Abstract
Regulated phosphorylation by protein tyrosine kinases (PTKs) such as c-Abl, is critical to cellular homeostasis. In turn, once deregulated as in the Chronic Myeloid Leukemia (CML) fusion protein Bcr-Abl, PTKs can promote cancer onset and progression. The dramatic success of the Bcr-Abl inhibitor imatinib as therapy for CML has inspired interest in other PTKs as targets for cancer drug discovery. Here we report a novel PTK activity and inhibition screening method using hydrogel-immobilized peptide substrates. Using acrylate crosslinkers, we tether peptides via terminal cysteines to thiol-presenting hydrogels in 96-well plates. These surfaces display low background and high reproducibility, allowing semi-quantitative detection of peptide phosphorylation by recombinant c-Abl or by Bcr-Abl activity in cell extracts using traditional anti-phosphotyrosine immunodetection and chemifluorescence. The capabilities of this assay were demonstrated by performing model screens for inhibition with several commercially available PTK inhibitors and a collection of pyridopyrimidine Src/Abl dual inhibitors. This assay provides a practical method to measure the activity of a single kinase present in a whole cell lysate with high sensitivity and specificity as a valuable means for efficient small molecule screening.
Keywords: kinase assay, imatinib mesylate (IM), peptide substrate, protein tyrosine kinase
Introduction
Phosphorylation by tyrosine kinases is a crucial signaling mechanism in pathways that control cellular development and proliferation. Consequently, these signals are normally tightly regulated by the cell and aberrant expression or activity of these kinases can have devastating results. For example, the c-Abl kinase signals at critical points in the growth and differentiation of hematopoetic cells. The Ph1 reciprocal translocation of chromosomes 9 and 22 [1] results in the fusion gene BCR-ABL [2]. In the Bcr-Abl gene product, the autoinhibitory domain of c-Abl is replaced with the amino terminal dimerization domain of Bcr [3,4], thereby disrupting the normal regulation of the kinase. As a result, Bcr-Abl displays constitutive protein tyrosine kinase activity towards c-Abl substrates. The resulting deregulation of proliferation and survival signals in hematopoietic stem cells predisposes to incomplete myeloid differentiation and Chronic Myeloid Leukemia (CML) [5].
The finding that Bcr-Abl activity underlies the CML phenotype rationalized a search for small molecule inhibitors of the Abl kinase as potential drugs. A pioneering research effort by Druker and colleagues revealed that imatinib mesylate (IM) (also called CGP57148B or STI571) could inhibit Bcr-Abl kinase activity, eliminate circulating CML cells and induce durable remission in chronic phase CML patients, leading to the first approved tyrosine kinase inhibitor drug (sold as Gleevec, Novartis) [6]. However, a significant fraction of chronic phase patients develop imatinib resistance and patients in advanced stages of CML typically fail to respond to imatinib at all. Chronic phase resistance is usually associated with point mutations in the Abl kinase catalytic domain that antagonize imatinib binding, but can also be a result of BCR-ABL gene amplification [7,8]. In the face of increased incidence of resistance to imatinib, two second generation agents have been approved, the 2-phenylaminopyrimidine AMN107 (Nilotinib, sold as Tasigna, Novartis) [9] and the 2-aminothiazole BMS-354825 (Dasatinib, sold as Sprycel, Bristol-Myers Squibb) [10]. Other agents remain under preclinical development and/or in clinical trials [11-17]. Several highly promising compounds that remain under study in the laboratory come from the pyrido-pyrimidine class of dual Src/Abl tyrosine kinase inhibitors, the best characterized of which are PD173955 [18], PD180970 [19] and PD166326 [20-22].
To expand CML drug development efforts and discover new lead compounds to combat or circumvent resistance requires efficient, biologically-relevant kinase assays, such as methods to detect Bcr-Abl activity in vivo. In turn, there is a clear need for Bcr-Abl kinase assays for clinical use where they could facilitate CML diagnosis, disease monitoring, detection of imatinib resistance, and selection of second-line therapy. Kinase assays designed to address these challenges must be reproducible, specific and robust. The ideal for these assays would be to measure the endogenous kinase activity in living cells or cell lysates. However, it can be challenging to follow the activity of a single kinase in such complex samples.
Conventional kinase assays based on detecting incorporation of the terminal phosphate from ATP-γ-32P, -33P or -35S (thiophosphate) are subject to artifacts such as nucleotide dilution and hydrolysis in cell lysate. While immunodetection with phospho-amino acid or phospho-peptide specific antibodies can obviate the need for radioactive tracers, phosphorylated proteins in cell extracts can produce confounding background signals. Here, solid-phase (heterogeneous) kinase assays with immobilized substrates have clear advantages. In typical solid-phase enzyme-linked immunosorbent assays (ELISA), an exogenously prepared protein or peptide substrate is captured on the surface of a 96-well plate either before or after being treated with a kinase. The phosphorylated substrate can be probed with a phospho-specific primary antibody and an enzyme-conjugated secondary antibody to detect phosphorylation [23]. ELISA-format kinase assays are particularly suitable for medium-throughput targeted-library screens, secondary screens, and as a follow-up to medicinal chemistry [24,25].
However, reproducibly tethering peptides or proteins onto surfaces so that they remain accessible to the kinase is not always straightforward, as their diverse chemical properties can lead to unanticipated interactions with surfaces [26-29]. Yet, the surfaces must be well passivated as non-specific adsorption of phosphorylated proteins from kinase reactions and/or cell lysate can cause background and obscure the signal. A number of approaches have been used to immobilize peptides and proteins to surfaces for kinase assays, including peptide synthesis in situ [30,31], direct covalent attachment to aldehydic surfaces [32], use of biotin or epitope tags to attach to functionalized surfaces [33], covalent attachment by native chemical ligation of amino-terminal cysteines to activated glass [34], covalent or adsorptive attachment to self-assembled monolayers of alkanethiols on gold surfaces [35], and co-polymerization into polyacrylamide gel pads [36]. While each of these approaches has advantages, as yet, no simple method that displays a high density and accessibility of substrate, resistance to non-specific protein adsorption, simple detection and high mechanical and chemical stability has been described. Each of these criteria needs to be met to ensure sensitivity, specificity and reproducibility in real-world applications such as multiwell screening.
In the method described here, we report a strategy for covalent immobilization of peptide kinase substrates in an acrylamide hydrogel in a 96-well plate format. Using the Abl kinase substrate “Abltide” (CEAIYAAPFAKKK) [37] attached to the acrylamide surface through conjugate Michael addition chemistry, we analyzed the activities and responses to small molecule drugs of recombinant c-Abl and Bcr-Abl present in the human erythroleukemia cell line K562. Phosphorylated Abltide was detected in an ELISA-style manner using an anti-phosphotyrosine antibody, a horseradish-peroxidase conjugated secondary antibody and chemifluorescent detection.
Materials and Methods
Chemicals were obtained from Sigma-Aldrich (St. Louis, MO, U.S.A.) unless otherwise specified.
Cell culture
K562 cells [38] were cultured at 37 C and 5% CO2 in RPMI-1640 medium containing 10% (v/v) heat-inactivated fetal bovine serum, 1% (v/v) penicillin/streptomycin, and 4 mM L-glutamine.
Cell extract preparation
Whole cell lysates were prepared in PhosphoSafe Extraction Reagent (Novagen, San Diego, CA, U.S.A.). Briefly, cells were washed with ice-cold PBS and incubated in 50 μl PhosphoSafe per 106 cells on ice for 20 min, briefly agitated with a vortex mixer, and centrifuged at 4°C for 10 min at 16,000 x g. The supernatant was collected and analyzed by Bradford assay using Coomassie Protein Assay Reagent (Bio-Rad Laboratories, Hercules, CA, U.S.A.).
Kinase inhibitors
Imatinib mesylate (Novartis, East Hanover, NJ, U.S.A.) was kindly provided by Dr. Wendy Stock (University of Chicago, Chicago, IL). AG957 and Genestein were purchased from Calbiochem (San Diego, CA, U.S.A.). The Screen-Well™ Kinase Inhibitor Library, a set of 80 ATP-competitive kinase inhibitors, was purchased from BioMol (Plymouth Meeting, PA, U.S.A.). All the compounds were dissolved and further diluted in dimethyl sulfoxide (DMSO).
PD166326, PD173955, and PD180970 and a focused compound library of 24 other pyrido[2,3-d]pyrimidine-7-one class multi-targeted kinase inhibitors were synthesized using methods developed by Klutchko et al. [39] and Boschelli et al. [40]. In short, a variety of aniline and amine derivatives were coupled with 6-(2,6-dichloro-phenyl)-2-methanesulfonyl-8-methyl-8H-pyrido [2,3-d] pyrimidin-7-one to generate a focused library.
Peptide synthesis and yield
The Cys-Abltide substrate peptide (Cys-AT), CEAIYAAPFAKKK, and the Cys-Abl SH3 ligand (Cys-AL), CGGAPTYSPPPPPLL, were synthesized using solid-phase Fmoc chemistry on a Prelude Parallel Peptide Synthesizer (Protein Technologies, Tucson, AZ, U.S.A.). The synthesized peptides were purified using a C18 reverse phase column on an Agilent 1200 series LC/MS (Santa Clara, CA, U.S.A.) and analyzed with an Applied Biosystems 4700 MALDI TOF/TOF MS (Foster City, CA, U.S.A.). The lyophilized peptides were dissolved in H2O and concentrations were determined using Beers’ Law from absorbance at 280 nm.
Immobilization of peptides
We adapted the ez-rays™ hydrogel multiwell plates from Matrix Technologies (Hudson, NH, U.S.A.) to immobilize our cysteine-labeled peptides on the surface. The ez-rays surface is a copolymer of acrylamide crosslinked with cystamine bisacrylamide which can be reduced to present reactive thiols for covalent immobilization of biomolecules [41]. Working at room temperature (24°C), the hydrogel surfaces were activated using 50 μl per well of 50 mM tris(2-carboxyethyl)phosphine (TCEP) for 30 min. The plates were washed three times with H2O in an ELx 405 plate washer (BioTek Instruments, Winooski, VT, U.S.A.). Activated hydrogel was next functionalized using 50 μl per well of 25 mM bisacrylamide or a Sartomer (Exton, PA, U.S.A.) water soluble acrylate linker in 100 mM NaHCO3, pH 8.4 for 30 min followed by three washes with H2O. The remaining unreacted thiol groups were blocked with 50 μl per well of 2 M sodium acrylate in 100 mM Na2CO3, pH 10.0, for 30 min and washed three times with H2O. The peptides were diluted in 100 mM NaHCO3 pH 8.4 at the indicated concentrations and 25 μl per well was added to the acrylamide-activated hydrogel surface for 1 h. Water soluble acrylates were kindly provided by Sartomer.
Kinase assays and immunodetection
The standard assay conditions are as follows. The surfaces were blocked against residual non-specific protein adsorption using 10% (w/v) BSA in 50 mM Tris, pH 7.5 for 1 h. The immobilized peptides were then subjected to kinase assays with 25 μg cell extract at 37 C or 0.005 U recombinant c-Abl (Upstate, Charlottesville, VA, U.S.A.) at 30 C in 50 μl kinase buffer (10 μM ATP, 50 mM Tris pH 7.5, 10 mM MgCl2, 1 mM DTT, 1 mM EDTA, 0.01% Brij 35 (v/v) and 1X complete protease inhibitor cocktail (Roche, Mannheim, Germany)). After the kinase reaction, the plate was washed six times with 2% SDS (w/v) in Tris-buffered saline (20 mM Tris, 137 mM NaCl, pH 7.6) in the ELx 405 plate washer.
Surfaces were further blocked for immunodetection with 10% BSA (w/v) in TBS-T (50 μl per well, 20 mM Tris, 137 mM NaCl, 0.1% Tween (v/v), pH 7.6) at 4°C for 1 h, followed by incubation with 50 μl per well of 1:1000 4G10 mouse anti-phosphotyrosine monoclonal antibody (Millipore, Billerica, MA, U.S.A.) in 5% BSA (w/v) in TBS-T for 1 h at 4°C. Plates were washed three times with TBS-Tand incubated with 50 μl per well of 1:2000 horseradish peroxidase (HRP) conjugated sheep anti-mouse IgG secondary antibody (Amersham, Piscataway, NJ, U.S.A.) in 5% BSA (w/v) in TBS-T for 30 min at 4°C followed by three washes with TBS-T.
For chemiluminiscent detection, SuperSignal West Pico chemiluminescent reagent (Pierce, Rockford, IL, U.S.A.) was added to the plates (50 μl per well) and the glass-bottom hydrogel plate was exposed to autoradiography film (Phenix, Hayward, CA, U.S.A.). The films were scanned using a ScanMaker 6800 (Microtek, Hsinchu, Taiwan) and quantitated with Quantity One software. For chemifluorescent detection, Amplex Red (Invitrogen, Carlsbad, CA, U.S.A.) was added to the plates (100 μl per well, 50 μM Amplex Red reagent, 1 mM H2O2 in 50 mM sodium phosphate, pH 7.5). Fluorescence of Amplex Red reactivity was scanned by a Bio-Rad Molecular Imager FX with excitation wavelength 532 nm and emission wavelength 590 nm and quantified with Quantity One software.
Statistical analysis of assay dynamic range and reproducibility
For statistical analysis of assay performance, the density of each well was determined with Quantity One software (BioRad) and the fluorescence reading was normalized by dividing by the mean of the negative controls (100 μM imatinib). The ratio of signal-to-background was calculated by: ((Xpos)/( Xneg)). The signal-to-noise ratio was calculated as: ((Xpos- Xneg)/(σneg)). The Z-factor, a dimensionless statistic representing both dynamic range and variability across multiple samples, was calculated with the formula: (1-((3σpos+3σneg)/(Xpos-Xneg))) [42]. A Z-factor of 1.0 represents ideal performance, but ones greater than 0.5 are considered adequate for screening assays.
Results
Development of kinase assays with hydrogel-immobilized peptide substrates in 96-well plates
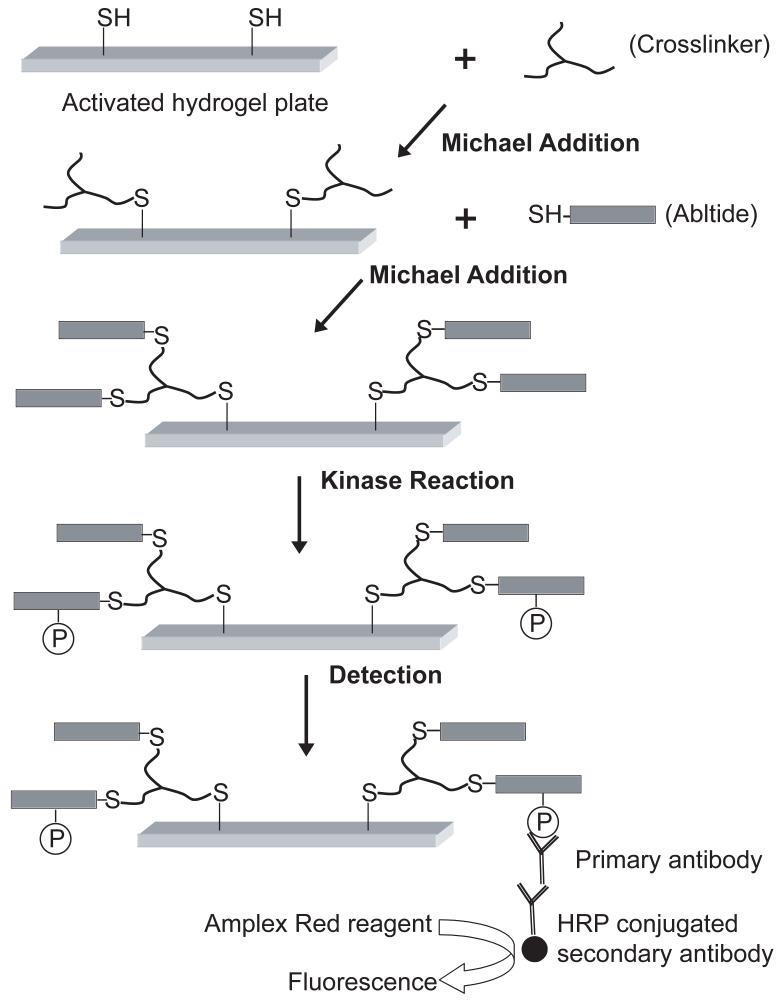
To detect c-Abl and Bcr-Abl activity, we covalently crosslinked Cys-AT (CEAIYAAPFAKKK), the synthetic substrate peptide Abltide [37] bearing an amino-terminal cysteine, to an ez-rays hydrogel surface by two Michael addition conjugations (Fig. 1). We used ez-rays, a glass substrate coated with a surface film of polyacrylamide crosslinked with cystamine bisacrylamide, yielding a hydrogel with a high density of embedded disulfide bonds. Reducing agents such as TCEP generate free thiol groups that can be functionalized via reaction with alkylating agents or conjugate (Michael) addition.
Fig. 1.
Basic strategy for immobilization, kinase reaction and immunodetection. Peptides are tethered to a hydrogel in multiwell plates via multivalent crosslinkers, subjected to phosphorylation by kinases, and then interrogated with anti-phosphotyrosine antibody and secondary antibody-horseradish peroxidase conjugate with detection by chemifluorescence.
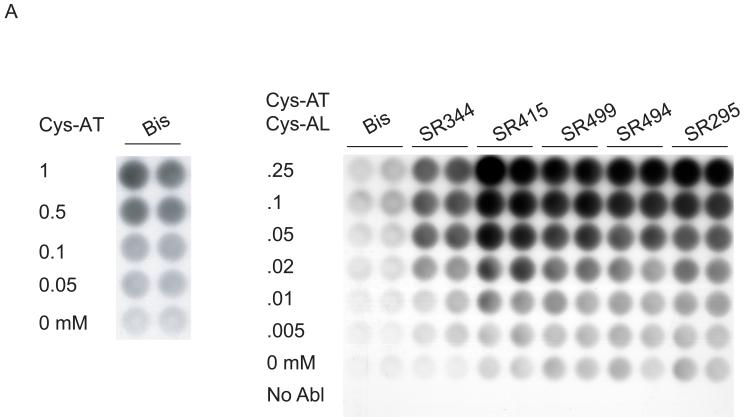
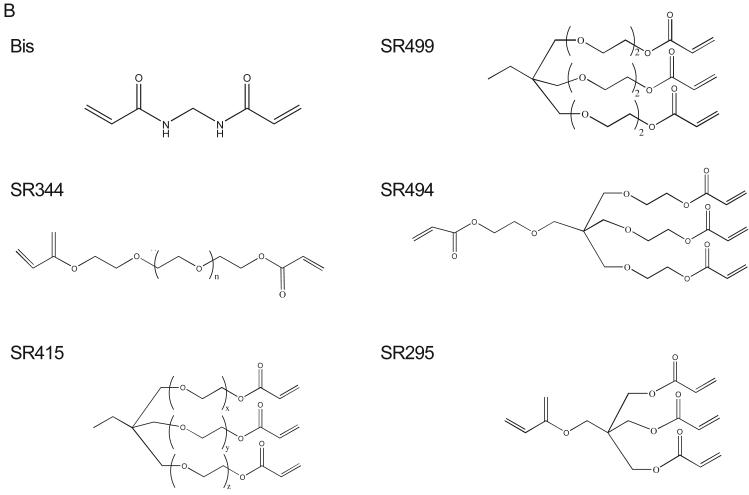
In our initial studies, Cys-AT was immobilized on the hydrogel surface via bisacrylamide and reacted with K562 lysate. The phosphorylated peptide was detected with anti-phosphotyrosine antibody, horseradish-peroxidase conjugated secondary antibody and chemiluminiscence. The background signal from no substrate peptide was very low and phosphorylation signal increased with peptide concentration (Fig. 2A, left). We then optimized a number of factors affecting sensitivity and background in the kinase assay. Combining Cys-AL (CGGAPTYSPPPPPLL), a high affinity peptide ligand for the Abl SH3 domain [43], with the Cys-AT improved assay sensitivity, presumably by recruiting the kinase to the surface (Fig. S1). We then evaluated several Sartomer bis-, tri-, and tetra-acrylate polyethylene glycol (PEG) crosslinkers (Fig. 2B) as a means to increase the concentration and/or accessibility of Cys-AT and Cys-AL on the hydrogel surface. The branched PEG linkers all gave superior results to bisacrylamide but SR415 yielded the strongest phosphorylation signal without compromising the apparent signal to noise ratio (Fig. 2A, right). Consistent with these results, compared to bisacrylamide, SR415 markedly increased the density of peptide on the hydrogel surface (Fig. S2). Comparison of detection methods demonstrated higher sensitivity for chemiluminiscence but superior reproducibility and linearity for chemifluorescence, particularly for Amplex Red, an HRP substrate that yields a red fluorescent product (data not shown).
Fig. 2.
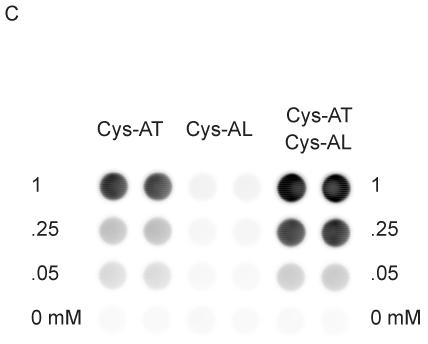
Phosphorylation of peptides by K562 lysates. (A, left) A titration of Cys-Abltide was attached to the surface in duplicate at the indicated concentrations, reacted with 25 μg K562 cell lysate, and detected by antibody and chemiluminiscent reagents as described in Materials and Methods. (A, right) The hydrogel surface was activated and then linked with different bis-, tri-, and tetra-acrylates. Cys-Abltide (Cys-AT) and Cys-Abl ligand (Cys-AL) were added in a 1:1 ratio at 0.25, 0.1, 0.05, 0.02, 0.01 and 0.005 mM. The kinase assay was performed as described in Materials and Methods with detection by chemiluminiscence. (B) Chemical structures of bisacrylamide and the Sartomer bis-, tri-, and tetra-acrylates: SR344, polyethylene glycol (400) diacrylate; SR415, ethoxylated (20) trimethylolpropane triacrylate; SR499, ethoxylated (6) trimethylolpropane triacrylate; SR494, ethoxylated (5) pentaerythritol tetraacrylate; SR295, pentaerythritol tetraacrylate. (C) After activation and SR415 attachment, the wells were treated with the peptides (Cys-AT alone, Cys-AL alone and Cys-AT/Cys-AL combined) in duplicate at the indicated concentrations. The kinase reactions and immunodetection with chemifluorescence were performed as described in Materials and Methods. Image shows accumulation of resorufin, the fluorescent Amplex Red reaction product, proportional to relative phosphorylation in each well.
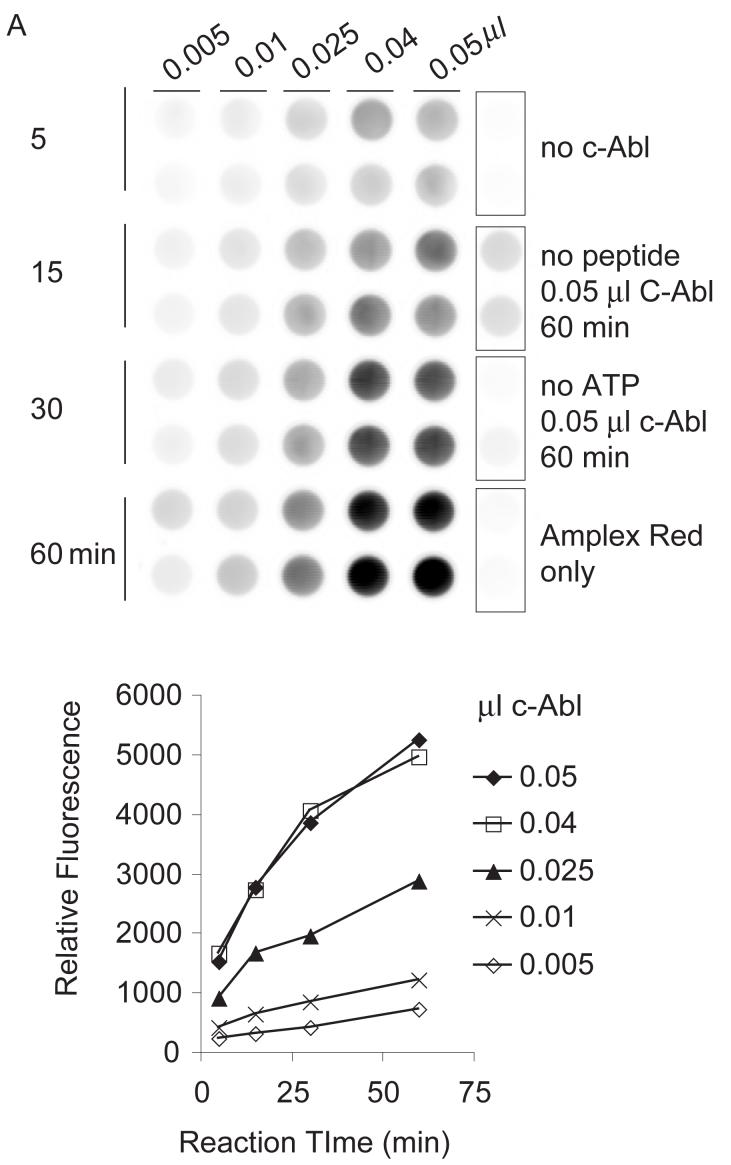
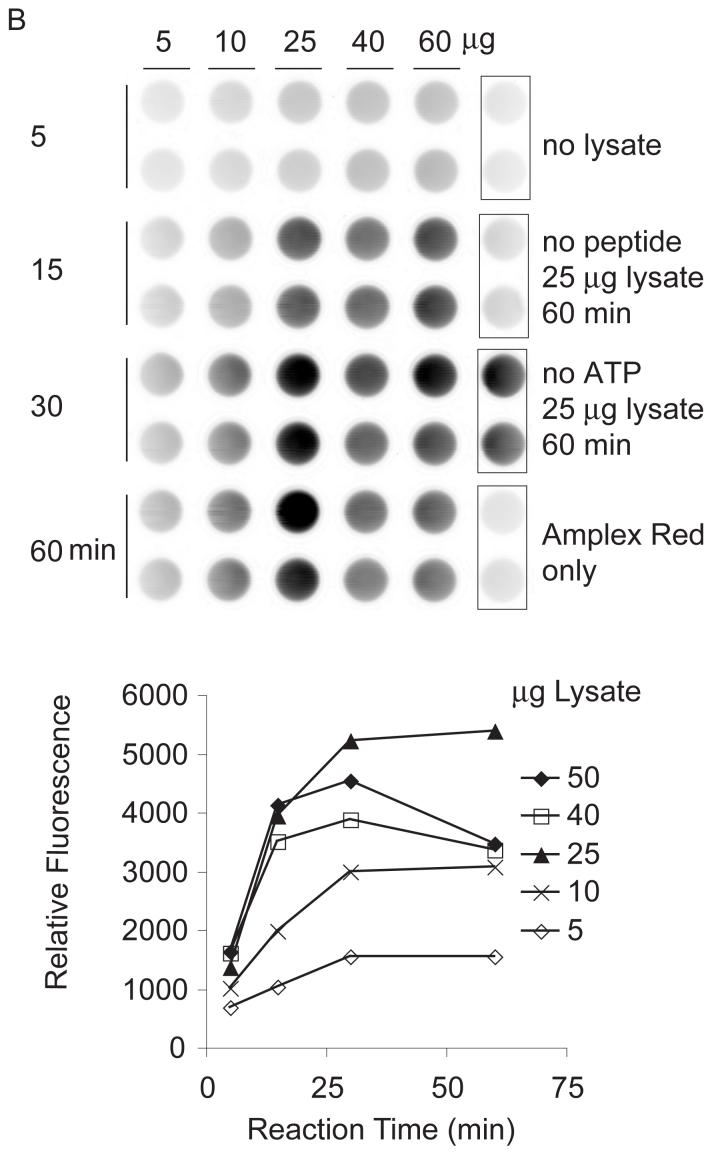
Combining SR415 tethering with Amplex Red detection, we observed that Cys-AT with Cys-AL yielded an increased phosphorylation signal over Cys-AT alone (Fig. 2C). Significantly, even though Cys-AL contains a tyrosine, little or no phosphorylation was measured after incubation with lysate. Under these conditions, recombinant c-Abl gave a maximum signal after 1 h with 0.005 U (0.05 μl) of enzyme at 30°C (Fig. 3A). K562 extracts (i.e. Bcr-Abl activity) gave the highest signal after 30 min with 25 μg of cell lysate at 37°C (Fig. 3B). The control wells lacking c-Abl or Bcr-Abl kinase, lacking peptides, or empty except Amplex Red all gave little or no signal. While c-Abl without added ATP displayed similarly low background, the negative control lacking additional ATP in the Bcr-Abl reaction displayed substrate phosphorylation, likely reflecting cellular ATP present in the cell extract. From these results 10 μM added ATP, 0.05 mM Cys-AT and Cys-AL, 30 min incubation, 0.05 μl c-Abl at 30°C or 25 μg K562 extract at 37°C, and Amplex Red detection were established as the standard conditions.
Fig. 3.
Dependence of kinase reaction on reaction time and kinase amount. Peptides (0.05 mM of Cys-AT and Cys-AL) were tethered to hydrogels in wells as describe in Materials and Methods. (A) Recombinant c-Abl: Kinase reactions were performed with 0.005, 0.01, 0.025, 0.04 and 0.05 μl c-Abl (0.1U/μl) in kinase buffer for 5, 15, 30, and 60 min. (B) K562 lysate: Kinase reactions were performed with 5, 10, 25, 40, or 50 μg of K562 cell lysate in kinase buffer for 5, 15, 30, or 60 min. Each panel shows a scan of Amplex Red reaction product on the top and a plot of total well fluorescence on the bottom. Control wells on the right are pairs of mock reactions lacking kinase (c-Abl or Bcr-Abl), lacking peptides, lacking ATP, or empty wells to which the Amplex Red detection reagent was added.
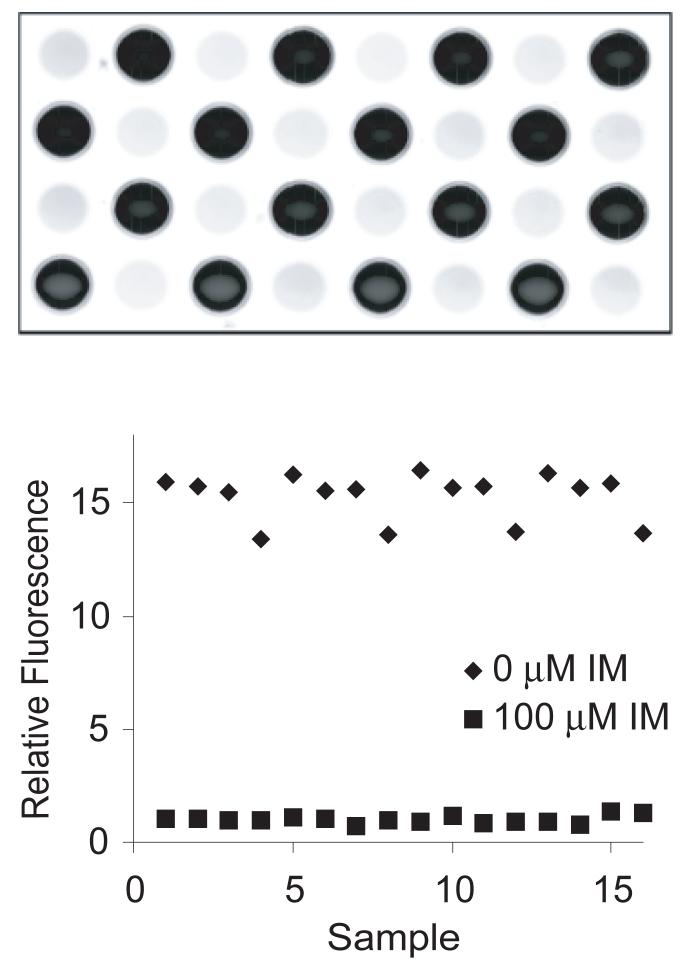
To characterize the assay, we examined the reproducibility of phosphorylation from well to well. Using imatinib to fully inhibit Bcr-Abl activity in K562 cell lysate, we quantitated fluorescence in 32 wells to which 0 (fully active) or 100 μM (fully inhibited) imatinib was added. These data yielded a signal to background of 15.3, a signal to noise of 82.8 and a Z-factor [42] of 0.74 (Fig. 4). These values represent satisfactory performance for a biochemical screening assay.
Fig. 4.
Evaluation of well-to-well consistency within the assay. (A) The kinase assay was performed using the standard assay conditions with or without 100 μM imatinib as negative and positive controls. (B) Plot of fluorescence intensity of wells after 30 min incubation with Amplex Red. Relative intensity was determined by dividing total well fluorescence by the mean intensity of the wells containing 100 μM imatinib. The signal to background, signal to noise, and Z-factor were calculated as described in Materials and Methods.
Model drug screens using this kinase assay
We examined this kinase assay as a tool to compare small molecule kinase inhibitors in a model screen using K562 cell lysate. The kinase reaction was found to be tolerant of DMSO up to a final concentration of 10% (v/v, not shown). The effects of six well-characterized tyrosine kinase inhibitors were assayed in a dilution series from 1 mM to 10 nM (Fig. 5). The highest potency was observed for the pyridopyrimidine dual Src/Abl inhibitors PD166326, PD173955 and PD180970. The apparent IC50, the concentration at which phosphorylation was decreased by 50%, was in the nanomolar range in each case. The first generation chronic myeloid leukemia therapeutic agent imatinib yielded a micromolar IC50 while the tyrphostin AG957 yielded an IC50 in the millimolar range. The general tyrosine kinase inhibitor genestein was comparable to AG957 in this assay. The relative potency and IC50 observed for the six compounds was generally consistent with literature values [20,44].
Fig. 5.
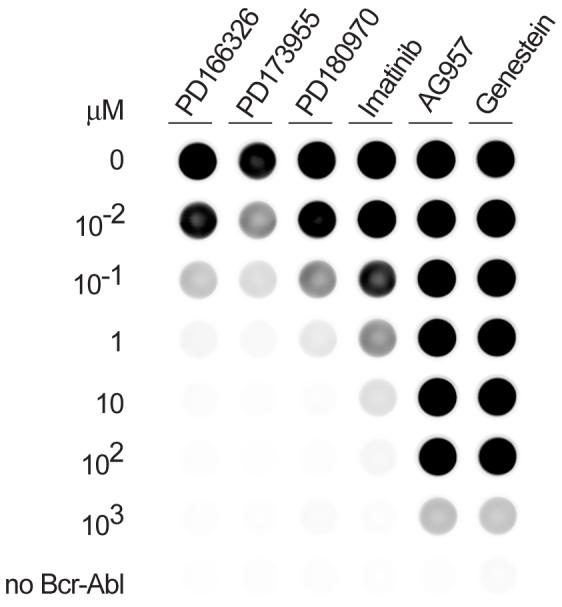
Model screen with known inhibitors. The kinase assay was performed with K562 lysates using the standard assay conditions in the presence of inhibitors at varying concentrations (10-2 to 103 μM).
As a further demonstration of the assay as a tool for compound screening, we examined representative compounds from a focused library of structurally-related pyridopyrimidine Src/Abl kinase inhibitors. Pyridopyrimidine compounds are well-characterized and their structure-activity relationships have been studied at length [21,45,46]. These analyses have shown that the substitution of the amino group on the carbon at position 2 (C-2) significantly affects the inhibitory activity. Several of the most potent pyridopyrimidine inhibitors such as PD166326, PD173955, PD173958, PD173956 and PD180970 all share a secondary aniline substitution at C-2. These compounds all yielded an apparent IC50 of 100 nM or less for inhibition of Bcr-Abl in K562 extracts in this assay (Figs. 5 and 6). From a focused library of pyrido[2,3-d]pyrimidine-7-ones that explored variously substituted anilines at C-2, five compounds, DV2-43, DV2-47, DV-87, DV2-273, and DV2-289, also displayed apparent IC50 values of 100 nM or less (Fig. 6). Of these DV2-43 and DV2-289 appeared to have the greatest activity. DV2-287, which lacks the aniline substitution at C-2, scored as a weaker inhibitor. DV2-103 which has a tertiary alkyl substituted amine, displayed no inhibition even at 1 μM. Further applications of this assay, including a screen of all 27 pyridopyrimidine compounds from this focused library (Fig. S3) and a screen of 80 kinase inhibitors in the presence of imatinib sensitization, are provided in Supplementary Data (Fig. S4 and Table S1).
Fig. 6.
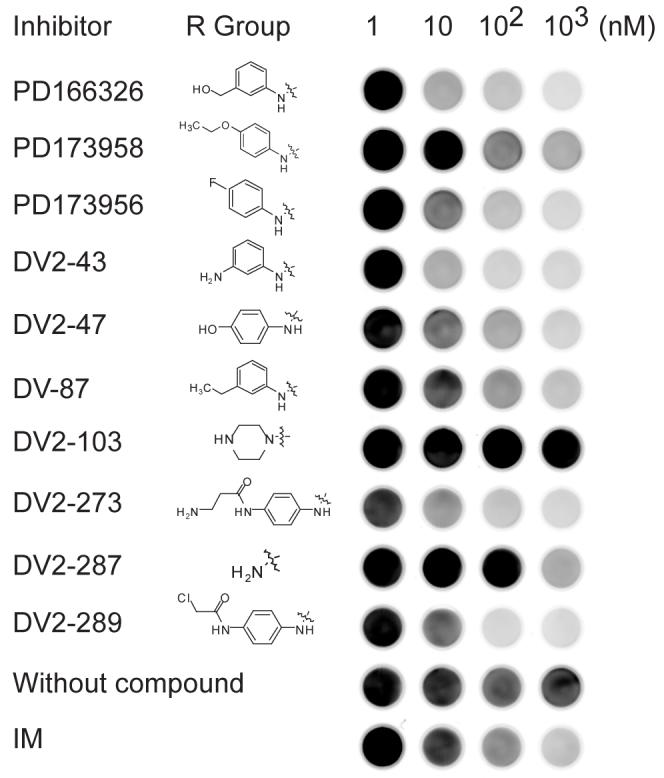
Model structure-activity screen of ten pyridopyrimidine compounds and controls. The kinase assay was performed with K562 lysates using the standard assay conditions with varying concentrations of each inhibitor (1 to 1000 nM).
Discussion
Our strategy for covalently tethering peptide substrates to acrylamide hydrogels offers several advantages. Background due to non-specific protein binding can be a significant confounding factor in antibody-based detection. Acrylamide hydrogels can passivate the glass surface and provide lower non-specific protein adsorption than poly-L-lysine or aldehydic surfaces [36,47]. Immobilization of substrates for solid-phase assays can also present challenges. The attachment chemistry described here is simple and reproducible, allowing straightforward, modular immobilization of peptides in a controlled orientation via cysteine. Multi-functional PEG linkers such as SR415 increased sensitivity by providing higher functionalization density without losing surface passivation.
We demonstrated that adding a recruitment peptide along with substrate enhanced substrate phosphorylation by Bcr-Abl kinase. We also showed that the assay provides a tool for rapidly and reproducibly screening small molecules for Bcr-Abl inhibition. This general approach should be readily adaptable to other protein tyrosine kinases for which sensitive and specific peptide substrates and affinity domain ligands have been identified.
In conclusion, we have demonstrated a kinase assay to measure the activity and inhibition of either Bcr-Abl from a cell extract or recombinant c-Abl using peptides immobilized in a hydrogel-coated 96-well plate. The chemistry employed for linking the peptide substrates to the hydrogel is simple and robust. The practicality of the method should make it useful for analysis of kinase activities in whole cell lysates and characterizing new small molecule tyrosine kinase inhibitors.
Supplementary Material
Acknowledgements
We thank Drs. Ezra Abrams and Jelena Janjic for helpful advice and discussion and Dr. Wendy Stock for kindly providing imatinib. We thank Won Jun Rhee and Stacey L. Kigar for synthesis of the peptides. Funding for this work was provided by NIH grants R33 CA10323 and R01 HG003864. L.L.P. was supported by Kirchstein NRSA F32 CA117672. D.R.V. and B.C. were generously supported by the MeadWestvaco Corporation and Leukemia & Lymphoma Society grant #6034. S.J.K. was a Fletcher Scholar of the Cancer Research Foundation and a Leukemia & Lymphoma Society Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Subject Category: Pharmacological and toxicological research techniques
References
- [1].Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- [2].Ben-Neriah Y, Daley GQ, Mes-Masson AM, Witte ON, Baltimore D. The chronic myelogenous leukemia-specific P210 protein is the product of the bcr/abl hybrid gene. Science. 1986;233:212–214. doi: 10.1126/science.3460176. [DOI] [PubMed] [Google Scholar]
- [3].Pluk H, Dorey K, Superti-Furga G. Autoinhibition of c-Abl. Cell. 2002;108:247–259. doi: 10.1016/s0092-8674(02)00623-2. [DOI] [PubMed] [Google Scholar]
- [4].Sawyers CL. Disabling Abl-perspectives on Abl kinase regulation and cancer therapeutics. Cancer Cell. 2002;1:13–15. doi: 10.1016/s1535-6108(02)00022-3. [DOI] [PubMed] [Google Scholar]
- [5].Warmuth M, Danhauser-Riedl S, Hallek M. Molecular pathogenesis of chronic myeloid leukemia: implications for new therapeutic strategies. Ann Hematol. 1999;78:49–64. doi: 10.1007/s002770050473. [DOI] [PubMed] [Google Scholar]
- [6].Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- [7].Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- [8].Nardi V, Azam M, Daley GQ. Mechanisms and implications of imatinib resistance mutations in BCR-ABL. Curr Opin Hematol. 2004;11:35–43. doi: 10.1097/00062752-200401000-00006. [DOI] [PubMed] [Google Scholar]
- [9].Weisberg E, Manley PW, Breitenstein W, Bruggen J, Cowan-Jacob SW, Ray A, Huntly B, Fabbro D, Fendrich G, Hall-Meyers E, Kung AL, Mestan J, Daley GQ, Callahan L, Catley L, Cavazza C, Azam M, Neuberg D, Wright RD, Gilliland DG, Griffin JD. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- [10].Das J, Chen P, Norris D, Padmanabha R, Lin J, Moquin RV, Shen Z, Cook LS, Doweyko AM, Pitt S, Pang S, Shen DR, Fang Q, de Fex HF, McIntyre KW, Shuster DJ, Gillooly KM, Behnia K, Schieven GL, Wityak J, Barrish JC. 2-aminothiazole as a novel kinase inhibitor template. Structure-activity relationship studies toward the discovery of N-(2-chloro-6-methylphenyl)-2-[[6-[4-(2-hydroxyethyl)-1- piperazinyl)]-2-methyl-4-pyrimidinyl]amino)]-1,3-thiazole-5-carboxamide (dasatinib, BMS-354825) as a potent pan-Src kinase inhibitor. J Med Chem. 2006;49:6819–6832. doi: 10.1021/jm060727j. [DOI] [PubMed] [Google Scholar]
- [11].Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- [12].Tauchi T, Ohyashiki K. The second generation of BCR-ABL tyrosine kinase inhibitors. Int J Hematol. 2006;83:294–300. doi: 10.1532/IJH97.06025. [DOI] [PubMed] [Google Scholar]
- [13].Burgess MR, Sawyers CL. Treating imatinib-resistant leukemia: the next generation targeted therapies. ScientificWorldJournal. 2006;6:918–930. doi: 10.1100/tsw.2006.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cortes J. Overcoming drug resistance in chronic myeloid leukemia. Curr Opin Hematol. 2006;13:79–86. doi: 10.1097/01.moh.0000208468.40991.f2. [DOI] [PubMed] [Google Scholar]
- [15].Walz C, Sattler M. Novel targeted therapies to overcome imatinib mesylate resistance in chronic myeloid leukemia (CML) Crit Rev Oncol Hematol. 2006;57:145–164. doi: 10.1016/j.critrevonc.2005.06.007. [DOI] [PubMed] [Google Scholar]
- [16].Martinelli G, Soverini S, Rosti G, Cilloni D, Baccarani M. New tyrosine kinase inhibitors in chronic myeloid leukemia. Haematologica. 2005;90:534–541. [PubMed] [Google Scholar]
- [17].Martinelli G, Soverini S, Rosti G, Baccarani M. Dual tyrosine kinase inhibitors in chronic myeloid leukemia. Leukemia. 2005;19:1872–1879. doi: 10.1038/sj.leu.2403950. [DOI] [PubMed] [Google Scholar]
- [18].Nagar B, Bornmann WG, Pellicena P, Schindler T, Veach DR, Miller WT, Clarkson B, Kuriyan J. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571) Cancer Res. 2002;62:4236–4243. [PubMed] [Google Scholar]
- [19].Huang M, Dorsey JF, Epling-Burnette PK, Nimmanapalli R, Landowski TH, Mora LB, Niu G, Sinibaldi D, Bai F, Kraker A, Yu H, Moscinski L, Wei S, Djeu J, Dalton WS, Bhalla K, Loughran TP, Wu J, Jove R. Inhibition of Bcr-Abl kinase activity by PD180970 blocks constitutive activation of Stat5 and growth of CML cells. Oncogene. 2002;21:8804–8816. doi: 10.1038/sj.onc.1206028. [DOI] [PubMed] [Google Scholar]
- [20].Huron DR, Gorre ME, Kraker AJ, Sawyers CL, Rosen N, Moasser MM. A novel pyridopyrimidine inhibitor of abl kinase is a picomolar inhibitor of Bcr-abl-driven K562 cells and is effective against STI571-resistant Bcr-abl mutants. Clin Cancer Res. 2003;9:1267–1273. [PubMed] [Google Scholar]
- [21].Wisniewski D, Lambek CL, Liu C, Strife A, Veach DR, Nagar B, Young MA, Schindler T, Bornmann WG, Bertino JR, Kuriyan J, Clarkson B. Characterization of potent inhibitors of the Bcr-Abl and the c-kit receptor tyrosine kinases. Cancer Res. 2002;62:4244–4255. [PubMed] [Google Scholar]
- [22].von Bubnoff N, Veach DR, Miller WT, Li W, Sanger J, Peschel C, Bornmann WG, Clarkson B, Duyster J. Inhibition of wild-type and mutant Bcr-Abl by pyrido-pyrimidine-type small molecule kinase inhibitors. Cancer Res. 2003;63:6395–6404. [PubMed] [Google Scholar]
- [23].Sadick MD, Sliwkowski MX, Nuijens A, Bald L, Chiang N, Lofgren JA, Wong WL. Analysis of heregulin-induced ErbB2 phosphorylation with a high-throughput Kinase receptor activation enzyme-linked immunosorbant assay. Anal Biochem. 1996;235:207–214. doi: 10.1006/abio.1996.0114. [DOI] [PubMed] [Google Scholar]
- [24].Mosier J, Olesen CE, Voyta JC, Bronstein I. Immunoassay protocol for quantitation of protein kinase activities. Methods Enzymol. 2000;305:410–416. doi: 10.1016/s0076-6879(00)05503-8. [DOI] [PubMed] [Google Scholar]
- [25].Schraag B, Staal GE, Adriaansen-Slot SS, Salden M, Rijksen G. Standardization of an enzyme-linked immunosorbent assay for the determination of protein tyrosine kinase activity. Anal Biochem. 1993;211:233–239. doi: 10.1006/abio.1993.1262. [DOI] [PubMed] [Google Scholar]
- [26].Marik J, Lam KS. Peptide and small-molecule microarrays. Methods Mol Biol. 2005;310:217–226. doi: 10.1007/978-1-59259-948-6_15. [DOI] [PubMed] [Google Scholar]
- [27].Reimer U, Reineke U, Schneider-Mergener J. Peptide arrays: from macro to micro. Curr Opin Biotechnol. 2002;13:315–320. doi: 10.1016/s0958-1669(02)00339-7. [DOI] [PubMed] [Google Scholar]
- [28].Schutkowski M, Reineke U, Reimer U. Peptide arrays for kinase profiling. Chembiochem. 2005;6:513–521. doi: 10.1002/cbic.200400314. [DOI] [PubMed] [Google Scholar]
- [29].Cretich M, Damin F, Pirri G, Chiari M. Protein and peptide arrays: recent trends and new directions. Biomol Eng. 2006;23:77–88. doi: 10.1016/j.bioeng.2006.02.001. [DOI] [PubMed] [Google Scholar]
- [30].Fodor SP, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- [31].Reineke U, Volkmer-Engert R, Schneider-Mergener J. Applications of peptide arrays prepared by the SPOT-technology. Curr Opin Biotechnol. 2001;12:59–64. doi: 10.1016/s0958-1669(00)00178-6. [DOI] [PubMed] [Google Scholar]
- [32].MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- [33].Aina OH, Sroka TC, Chen ML, Lam KS. Therapeutic cancer targeting peptides. Biopolymers. 2002;66:184–199. doi: 10.1002/bip.10257. [DOI] [PubMed] [Google Scholar]
- [34].Falsey JR, Renil M, Park S, Li S, Lam KS. Peptide and small molecule microarray for high throughput cell adhesion and functional assays. Bioconjug Chem. 2001;12:346–353. doi: 10.1021/bc000141q. [DOI] [PubMed] [Google Scholar]
- [35].Houseman BT, Huh JH, Kron SJ, Mrksich M. Peptide chips for the quantitative evaluation of protein kinase activity. Nat Biotechnol. 2002;20:270–274. doi: 10.1038/nbt0302-270. [DOI] [PubMed] [Google Scholar]
- [36].Brueggemeier SB, Wu D, Kron SJ, Palecek SP. Protein-acrylamide copolymer hydrogels for array-based detection of tyrosine kinase activity from cell lysates. Biomacromolecules. 2005;6:2765–2775. doi: 10.1021/bm050257v. [DOI] [PubMed] [Google Scholar]
- [37].Songyang Z, Carraway KL, 3rd, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, et al. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- [38].Uphoff CC, Habig S, Fombonne S, Matsuo Y, Drexler HG. ABL-BCR expression in BCR-ABL-positive human leukemia cell lines. Leuk Res. 1999;23:1055–1060. doi: 10.1016/s0145-2126(99)00131-9. [DOI] [PubMed] [Google Scholar]
- [39].Klutchko SR, Hamby JM, Boschelli DH, Wu Z, Kraker AJ, Amar AM, Hartl BG, Shen C, Klohs WD, Steinkampf RW, Driscoll DL, Nelson JM, Elliott WL, Roberts BJ, Stoner CL, Vincent PW, Dykes DJ, Panek RL, Lu GH, Major TC, Dahring TK, Hallak H, Bradford LA, Showalter HD, Doherty AM. 2-Substituted aminopyrido[2,3-d]pyrimidin-7(8H)-ones. structure-activity relationships against selected tyrosine kinases and in vitro and in vivo anticancer activity. J Med Chem. 1998;41:3276–3292. doi: 10.1021/jm9802259. [DOI] [PubMed] [Google Scholar]
- [40].Boschelli DH, Wu Z, Klutchko SR, Showalter HD, Hamby JM, Lu GH, Major TC, Dahring TK, Batley B, Panek RL, Keiser J, Hartl BG, Kraker AJ, Klohs WD, Roberts BJ, Patmore S, Elliott WL, Steinkampf R, Bradford LA, Hallak H, Doherty AM. Synthesis and tyrosine kinase inhibitory activity of a series of 2-amino-8H-pyrido[2,3-d]pyrimidines: identification of potent, selective platelet-derived growth factor receptor tyrosine kinase inhibitors. J Med Chem. 1998;41:4365–4377. doi: 10.1021/jm980398y. [DOI] [PubMed] [Google Scholar]
- [41].Abrams ES, Zhang T, MIelewczyk S, Patterson BC. Methods of immobilizing ligands on solid supports. 6292118. U.S. Patent. 2000 Aug 28;
- [42].Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- [43].Pisabarro MT, Serrano L. Rational design of specific high-affinity peptide ligands for the Abl-SH3 domain. Biochemistry. 1996;35:10634–10640. doi: 10.1021/bi960203t. [DOI] [PubMed] [Google Scholar]
- [44].Sun X, Layton JE, Elefanty A, Lieschke GJ. Comparison of effects of the tyrosine kinase inhibitors AG957, AG490, and STI571 on BCR-ABL--expressing cells, demonstrating synergy between AG490 and STI571. Blood. 2001;97:2008–2015. doi: 10.1182/blood.v97.7.2008. [DOI] [PubMed] [Google Scholar]
- [45].Trumpp-Kallmeyer S, Rubin JR, Humblet C, Hamby JM, Hollis HD. Showalter, Development of a binding model to protein tyrosine kinases for substituted pyrido[2,3-d]pyrimidine inhibitors. J Med Chem. 1998;41:1752–1763. doi: 10.1021/jm970634p. [DOI] [PubMed] [Google Scholar]
- [46].Hamby JM, Connolly CJ, Schroeder MC, Winters RT, Showalter HD, Panek RL, Major TC, Olsewski B, Ryan MJ, Dahring T, Lu GH, Keiser J, Amar A, Shen C, Kraker AJ, Slintak V, Nelson JM, Fry DW, Bradford L, Hallak H, Doherty AM. Structure-activity relationships for a novel series of pyrido[2,3-d]pyrimidine tyrosine kinase inhibitors. J Med Chem. 1997;40:2296–2303. doi: 10.1021/jm970367n. [DOI] [PubMed] [Google Scholar]
- [47].Suzawa T, Shirahama H. Adsorption of plasma proteins onto polymer latices. Adv Colloid Interface Sci. 1991;35:139–172. doi: 10.1016/0001-8686(91)80021-b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.