cryptococcus neoformans. Localization hypothesized. EC number obtained from ExPASy ENZYME database |
| LmjF31.2920 | Hypothetical protein, conserved | Unknown | Unknown | Inositol polyphosphate 1-phosphatase | Cytosol | EC: 3.1.3.57 | BLAST search shows amino-acid sequence similarity with gene from T. brucei. Localization hypothesized. EC number obtained from ExPASy ENZYME database |
| LmjF32.1530 | Hypothetical protein, conserved | Unknown | EC: 3.6.1.- | NUDIX hydrolase dihydroneopterin triphosphate pyrophosphohydrolase/hydrolase | Nucleus | EC: 3.6.1.- | BLAST search shows amino-acid sequence similarity with two annotated genes each from T. cruzi and T. brucei. Localization hypothesized |
| LmjF32.2290 | Hypothetical protein, conserved | Unknown | Unknown | Sphingosine-1-phosphate phosphatase | Endoplasmic reticulum | EC: 3.1.3.- | BLAST search shows amino-acid sequence similarity with genes from C. neoformans, Aspergillus oryzae, S. pombe, A. clavatus and H. sapiens. Localization and EC number hypothesized |
| LmjF32.3310 | Dihydrolipoamide dehydrogenase, putative | Cytoplasm | EC: 1.8.1.4 | Dihydrolipoamide dehydrogenase | Mitochondria | EC: 1.8.1.4 | See Hannaert et al (2003b) (part of the pyruvate dehydrogenase complex) |
| LmjF32.3760 | Hypothetical protein, conserved | Unknown | Unknown | Enolase | Cytosol | EC: 4.2.1.11 | See Hannaert et al (2003a). BLAST search shows amino-acid sequence similarity with two T. cruzi enolase genes and one T. brucei enolase gene. EC number obtained from ExPASy ENZYME database |
| LmjF33.3270 | Fatty acid desaturase, putative | Membrane | Unknown | Delta-12 oleate desaturase | Cytosol | EC: 1.3.1.35 | BLAST search shows amino acid sequence similarity with one functionally annotated gene each from T. cruzi and T. brucei. Localization hypothesized. EC number obtained from ExPASy ENZYME database |
| LmjF34.4600 | Hypothetical protein, conserved | Unknown | Unknown | 1,2-Dihydroxy-3-keto- 5-methylthiopentene dioxygenase | Cytosol | EC: 1.13.11.54 | BLAST search shows amino-acid sequence similarity with genes from S. pombe, B. taurus, R. norvegicus, M. musculus, A. terreus, A. thaliana, H. sapiens and G. gallus. Localization hypothesized. EC number obtained from ExPASy ENZYME database |
| LmjF35.2740 | Galactokinase-like protein | Cytoplasm | EC: 2.7.1.6 | Galactokinase | Glycosome | EC: 2.7.1.6 | PTS detected |
| LmjF36.1960 | Phosphomannomutase, putative | Cytoplasm | EC: 3.1.3.11 | Phosphomannomutase | Cytosol | EC: 5.4.2.8 | EC number obtained from ExPASy ENZYME database |
| LmjF36.2540 | Hypothetical protein, unknown function | Unknown | Unknown | C-4 sterol methyl oxidase | Endoplasmic reticulum | EC: 1.14.13.72 | BLAST search shows amino-acid sequence similarity with genes from A. oryzae, D. rerio, G. gallus, N. fischeri, Ictalurus punctatus and X. tropicalis. Localization hypothesized. EC number obtained from ExPASy ENZYME database |
| LmjF36.2950 | Succinyl-CoA ligase (GDP forming) beta-chain, putative | Mitochondria | EC: 6.2.1.5 | Succinyl-CoA ligase (GDP forming) | Mitochondria | EC: 6.2.1.4 | EC: 6.2.1.5 is for ADP-forming succinyl-CoA ligase. EC number obtained from ExPASy ENZYME database |
| LmjF36.4930 | Translation initiation factor 2 subunit, putative | Unknown | Unknown | Methylthioribose-1-phosphate isomerase | Cytosol | EC: 5.3.1.23 | BLAST search shows amino-acid sequence similarity with genes from Syntrophomonas wolfei, Pseudomonas syringae, P. aeruginosa and Xylella fastidiosa. Localization hypothesized. EC number obtained from ExPASy ENZYME database |
| LmjF36.5910 | Hypothetical protein, conserved | Unknown | Unknown | Enolase- phosphatase E1 | Cytosol | EC: 3.1.3.77 | BLAST search shows amino-acid sequence similarity with genes from Leptospira spp., B. taurus, M. musculus, H. sapiens and Prochlorococcus marinus. Localization hypothesized. EC number obtained from ExPASy ENZYME database |
Current gene annotations, including putative identification, localization, and EC classification were compiled from KEGG and GeneDB. The new proposed iAC560 annotations for gene identification, localization or EC classification are shown in bold and italics. The corresponding evidence for the refined annotation is provided to the right.
As an example of a hypothetical gene annotation refinement, consider LmjF23.1480, which this study characterizes as an ‘alanine racemase.' D-Alanine was included as a substrate in the experimentally defined medium provided in Merlen et al (1999); Schuster and Sullivan (2002). This was the first indication that the L. major genome coded for a racemase that converted D-alanine into L-alanine, the latter isomer required for protein synthesis (Panizzutti et al, 2006). However, there were no L. major genes in KEGG (http://www.genome.jp/kegg/) or GeneDB (http://www.genedb.org/) annotated as ‘alanine racemase.' Moreover, two Trypanosoma cruzi genes that coded for alanine racemase were found. A BLAST (http://www.expasy.org/tools/blast/) search using T. cruzi genes resulted in very poor amino-acid sequence similarity against L. major genes (E-value: >2.0). The existence of a cytosolic alanine racemase in Leishmania amazonensis was previously described (Panizzutti et al, 2006). Also, the presence of a ‘hypothetical protein' in L. major with possible alanine racemase activity was stated in Panizzutti et al (2006) and a GenBank (http://www.ncbi.nlm.nih.gov/Genbank/index.html) identification number was provided, which was associated with LmjF23.1480, annotated as a ‘hypothetical protein, conserved' in KEGG and GeneDB. However, there was no localization or EC information provided in the databases. A BLAST search was conducted, which showed strong amino-acid sequence similarity with alanine racemase from several organisms, including Ostreococcus tauri (E-value: 4e−61), Aspergillus fumigatus (E-value: 3e−39), Aspergillus clavatus (E-value: 3e−39) and Schizosaccharomyces pombe (E-value: 1e−37). EC information was obtained from the ExPASy ENZYME database (http://ca.expasy.org/enzyme/). Hence, the gene annotation for LmjF23.1480 was refined as a ‘cytosolic alanine racemase' with EC: 5.1.1.1.
As an example of an incorrect annotation in public databases, the gene LmjF04.1160 was annotated as ‘fructose-1,6-bisphosphatase, cytosolic' in KEGG and GeneDB. This is a gluconeogenic enzyme whose subcellular localization has been characterized as the glycosome rather than the cytosol (Naderer et al, 2006). Additionally, GeneDB stated that the isoelectric point (pI) of the predicted peptide for LmjF04.1160 was 9.1. Enzymes in the glycosome are associated with highly alkaline isoelectric points (Umasankar et al, 2005). The existence of a peroxisomal-targeting signal 1 (PTS1) at the extreme carboxyl terminus of the amino-acid sequence for LmjF04.1160 was also discovered (Opperdoes and Szikora, 2006). The targeting signal was a three amino-acid peptide consisting of serine, lysine and leucine (-SKL-), which is used to characterize a glycosomal subcellular localization (Keller et al, 1991; Opperdoes and Szikora, 2006). Hence, based on literature, isoelectric point and targeting signal evidence, the gene annotation for LmjF04.1160 was refined as a ‘glycosomal fructose-1,6-bisphosphatase.' This type of model-driven discovery would be impossible without a systematic model-building approach that this reconstruction entails.
Refinements of gene annotation typically surface whenever a gap or dead end is identified in the in silico metabolic network and there is no gene specified in the organism's genome to fill in for that required metabolic function. Figure 3 illustrates the process employed in the construction of a GPR when there is limited gene or EC information for a particular reaction. A dead end occurs when a metabolite is either consumed or produced only, hence violating the rule of mass balance, the principal physiochemical constraint (Lee et al, 2006). Currently, there are 261 dead-end metabolites in the L. major metabolic network. Consequently, there are 210 reactions with at least one participating dead-end metabolite that will always have a flux of zero. It is likely that in future iterations of the reconstruction, more functional annotation refinements will surface as more gaps and dead ends are resolved. See Supplementary Information III for all dead-end metabolites belonging to various subcellular compartments within the metabolic network.
Figure 3.
Construction of a gene–protein reaction (GPR) relationship. A flowchart highlighting the process of constructing a GPR based on the amount of gene or EC information available for a particular reaction in public databases or literature is illustrated.
In silico minimal medium prediction
An in silico minimal medium composed of arginine, cysteine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, valine, hypoxanthine, phosphate and oxygen was predicted (see Figure 4A). In addition, Figure 4A delineates non-essential and essential substrates in the defined medium.
Figure 4.
Minimal medium prediction and effects of non-essential substrates in defined medium on growth. (A) Minimal medium and non-essential substrates in defined medium are delineated. The defined medium was obtained from Merlen et al (1999); Schuster and Sullivan (2002). (B) The effects of in silico minimal medium on production of biomass constituents are summarized. Gray- and red-colored boxes are indicative of particular biomass constituents that are and are not produced, respectively, when the corresponding substrate in the minimal medium is prevented from entering the metabolic system. The dotted ‘X' indicates biomass constituents that cannot be produced when oxygen and another substrate in minimal medium are restricted from entering the metabolic system. (C) The effects of non-essential substrates in defined medium on growth rate are shown. The vertical bars represent normalized growth rate when each substrate is removed from the environment. †See Supplementary Information VII for all metabolite abbreviations. ‡Non-essential substrates in defined medium belonging to the ‘other' category include a-D-glucose, b-D-glucose, ascb, asn-L, asp-L, btn, ca2, chsterol, co2, fol, gln-L, glu-L, gly, gthrd, gua, h, h2o, inost, nac, ncam, nh3, nh4, pnto-R, pydx, pydxn, rib-D, so4, thym, tyr-L and xan.
The list of essential amino acids contained in the in silico minimal medium differed from those specified in the experimentally defined medium (see Merlen et al, 1999; Schuster and Sullivan, 2002). Specifically, the experimental medium listed three additional amino acids as being essential, namely cystine, glutamine and tyrosine. Cystine, which is a dimer of cysteines, was not included as a drain in biomass. L. major has a gene for glutamine synthetase (LmjF06.0370), which synthesizes glutamine from glutamate in the presence of ATP and ammonia. Therefore, the model predicted that glutamine was not essential for growth. In addition, L. major has a gene for tyrosine aminotransferase (LmjF36.2360), which can synthesize tyrosine from glutamate. ExPASy's ENZYME database noted that aspartate aminotransferase (LmjF35.0820) can act on tyrosine as well.
In addition, bioinformatics analyses have been performed to predict essential and non-essential amino acids in L. major (Payne and Loomis, 2006). In prior work, if one or more genes in an amino-acid pathway were missing in the organism, that particular amino acid was characterized as being essential. Through bioinformatics, cysteine was predicted as non-essential and serine as an essential amino acid (Payne and Loomis, 2006). Our minimal medium prediction indicated just the inverse that cysteine was essential and serine was non-essential. The model was still able to grow if serine and hydrogen sulfide were allowed to enter the metabolic system from the environment instead of cysteine, thus validating the prediction of essential amino acids to that using solely bioinformatics analyses (Payne and Loomis, 2006). There can be several combinations of metabolites that can support growth; however, the one particular combination of essential medium with the fewest number of metabolites was chosen. Hence, the reconstruction was used to predict essential components of a defined medium and aided in reconciling discrepancies between published data sources (on essential amino acids).
Since Leishmania spp. can grow under anaerobic conditions (Darling et al, 1987), oxygen in the minimal medium was replaced with tyrosine, uracil, ergosterol and zymosterol. Additionally, 5-methylthioadenosine, an intermediate in spermidine synthesis, needed to be excreted out of the system for the model to grow anaerobically. 5-Methylthioadenosine is a by-product of the spermidine synthase reaction. An enzymatic reaction downstream of spermidine synthase in the methionine salvage cycle, namely 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase requires oxygen to produce 2-keto-4-methylthiobutyrate. Subsequently, tyrosine transaminase converts this metabolite and L-glutamate into L-methionine and 2-oxoglutarate. For organisms that undergo anaerobic respiration, the reaction that synthesizes 2-keto-4-methylthiobutyrate has not been identified (Sekowska et al, 2004).
Contribution of minimal medium components to biomass
The contribution of each component of minimal medium to the generation of biomass precursors was computed (see Figure 4B). After each minimal medium component was eliminated, the red boxes denote the corresponding biomass precursors that could not be generated, while the gray boxes indicate the biomass precursors that were produced. For example, if arginine was prevented from entering the metabolic system, then arginine, putrescine and spermidine were not produced. Likewise, if hypoxanthine was removed, adenosine monophosphate (AMP), deoxyAMP (dAMP), deoxyguanosine monophosphate (dGMP) and GMP could not be synthesized. Interestingly, this result validated a key point of L. major metabolism. L. major is unable to synthesize purines de novo (Berriman et al, 2005). In total, 9 out of 10 genes coding for a pathway beginning with phosphoribosyl pyrophosphate and ending in inosine monophosphate are not present in trypanosomatids (Berriman et al, 2005). Therefore, a purine source is required from the environment to fulfill purine requirements in biomass. The minimal medium prediction demonstrated that hypoxanthine, a purine derivative, was the essential purine source. Also, in Figure 4B, a dotted ‘X' denotes a biomass precursor that could not be generated when oxygen and another component of minimal medium were restricted from entering the metabolic system. Cysteine and oxygen had a severe impact on several biomass constituents. With cysteine and oxygen absent, metabolites such as S-adenosylhomocysteine, L-homocysteine, S-adenosylmethionine, O-succinyl-L-homoserine and L-cystathionine among others were not synthesized. These have important roles for the synthesis of biomass constituents. For example, S-adenosylmethionine plays a role in spermidine, zymosterol, ergosterol and phosphatidylcholine synthesis, and is known to be one of the most ubiquitous metabolites in metabolic networks paralleling ATP (Reguera et al, 2007). The relationships between minimal medium components and biomass constituents cannot be ascertained intuitively without these types of network analyses.
Contribution of non-essential defined medium components to biomass
The effects of the non-essential defined medium components on growth rate were investigated. Each of the non-essential substrates in defined medium (see Figure 4A) was prevented from entering the system and allowed only to be excreted (flux restricted to positive flux space). Figure 4C depicts the reduction in growth rate caused by removing each of the non-essential substrates. Interestingly, the majority of the substrates had absolutely no effect. Only 7 out of 36 metabolites had any effect on the growth rate. Preventing the entry of deoxyribose into the metabolic system had the most drastic effect of all, by causing a drop in growth rate by 16.8%. The flux through deoxyribokinase, which converts deoxyribose to 2-deoxy-D-ribose 5-phosphate with the help of ATP, was reduced to zero. Consequently, the flux through deoxyribose-phosphate aldolase, which converts 2-deoxy-D-ribose 5-phosphate to acetaldehyde and glyceraldehyde 3-phosphate, was reduced. In the reconstruction, acetaldehyde was transported into the mitochondria where it participated in a reaction catalyzed by aldehyde dehydrogenase. The by-product of aldehyde dehydrogenase reaction, acetate, in turn was used for synthesis of acetyl-CoA via the acetyl-CoA synthetase reaction. Acetyl-CoA, an important intermediate in the citrate cycle, is a well-known precursor to fatty acid synthesis and several biomass constituents (Wakil et al, 1964). Hence, due to absence of deoxyribose in the medium, the flux through the acetyl-CoA synthetase reaction was reduced, which in turn caused a reduction in growth rate.
F0F1-ATP synthase robustness analysis
The robustness of the metabolic network can be evaluated by constraining the flux of any reaction from its wild-type value to zero and calculating the resultant effect on the growth rate. As an example, oligomycin is an inhibitor of mitochondrial F0F1-ATP synthase (Schnaufer et al, 2005). Therefore, the robustness of the metabolic network in response to varying the flux of mitochondrial F0F1-ATP synthase-catalyzed reaction was investigated. Figure 5 depicts the normalized growth rate response to changing the flux through mitochondrial F0F1-ATP synthase reaction. At different ATP synthase flux values, a single optimization was run and growth rate (i.e. the flux through the biomass reaction) was measured. The graph illustrates that growth rate is reduced to about 50% when the flux through ATP synthase is reduced to 8–9% of wild-type flux value. Reducing ATP synthase flux to 40% of its wild-type causes only a 3% change in growth rate. Robustness analysis provides an optimal flux value for a particular reaction that corresponds to the optimal growth rate. This is a proof-of-concept rather than a validation study of the in silico metabolic network. Particular enzymes with known inhibitors can be targeted to help reduce the proliferation of Leishmania spp. in mammalian hosts and subsequently slow the spread of infection.
Figure 5.
F0F1-ATP synthase robustness analysis. The normalized growth rate corresponding to F0F1-ATP synthase flux is plotted. The white circles indicate single optimization simulations that are run at different ATP synthase flux values to measure flux through biomass (growth rate).
Amastigote or promastigote stage-specific metabolism
Differential protein expression data from Leishmania infantum suggested hexokinase was preferentially expressed in the amastigote stage, while alcohol dehydrogenase, enolase and ATP synthase were preferentially expressed in the promastigote stage (Leifso et al, 2007). Figure 6 depicts the reactions catalyzed by the differentially expressed enzymes. A red ‘X' indicates that the flux through a particular reaction was reduced to 10% of its wild-type stage-independent value. The value of 10% was arbitrarily chosen to emphasize that these enzymes are not solely present in one stage over another, but rather show higher abundance in a particular morphological stage. Figure 7 illustrates the changes in normalized growth rate as the fluxes through the hexokinase, alcohol dehydrogenase, enolase and ATP synthase reactions were reduced. Since hexokinase was more highly abundant in the amastigote, knocking down flux of hexokinase would partially represent a promastigote metabolic network. Likewise, since alcohol dehydrogenase, enolase and ATP synthase had higher abundances in the promastigote, knocking down their fluxes in combination would imply an amastigote metabolic network. As evidenced from the robustness analysis described above, the mitochondrial F0F1-ATP synthase had the greatest impact on the growth rate. The motivation for this analysis was not to make predictions regarding growth rates in the two morphological forms of the parasite, but rather to demonstrate the utility of integrating protein expression data with a genome-scale metabolic network. Identifying pathways upregulated in the amastigote, which faces a drastically different environment in the macrophage as opposed to a promastigote in the sandfly, is critical when exploring novel therapeutic targets.
Figure 6.
Reactions differentially expressed in amastigote or promastigote stages. The metabolic reactions that distinguish the morphological stages of L. major are highlighted. These include reactions catalyzed by hexokinase, ATP synthase, alcohol dehydrogenase and enolase. The red ‘X' indicates that the flux through each reaction is reduced to 10% of its wild-type stage-independent flux value to characterize morphological stage-specific metabolism.
Figure 7.
Growth effects in amastigote and promastigote stages. The growth rate is assessed after accounting for the flux through reactions catalyzed by differentially expressed enzymes in amastigote and promastigote stages. Hexokinase is preferentially expressed in amastigotes, while ATP synthase, alcohol dehydrogenase and enolase are preferentially expressed in promastigotes (Leifso et al, 2007). As a proof-of-concept, reducing the flux through hexokinase represents a promastigote metabolic network, and reducing the flux through ATP synthase, alcohol dehydrogenase and enolase in combination implies an amastigote metabolic network. Vertical bars indicate normalized growth rate. **The flux through each reaction is reduced to 10% of its wild-type stage-independent flux value.
Model refinement and iterative design
The step of model refinement and iterative design (see Figure S2 in Supplementary Information I) is an important facet of the model-building process. According to the literature, removing the function of adenosylmethionine decarboxylase, spermidine synthase or ornithine decarboxylase was lethal for Leishmania donovani (Jiang et al, 1999; Roberts et al, 2001, 2002, 2004). Knocking out arginase was lethal for Leishmania mexicana (Roberts et al, 2004). After the first iteration of the metabolic model, these four reaction knockouts were non-lethal for the in silico growth of L. major under defined medium conditions. These four enzymes are involved in polyamine (spermidine and putrescine) synthesis. Additionally, it has been suggested that polyamine synthesis is important for growth of Leishmania parasites (Bachrach et al, 1979a, 1979b, 1981). Hence, spermidine and putrescine were included in biomass to provide a drain for these essential metabolites. Previous metabolic reconstructions for H. pylori and Methanosarcina barkeri have also included polyamines as part of biomass (Schilling et al, 2002; Feist et al, 2006).
After accounting for polyamines, a single optimization simulation (maximization for biomass) was implemented. However, there was no flux through the biomass reaction, and hence, no growth. This indicated that there existed a gap or a dead end within the resulting network. Upon further investigation, it was concluded that there was a gap in the methionine salvage cycle, downstream of the spermidine synthase reaction. Only the metabolite 5-methylthio-5-deoxy-D-ribose 1-phosphate was produced, violating the principal physiochemical constraint of mass balance. We subsequently added four enzymatic reactions to the model, namely 5-methylthioribose-1-phosphate isomerase, 5-methylthio-5-deoxy-D-ribulose 1-phosphate dehydratase, enolase-phosphatase E-1 and 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase to complete the methionine salvage cycle. KEGG and GeneDB did not have functionally annotated genes for these enzymes. LmjF28.1840, LmjF36.5910, LmjF20.0970 and LmjF34.4600, previously uncharacterized genes, received new functional annotation assignments (see Table II). LmjF36.4930 was annotated in KEGG and GeneDB as ‘translation initiation factor 2 subunit.' However, a BLAST search showed strong amino-acid sequence similarity with 5-methylthioribose-1-phosphate isomerase from several organisms, including S. pombe (E-value: 2e−60), Syntrophomonas wolfei (E-value: 2e−57), Pseudomonas syringae (E-value: 6e−57), Pseudomonas aeruginosa (E-value: 2e−56) and Xylella fastidiosa (E-value: 2e−56) (see Table II). Therefore, in iAC560, this gene was annotated as ‘5-methylthioribose-1-phosphate isomerase.' Whether this gene is involved in dual function requires further investigation. After filling in the gaps in the methionine salvage cycle and adding polyamines to biomass, iAC560 predicted adenosylmethionine decarboxylase, spermidine synthase, ornithine decarboxylase and arginase as lethal knockouts in all in silico medium conditions (see Table I).
Discussion
We present the first constraint-based model for a protozoan accounting for 560 genes, 1112 reactions, 1101 metabolites and 8 unique subcellular localizations. Among unicellular eukaryotes, Leishmania spp. are unique in their capability for polycistronic transcription (Campbell et al, 2003), the presence of glycosome, acidocalcisome and flagellar compartments (Hart and Opperdoes, 1984; Opperdoes and Michels, 1993; de Souza, 2002; Docampo et al, 2005), their use of trypanothione as an analog of glutathione (Fairlamb et al, 1985), the occurrence of novel fatty acid synthesis mechanisms (Lee et al, 2007) and the presence of kinetoplast DNA (kDNA) network (Simpson and Da Silva, 1971; Stuart, 1983).
Using a systems-based approach, we identified 12% of single gene knockouts in the network as lethal and 10% as growth reducing, and proposed 56 non-trivial lethal double deletions. Each one of these deletions constitutes a promising drug target and an experimentally testable hypothesis. We validated the metabolic network with experimental knockout data from related Leishmania and Trypanosoma species. In silico predictions in minimal medium had the best overall compliance with experimental data (72.4%). Predictions in minimal medium–glucose, minimal medium–glucose–other amino acids and defined medium had overall compliances of 69.0, 69.0 and 65.5%, respectively. Through the network reconstruction and associated analyses, we demonstrated systematic refinement of genome annotation. A total of 17 out of 25 new annotation refinements were proposed for ‘hypothetical proteins.' The rest of the proposed annotations were for genes that had an incorrect annotation, localization or EC classification.
In addition, we proposed a novel in silico minimal medium that supports growth of L. major and evaluated growth response to non-essential components in defined medium. The in silico medium consisted of arginine, cysteine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, valine, hypoxanthine, phosphate and oxygen. The essential amino acids in our minimal medium matched with those predicted by bioinformatics analyses with one exception (Payne and Loomis, 2006). Permitting serine and hydrogen sulfide to enter from the environment instead of cysteine allowed the model to grow, and further validated our prediction of essential amino acids with that in Payne and Loomis (2006). However, we chose the minimal medium composition based on the fewest number of metabolites required for in silico growth.
The presence of hypoxanthine in the minimal medium highlighted the fact that L. major is unable to synthesize purines de novo (Berriman et al, 2005). If hypoxanthine is removed, AMP, dAMP, dGMP and GMP cannot be synthesized. Hypoxanthine, a purine derivative, fulfills the purine source requirement from the environment. Moreover, the majority of non-essential components of defined medium have no effect on growth. Only 7 out of 36 metabolites had any effect, and deoxyribose had the most drastic effect of all, causing a drop in growth rate by 16.8%. Preventing the entry of deoxyribose affected synthesis of acetyl-CoA due to a reduction in flux through the acetyl-CoA synthetase reaction. Acetyl-CoA is an important precursor to fatty acid synthesis and several biomass constituents (Wakil et al, 1964). Finally, we provided two proof-of-concept examples yielding insight into the utility of network reconstruction; the robustness of the metabolic network was evaluated by varying the flux through F0F1-ATP synthase reaction, and pathways differentially upregulated in the morphological stages of the parasite were identified by the use of protein expression data.
A comprehensive metabolic model such as iAC560 is an ideal forum for continual integration and interpretation of high-throughput data. Systems analysis of L. major is particularly challenging because, unlike organisms like E. coli and S. cerevisiae, L. major is not as well-characterized experimentally. As more information on L. major becomes available, the model will be refined to better match experimental validating data. As an example of the model refinement process, we identified four gaps in the methionine salvage cycle and provided functional annotation to previously uncharacterized genes. Consequently, we improved our compliance of knockout predictions with experimental data from 51.7 to 65.5% agreement under defined medium conditions.
Limitations of the model include poor gene associations for inter-compartment transport reactions and transporters to the environment. Most of the metabolites in iAC560 that needed to be transported across membranes were allowed to do so via passive diffusion. It was previously published that metabolites and coenzymes cannot permeate through the glycosomal membrane (Bakker et al, 2000). A publicly accessible membrane transport database, TransportDB (http://www.membranetransport.org/), has gene listings for transporters in L. major that will need to be investigated. The metabolic network also did not account for kDNA. L. major maxicircle kDNA plays an important role in energy metabolism, encoding for subunits of NADH-ubiquinone oxidoreductase, for example (Akopyants et al, 2004).
Comparing the metabolic network of L. major with that of humans can aid in identifying unique features of trypanosomatid metabolism. Focusing on metabolic causes of pathogenicity and virulence will lead to discovery of unique therapeutic targets dissimilar from human metabolism. Ultimately, the genome-scale metabolic reconstruction of L. major presented here provides a framework for the interrogation of human pathogens and a platform for integration of high-throughput data and generation of experimental hypotheses. These types of novel network analyses may be relevant to treating infectious diseases, particularly when diseases like leishmaniasis are considered emerging and uncontrolled, necessitating improvements in therapeutics and treatment strategies (Remme et al, 2002).
Materials and methods
Metabolic network reconstruction
The process of reconstructing the metabolic network of an organism has been thoroughly described in the literature (Edwards and Palsson, 2000; Schilling et al, 2002; Forster et al, 2003; Reed et al, 2003, 2006a; Borodina and Nielsen, 2005; Francke et al, 2005; Feist et al, 2006, 2007; Duarte et al, 2007; Jamshidi and Palsson, 2007; Kim et al, 2007). The basis of a metabolic reconstruction is the formulation of GPR relationships and organization of reactions into a S-matrix for subsequent mathematical analyses (Reed et al, 2006a). The S-matrix, with rows of metabolites and columns of reactions, is comprised of stoichiometric coefficients corresponding to the chemical transformations in the network. For example, a reaction where 1 mol of ‘A' yields 1 mol of ‘B' may be represented as a single column in the S-matrix, with the row corresponding to ‘A' having a ‘−1' denoting consumption of 1 mol of ‘A' and the row corresponding to ‘B' having a ‘+1' denoting production of 1 mol of ‘B.'
Using a variety of data sources such as gene and protein databases, biochemistry textbooks and published literature, information relating to gene locus, enzyme biochemistry, reaction stoichiometry and subcellular localization was compiled (see Figure S2 in Supplementary Information I). Further, rules on reaction reversibility and promastigote/amastigote protein expression data were used to delineate the lower and upper bounds on reaction fluxes for every reaction in the S-matrix. With the ability of trypanosomes, unlike other eukaryotic organisms, to perform polycistronic transcription (Campbell et al, 2003), one transcript can potentially code for multiple proteins. However, a simplifying assumption was made at the start of the reconstruction process to allow one gene to code for a single protein. The single protein, however, was allowed to catalyze multiple reactions in different subcellular compartments. Two or more proteins arising from independent genes that catalyze the same reaction (e.g. isozymes) were characterized with an ‘OR' relationship. Two or more proteins that are needed to work in conjunction for a reaction to occur (e.g. as subunits in a protein complex) were characterized with an ‘AND' relationship (Reed et al, 2006a; Duarte et al, 2007).
Published literature was explored extensively for assigning subcellular localizations to reactions. In cases where literature evidence was limited, the presence of signaling peptides was taken into account. Several metabolic genes containing a PTS were listed in Opperdoes and Szikora (2006), and this list was used for locating particular reactions to the glycosome, a unique trypanosomatid peroxisome-like organelle. In some cases, an isoelectric point (pI) of above 8.4 was also used to place reactions into the glycosome (Umasankar et al, 2005). In addition, the PSORT prediction tool (http://psort.nibb.ac.jp/) was used to locate targeting signals for compartments other than the glycosome. When no literature evidence or presence of targeting signals was available, the default subcellular compartment was the cytosol.
Reactions in the metabolic network were classified as gene associated or non-gene associated. Those reactions that occurred spontaneously without the presence of an enzyme and in silico ‘model' reactions comprised the non-gene-associated category. Several subcellular transport reactions and transporters to the extracellular environment were added to the model as in silico ‘model' reactions due to lack of gene annotations and literature evidence. Various types of network analyses are possible following a metabolic network reconstruction. As illustrated in Figure S2 in Supplementary Information I, the iterative model-building process is vital for subsequent model refinement. See Supplementary Information IV for the complete metabolic reconstruction in Microsoft Excel format, Supplementary Information V for complete metabolic reconstruction in SBML format, Supplementary Information VI for a comprehensive reaction map and Supplementary Information VII for all metabolite abbreviations.
Naming convention
The L. major metabolic reconstruction was named following the in silico labeling convention first introduced in Reed et al (2003). The computational model was named ‘iAC560,' where ‘i' denotes ‘in silico,' ‘AC' indicates the initials of the first author and ‘560' represents the number of L. major genes accounted for in the model.
Fatty acid composition
The fatty acid composition for L. major was obtained from Vessal et al (1974). Chain lengths with less than 2% prevalence were not taken into account and the percentages of the remaining fatty acids were normalized. Subsequently, based on the percentages of individual chain lengths, an average fatty acid chain length was derived. It was assumed that all metabolites with fatty acyl side chains in Leishmania had equivalent chain lengths. See Supplementary Information VIII for the average fatty acid composition and the molecular formulas of all metabolites in the model with fatty acyl side chains.
Biomass calculations
The purpose of the biomass equation is to provide a drain of essential metabolites that are needed to support growth of the metabolic system (Varma and Palsson, 1993; Edwards and Palsson, 2000). The equation is an approximation as the composition is likely to vary under different physiological conditions (Varma and Palsson, 1993). However, it has been shown that perturbations in the coefficients of the metabolites present in biomass do not affect overall biomass yield significantly (Varma and Palsson, 1993, 1994). Additionally, model predictions have been shown to be insensitive to various factors including ATP maintenance and biomass scaling (Varma and Palsson, 1995). Due to the scarcity of data on cellular composition for L. major, data from related Leishmania species and other protozoans (e.g. Tetrahymena) was used. Therefore, the biomass equation used is only an estimate and mainly serves to provide a demand for essential metabolites. See Supplementary Information I for detailed explanation on the derivation of the biomass equation used in the computational analysis.
Flux balance analysis
Flux balance analysis (FBA) has been extensively used to interrogate metabolic networks (Edwards and Palsson, 2000; Schilling et al, 2002; Forster et al, 2003; Reed et al, 2003; Feist et al, 2006, 2007; Duarte et al, 2007; Jamshidi and Palsson, 2007; Kim et al, 2007). FBA is used to generate a set of steady-state fluxes for all the reactions in the biochemical network upon the optimization of an objective under a set of constraints (see Kauffman et al (2003); Lee et al (2006) for more detailed review of FBA). Constraints can be categorized into four different types: physicochemical, topological, thermodynamic and environmental (Kauffman et al, 2003; Lee et al, 2006). The principal physicochemical constraint used in FBA is conservation of mass. Topological constraints include restrictions placed on metabolites due to subcellular compartmentalization, thermodynamic constraints are applied by defining effective irreversibility of a particular reaction, and environmental constraints are defined by the medium in which the cell is cultivated (Lee et al, 2006).
The change in concentration of a metabolite in the network can be represented in terms of the fluxes through the reactions in which the metabolite is participating.
 |
In equation (1), ‘C' is a vector of concentrations of metabolites, ‘t' is time, ‘S' is the S-matrix with m rows of metabolites and n columns of reactions and ‘v' is a vector of fluxes through the corresponding reactions. At steady state, the change in concentration of any metabolite over time throughout the cellular network is zero. The steady-state assumption, frequently applied in the analysis of metabolic networks, is reasonable for a metabolic network as individual reactions are fast in comparison to changes in the overall cellular objective, such as maximization of growth rate (Lee et al, 2006).
Therefore,
 |
And,
Because of the under-determined nature of the system (i.e. the number of equations is less than the number of unknown fluxes), a linear programming (LP) formulation was used to generate a flux distribution for the entire network. Typical objective functions include maximizing for biomass production, maximizing or minimizing for ATP or maximizing for certain by-products of the metabolic system (Lee et al, 2006). In this study, the biomass equation was the objective function for simulating growth of L. major. In addition to conservation of mass, other constraints included reaction reversibility rules, promastigote/amastigote protein expression data, various medium conditions and prevalence of transport reactions across cellular compartments. All intracellular reversible reactions were allowed to traverse the entire flux space from −∞ to +∞ (mmol/gDW/h). Fluxes on irreversible intracellular reactions (mmol/gDW/h) and the biomass demand (h−1) were restricted to the positive flux space from 0 to +∞. The fluxes on substrates (mmol/h) present in the experimentally defined medium (see Merlen et al (1999); Schuster and Sullivan (2002)) were constrained between −100 and +100, i.e. these substrates were free to enter or leave the system. Reactions that allowed entry of metabolites not present in the defined medium had their fluxes constrained from 0 to 100, i.e. these metabolites were only allowed to be excreted out of the system. For minimal medium simulations, the fluxes on substrates present in minimal medium were constrained between −∞ to +∞ and remaining substrate fluxes were constrained to the positive flux space (biomass flux was constrained to+100). All LP simulations were carried out in Simpheny™ (Genomatica, San Diego, CA).
Single and double gene deletion predictions
Gene deletion predictions allowed for identification of genes that were lethal for growth of L. major. Each of the 560 genes in the metabolic network was ‘knocked out' and grouped into one of the following categories with respect to wild type: lethal (0%), growth reducing (>0% and <90%), growth reducing (>90% and <100%) and no effect (100%). For each case, the percentage of growth is indicated in the parenthesis. All possible double deletions were also simulated and classified as trivial lethal or non-trivial lethal. In trivial lethal deletions, at least one of the genes involved is lethal in a single deletion, and in non-trivial lethal deletions, both genes involved are not lethal individually as single deletions but are lethal only in combination. Single and double gene deletion predictions were performed under defined medium conditions (see Merlen et al (1999); Schuster and Sullivan (2002)). The substrates included in defined medium are listed in Figure 4A.
Validation of reaction knockout predictions
Experimental data on gene essentiality in L. major were not readily available, and RNA interference has not been effectively demonstrated in Leishmania spp. (Beverley, 2003; Opperdoes and Michels, 2008). Therefore, gene knockout data were obtained from published literature for related Leishmania and Trypanosoma species. Data for six knockouts were obtained from TrypanoFAN (http://trypanofan.path.cam.ac.uk/trypanofan/main/), the Trypanosoma brucei functional genomics project. A reaction knockout strategy (as opposed to individual gene knockouts) was pursued to completely remove function encoded by particular genes obtained from literature. The in silico reaction knockouts were carried out under four different medium conditions: (a) minimal medium, (b) minimal medium and glucose, (c) minimal medium, glucose and other amino acids and (d) defined medium (see Merlen et al (1999); Schuster and Sullivan (2002)). The model results were compared with experimental knockout studies.
In silico minimal medium prediction
The completely defined medium for the continuous cultivation and growth of Leishmania promastigotes has been experimentally characterized (Merlen et al, 1999; Schuster and Sullivan, 2002). With the exception of a few vitamins and salts, the majority of the metabolites in the experimentally defined medium were accounted for in the L. major metabolic network reconstruction. To predict an in silico minimal medium for growth, each of the extracellular metabolite fluxes was prevented from entering the metabolic system. This yielded a combination of essential metabolites necessary to support growth. Constraining the flux of any one of these essential metabolites resulted in no growth (the flux of biomass was 0 h−1).
Characterizing stage-specific metabolism
L. major exists as an amastigote within macrophages in the human host. Therefore, identifying specific pathways of L. major metabolism that are differentially upregulated in amastigotes as opposed to promastigotes is necessary. With the help of mRNA or protein expression data, morphological stage-specific metabolic networks can be characterized. The mRNA expression may not directly correspond to protein expression in Leishmania spp., and obtaining protein expression data from L. major lesion-derived amastigotes can be challenging as they are difficult to culture in vitro (Leifso et al, 2007). Therefore, as a proof-of-concept, information on differential protein expression from L. infantum (a related Leishmania spp.) was integrated with the in silico L. major metabolic network. According to the thresholds set in Leifso et al (2007), hexokinase was preferentially expressed in the amastigote stage, while alcohol dehydrogenase, enolase and ATP synthase were preferentially expressed in the promastigote stage. The fluxes through each of these reactions were reduced to 10% of their wild-type stage-independent values and the effects on growth rate were observed.
Supplementary Material
Supplementary Information 1
Supplementary Information 2
Supplementary Information 3
Supplementary Information 4
Supplementary Information 5
Supplementary Figure 6
Supplementary Information 7
Supplementary Information 8
Acknowledgments
We acknowledge Erwin Gianchandani, Monica Lee, Kyle Singleton, Jennifer Reed, Natalie Duarte, Matthew Oberhardt, Corinne Locke, Jennifer Robichaux, Jong Min Lee, Richard Pearson and Fred Opperdoes for insightful comments and feedback. We also thank the National Science Foundation CAREER program (grant no. 0643548) for financial support.
References
- Akopyants NS, Matlib RS, Bukanova EN, Smeds MR, Brownstein BH, Stormo GD, Beverley SM (2004) Expression profiling using random genomic DNA microarrays identifies differentially expressed genes associated with three major developmental stages of the protozoan parasite Leishmania major. Mol Biochem Parasitol 136: 71–86 [DOI] [PubMed] [Google Scholar]
- Alphey MS, Bond CS, Tetaud E, Fairlamb AH, Hunter WN (2000) The structure of reduced tryparedoxin peroxidase reveals a decamer and insight into reactivity of 2Cys-peroxiredoxins. J Mol Biol 300: 903–916 [DOI] [PubMed] [Google Scholar]
- Bachrach U, Abu-Elheiga L, Talmi M, Schnur LF, El-On J, Greenblatt CL (1981) Polyamines and the growth of leishmanial parasites. Med Biol 59: 441–447 [PubMed] [Google Scholar]
- Bachrach U, Brem S, Wertman SB, Schnur LF, Greenblatt CL (1979a) Leishmania spp.: cellular levels and synthesis of polyamines during growth cycles. Exp Parasitol 48: 457–463 [DOI] [PubMed] [Google Scholar]
- Bachrach U, Brem S, Wertman SB, Schnur LF, Greenblatt CL (1979b) Leishmania spp.: effect of inhibitors on growth and on polyamine and macromolecular syntheses. Exp Parasitol 48: 464–470 [DOI] [PubMed] [Google Scholar]
- Bakker BM, Mensonides FI, Teusink B, van Hoek P, Michels PA, Westerhoff HV (2000) Compartmentation protects trypanosomes from the dangerous design of glycolysis. Proc Natl Acad Sci USA 97: 2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson TJ, McKie JH, Garforth J, Borges A, Fairlamb AH, Douglas KT (1992) Rationally designed selective inhibitors of trypanothione reductase. Phenothiazines and related tricyclics as lead structures. Biochem J 286 (Pt 1): 9–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Böhme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ (2005) The genome of the African trypanosome Trypanosoma brucei. Science 309: 416–422 [DOI] [PubMed] [Google Scholar]
- Beverley SM (2003) Protozomics: trypanosomatid parasite genetics comes of age. Nat Rev Genet 4: 11–19 [DOI] [PubMed] [Google Scholar]
- Bochud-Allemann N, Schneider A (2002) Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J Biol Chem 277: 32849–32854 [DOI] [PubMed] [Google Scholar]
- Boitz JM, Ullman B (2006) Leishmania donovani singly deficient in HGPRT, APRT or XPRT are viable in vitro and within mammalian macrophages. Mol Biochem Parasitol 148: 24–30 [DOI] [PubMed] [Google Scholar]
- Boone C, Bussey H, Andrews BJ (2007) Exploring genetic interactions and networks with yeast. Nat Rev Genet 8: 437–449 [DOI] [PubMed] [Google Scholar]
- Borodina I, Nielsen J (2005) From genomes to in silico cells via metabolic networks. Curr Opin Biotechnol 16: 350–355 [DOI] [PubMed] [Google Scholar]
- Braga MV, Urbina JA, de Souza W (2004) Effects of squalene synthase inhibitors on the growth and ultrastructure of Trypanosoma cruzi. Int J Antimicrob Agents 24: 72–78 [DOI] [PubMed] [Google Scholar]
- Campbell DA, Thomas S, Sturm NR (2003) Transcription in kinetoplastid protozoa: why be normal? Microbes Infect 5: 1231–1240 [DOI] [PubMed] [Google Scholar]
- Chan C, Yin H, McKie JH, Fairlamb AH, Douglas KT (2002) Peptoid inhibition of trypanothione reductase as a potential antitrypanosomal and antileishmanial drug lead. Amino Acids 22: 297–308 [DOI] [PubMed] [Google Scholar]
- Chen M, Zhai L, Christensen SB, Theander TG, Kharazmi A (2001) Inhibition of fumarate reductase in Leishmania major and L.donovani by chalcones. Antimicrob Agents Chemother 45: 2023–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou V, Besteiro S, Biran M, Diolez P, Bouchaud V, Voisin P, Michels PA, Canioni P, Baltz T, Bringaud F (2003) ATP generation in the Trypanosoma brucei procyclic form: cytosolic substrate level is essential, but not oxidative phosphorylation. J Biol Chem 278: 49625–49635 [DOI] [PubMed] [Google Scholar]
- Darling TN, Davis DG, London RE, Blum JJ (1987) Products of Leishmania braziliensis glucose catabolism: release of D-lactate and, under anaerobic conditions, glycerol. Proc Natl Acad Sci USA 84: 7129–7133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies CR, Kaye P, Croft SL, Sundar S (2003) Leishmaniasis: new approaches to disease control. BMJ 326: 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza W (2002) Special organelles of some pathogenic protozoa. Parasitol Res 88: 1013–1025 [DOI] [PubMed] [Google Scholar]
- Docampo R, de Souza W, Miranda K, Rohloff P, Moreno SN (2005) Acidocalcisomes—conserved from bacteria to man. Nat Rev Microbiol 3: 251–261 [DOI] [PubMed] [Google Scholar]
- Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BO (2007) Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA 104: 1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas C, Ouellette M, Tovar J, Cunningham ML, Fairlamb AH, Tamar S, Olivier M, Papadopoulou B (1997) Disruption of the trypanothione reductase gene of Leishmania decreases its ability to survive oxidative stress in macrophages. EMBO J 16: 2590–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JS, Palsson BO (2000) The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proc Natl Acad Sci USA 97: 5528–5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairlamb AH (2003) Target discovery and validation with special reference to trypanothione. In Drugs Against Parasitic Diseases: R&D Methodologies and Issues, Fairlamb AH, Ridley RG, Vial HJ (eds), pp 107–118. World Health Organization on behalf of the Special Programme for Research and Training in Tropical Diseases: Geneva, Switzerland [Google Scholar]
- Fairlamb AH, Blackburn P, Ulrich P, Chait BT, Cerami A (1985) Trypanothione: a novel bis(glutathionyl)spermidine cofactor for glutathione reductase in trypanosomatids. Science 227: 1485–1487 [DOI] [PubMed] [Google Scholar]
- Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BO (2007) A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol 3: 121 10.1038/msb4100155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist AM, Scholten JC, Palsson BO, Brockman FJ, Ideker T (2006) Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol Syst Biol 2: 2006.0004 10.1038/msb4100046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forst CV (2006) Host–pathogen systems biology. Drug Discov Today 11: 220–227 [DOI] [PubMed] [Google Scholar]
- Forster J, Famili I, Fu P, Palsson BO, Nielsen J (2003) Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res 13: 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francke C, Siezen RJ, Teusink B (2005) Reconstructing the metabolic network of a bacterium from its genome. Trends Microbiol 13: 550–558 [DOI] [PubMed] [Google Scholar]
- Garami A, Ilg T (2001a) Disruption of mannose activation in Leishmania mexicana: GDP-mannose pyrophosphorylase is required for virulence, but not for viability. EMBO J 20: 3657–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garami A, Ilg T (2001b) The role of phosphomannose isomerase in Leishmania mexicana glycoconjugate synthesis and virulence. J Biol Chem 276: 6566–6575 [DOI] [PubMed] [Google Scholar]
- Hannaert V, Albert MA, Rigden DJ, da Silva Giotto MT, Thiemann O, Garratt RC, Van Roy J, Opperdoes FR, Michels PA (2003a) Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase. Eur J Biochem 270: 3205–3213 [DOI] [PubMed] [Google Scholar]
- Hannaert V, Bringaud F, Opperdoes FR, Michels PA (2003b) Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetoplastid Biol Dis 2: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart DT, Opperdoes FR (1984) The occurrence of glycosomes (microbodies) in the promastigote stage of four major Leishmania species. Mol Biochem Parasitol 13: 159–172 [DOI] [PubMed] [Google Scholar]
- Hidalgo-Zarco F, Gonzalez-Pazanowska D (2001) Trypanosomal dUTPases as potential targets for drug design. Curr Protein Pept Sci 2: 389–397 [DOI] [PubMed] [Google Scholar]
- Hwang HY, Ullman B (1997) Genetic analysis of purine metabolism in Leishmania donovani. J Biol Chem 272: 19488–19496 [DOI] [PubMed] [Google Scholar]
- Jamshidi N, Palsson BO (2007) Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets. BMC Syst Biol 1: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardim A, Bergeson SE, Shih S, Carter N, Lucas RW, Merlin G, Myler PJ, Stuart K, Ullman B (1999) Xanthine phosphoribosyltransferase from Leishmania donovani. Molecular cloning, biochemical characterization, and genetic analysis. J Biol Chem 274: 34403–34410 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Roberts SC, Jardim A, Carter NS, Shih S, Ariyanayagam M, Fairlamb AH, Ullman B (1999) Ornithine decarboxylase gene deletion mutants of Leishmania donovani. J Biol Chem 274: 3781–3788 [DOI] [PubMed] [Google Scholar]
- Kauffman KJ, Prakash P, Edwards JS (2003) Advances in flux balance analysis. Curr Opin Biotechnol 14: 491–496 [DOI] [PubMed] [Google Scholar]
- Keller GA, Krisans S, Gould SJ, Sommer JM, Wang CC, Schliebs W, Kunau W, Brody S, Subramani S (1991) Evolutionary conservation of a microbody targeting signal that targets proteins to peroxisomes, glyoxysomes, and glycosomes. J Cell Biol 114: 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TY, Kim HU, Park JM, Song H, Kim JS, Lee SY (2007) Genome-scale analysis of Mannheimia succiniciproducens metabolism. Biotechnol Bioeng 97: 657–671 [DOI] [PubMed] [Google Scholar]
- Krauth-Siegel RL, Meiering SK, Schmidt H (2003) The parasite-specific trypanothione metabolism of Trypanosoma and Leishmania. Biol Chem 384: 539–549 [DOI] [PubMed] [Google Scholar]
- Lee JM, Gianchandani EP, Papin JA (2006) Flux balance analysis in the era of metabolomics. Brief Bioinform 7: 140–150 [DOI] [PubMed] [Google Scholar]
- Lee SH, Stephens JL, Englund PT (2007) A fatty-acid synthesis mechanism specialized for parasitism. Nat Rev Microbiol 5: 287–297 [DOI] [PubMed] [Google Scholar]
- Leifso K, Cohen-Freue G, Dogra N, Murray A, McMaster WR (2007) Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: the Leishmania genome is constitutively expressed. Mol Biochem Parasitol 152: 35–46 [DOI] [PubMed] [Google Scholar]
- Lipoldova M, Demant P (2006) Genetic susceptibility to infectious disease: lessons from mouse models of leishmaniasis. Nat Rev Genet 7: 294–305 [DOI] [PubMed] [Google Scholar]
- Martin KL, Smith TK (2005) The myo-inositol-1-phosphate synthase gene is essential in Trypanosoma brucei. Biochem Soc Trans 33: 983–985 [DOI] [PubMed] [Google Scholar]
- Merlen T, Sereno D, Brajon N, Rostand F, Lemesre JL (1999) Leishmania spp: completely defined medium without serum and macromolecules (CDM/LP) for the continuous in vitro cultivation of infective promastigote forms. Am J Trop Med Hyg 60: 41–50 [DOI] [PubMed] [Google Scholar]
- Molyneux D, Killick-Kendrick R (1987) Morphology, ultrastructure and life cycles. In The Leishmaniases in Biology and Medicine, Peters W, Killick-Kendrick R (eds), Vol. v. 1, pp 121–176. Academic Press Inc. (London) Limited: London [Google Scholar]
- Naderer T, Ellis MA, Sernee MF, De Souza DP, Curtis J, Handman E, McConville MJ (2006) Virulence of Leishmania major in macrophages and mice requires the gluconeogenic enzyme fructose-1,6-bisphosphatase. Proc Natl Acad Sci USA 103: 5502–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhardt MA, Puchalka J, Fryer KE, Dos Santos VA, Papin JA (2008) Genome-scale metabolic network analysis of the opportunistic pathogen Pseudomonas aeruginosa PAO1. J Bacteriol 8 (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperdoes FR, Michels PA (1993) The glycosomes of the Kinetoplastida. Biochimie 75: 231–234 [DOI] [PubMed] [Google Scholar]
- Opperdoes FR, Michels PAM (2008) The metabolic repertoire of Leishmania and implications for drug discovery. In Leishmania: After The Genome, Myler PJ, Fasel N (eds), Norfolk, UK: Caister Academic Press (in press) [Google Scholar]
- Opperdoes FR, Szikora JP (2006) In silico prediction of the glycosomal enzymes of Leishmania major and trypanosomes. Mol Biochem Parasitol 147: 193–206 [DOI] [PubMed] [Google Scholar]
- Panizzutti R, de Souza Leite M, Pinheiro CM, Meyer-Fernandes JR (2006) The occurrence of free D-alanine and an alanine racemase activity in Leishmania amazonensis. FEMS Microbiol Lett 256: 16–21 [DOI] [PubMed] [Google Scholar]
- Payne SH, Loomis WF (2006) Retention and loss of amino acid biosynthetic pathways based on analysis of whole-genome sequences. Eukaryot Cell 5: 272–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JL, Famili I, Thiele I, Palsson BO (2006a) Towards multidimensional genome annotation. Nat Rev Genet 7: 130–141 [DOI] [PubMed] [Google Scholar]
- Reed JL, Patel TR, Chen KH, Joyce AR, Applebee MK, Herring CD, Bui OT, Knight EM, Fong SS, Palsson BO (2006b) Systems approach to refining genome annotation. Proc Natl Acad Sci USA 103: 17480–17484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JL, Vo TD, Schilling CH, Palsson BO (2003) An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol 4: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera RM, Redondo CM, Perez-Pertejo Y, Balana-Fouce R (2007) S-Adenosylmethionine in protozoan parasites: functions, synthesis and regulation. Mol Biochem Parasitol 152: 1–10 [DOI] [PubMed] [Google Scholar]
- Remme JH, Blas E, Chitsulo L, Desjeux PM, Engers HD, Kanyok TP, Kengeya Kayondo JF, Kioy DW, Kumaraswami V, Lazdins JK, Nunn PP, Oduola A, Ridley RG, Toure YT, Zicker F, Morel CM (2002) Strategic emphases for tropical diseases research: a TDR perspective. Trends Parasitol 18: 421–426 [DOI] [PubMed] [Google Scholar]
- Roberts SC, Jiang Y, Jardim A, Carter NS, Heby O, Ullman B (2001) Genetic analysis of spermidine synthase from Leishmania donovani. Mol Biochem Parasitol 115: 217–226 [DOI] [PubMed] [Google Scholar]
- Roberts SC, Scott J, Gasteier JE, Jiang Y, Brooks B, Jardim A, Carter NS, Heby O, Ullman B (2002) S-Adenosylmethionine decarboxylase from Leishmania donovani. Molecular, genetic, and biochemical characterization of null mutants and overproducers. J Biol Chem 277: 5902–5909 [DOI] [PubMed] [Google Scholar]
- Roberts SC, Tancer MJ, Polinsky MR, Gibson KM, Heby O, Ullman B (2004) Arginase plays a pivotal role in polyamine precursor metabolism in Leishmania. Characterization of gene deletion mutants. J Biol Chem 279: 23668–23678 [DOI] [PubMed] [Google Scholar]
- Roper JR, Guther ML, Milne KG, Ferguson MA (2002) Galactose metabolism is essential for the African sleeping sickness parasite Trypanosoma brucei. Proc Natl Acad Sci USA 99: 5884–5889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling CH, Covert MW, Famili I, Church GM, Edwards JS, Palsson BO (2002) Genome-scale metabolic model of Helicobacter pylori 26695. J Bacteriol 184: 4582–4593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A, Clark-Walker GD, Steinberg AG, Stuart K (2005) The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J 24: 4029–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster FL, Sullivan JJ (2002) Cultivation of clinically significant hemoflagellates. Clin Microbiol Rev 15: 374–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekowska A, Denervaud V, Ashida H, Michoud K, Haas D, Yokota A, Danchin A (2004) Bacterial variations on the methionine salvage pathway. BMC Microbiol 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L, Da Silva A (1971) Isolation and characterization of kinetoplast DNA from Leishmania tarentolae. J Mol Biol 56: 443–473 [DOI] [PubMed] [Google Scholar]
- Stuart K (1983) Kinetoplast DNA, mitochondrial DNA with a difference. Mol Biochem Parasitol 9: 93–104 [DOI] [PubMed] [Google Scholar]
- Sutterwala SS, Creswell CH, Sanyal S, Menon AK, Bangs JD (2007) De novo sphingolipid synthesis is essential for viability, but not for transport of glycosylphosphatidylinositol-anchored proteins, in African trypanosomes. Eukaryot Cell 6: 454–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar J, Wilkinson S, Mottram JC, Fairlamb AH (1998) Evidence that trypanothione reductase is an essential enzyme in Leishmania by targeted replacement of the tryA gene locus. Mol Microbiol 29: 653–660 [DOI] [PubMed] [Google Scholar]
- Umasankar PK, Jayakumar PC, Shouche YS, Patole MS (2005) Molecular characterization of the hexokinase gene from Leishmania major. J Parasitol 91: 1504–1509 [DOI] [PubMed] [Google Scholar]
- Urbina JA, Concepcion JL, Rangel S, Visbal G, Lira R (2002) Squalene synthase as a chemotherapeutic target in Trypanosoma cruzi and Leishmania mexicana. Mol Biochem Parasitol 125: 35–45 [DOI] [PubMed] [Google Scholar]
- Urbina JA, Docampo R (2003) Specific chemotherapy of Chagas disease: controversies and advances. Trends Parasitol 19: 495–501 [DOI] [PubMed] [Google Scholar]
- Varma A, Palsson BO (1993) Metabolic capabilities of Escherichia coli 0.2. Optimal-growth patterns. J Theor Biol 165: 503–522 [DOI] [PubMed] [Google Scholar]
- Varma A, Palsson BO (1994) Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl Environ Microbiol 60: 3724–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, Palsson BO (1995) Parametric sensitivity of stoichiometric flux balance models applied to wild-type Escherichia coli metabolism. Biotechnol Bioeng 45: 69–79 [DOI] [PubMed] [Google Scholar]
- Vessal M, Rezai HR, Pakzad P (1974) Leishmania species: fatty acid composition of promastigotes. Exp Parasitol 36: 455–461 [DOI] [PubMed] [Google Scholar]
- Wakil SJ, Pugh EL, Sauer F (1964) The mechanism of fatty acid synthesis. Proc Natl Acad Sci USA 52: 106–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Showalter M, Revollo J, Hsu FF, Turk J, Beverley SM (2003) Sphingolipids are essential for differentiation but not growth in Leishmania. EMBO J 22: 6016–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information 1
Supplementary Information 2
Supplementary Information 3
Supplementary Information 4
Supplementary Information 5
Supplementary Figure 6
Supplementary Information 7
Supplementary Information 8