Abstract
Transcription consists of a series of highly regulated steps: assembly of the preinitiation complex (PIC) at the promoter, initiation, elongation, and termination. PIC assembly is nucleated by TFIID, a complex composed of the TATA-binding protein (TBP) and a series of TBP-associated factors (TAFs). One component, TAF7, is incorporated in the PIC through its interaction with TFIID but is released from TFIID upon transcription initiation. We now report that TAF7 interacts with the transcription factors, TFIIH and P-TEFb, resulting in the inhibition of their Pol II CTD kinase activities. Importantly, in in vitro transcription reactions, TAF7 inhibits steps after PIC assembly and formation of the first phosphodiester bonds. Further, in vivo TAF7 coelongates with P-TEFb and Pol II downstream of the promoter. We propose a model in which TAF7 contributes to the regulation of the transition from PIC assembly to initiation and elongation.
Keywords: MHC class I genes, regulation, transcription initiation
In eukaryotic cells, expression of protein-encoding genes depends on the ordered recruitment of the general transcription factors (GTF) TFIID, TFIIB, and TFIIA to the promoter, followed by association of Pol II, the mediator, and the remaining GTFs, TFIIF, TFIIE, and TFIIH, to form a preinitiation complex (PIC) (1). Once PIC assembly is complete, transcription initiation ensues; Pol II with the elongation complex dissociate from the PIC (2, 3). A required step during transcription initiation is the phosphorylation of serine 5 in the carboxy terminal domain (CTD) heptad repeat of Pol II by the kinase subunit of TFIIH, CDK7 (4, 5), after which Pol II pauses to ensure proper pre-mRNA capping (6–9). The transition from pausing to elongation is facilitated by the P-TEFb elongation complex, which also mediates efficient elongation (10). P-TEFb consists of two subunits, cyclin T1 and the kinase CDK9, which phosphorylates serine 2 of the CTD, required for productive elongation and the recruitment of complexes involved in mRNA processing (splicing and polyadenylation) (10–15).
Although the general mechanics of transcription have been characterized, relatively little is known about how the transitions from PIC assembly to initiation/pausing to elongation are regulated. Promoter recognition is largely mediated by TFIID, which is composed of the TATA binding protein (TBP) and over a dozen TBP associated factors (TAFs) (16, 17, 18). The largest TFIID component, TAF1, has both acetyltransferase (AT) and kinase activities (19, 20). We demonstrated that TAF1 and its intrinsic acetyltransferase activity are essential for transcription of an MHC class I gene (21). Importantly, MHC class I transcription is inhibited both in vitro and in vivo by the viral transactivator, HIV Tat, which binds to the TAF1 AT domain, inhibiting its enzymatic activity (22, 23). TAF7, a cellular 55-kDa TFIID component (24, 25), also binds to TAF1 inhibiting its AT activity and repressing MHC class I transcription (26) Significantly, we have demonstrated that TAF7 remains bound to TAF1/TFIID until PIC assembly is complete, whereupon it is released enabling transcription initiation and elongation (27). Thus, TAF7 is an intrinsic regulator of transcription.
These studies were designed to determine the fate of TAF7 after its dissociation from the PIC. We report that TAF7 functionally interacts with both the general transcription factor TFIIH and the elongation factor P-TEFb. Association of TAF7 with TFIIH inhibits its CDK7 kinase thereby inhibiting TFIIH-mediated phosphorylation of the Pol II CTD Ser-5; binding of TAF7 to the P-TEFb elongation complex inhibits its CDK9-mediated phosphorylation of Pol II CTD Ser-2. Importantly, we show that TAF7 functions in vitro to inhibit transcription at steps after PIC assembly and in vivo colocalizes with P-TEFb and Pol II downstream of the promoter. Thus, in addition to its role in transcription initiation as a TFIID component, TAF7 also functions in the transition from PIC assembly to initiation and elongation. We propose a model in which TAF7 regulates the orderly progression of events in transcription, preventing transcription elongation until the steps of transcription initiation are completed and the transcription elongation complex (TEC) is fully assembled.
Results
TAF7 Functionally Interacts with TFIIH and P-TEFb and Inhibits Their Phosphorylation of the Pol II CTD.
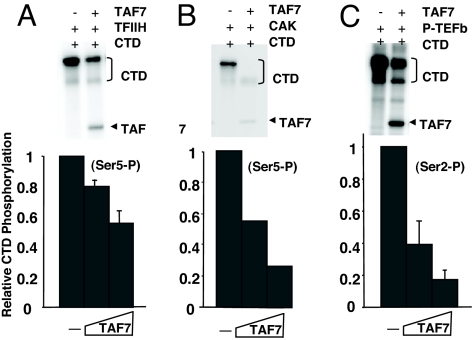
We demonstrated that TAF7 is released from the TFIID complex upon transcription initiation (27). The present studies were designed to determine whether TAF7 associates with other components of the transcription complex after its dissociation from TFIID. Because TAF7 was released from a partially assembled preinitiation complex with TFIIB, TFIIF, and Pol II (27), none of these factors is likely to be a TAF7 interacting partner. Therefore, we considered the possibility that TAF7 functionally interacts with TFIIH and/or P-TEFb (7). To assess the functional interaction of TAF7 with TFIIH or P-TEFb, its effect on their Pol II CTD kinase activities was determined. TAF7 inhibited the CTD kinase activities of TFIIH (Fig. 1A). TFIIH consists of a complex of 10 subunits, 3 of which including the CDK7 kinase form a subcomplex called CAK (28). TAF7 inhibited the phosphorylation of the Pol II CTD by the CAK complex, demonstrating that TAF7 acts directly on CAK (Fig. 1B). Remarkably, TAF7 also inhibited P-TEFb phosphorylation of the Pol II Ser-2 CTD (Fig. 1C). In contrast, recombinant CREB has no effect on CDK9 kinase phosphorylation of the CTD (29) [supporting information (SI) Fig. S1D]. Thus, TAF7 functionally interacts with both TFIIH and P-TEFb, inhibiting their respective CTD kinase activities.
Fig. 1.
TAF7 inhibits the kinase activities of TFIIH and P-TEFb. (A) TAF7 inhibits TFIIH-mediated phosphorylation of the Pol II CTD Ser-5. (Upper) Purified TFIIH and 100 ng of GST-tagged Pol II CTD were incubated in the presence or absence of recombinant Flag-tagged TAF7 (F-TAF7) in an in vitro kinase assay using [32P]ATP. Phosphorylation was determined by autoradiography after gel electrophoresis; positions of TAF7 and CTD are indicated based on mobility markers (data not shown). (Lower) Purified TFIIH and 100 ng of GST-tagged Pol II CTD were incubated in the presence of increasing amount of recombinant F-TAF7 (125 and 250 ng), in an in vitro kinase assay. Ser-5 CTD phosphorylation was determined by Western blot analysis, using H14 anti-phosphoSer5 antibody, and quantified by densitometry. (B) TAF7 inhibits the CAK-mediated phosphorylation of the Pol II CTD. (Upper) In vitro kinase assay with purified CAK, 100 ng of GST-tagged Pol II CTD, and F-TAF7 as in A Upper. (Lower) In vitro kinase assay with purified CAK, 100 ng of GST-tagged Pol II CTD, and increasing F-TAF7 as in A Lower. (C) TAF7 inhibits P-TEFb-mediated phosphorylation of the Pol II CTD Ser-2. (Upper) In vitro kinase assay with 50 ng of purified P-TEFb, 100 ng of GST-tagged Pol II CTD, and F-TAF7 as in A Upper. (Lower) In vitro kinase assay with 50 ng of P-TEFb, 100 ng of GST-tagged Pol II CTD, and increasing F-TAF7 as in A Lower. CTD Ser-2 phosphorylation was determined by Western blot analysis, using H5 anti-phosphoSer2 antibody.
Because both TFIIH and P-TEFb also phosphorylate TAF7 (Fig. 1), we considered the possibility that TAF7 acts as a competitive inhibitor. However, phosphorylation of SPT5, another P-TEFb substrate (30), is enhanced in the presence of TAF7 (Fig. S1A). Furthermore, TAF7 does not inhibit the activity of CDK2, another CDK family member (Fig. S1B). Finally, neither CTD nor TAF7 alone has any kinase activity (Fig. S1C). Thus, TAF7 inhibition of TFIIH and P-TEFb CTD kinase activities is specific. Thus, TAF7 regulates TAF1 acetyl transferase activity and function and the CTD kinase activities of both TFIIH and P-TEFb.
TAF7 Physically Interacts with P-TEFb.
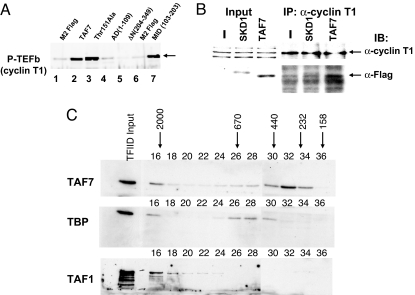
The experiments described above demonstrate a functional association between TAF7 and both TFIIH and P-TEFb. To examine the physical interactions, we performed in vitro pull-down assays with recombinant Flag-tagged TAF7 and either purified P-TEFb or purified TFIIH. A weak association of TAF7 with the CAK subunit of TFIIH was observed in the pull-down assay (data not shown). TAF7 efficiently recovered P-TEFb as revealed by anti-cyclin T1 immunoblotting, indicating that TAF7 stably interacts with P-TEFb in vitro (Fig. 2A; compare lanes1 and 2). Similar results were obtained in an anti-CDK9 immunoblot (data not shown). The interaction domain of TAF7 with P-TEFb was mapped using a set of deletion mutants to the central 103- to 203-aa segment of TAF7 [MID (103–203)] (Fig. 2A, compare lanes 6 and 7), which is within the same domain that interacts with TAF1 (24). However, mutation of Thr-151 in the TAF1 binding domain (24) has no effect on TAF7 binding to P-TEFb. Thus, TAF7 binds P-TEFb in vitro.
Fig. 2.
TAF7 interacts with P-TEFb in vitro and in vivo and exists in a TFIID-independent complex. (A) TAF7 binds to P-TEFb through its central domain in in vitro pull-down assays. Purified recombinant P-TEFb (200 ng) was incubated with F-TAF7 or each of the F-TAF7 mutants immobilized on Sepharose beads. TAF7-bound fractions were assayed for retention of P-TEFb by immunoblotting, using anti-cyclin T1 antibody. (B) TAF7 binds to P-TEFb in vivo. Extracts from cells transiently transfected with either F-TAF7, pcDNA3, or Flag-SKD1, an irrelevant control protein, were immunoprecipitated with anti-cyclin T1 antibody. Immunoprecipitates were analyzed for cyclin T1 (Upper) or F-TAF7 (Lower) by Western blot. (Upper) Anti-cyclin T1. Total extract from cells transfected with vector (lane 1), Flag-SKD1 (lane 2), or F-TAF7 (lane 3) before immunoprecipitation and immunoprecipitated cyclin T1 (lanes 4–6). (Lower) Anti-Flag M2 antibody, detection of F-TAF7 coimmunoprecipitated from extracts of cells transfected with vector control (lane 4), Flag-SKD1 (lane 5), or F-TAF7 expression vector (lane 6). (C) TAF7 exists in a TFIID-independent form. C8166 whole cell extracts were fractionated on an FPLC column and fractions analyzed by Western blotting as indicated. Arrows refer to the indicated molecular sizes. Purified TFIID was included as a marker. TFIID consistently appears in the excluded volume in column fractionation of whole cell extracts. Fractions 16–28 and 30–36 were run in parallel but on two separate SDS/PAGEs.
The in vivo interaction of TAF7 with P-TEFb was determined by immunoprecipitation after transfection of Flag-TAF7 into 293 cells. Consistent with the in vitro findings, immunoprecipitation of endogenous cyclin T1 coimmunoprecipitated TAF7 from Flag-TAF7-transfected 293 extracts but not control protein from control extracts (Fig. 2B Lower, lanes 5 and 6).
The observed TAF7 inhibition of P-TEFb and TFIIH kinase activities predicted that TAF7 exists in a TFIID-independent form in vivo. To test this prediction, native cell extracts were fractionated by FPLC gel filtration and assayed for the elution pattern of TAF7, TBP, and TAF1 (Fig. 2C). As expected, TFIID components (TBP, TAF1, and TAF7) were detected in high molecular weight fractions (fractions 16–18); a second peak of TBP was detected in fractions 26–30, corresponding to free TFIID. Importantly, TAF7 eluted in a distinct peak in fractions corresponding to molecular mass between 230 and 440 kDa (fractions 30–32). Thus, TAF7 is found in a form independent of TFIID. Detecting the 55-kDa TAF7 in fractions corresponding to 230 kDa suggests that it is in a complex with other proteins. A TFIID-independent TAF7 fraction was also observed in MOLT4 whole cell extracts (Fig. S2). Whether TAF7 is associated with cyclin T1 or CDK9 or other proteins in the 230-kDa fraction remains to be determined.
These experiments provide the first demonstration that TAF7 exists in at least two distinct forms in vivo—a TFIID-independent form and the well known TFIID complex.
TAF7 Regulates Both PIC Assembly and Transcription Initiation in Vitro.
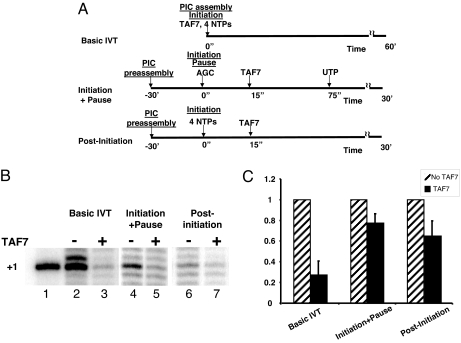
TAF1, TFIIH and P-TEFb are critical for PIC assembly, transcription initiation, and elongation. We have shown that TAF7 functionally interacts with all three. Because TFIIH and P-TEFb are involved in transcription initiation and elongation, we postulated that TAF7 functions in the regulation of transcription at steps after PIC assembly. To address this question, in vitro transcription assays were performed in which TAF7 was added at one of three times: before PIC assembly, at transcription initiation, or after initiation (schematized in Fig. 3A). The presence of TAF7 during the entire in vitro transcription assay is predicted to block transcription at all steps of transcription initiation: inhibition of TAF1 AT activity and of TFIIH and P-TEFb kinase activities (Basic IVT). Indeed, addition of TAF7 before PIC assembly significantly reduced transcription (Fig. 3B, basic IVT, lanes 2 and 3). Transcriptional repression in the presence of TAF7 is specific; addition of irrelevant proteins to the in vitro transcription reaction had no effect (Fig. S3).
Fig. 3.
TAF7 regulates multiple distinct steps in transcription initiation. (A) Scheme of experimental protocol. Basic IVT, TAF7, and the four NTPs are present during PIC assembly and initiation; Initiation+Pause, PIC preassembly occurs in the absence of NTPs or TAF7. TAF7 is added after transcription is initiated with AGC; UTP is added at 75 sec to permit clearance and elongation. Post-Initiation, PIC preassembly occurs in the absence of NTPs or TAF7, which is added after transcription is initiated with all four NTPs. (B) TAF7 regulates in vitro transcription. Lane 1, marker; lanes 2–3, HeLa nuclear extract in the presence or absence of purified TAF7 preincubated for 15 min in presence of rNTPs. Transcription was initiated by the addition of the MHC class I −313CAT plasmid. In lanes 4–7, −313CAT DNA and extracts were preincubated for 30 min in presence of 40 μM ATP to permit PIC assembly. Transcription was initiated by addition of ATP/CTP/GTP mix (lanes 4 and 5) or rNTP (lanes 6 and 7). Control TKEG buffer (−) or purified F-TAF7 (+) were added 15 seconds after initiation of transcription and incubated for 30min. Analysis of −313CAT transcripts was by primer extension. [In previous studies (27), addition of TAF7 was after promoter clearance so did not affect transcription.] (C) Quantitation of results of three experiments, showing averages and SEM. Addition of TAF7 in each of these conditions results in significant inhibition of transcription: IVT, P < 0.003; pause, P < 0.03; initiation, P < 0.04.
The effect of TAF7 on transcription after initiation was determined as follows: After PIC assembly, transcription was initiated by the addition of ATP, GTP, and CTP to allow the incorporation of the first three nucleotides in the nascent RNA and pausing of Pol II. (The sequence at the transcription start site is gctcAGCTTCT; transcription initiates at A). Then, TAF7 was added, followed by UTP to allow promoter clearance and elongation (initiation plus pause). Consistent with a role for TAF7 after initiation of transcription, TAF7 significantly reduced transcription even when added after phosphodiester bond formation (Fig. 3B, Initiation+Pause, lanes 4 and 5). Finally, to examine the effect of TAF7 after promoter clearance, transcription was initiated with the 4NTPs after PIC assembly, followed by addition of TAF7 (Fig. 3A, Post-Initiation). Even added after PIC assembly and promoter clearance, TAF7 is still able to significantly reduce transcription (Fig. 3B, lanes 6 and 7). The results of three independent experiments are summarized in Fig. 3C. The effects of TAF7 after transcription initiation, although modest, are reproducible and significant. Thus, TAF7 functionally targets components of the transcription machinery other than TAF1/TFIID, consistent with a functional role for its inhibition of the kinase activities of TFIIH and P-TEFb.
TAF7 Regulates Promoter Activity in Vivo.
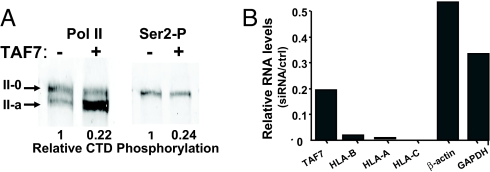
The biological implications of TAF7 inhibition of Pol II CTD phosphorylation were examined in vivo in 293T cells stably transfected with TAF7 (293/F-TAF7). Both TAF7 protein and RNA levels were increased in the 293/F-TAF7 cells relative to control 293 cells (Fig. S4 A and B). Importantly, the fraction of total phosphorylated Pol II species was markedly reduced in the 293/F-TAF7 cells relative to controls (Fig. 4A; compare two left lanes); CTD Ser-2 Pol II was also reduced by 75% in the TAF7-transfected cells (Fig. 4A; compare Ser-2 in Right to total Pol II in Left). These results clearly demonstrate that over-expression of TAF7 leads to inhibition of CTD phosphorylation in vivo and in vitro.
Fig. 4.
TAF7 regulates promoter activity in vivo. (A) TAF7 inhibits Pol II CTD phosphorylation in vivo. Nuclear extracts from either 293/F-TAF7 (+) or control 293T (−) cells were analyzed by Western blotting with either an antibody specific for the Ser-2 CTD phosphorylated form of pol II (Right) or an antibody reactive with all forms of Pol II CTD (Left). (Quantitation) (Left) Phosphorylated Pol II (II-0) is expressed relative to nonphosphorylated Pol II (II-a). (Right) The amount of Ser-2-P is expressed relative to the total amount of Pol II in the same sample (Left). (B) Depletion of TAF7 results in reduced levels of endogenous MHC class I RNA. Human 293T cells were transiently transfected with TAF7 siRNA or scrambled siRNA, as described; levels of TAF7, HLA class I RNAs, actin, and GAPDH RNAs were determined by real time PCR. Results are presented relative to control siRNA.
Because TAF7 regulates the functions of TAF1, TFIIH and P-TEFb and has a major effect on MHC class I expression in vitro (Fig. 3) (26, 27), we predicted that both overexpression and underexpression of TAF7 would perturb transcription in vivo: Overexpression would sustain inhibition of TAF1, TFIIH, and P-TEFb functions, whereas under-expression would allow premature initiation and promoter clearance, resulting in abortive transcripts. In either case, MHC class I expression would be reduced. Indeed, overexpression of TAF7 decreased MHC class I promoter activity in a transient transfection assay (Fig. S5). To determine whether ablated expression of TAF7 affected transcription, human 293T cells were transiently transfected with TAF7 siRNA, resulting in a reduction of TAF7 RNA to 19% of the control level (Fig. 4B). As predicted, RNA levels of the endogenous MHC class I genes HLA-A,B,C were dramatically reduced after TAF7 siRNA treatment (Fig. 4B); siRNA to a control transcript did not affect HLA-A RNA levels (Fig. S6B). A TAF7 shRNA construct stably introduced into HeLa cells similarly decreased TAF7, HLA-A, and HLA-C RNA levels (Fig. S6). Because the siRNA and shRNA sequences and the transfected cell lines were different, the observed effects of TAF7 knock-down on HLA class I RNA levels are unlikely due to off-target effects. Taken together, these findings demonstrate that TAF7 actively contributes to regulating transcription of MHC class I genes in vivo.
Interestingly, the depletion of TAF7 did not result in a global inhibition of transcription, because the levels of actin and GAPDH were only modestly reduced, suggesting that TAF7 regulates a subset of mammalian promoters, consistent with the observation in yeast that loss of TAF7 affects only a subset of genes (31).
TAF7 Plays a Critical Role in Cell Viability and Growth.
Because transcription of many cell cycle genes depends on TFIID, we considered the possibility that TAF7 levels may play a critical role in cell viability and growth. As assessed by limiting dilution analysis in HeLa tet-off cells stably transfected with an inducible TAF7 shRNA, TAF7 depletion reduced both cell viability and growth (Table 1). Thus, TAF7 contributes to the regulation of sustained cell viability and growth.
Table 1.
TAF7 deficiency compromises cell growth
| Construct | Absence of colonies, % seeded wells | Avg cells per well, no. |
|---|---|---|
| Control | 4.2 | 21.8 ± 1.9 |
| shRNA1 | 22.9 | 10.6 ± 1.6, P = 1.4 × 10−4 |
| shRNA2 | 17.8 | NT |
Human 293 cells, stably transfected with two different TAF7 shRNA retroviral vectors, shRNA1 or shRNA2, or the control vector, were seeded into 100 microtitre wells at an average density of five cells per well. After TAF7 shRNA induction, the number of wells with colonies and the size of each colony were determined by microscopic examination. In the absence of induction of TAF7 shRNA, transfectant growth was comparable to a control cell line transfected with an empty vector (data not shown).
TAF7 Is Associated with Both the PIC and Transcription Initiation/Elongation Complex in Vivo.
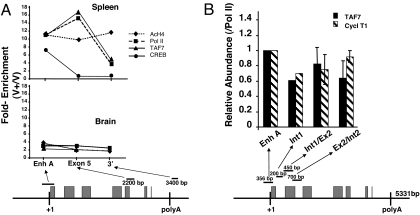
Because TAF 7 interacts with P-TEFb, an elongation factor that moves with the elongation complex, we postulated that, in vivo, TAF7 would be associated with coding regions of the gene. To test this prediction, we examined the in situ localization of TAF7 by in vivo ChIP assays of the class I gene in spleen and brain (Fig. 5). These two tissues differ in MHC class I expression by >2 orders of magnitude (32). In the high-expressing spleen, TAF7 was associated with both the promoter and downstream sequences as far as exon 5 (Fig. 5A). The extent of TAF7 association paralleled that of Pol II. In contrast, the transcription factors CREB and c-jun were found only at the promoter (Fig. 5A and data not shown). Consistent with the high level of transcription of the class I gene in spleen, acetylated H4 histone—a marker of active chromatin—was detected throughout the gene. In brain, where class I transcription is barely detectable, the levels of TAF7, Pol II, and acetylated H4 histone were correspondingly low. No TAF7 was detected either to sequences beyond the poly(A) addition site or to sequences 700 bp 5′ to the promoter in either tissue (data not shown). Thus, association of TAF7 with the promoter and coding sequences is specific and dependent on active transcription.
Fig. 5.
TAF7 remains associated with the transcription elongation complex in vivo. (A) TAF7 is associated with MHC class I coding sequences in spleen but not brain. A representative ChIP assay out of four performed by using chromatin from spleen cells and brain of mice transgenic for the pig MHC class I gene, PD1 (B10.PD1). The fold enrichments of TAF7, Pol II, AcHistone H4, and CREB were calculated for different regions of the PD1 gene as described in Methods. The PD1 gene segments analyzed and primer sets in the ChIP assays are shown diagrammatically. Boxes indicate exons. PCR fragments are nonoverlapping. (B) TAF7 and Cyclin T1 remain associated with Pol II through exon 2 of the class I gene. ChIP assays were performed on B10.PD1 spleen cells. The amounts of TAF7 or cyclin T1 were determined relative to Pol II for different regions of the PD1 gene and are expressed as “relative abundance”: the fold enrichment (V+/V−) of TAF7 and cyclin T1 relative to the fold enrichment of Pol II, to normalize for variations in efficiency of PCR amplification of primer sets across the gene. The PD1 gene segments analyzed and primer sets in the ChIP assays are shown diagrammatically. Gray boxes indicate exons. PCR fragments are nonoverlapping. The graph represents the composite of three independent experiments.
To examine the relationship between TAF7 and the elongation complex, the correlation between TAF7 and P-TEFb migration along the gene was determined by ChIP analysis of spleen cells with antibodies to TAF7, cyclin T1, and Pol II. Because the abundance of the elongation complex varies along the gene, the abundances of TAF7 and cyclin T1 were normalized to the level of Pol II association at each point along the gene. This normalization also corrects for differences between the antibodies and efficiency of PCR primers at the various positions within the gene. As shown in Fig. 5B, the relative abundance of TAF7 paralleled that of cyclin T1 through the exon 2/intron 2 junction. Thus, the occurrence of TAF7 within the class I coding sequences correlates with the abundance of Pol II and the level of transcription. From these experiments, we conclude that TAF7 is associated with initiation/elongation complexes in situ in tissues.
Discussion
Transcription depends on the temporal and spatial integration of preinitiation complex assembly, initiation, elongation, and termination. Regulation of the transitions from PIC assembly to initiation to elongation is necessary to assure proper rates of productive transcription. Here, we report that the TFIID component, TAF7, functionally associates with other components of the transcription machinery both in vitro and in vivo. We demonstrate that TAF7 interacts with both the general transcription factor TFIIH and the elongation factor P-TEFb, inhibiting phosphorylation of the Pol II CTD. Importantly, as a presumed functional consequence of its modulation of the CTD kinase activities of TFIIH/CDK7 and P-TEFb/CDK9, TAF7 regulates transcription subsequent to PIC assembly and initiation (i.e., promoter clearance and elongation). Finally, we demonstrate that TAF7 regulates MHC class I expression in vivo and is associated with MHC class I DNA coding sequences. These studies demonstrate that TAF7 plays a critical role in regulating distinct steps in transcription initiation.
We propose a model in which TAF7 contributes to the regulation of early transitions in transcription (Fig. S7) as follows. As a component of TFIID, TAF7 inhibits the TAF1 AT activity until PIC assembly is complete, thereby preventing premature initiation (27). Upon completion of PIC assembly, TAF7 is released from TAF1, allowing transcription to initiate with the first phosphodiester bond formation. TAF7 inhibition of TFIIH phosphorylation of Ser-5 CTD would delay 5′ cap formation and the recruitment of CTD binding factors until formation of the first phosphodiester bonds and separation of the nascent elongation complex from the PIC, thus effectively pausing initiation. This model is consistent with the known pausing of Pol II ≈20–60 bp downstream of the transcription start site. For genes, like the MHC class I gene, whose expression is dynamically regulated by extrinsic signaling events (i.e., hormonal or cytokine), Pol II pausing could provide a mechanism to regulate the rate of transcription, either by modulating the length of pausing or by maintaining the promoter in an open conformation that is “poised” for rapid response but not actively transcribed (33).
Release of TAF7 from TFIIH/CDK7 would overcome pausing, whereas its inhibition of P-TEFb phosphorylation of Ser-2 CTD would delay elongation to allow recruitment of CTD-associated complexes necessary for productive elongation. Overcoming pausing and resumption of elongation are known to require activation by additional factors, such as NELF, TFIIF, Elongins, and members of the ELL family (7). The finding that TAF7 does not affect P-TEFb phosphorylation of Spt5 raises the intriguing possibility that TAF7 functions to alter the substrate specificity of P-TEFb.
In vivo, TAF7 remains associated with the elongation complex within MHC class I coding sequences, suggesting that, like TFIIF, it may modulate the late pauses of transcription (2). The extent of TAF7 association with the class I gene in different tissues from the same animal parallels the extent of association of Pol II and the level of class I expression in those tissues.
Although the multiplicity of TAF7 interactions we have described are unusual, they are not unique. A well documented example, with a strikingly similar pattern of interactions, is the HIV transactivator protein Tat. Like TAF7, Tat (i) binds TAF1 and inhibits its acetyl transferase activity (23), (ii) binds P-TEFb and activates its autophosphorylation (34), and (iii) modulates the phosphorylation of RNAP CTD by TFIIH and P-TEFb (35). Although there is no structural similarity between these two proteins, the remarkable parallels between TAF7 and Tat function suggest that Tat has subverted the cellular processes normally regulated by TAF7.
In conclusion, the studies reported here define important new roles for TAF7 in regulating the transition from PIC assembly to productive elongation and demonstrate that transcription factor complexes, such as TFIID, TFIIH, and P-TEFb, are not static entities but rather dynamic ones with constituent proteins entering and leaving at various points during the transcription process.
Methods
Kinase activities were measured with Flag-tagged TAF7 (125 or 250 ng) in the presence of 100-ng GST-Pol II CTD, 10 μCi of [32P] μATP (6,000 Ci/mM) in 20 μl of 50 mM Tris (pH 6.8 for P-TEFb, pH 7.9 for TFIIH), 5 mM DTT, 10 μM ZnSO4, 5 mM MnCl, 4 mM MgCl2 and 1 mM ATP. In vitro transcription assays, immunoprecipitations, and Western blot anlyses were performed as described in ref. 27. Column fractionation of C8166 T whole cells extracts was on a Superose 6 column in 150 mM Hepes (pH 7.9), 1.5 mM MgCl2, 1 mM EGTA (pH 8), 10% glycerol, 150 mM NaCl, 10 mM B-glycerophosphate, 1 mM DTT, 50 mM NaF, 0.2 mM sodium vanadate, and protease inhibitors. TAF7 siRNA transient transfections were performed with Hiperfect (Qiagen) according to the manufacturer's protocol. siRNA duplexes specifically targeting TAF7 expression (Dharmacon), a control target (AP2 RNA sequence GUGGAUGCCUUUCGGGUCA), and a scrambled duplex control (Dharmacon; catalog no. D-001810-10-05) were transfected in parallel experiments to a final concentration of 25 nM. ChIP assays were performed using a modification of the method of Oberley and Farnham (36) on tissues from the B10.PD1 transgenic mouse strain (32). Further experimental details can be found in SI Methods.
Acknowledgments.
We thank Dr. Greg Hannon (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) for providing TAF7 shRNA constructs and helpful discussions; Drs. Danny Reinberg (New York University School of Medicine, New York, NY), David Levens (National Cancer Institute, Bethesda, MD), and Hui Ge (Protein One, College Park, MD) for providing TFIIH and CAK; Drs. Kevin Howcroft, Helit Cohen, Aparna Kotekar, and Steve Shaw for helpful discussions; and Drs. Brian Lewis, David Levens, Keji Zhao for helpful discussions and critical review of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801637105/DCSupplemental.
References
- 1.Woychik NA, Hampsey M. The RNA polymerase II machinery: Structure illuminates function. Cell. 2002;108:453–463. doi: 10.1016/s0092-8674(02)00646-3. [DOI] [PubMed] [Google Scholar]
- 2.Zawel L, Kumar KP, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 3.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz BE, Larochelle S, Suter B, Lis JT. Cdk7 is required for full activation of Drosophila heat shock genes and RNA polymerase II phosphorylation in vivo. Mol Cell Biol. 2003;23:6876–6886. doi: 10.1128/MCB.23.19.6876-6886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Sims RJ, III, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: The short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 8.Ho CK, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 9.Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou M, et al. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol. 2000;20:5077–5086. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 12.Peng J, Zhu Y, Milton JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carty SM, Greenleaf AL. Hyperphosphorylated C-terminal repeat domain-associating proteins in the nuclear proteome link transcription to DNA/chromatin modification and RNA processing. Mol Cell Proteomics. 2002;1:598–610. doi: 10.1074/mcp.m200029-mcp200. [DOI] [PubMed] [Google Scholar]
- 14.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol Cell. 2004;13:55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- 16.Goodrich JA, Tjian R. TBP-TAF complexes: Selectivity factors for eukaryotic transcription. Curr Opin Cell Biol. 1994;6:403–409. doi: 10.1016/0955-0674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 17.Hahn S. The role of TAFs in RNA polymerase II transcription. Cell. 1998;95:579–582. doi: 10.1016/s0092-8674(00)81625-6. [DOI] [PubMed] [Google Scholar]
- 18.Muller F, Demeny MA, Tora L. New problems in RNA polymerase II transcription initiation: Matching the diversity of core promoters with a variety of promoter recognition factors. J Biol Chem. 2007;282:14685–14689. doi: 10.1074/jbc.R700012200. [DOI] [PubMed] [Google Scholar]
- 19.Dikstein R, Ruppert S, Tjian R. TAFII250 is a bipartite protein kinase that phosphorylates the base transcription factor RAP74. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 20.Mizzen CA, et al. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 21.Weissman JD, Howcroft TK, Singer DS. TAF(II)250-independent transcription can be conferred on a TAF(II)250-dependent basal promoter by upstream activators. J Biol Chem. 2000;275:10160–10167. doi: 10.1074/jbc.275.14.10160. [DOI] [PubMed] [Google Scholar]
- 22.Howcroft TK, Strebel K, Martin M, Singer DS. Repression of MHC class I gene promoter activity by two-exon tat of HIV. Science. 1993;260:91–93. doi: 10.1126/science.8493575. [DOI] [PubMed] [Google Scholar]
- 23.Weissman J, et al. HIV-1 Tat binds TAFII250 and represses TAFII250-dependent transcription of MHC class I genes. Proc Natl Acad Sci. 1998;95:11601–11606. doi: 10.1073/pnas.95.20.11601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiang CM, Roeder RG. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 25.Lavigne AC, et al. Multiple interactions between hTAFII55 and other TFIID subunits. Requirements for the formation of stable ternary complexes between hTAFII55 and the TATA-binding protein. J Biol Chem. 1996;271:19774–19780. doi: 10.1074/jbc.271.33.19774. [DOI] [PubMed] [Google Scholar]
- 26.Gegonne A, Weissman JD, Singer DS. TAFII55 binding to TAFII250 inhibits its acetyltransferase activity. Proc Natl Acad Sci USA. 2001;98:12432–12437. doi: 10.1073/pnas.211444798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gegonne A, Weissman JD, Zhou M, Brady JN, Singer DS. TAF7: A possible transcription initiation check-point regulator. Proc Natl Acad Sci USA. 2006;103:602–607. doi: 10.1073/pnas.0510031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tassan JP, Schultz SJ, Bartek J, Nigg EA. Cell cycle analysis of the activity, subcellular localization, and subunit composition of human CAK (CDK-activating kinase) J Cell Biol. 1994;127:467–478. doi: 10.1083/jcb.127.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou M, et al. Tax interacts with P-TEFb in a novel manner to stimulate human T-lymphotropic virus type 1 transcription. J Virol. 2006;80:4781–4791. doi: 10.1128/JVI.80.10.4781-4791.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JB, Sharp PA. Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J Biol Chem. 2001;276:12317–12323. doi: 10.1074/jbc.M010908200. [DOI] [PubMed] [Google Scholar]
- 31.Shen WC, et al. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 2003;22:3395–3402. doi: 10.1093/emboj/cdg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singer DS, Maguire J. Regulation of expression of MHC class I genes. CRC Rev Immunol. 1990;10:235–257. [PubMed] [Google Scholar]
- 33.Jones KA. Taking a new TAK on tat transactivation. Genes Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- 34.Zhou M, et al. TFIIH inhibits CDK9 phosphorylation during human immunodeficiency virus type 1 transcription. J Biol Chem. 2001;276:44633–44640. doi: 10.1074/jbc.M107466200. [DOI] [PubMed] [Google Scholar]
- 35.Kobor MS, Greenblatt J. Regulation of transcription elongation by phosphorylation. Biochim Biophys Acta. 2002;1577:261–275. doi: 10.1016/s0167-4781(02)00457-8. [DOI] [PubMed] [Google Scholar]
- 36.Oberley MJ, Farnham PJ. Probing chromatin immunoprecipitates with CpG-island microarrays to identify genomic sites occupied by DNA-binding proteins. Methods Enzymol. 2003;371:577–596. doi: 10.1016/S0076-6879(03)71043-X. [DOI] [PubMed] [Google Scholar]