Abstract
Two directed evolution experiments on p-nitrobenzyl esterase yielded one enzyme with a 100-fold increased activity in aqueous-organic solvents and another with a 17°C increase in thermostability. Structures of the wild type and its organophilic and thermophilic counterparts are presented at resolutions of 1.5 Å, 1.6 Å, and 2.0 Å, respectively. These structures identify groups of interacting mutations and demonstrate how directed evolution can traverse complex fitness landscapes. Early-generation mutations stabilize flexible loops not visible in the wild-type structure and set the stage for further beneficial mutations in later generations. The mutations exert their influence on the esterase structure over large distances, in a manner that would be difficult to predict. The loops with the largest structural changes generally are not the sites of mutations. Similarly, none of the seven amino acid substitutions in the organophile are in the active site, even though the enzyme experiences significant changes in the organization of this site. In addition to reduction of surface loop flexibility, thermostability in the evolved esterase results from altered core packing, helix stabilization, and the acquisition of surface salt bridges, in agreement with other comparative studies of mesophilic and thermophilic enzymes. Crystallographic analysis of the wild type and its evolved counterparts reveals networks of mutations that collectively reorganize the active site. Interestingly, the changes that led to diversity within the α/β hydrolase enzyme family and the reorganization seen in this study result from main-chain movements.
Directed evolution allows the rapid and simultaneous exploration of sequence, structure, and function space and thus provides a powerful tool to address biochemical questions involving protein stability and function. Using low mutation rates and recombination to minimize the number of silent or deleterious mutations (1, 2), directed evolution delivers protein variants that are instructive solutions to defined biochemical or environmental challenges. Unlike proteins from different species, in which the bulk of the mutations arise from genetic drift (3), sequence or structural alignments of proteins obtained by directed evolution allow immediate identification of accessible sequence changes that result in the desired phenotypic changes. Additionally, mechanisms of response to different, defined selection pressures can be investigated. This work represents a structural study of evolutionary divergence in a setting where the selection pressure, start point, pathway, and endpoint all are well defined.
The enzyme para-nitrobenzyl (pNB) esterase has been evolved for increased activity in organic solvents (mutant 5–6c8) (4, 5) and higher thermal stability (mutant 8g8) (6). The organophile 5–6c8 was evolved by five generations of error-prone PCR and gene shuffling. Seven amino acid mutations (Table 1) yielded an enzyme whose activity toward the pNB ester of the antibiotic loracarbef (pNB-LCN) in 25% dimethylformamide is 100-fold greater than the wild type (WT) (5). Thermophile 8g8, the product of eight generations of laboratory evolution (the first six of which are described in ref. 6), has 13 amino acid mutations (Table 2) that increase the melting temperature by 17°C (Tm = 69.5°C), increase the optimal temperature for activity by 15°C (Topt = 60°C), and increase activity toward p-nitrophenyl acetate. Here we compare the crystal structures of the WT, organophile 5–6c8, and thermophile 8g8. The organophile 5–6c8 was determined to 1.6-Å resolution by a three-wavelength mercury multiple anomalous dispersion experiment and was used to determine the WT and thermophile 8g8 structures to 1.5 Å and 2.0 Å, respectively. The WT structure shows high structural similarity to acetylcholine esterase (rms deviation = 2.6 Å for the Cα of 460 aligned residues) and bile salt-activated lipase (rms deviation = 2.5 Å for the Cα of 444 residues) (7). As with acetylcholine esterase, a large fraction of the pNB esterase has no defined secondary structure (52% random coil, 33% α-helix, 14% β-sheet). The structures show how complex networks of interacting mutations increase the stability and alter the specificity of pNB esterase.
Table 1.
Proposed role of each mutation introduced into 5-6c8
| 56c8 mutations | Generation | Distance to S189 Oγ | Proposed role for 5-6c8 mutations |
|---|---|---|---|
| H322R | 1 | 21.8 Å | Stabilizes 314–324 loop. Moves 265-275 active site forming loop. Interacts with M358V, L144M, and L334S. |
| Y370F | 1 | 16.3 Å | Perhaps results in more favorable solvation. |
| M358V | 2 | 11.4 Å | Reorganizes 265–275 and 315–325 loops. Interacts with L144M. H322R, and L334S. |
| L144M | 3 | 18.5 Å | Reorients helix 9, which stabilizes 265-275 loop. Interacts with M358V, H322R, and L334S. |
| 160V | 4 | 18.1 Å | Stabilizes 66–74 loop, allows 10 intramolecular H bonds. |
| L334S | 4 | 17.1 Å | Stabilizes movement of helix 9 and 265-275 loop. Interacts with H322R, M358V, and L144M. |
| P317S | 5 | 20.8 Å | Perhaps stabilizes new orientation of 315-324 loop. |
Table 2.
Proposed role of each mutation introduced into 8g8
| 8g8 mutations | Generation | Mutation group | Distance to S189 Oγ | Proposed role for 8g8 mutations |
|---|---|---|---|---|
| 160V | 1 | 2 | 17.0 Å | Stabilizes 64–74 and 414–420 loops. Allows 10 intramolecular H bonds. |
| L144M | 1 | 1 | 17.8 Å | Stabilizes 265–275 loop. Interacts with M358V, H322Y, and L343. |
| M358V | 1 | 1 | 11.9 Å | Shifts 265-275 and 315-324 loops (smaller shift than 56c8). Interacts with H322Y, L144M, and A343V. |
| Y370F | 1 | 3 | 16.5 Å | Perhaps results in more favorable solvation. |
| A343V | 2 | 1 | 25.0 Å | Alters packing among helices 9, 10, 11, and 15. |
| 1437T | 3 | 3 | 21.7 Å | Forms intramolecular H bond, stabilizing helix 17. |
| H322Y | 4 | 1 | 20.8 Å | Positions 315–324 loop. Determines the interaction between 265-275 active site loop and 315-324 loop. Interacts with M358V, L144M, and A343V. |
| L313F | 5 | 3 | 10.7 Å | Forms face-to-edge interaction with F314. |
| G412E | 5 | 2 | 17.0 Å | Precedes 413–421 loop, which is unstructured in WT. Forms salt bridge with R415. |
| T73K | 7 | 2 | 21.1 Å | In 65–75 loop, which is unstructured in WT. Forms salt bridge with E74. |
| T459S | 7 | 4 | 28.4 Å | Thr and Ser both H bond to H456. Small cost of exposing a methyl group is saved. |
| A56V | 8 | 4 | 25.8 Å | No observable changes. |
| A400T | 8 | 3 | 7.8 Å | Thr-400 is in the active site. |
Materials and Methods
Purification and Crystallization.
pNB esterases were expressed by using a modification of the protocol described by Zock et al. (8). Cells were grown at 32°C in 2× super broth liquid media in a 10-liter high-density bioreactor (Braun, Melsungen, Germany) and induced by raising the temperature to 42°C. Esterases were purified by using a modification of the protocol of Chen et al. (9). Cells were lysed by French press and pelleted by centrifugation. The supernatant then was titrated to pH 5.0 and centrifuged. Ammonium sulfate cuts (45% and 85%) were taken, and the second pellet was dialyzed against 10 mM Tris (pH 8.5), 50 mM NaCl, 0.5 mM EDTA, and 0.5 mM DTT. The protein was loaded onto a 25-ml FF Sepharose DEAE column (Amersham Pharmacia) and eluted with a salt gradient (50–500 mM NaCl). The peak containing pNB esterase was concentrated, and buffer was exchanged into low salt (50 mM NaCl), loaded on a 45-ml Poros HQ column, and eluted with a salt gradient (50–500 mM NaCl). pNB esterase eluted as a single peak, which was concentrated and loaded onto a 150-ml HR Sephacryl S-100 column equilibrated with 10 mM Tris (pH 7.5), 100 mM NaCl, and 1 mM methionine. Purified pNB esterase was analyzed by MS and dynamic light scattering. MS showed a single major species, and dynamic light scattering showed no measurable polydispersity. Organophile 5–6c8 crystallized by using the hanging drop method at 18°C in the optimized conditions of 5 μl of 20 mg/ml 5–6c8 and 5 μl of mother liquor [29% PEG 4000, 100 mM Tris (pH 8.4), 160–185 mM LiSO4, and 8–12% dimethylformamide]. WT and 8g8 crystallized in slightly modified conditions from 5–6c8 [WT: 15 mg/ml protein, 28% PEG 4000, 100 mM Tris (pH 8.25), 150 mM LiSO4; 8g8: 15 mg/ml protein, 20% PEG 4000, 100 mM Tris (pH 7.5), 200 mM MgOAc2]. All three enzymes crystallized in the space group P212121, a = 67 Å, b = 83 Å, c = 99 Å.
Data Collection and Structure Determination.
Crystals were frozen after a 30-sec soak in fresh mother liquor. Native and derivative data sets were collected at the Advance Light Source beamline 5.0.2. The structure of 5–6c8 was determined by a three-wavelength Hg multiple anomalous dispersion experiment (see Table 4, which is published as supplementary material on the PNAS web site, www.pnas.org). Data were processed with denzo and scalepack (10). Scaling and phase determination were done with solve (11). The phasing figure of merit (FOM) was 0.54 before solvent flattening and 0.75 after solvent flattening with the program dm (12). Density modification in ccp4 (12) produced a very good map into which ≈400 aa were built with side chains. Phased restrained refinement in cns (13) and phase combination in ccp4 (12) allowed the remainder of the structure to be built. The model was refined into the native data, by using rigid body refinement, followed by Powell minimization, and individual B-factor refinement, resulting in an Rfree of 42% and Rcryst of 38%. Alternate rounds of model building in o (14) with ccp4 (12) generated omit maps, and refinement with cns (13) resulted in the final model summarized in Table 3. WT and 8g8 crystallized in isomorphous cells and were solved with direct replacement by using the 5–6c8 model. All native data were processed and scaled with denzo and scalepack (10). In both cases initial R factors were in the low 50s but dropped into the low 30s after rigid body refinement (6.0–3.0 Å). Further refinement using all data (25 Å-dmin) and building into omit maps produced the models described in Table 3. For all models, refinement using cns included simulated annealing, Powell minimization, and individual B-factor refinement (13).
Table 3.
Structure refinement statistics for pNB esterases
| Protein | Resolution Å | Rfree | Rcryst | rms deviation
|
Amino acids | #Waters | #Ions | |
|---|---|---|---|---|---|---|---|---|
| Bonds | Angles | |||||||
| 5-6c8 | 25–1.6 | 23.2 | 21.8 | 0.0055 | 1.470 | 2–484 | 272 | 2 |
| WT | 25–1.5 | 24.0 | 22.0 | 0.0049 | 1.348 | 2–65, 75–413, 421–484 | 380 | 2 |
| 8g8 | 25–2.0 | 24.9 | 21.1 | 0.0059 | 1.313 | 2–484 | 161 | 1 |
Rfree = Rcryst calculated with 10% of the data chosen at random and not used in refinement. Rcryst = 100*Σ|Fobs − Fcalc|/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors.
Overall Structure Description
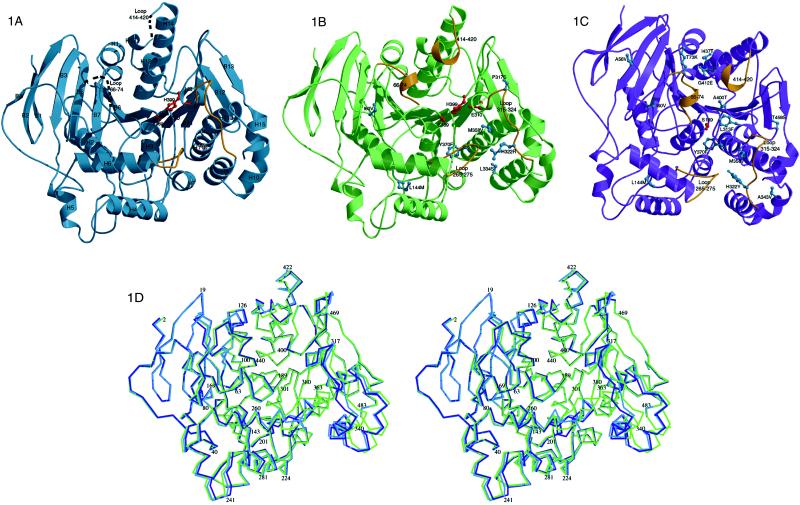
Bacillus subtilis pNB esterase (Fig. 1) is an α/β protein of 489 aa composed of a central 13-stranded β-sheet surrounded by 15 α-helices. Residues 66–74 and 414–420 in the WT structure are not observed in the electron density. However, analysis by MS identified only the N-terminal methionine as deleted. These residues therefore are presumably unstructured or in multiple conformations and contribute only weakly to the measured diffraction intensities.
Figure 1.
molscript diagrams (15, 16) of pNB esterases looking into the active site cavity, showing loops that are not visible in the electron density as dashed lines and loops that reorganize most significantly in gold. The catalytic triad is shown in red and mutations are shown in blue. (A) The WT structure with secondary structural elements labeled. (B) The 5–6c8 structure. (C) The 8g8 structure, rotated slightly from the others to clarify the location of the mutations. (D) A wall-eyed stereo overlay of the Cα positions of the three structures, oriented as in A and B. WT is shown in blue, organophile 5–6c8 in green, and thermophile 8g8 in purple.
Based on sequence and structural homology, pNB esterase belongs to the α/β hydrolase family (17) and contains a catalytic triad formed by Ser-189, His-399, and Glu-310 (Fig. 1). The active site is located at the base of a cavity with dimensions 20 Å by 13 Å by 18 Å. pNB esterase shares with acetylcholine esterase a rare use of Glu instead of Asp as the active site carboxylate (18). The entrance to the active site is formed by four loops that reorganize substantially in response to selection pressure. Residues 64–71 and 413–417, which are in the unstructured WT loops, form one side of the entrance. The rest of the entrance is formed by residues 316–320 (poorly structured electron density in WT) and 260–268. The active site cavity, which also changes structure substantially in the evolved esterases, is comprised of residues 105–108, 193, 215–216, 268–275, 310–314, 362–363, and 399–400.
Structural Evolution of an Organophile
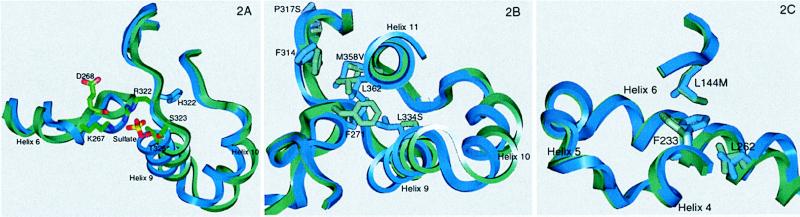
The organophile 5–6c8 structure is overall quite similar to WT, with a rms deviation of 0.67 Å for all C-α atoms (467 residues). The positions and orientations of the catalytic triad are unaltered by the mutations. However, the mutations have dramatic effects on the surface loops that form the opening to the active site and on residues that form the sides of the active site. None of the seven substitutions in 5–6c8 (Table 1 and Fig. 1B) is in the active site, and only one is in the entrance. These substitutions serve, however, to decrease the flexibility of surface loops and reorganize the active site for substrate pNB-LCN (the pNB ester of the antibiotic loracarbef). The 315–324 loop, poorly structured in the WT, is well defined in both organophile 5–6c8 and thermophile 8g8 (Fig. 1). As shown in Fig. 2A, the His-322 → Arg mutation from the first generation plays a major role. The side chain of 322 in 5–6c8 points ≈95° away from its position in WT and directly toward the 265–275 loop. In 5–6c8, the guanidino group of Arg-322 H-bonds to the main-chain oxygen of Asp-268 and contributes to a binding pocket for a well-defined sulfate, which in turn H-bonds to Lys-267 (the sulfate binding pocket is formed by Lys-267, Arg-322, Ser-323, and Thr-326). This hydrogen bonding network reorganizes the 265–275 loop that forms a face of the active site and stabilizes the 265–275 and 315–324 loops by introducing hydrogen bonds between them.
Figure 2.
Comparison between organophile 5–6c8 (green) and WT (blue). (A) A superposition of the 315–324 and 265–275 loops. The His-322 → Arg mutation results in a hydrogen bond to the main-chain oxygen of Asp-268. Arg-322 also contributes to a binding pocket for a well-defined sulfate. The sulfate is oriented by hydrogen bonds to Ser-323 and Thr-326, as well as salt bridges to Lys-267 and Arg-322. The His-322 → Arg mutation results in two interactions between loop 265–275 and loop 315–324. (B) A superposition of the main chain in the vicinity of changes introduced by the Met-358 → Val and Leu-334 → Ser mutations. The mutation to a smaller side chain at 358 allows Phe-314 to rotate into the area vacated, shifting loop 315–324. Leu-362 also rotates into the vacated space, as does Phe-271, resulting in a shift of residues 258–275, which form a face of the active site. The changes caused by the third-generation Met-358 → Val mutation are stabilized by the fourth-generation Leu-334 → Ser mutation, which accommodates the new orientation of Phe-271. (C) The new orientation of 265–275 caused by the second-generation Met-358 → Val mutation is stabilized by the fourth-generation Leu-144 → Met mutation. Leu-144 → Met allows Phe-233 to shift, accommodating the new position of Leu-262 and stabilizing the new orientation of 258–275.
Reorganization of the active site is further affected by a group of three interacting mutations: Met-358 → Val, Leu-144 → Met, and Leu-334 → Ser. As shown in Fig. 2B, Met-358 → Val is located in helix 11, which packs against loop 315–324. The Met-358 → Val mutation allows Phe-314 (at the end of helix 8) to shift toward the space previously occupied by the longer Met side chain, shifting loop 315–324 and the following helix (helix 8). This rearrangement accommodates the Pro-317 → Ser mutation. Phe-314 in either orientation forms a part of the active site pocket. Additionally, Leu-362 rotates ≈70° toward the active site. The largest change allowed by the Met-358 → Val mutation is a 2-Å movement of residue Phe-271 into the area previously occupied by Met-358. This movement shifts a 17-aa stretch from 258–275, which forms the entrance to the active site as well as forming a face of the active site. The new loop orientation is further stabilized by Leu-144 → Met (Fig. 2C), which allows Phe-233 to rotate 50° around χ1 and 30° around χ2. This allows Leu-262 to shift into the vacated space, resulting in a reorientation of helix 9 that stabilizes the new orientation of the 265–275 loop. Finally, the last of these three mutations, the fourth-generation Leu-334 → Ser mutation, accommodates the movement of Phe-271 (Fig. 2B).
Ile-60 → Val, found in the fourth generation, stabilizes a surface loop that is unstructured in the WT enzyme. The replacement of Ile-60 with the smaller valine allows the 66–74 loop to pack against residues 60–63 and 110–116 in a manner that would cause steric clashes between side chains of residues 60 and Leu-68 if both adopted conformations similar to the WT. The alternate loop structure of residues 66–74 in 5–6c8 provides 10 additional H bonds.
Mutation Tyr-370 → Phe is buried and results in no structural changes. An H bond, from the side chain or Tyr-370 to the main-chain carbonyl of 196, formed in the WT is lost in 5–6c8. However, this may not compensate the higher cost of desolvating Tyr versus Phe.
Structural Evolution of a Thermophile
Currently, at least 13 pairs of mesophile and thermophile structures have been determined (for a recent review see ref. 19). However, these structures are of significantly divergent proteins with sequence identities as low as 36% and in no cases exceeding 75% (19), a fact that hinders the assignment of stabilizing mechanisms (19–21). By contrast, the most thermophilic variant of pNB esterase, 8g8, with only 13 mutations is 97% identical to WT and has a rms deviation of 0.44 Å for C-α atoms (467 residues). As with 5–6c8, the catalytic triads are superimposable. This high sequence identity, in conjunction with the crystal structures of both WT and thermophile, affords an interesting opportunity to study the structural basis for thermostability. Thermophile 8g8 is the product of eight generations of directed evolution for retention of activity toward p-nitrophenyl acetate after incubation at high temperature (ref. 6 and data not shown). The starting point was a thermophilic esterase from a pre-existing library (generated for the organophile study) that already contained Ile-60 → Val, Leu-144 → Met, His-322 → Arg, Tyr-370 → Phe, and Met-358 → Val.
As with organophile 5–6c8, the mutations in thermophile 8g8 serve to both reorganize the active site and stabilize surface loops. The 13 mutations in 8g8 fall into four groups. Mutations in the first group (Leu-144 → Met, Met-358 → Val, His-322 → Tyr, and Ala-343 → Val) all interact to reorganize the 265–275 and 315–324 loops. A second group (Ile-60 → Val, Gly-412 → Glu, and Thr-73 → Lys) stabilizes the 66–74 and 414–420 loops. These loops, which do not contribute to the lattice structure in any of these isomorphous crystals, are unstructured in WT. Mutations Tyr-370 → Phe, Leu-313 → Phe, Ile-437 → Thr, and Ala-400 → Thr have isolated, interpretable effects, whereas mutations Thr-459 → Ser and Ala-56 → Val cannot be explained. The 13 mutations and their putative stabilization mechanisms are summarized in Table 2.
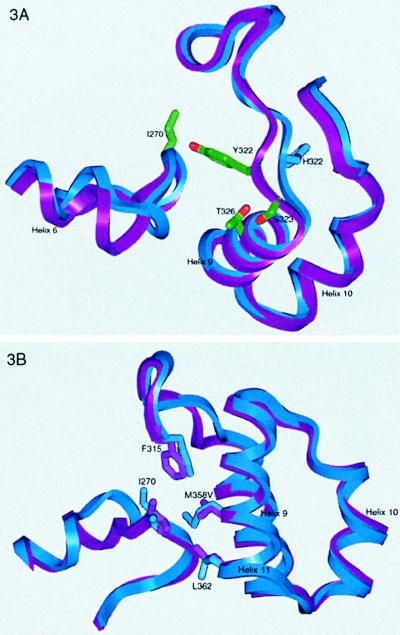
The first group contains two mutations, Leu-144 → Met and Met-358 → Val, that are found in organophile 5–6c8. Although they make similar contacts in both structures, the overall effect in the thermophile is greatly influenced by a third mutation, Arg-322 → Tyr. Met-358 → Val allows Ile-270, Leu-362, and Phe-315 to move, but the large reorganization of the 258–275 stretch in the organophile is not observed in the thermophile. Likewise, the movement in the 315–348 stretch is much less substantial (compare Figs. 2 and 3). The explanation for why identical mutations have different structural (and functional) results involves the role of residue 322 (Figs. 2A and 3A), which differs in all three proteins. This site dictates the nature of the interaction between the 315–324 loop and the 265–275 loop. In the evolution of thermophile 8g8, residue 322 starts as an arginine, then mutates to a cysteine and finally to a tyrosine (6). In thermophile 8g8 the 265–275 loop follows WT much more closely than in organophile 5–6c8, but the 315–324 loop is pulled closer to the active site. Residue 322 moves 2.5 Å (measured at Cα) from its WT position, and Cβ points ≈90° away from its WT position and toward the 265–275 loop. Unlike Arg-322, Tyr-322 does not interact directly with the adjacent 265–275 loop (except for a 3.5-Å H bond to the backbone amide of residue 270). The new position of the 315–324 loop allows Ser-323 to H-bond to Thr-326, stabilizing the loop and helix 8. A fourth mutation, Ala-343 → Val, is in helix 10, which packs against helices 9, 11, and 15. To accommodate the movement of loop 315–324, helix 10 moves closer to helix 9 and further from helix 11, resulting in space nicely filled by the Ala-343 → Val mutation.
Figure 3.
Comparison between the thermophile 8g8 (purple) and WT (blue). (A) A superposition of the 315–324 and 265–275 loops. The His-322 → Tyr mutation introduces a direct interaction between the loops. The 315–324 loop is pulled closer to the active site to accommodate the smaller substrate, allowing a 3.5-Å H bond between Tyr-322 and the main-chain N of Ile-270. Additionally, the new orientation of 315–324 allows an H bond between the side chains of Ser-323 and Thr-326, stabilizing helix 12 and the 315–324 loop. (B) A superposition of WT and 8g8 shows the effect of the Met-358 → Val mutation. In the absence of the His-322 → Arg mutation, the large reorganization of 265–275 is not seen. Leu-362 and Ile-270 move to fill the cavity created by the Met-358 → Val mutation.
The second group of interacting mutations (Ile-60 → Val, Gly-412 → Glu, and Thr-73 → Lys) stabilizes the loops that form the opening to the active site cavity (Fig. 1C). These loops are also well defined in organophile 5–6c8; therefore, the stabilization of 66–74 by Ile-60 → Val appears to also fix the adjacent 414–420 loop. Gly-412 → Glu is not necessary for the stabilization of 413–421, but it forms a salt bridge with Arg-415, which stabilizes helix 12. This interaction has been shown by mutational analysis to contribute to thermostability (6). The Thr-73 → Lys mutation is also in the loop stabilized by Ile-60 → Val, and its WT position is therefore unknown. In the thermophile 8g8 Lys-73 forms a salt bridge with Glu-74. Both Gly-412 → Glu and Thr-73 → Lys occur after the 66–74 and 414–420 loops are fixed (the loops are fixed in the first generation) and are thus clear examples of stabilizing salt bridges that could not be designed into the WT background, illustrating the power of iterative methods in protein engineering.
Mutations in the third group, Tyr-370 → Phe, Leu-313 → Phe, Ile-437 → Thr, and Ala-400 → Thr all have been shown by biochemical studies to be thermostabilizing (6). Tyr-370 → Phe results in no structural changes other than the loss of an H bond between Tyr-370 and the main-chain carbonyl of residue 196. As mentioned above, the higher cost of desolvating Tyr versus Phe may explain how this mutation stabilizes the enzyme. Leu-313 → Phe results in a stabilizing face-to-edge interaction (22, 23) with Phe-314. Ile-437 → Thr is in helix 13 and results in an intra-helix H bond to the main-chain oxygen of 433, stabilizing the helix. Ala-400 → Thr is not thermostabilizing when introduced into WT as a double mutant with Ala-56 → Val, the other eighth-generation mutation (data not shown). However, it may be thermostabilizing in the seventh-generation background or may be responsible for the increased activity of 8g8. Although residue 400 is visible in all structures, Thr-400 packs against Met-416, which is not visible in the WT structure. Met-416 is shifted away from residue 400 in the thermophile relative to the organophile, because of the salt bridge introduced by the fifth-generation Gly-412 → Glu mutation. The movement of Met-416 is accommodated by the larger Thr side chain. Furthermore, the Thr-400 side chain is in the active site and may help stabilize His-399.
As seen in antibody maturation structural studies, not all mutations can be rationalized even with structures for comparison (24, 25). Thus although screening mutant libraries often can identify interesting proteins, neutral mutations do arise. The ability of recombination to remove neutral mutations (1, 2) (performed most recently after the seventh generation in the evolution of 8g8) presents a tool to be enlisted in the study of somatic mutation in the immune system. Thr-459 → Ser and Ala-56 → Val are two neutral mutations in thermophile 8g8. Both are exposed to solvent and are more than 25 Å from the active site. The mutation at 459 appeared in the same generation with a known stabilizing mutations and therefore may not be stabilizing itself.
Relevance to Protein Engineering
Directed evolution offers an efficient route to redesigning proteins for new functional characteristics. Adaptive mutations and well-defined selection pressures allow structural analysis of the evolved products to provide insights into the molecular basis of protein function. Here, the stabilization of a monomeric protein is accompanied by altered core packing, helix stabilization, introduction of surface salt bridges and reduction of flexibility in surface loops, all of which are proposed mechanisms by which proteins from thermophiles are stabilized (19–21, 26). The thermostabilizing mechanisms that were recruited in 8g8 represent a far from exhaustive list of the mechanisms identified in naturally thermostable proteins. A reduction of internal cavity volume, introduction of salt bridge clusters, decreased surface area to volume ratio, removal of redox active residues, burial of exposed hydrophobic residues, elimination of β-branched amino acids, and removal of amino acids that can deamidate, all proposed routes to enhanced thermostability (19), were not observed in this experiment. Some of these mechanisms may not be accessible via a single amino acid mutation pathway, requiring instead multiple simultaneous and compensating mutations. Others may not become apparent under heavy selection pressure that requires measurable changes (1–2°C) in thermostability with each generation. Still others will be associated with deactivation mechanisms that are not important at the temperature (≈70°C) and time scale of the directed evolution experiment (e.g., chemical modification of amino acids).
The membership of pNB esterase in the diverse group of α/β hydrolases invites comparisons of how directed and natural evolution solve challenges requiring adaptation to new biochemical tasks. The two enzymes with which pNB esterase shares the greatest structural identity are acetylcholine esterase (AChE) and bile-salt activated lipase (7), both of which differ from pNB esterase in the loops affected by directed evolution. Both directed and natural evolution introduce main-chain movements into the loops around the active site. The changes introduced by directed evolution are similar to the main-chain movements seen in members of the α/β hydrolase family. In AChE, recognition of the small acteylcholine substrate is aided by the loop homologous to the 64–78 loop in pNB esterase. In AChE the 64–78 loop is replaced by a longer (21 aa) loop that shrinks the active site, which is advantageous for hydrolyzing the smaller acetylcholine substrate, by inserting large residues, Met-83 and Trp-84, into the binding cleft. While the experiments conducted on pNB esterase produced variants with conserved loop lengths, natural evolution and V(D)J recombination (the combinatorial gene shuffling procedure the immune system uses to produce large libraries of antibodies) use loop length variation to enhance diversity. It should be possible to introduce this feature into directed evolution libraries by mixing parent sequences from different species (27).
The complex changes brought about by directed evolution underscore the difficulty of the rational design problem. Some efforts to simplify the rational design problem have focused on the additivity of single site mutations (28, 29). Mutations are considered additive when the effect of multiple mutations and the sum of the effects of single site mutants are equal, and the effect being measured is a state function. Such additivity is observed when the individual mutation sites do not interact with each other or do not alter the rate-determining step in catalysis (29). However, in addition to these limitations, the reported precision of such results may not be adequate to have broadly predictive value (30). Directed evolution, in which mutations in the later generations interact with the results of previous mutations, can reveal nonadditive mutation pathways. The introduction of interacting mutations in successive generations indicates that rational design will benefit from efforts to predict the effects of single mutations and then introduce later-generation mutations in an iterative manner.
However, even given a high-resolution crystal structure, it is unlikely that the mutations discovered in these experiments could have been predicted. Mutations that fix disordered regions of pNB esterase were found in both evolutionary experiments. The lack of defined electron density for these regions in the WT crystal structure certainly would have precluded rational design to stabilize them. Three of the mutations in thermophile 8g8, Ile-60 → Val, Gly-412 → Glu, and Thr-73 → Lys, interact directly with regions that are unstructured in the WT. Additionally, these mutations interact with Met-358 → Val, Ala-343 → Val, Leu-144 → Met, and His-322 → Tyr, thus a majority of the stabilizing mutations are functionally related to regions that are unstructured in the WT enzyme. Some mutations that affect the active site were found to do so by transmitting subtle changes into more significant active site perturbations. The increased activity of the organophile is a result of mutations outside the active site, which nonetheless affect significant reorganization of the active site cavity. The Leu-144 → Met, Ala-343 → Val, and Met-358 → Val mutations shift α-helices, which, in turn, reorient loops forming the active site. Recently, the structure of an amino-transferase directed evolution product was reported (31). Those authors also found clusters of interacting mutations distant from the active site. The ability of directed evolution to identify structurally and functionally important amino acid clusters in proteins is similar to the immune system’s ability to control the shapes of the complementarity determining regions of antibodies with a small number of critical sites (32). Both methods point to the existence of mutational hotspots that influence function across long distances.
Supplementary Material
Acknowledgments
We thank Dan Koshland, Judith Klinman, Andrew Mesecar, Melanie Gee, Emily Munsdorff, and D. Borden Lacy for reviewing the manuscript, T. Earnest and L.W. Hung for help at beamline 5.0.2 of the Advance Light Source, P. Kuhn and M. Solstice for help at beamline 7–1 of Stanford Synchrotron Radiation Laboratory. B. Jap and P. Walian for access to in-house x-ray equipment, and D. King for MS. This work was supported in part by the Army Research Office (F.H.A. and A.G.) and by Department of Energy Contract No. DE-AC03-76SF0098 (R.C.S. and B.S.).
Abbreviations
- pNB
para-nitrobenzyl
- WT
wild type
Footnotes
Data deposition: The coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID code 1QE3 (WT), 1QE8 (organophile 5-c68), and 1C00 (thermophile 8g8)].
References
- 1.Stemmer W P C. Nature (London) 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 2.Arnold F H. Acc Chem Res. 1998;31:125–131. [Google Scholar]
- 3.Kimura M. Molecular Evolution, Protein Polymorphism, and the Neutral Theory. Tokyo: Japan Scientific Societies Press; 1982. [Google Scholar]
- 4.Moore J C, Arnold F H. Nat Biotechnol. 1996;14:458–467. doi: 10.1038/nbt0496-458. [DOI] [PubMed] [Google Scholar]
- 5.Moore J C, Jin H M, Kuchner O, Arnold F H. J Mol Biol. 1997;272:336–347. doi: 10.1006/jmbi.1997.1252. [DOI] [PubMed] [Google Scholar]
- 6.Giver L, Gershenson A, Freskgard P O, Arnold F H. Proc Natl Acad Sci USA. 1998;95:12809–12813. doi: 10.1073/pnas.95.22.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holm L, Sander C. Nucleic Acids Res. 1994;22:3600–3609. [PMC free article] [PubMed] [Google Scholar]
- 8.Zock J, Cantwell C, Swartling J, Hodges R, Pohl T, Sutton K, Rosteck P, Jr, McGilvray D, Queener S. Gene. 1994;151:37–43. doi: 10.1016/0378-1119(94)90630-0. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y R, Usui S, Queener S W, Yu C A. J Ind Microbiol. 1995;15:10–18. [Google Scholar]
- 10.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 11.Terwilliger T C, Berendzen J. Acta Crystallogr D. 1999;55:501–505. doi: 10.1107/S0907444998012657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collaborative Computational Project 4. ccp4: A Suite of Programs for Protein Crystallography. Warrington, U.K.: SERC Daresbury Laboratory; 1979. [Google Scholar]
- 13.Adams P D, Pannu N S, Read R J, Brunger A T. Proc Natl Acad Sci USA. 1997;94:5018–5023. doi: 10.1073/pnas.94.10.5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 15.Merrit E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 16.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 17.Ollis D L, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken S M, Harel M, Remington S J, Silman I, Schrag J, et al. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 18.Sussman J L, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Science. 1991;253:872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- 19.Facchiano A M, Colonna G, Ragone R. Protein Eng. 1998;11:753–760. doi: 10.1093/protein/11.9.753. [DOI] [PubMed] [Google Scholar]
- 20.Jaenicke R, Böhm G. Curr Opin Struct Biol. 1998;8:738–748. doi: 10.1016/s0959-440x(98)80094-8. [DOI] [PubMed] [Google Scholar]
- 21.Vogt G, Woell S, Argos P. J Mol Biol. 1997;269:631–643. doi: 10.1006/jmbi.1997.1042. [DOI] [PubMed] [Google Scholar]
- 22.Hunter C A, Singh J, Thornton J M. J Mol Biol. 1991;218:837–846. doi: 10.1016/0022-2836(91)90271-7. [DOI] [PubMed] [Google Scholar]
- 23.Serrano L, Bycroft M, Fersht A R. J Mol Biol. 1991;218:465–475. doi: 10.1016/0022-2836(91)90725-l. [DOI] [PubMed] [Google Scholar]
- 24.Romesberg F E, Spiller B, Schultz P G, Stevens R C. Science. 1998;279:1929–1933. doi: 10.1126/science.279.5358.1929. [DOI] [PubMed] [Google Scholar]
- 25.Wedemayer G J, Patten P A, Wang L H, Schultz P G, Stevens R C. Science. 1997;276:1665–1669. doi: 10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- 26.Zavodszky P, Kardos J, Svingor A, Petsko G A. Proc Natl Acad Sci USA. 1998;95:7406–7411. doi: 10.1073/pnas.95.13.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crameri A, Raillard S A, Bermudez E, Stemmer W P C. Nature (London) 1998;391:288–291. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]
- 28.Skinner M M, Terwilliger T C. Proc Natl Acad Sci USA. 1996;93:10753–10757. doi: 10.1073/pnas.93.20.10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wells J A. Biochemistry. 1990;29:8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- 30.Dill K A. J Biol Chem. 1997;272:701–704. doi: 10.1074/jbc.272.2.701. [DOI] [PubMed] [Google Scholar]
- 31.Oue S, Okamoto A, Yano T, Kagamiyama H. J Biol Chem. 1999;274:2344–2349. doi: 10.1074/jbc.274.4.2344. [DOI] [PubMed] [Google Scholar]
- 32.Chothia C, Lesk A M. J Mol Biol. 1987;196:901–917. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.