Abstract
Although it has been established how CD1 binds a variety of lipid antigens (Ag), data are only now emerging that show how αβ T cell receptors (TCRs) interact with CD1-Ag. Using the structure of the human semiinvariant NKT TCR–CD1d–α-galactosylceramide (α-GalCer) complex as a guide, we undertook an alanine scanning mutagenesis approach to define the energetic basis of this interaction between the NKT TCR and CD1d. Moreover, we explored how analogues of α-GalCer affected this interaction. The data revealed that an identical energetic footprint underpinned the human and mouse NKT TCR–CD1d–α-GalCer cross-reactivity. Some, but not all, of the contact residues within the Jα18-encoded invariant CDR3α loop and Vβ11-encoded CDR2β loop were critical for recognizing CD1d. The residues within the Vα24-encoded CDR1α and CDR3α loops that contacted the glycolipid Ag played a smaller energetic role compared with the NKT TCR residues that contacted CD1d. Collectively, our data reveal that the region distant to the protruding Ag and directly above the F′ pocket of CD1d was the principal factor in the interaction with the NKT TCR. Accordingly, although the structural footprint at the NKT TCR–CD1d–α-GalCer is small, the energetic footprint is smaller still, and reveals the minimal requirements for CD1d restriction.
αβ TCRs interact with peptide- and lipid-laden MHC and CD1 molecules, respectively. The MHC is highly polymorphic, and distinct features within the peptide-binding groove enable the MHC to present a wide array of peptides to the T cells. Nevertheless, TCRs are highly specific and genetically restricted to recognize MHC molecules of the individual from which they were derived (1). In contrast, CD1 family members are monomorphic glycoproteins that are recognized by T cells from different individuals, and even across species in the case of CD1d. On the basis of their structural and functional features, the CD1 family is predominantly divided into group I CD1a, CD1b, and CD1c molecules, and the group II CD1d molecule, the latter of which is the only member of the family expressed in mice and rats. The Ag-binding cleft of the CD1 family contains large hydrophobic pockets that are suited to bind lipid-based antigens. Recent structural studies have highlighted how different-sized cavities among the CD1 family enable it to bind defined lipids (2, 3, 4). For example, the CD1d family binds a restricted repertoire of glycolipids, which includes foreign glycolipids such as the glycosphingolipid α-galactosylceramide (α-GalCer) (5), which is an archetypal CD1d ligand that binds well to both human (hCD1d) and mouse CD1d (mCD1d) molecules (6, 7).
Structural studies on TCR–peptide–MHC complexes have revealed markedly differing docking strategies in which TCRs can interact with peptide-MHC ligands (pMHC) (8, 9). Nevertheless, a rough docking mode is preserved, in which the Vα domain is positioned over the α2-helix and the N-terminal end of the peptide, while the Vβ domain is positioned over the α1-helix and C-terminal end of the peptide. These studies have been complemented by biophysical studies (e.g., surface plasmon resonance) of the TCR–pMHC interaction, which have demonstrated that the TCR–pMHC interaction is dominated by weak intermolecular interactions (low micromolar range), with slow association rates and fast dissociation rates (10, 11). Moreover, alanine-scanning mutagenesis has been important in understanding the energetic basis of the TCR–pMHC interaction, and to date, has been conducted in several TCR–pMHC systems (12–15). These studies have revealed that the energetic contributions of the CDR loops can vary quite considerably at the TCR–pMHC interface, as well as between different TCR–pMHC systems. For example, in the 2C TCR system, the CDR1 and CDR2 loops were shown to be critically important (13), whereas in the LC13 TCR system, the CDR3 loops contributed mostly to the energetic landscape (14). Moreover, studies on the I-Ek-moth cytochrome c system has suggested a two-step mechanism for TCR recognition of the pMHC complex, whereby the CDR1 and CDR2 loops initially contact the MHC, followed by the CDR3 loops contacting the peptide (15).
NKT cell activation is implicated in many aspects of immunity and can enhance the response to some bacterial, viral, and parasitic infections, and some types of cancer, yet can suppress autoimmune disease, allograft rejection, and graft-versus-host disease (16). Type I NKT cells typically express a semiinvariant αβ TCR (NKT TCR) that comprises an invariant α chain, and a limited TCRβ repertoire. The NKT TCR binds to CD1d, which can present self- or foreign glycolipid to NKT cells. The human invariant NKT TCR α chain uses a Vα24-Jα18 (TRAV10-TRAJ18) rearrangement that encodes a germline-encoded junctional sequence, preserving amino acid sequence identity among human NKT TCR α chains. Moreover, most type I human NKT cells express Vβ11 (TRBV25-1) (17) rearranged to form variable Dβ−Jβ combinations with N-region additions or deletions (18–21). In addition, some CD1d-restricted, Vα24-independent NKT TCRs have been described that nevertheless maintain Jα18 and Vβ11 usage (18, 22), but these are less well studied. In addition to being highly selected, the human invariant NKT TCR (Vα24-Jα18; Vβ11) and the mouse NKT TCR homologue (Vα14-Jα18; Vβ8.2) (17) are cross-species reactive with mouse and human CD1d, respectively (23, 24). Such evolutionary conserved recognition is contrary to the highly restricted syngeneic TCR recognition of MHC class I and II molecules.
It is unclear, however, whether the principles underlying αβ TCR–pMHC recognition will be applicable to that of αβ TCR–CD1 interactions, and this consideration has been hampered because of a lack of structural information on αβ TCR–CD1–Ag complexes. Nevertheless, recent insight has been gained into lipid-mediated recognition, with the structure determination of the NKT TCR in the nonliganded state (25, 26) and in complex with CD1d–α-GalCer (27). Moreover, a recent study has highlighted the important mouse NKT TCR residues that are critical for mouse CD1d-Ag recognition, although the structure of the mouse NKT TCR–CD1d–Ag complex is unknown (28). To establish the underlying energetic basis of this human NKT TCR–CD1d–α-GalCer interaction, we undertook an alanine scanning approach using surface plasmon resonance, which has revealed the minimal requirements to enable CD1d-Ag recognition.
RESULTS
Experimental rationale
To define the underlying energetic basis of the interaction between the NKT TCR and CD1d–α-GalCer, we undertook an alanine scanning approach on both the NKT TCR and CD1d binding partners. We analyzed the effect of the mutations on the interaction using surface plasmon resonance. Given that the structures of the NKT TCR and CD1d–α-GalCer are available in the nonliganded state and in complex with CD1d–α-GalCer, we were able to rationalize the NKT TCR and CD1d residues selected for mutational analysis. Solvent-exposed NKT TCR residues whose sidechains interacted with either CD1d and/or α-GalCer were selected for substitution to alanine (Fig. 1 A). CD1d residues selected for substitution were those that made contact with the NKT TCR, but were not involved in contacting α-GalCer, as mutation of these latter residues (Tyr73, Ser76, Phe77, Asp80, Phe84, Asp151, Trp153, and Thr154) were considered likely to impact on the presentation of the ligand.
Figure 1.
A NKT TCR CDR3α and CDR2β loop contacts dominate CD1d-mediated interactions. (A) Residues on the NKT TCR important for recognizing CD1d–α-GalCer. (B) CDR1α loop interacts solely with α-GalCer galactose head group. (C) CDR3α loop mediates multiple contacts between CD1d α-helices and α-GalCer. (D) CDR3β loop contacts with the CD1d α2-helix. (E) Residues within and next to the CDR2β loop make polar interactions with the CD1d α1-helix. Pink, α-GalCer; yellow, CDR1α loop; cyan, CDR3α; orange; CDR2β loop; blue, CDR3β loop; grey, CD1d α-helices. H-bond or salt-bridge interactions, dotted lines. In Fig. 1 (B–E), substituted residues with a >10-fold reduction in affinity are shown in red; with a 4–6-fold reduction in affinity are shown in green; and with a <4-fold reduction in affinity are shown in blue.
NKT TCR substitutions
In brief, the NKT TCR–CD1d–α-GalCer complex revealed that the CDR1α loop solely contacted the Ag, whereas the CDR3α loop contacted CD1d and the Ag, whereas contributions from the Vβ chain predominantly arose via the CDR2β loop contacting CD1d (Fig. 1 A). In the original study on the 3.2 Å crystal structure of NKT TCR–CD1d–α-GalCer complex, there were two ternary complexes within the asymmetric unit. Subtle differences in the NKT TCR-CD1d contacts between these two ternary complexes were observed (Table I), which could potentially help to further refine the residues important in the interaction; alternatively, these differences may either be a consequence of crystal-packing effects or be a function of refining a ternary complex at 3.2 Å resolution. As such, we decided to mutate all NKT TCR residues implicated in the interaction with CD1d–α-GalCer and in total, 14 NKT TCR amino acid substitutions were made and included CDR1α (Pro28α and Ser30α); CDR3α (Asp94α, Arg95α, Gly96α, Ser97α, Thr98α, Leu99α, and Arg103α); CDR2β framework (Tyr48β and Glu56β); CDR2β (Tyr50β and Asn53β); and CDR3β (Tyr103β). All of the mutant NKT TCR proteins expressed and refolded with similar yield to WT NKT TCR, except for Asp94α, which produced half the amount of protein during expression but has the same refold efficiency as the WT. These mutant NKT TCR proteins behaved similarly to the WT NKT TCR in gel filtration and analysis under reducing and nonreducing SDS-PAGE. Furthermore, the NKT TCR mutant proteins were as reactive as the WT NKT TCR to a panel of conformation-sensitive mAbs reactive against regions of the Vα24 (A2G10) and Vβ11 (1A6 and A2G6) variable domains and the TCR constant domain (12H8). However, the Pro28αAla mutant was the exception, as there was no reactivity with the Vα24 (A2G10) mAb, but equal reactivity with mAbs toward the Vβ11 (1A6 and A2G6) and constant domain (12H8). Accordingly, the controls were consistent with the NKT TCR mutants; with the exception of Pro28α, they all retained their native conformation (unpublished data).
Table I.
The effect of NKT TCR alanine substitutions on CD1d–α-GalCer binding
| Residue | mouse CD1d K deq | human CD1d K deq | human CD1d–α-GalCer contact | Effect on affinity |
|---|---|---|---|---|
| μM | μM | |||
| NKT WT | 0.94 ± 0.13 | 0.46 ± 0.05 | – | – |
| CDR1α | ||||
| Pro28α | 4.75 ± 0.13 | 5.44 ± 0.23 | α-GalCer Galactose; 6′-OH, 5′-O | *** |
| Ser30α | 1.16 ± 0.05 | 0.70 ± 0.07 | α-GalCer Galactose; 3′-OH, 4′-OH | * |
| CDR3α | ||||
| Asp94αa | >58 | >58 | α1-helix; Arg79 | *** |
| Arg95αa | >58 | >58 | α1-helix; Ser76, Arg79, Asp80 | *** |
| α-GalCer Galactose; 2′-OH, C-2 | ||||
| Sphingosine; 3′-OH, 4′-OH, C-3, C-4 | ||||
| Gly96αa | 1.19 ± 0.12 | 1.07 ± 0.13 | α2-helix; Gln150, Asp151 | * |
| α-GalCer Galactose; 2′-OH, 3′-OH, C-2 | ||||
| Ser97αa | >58 | >58 | α2-helix; Val147, Gln150 | *** |
| Thr98α | 0.90 ± 0.05 | 0.81 ± 0.07 | α2-helix; Gln150 | * |
| Leu99αa | >58 | >58 | α1-helix; Asp80, Phe84; | *** |
| α2-helix; Val147 | ||||
| Arg103αa | 2.56 ± 0.24 | 0.39 ± 0.06 | α1-helix; Arg79c | * |
| CDR2β | ||||
| Tyr48βb | >30 | >30 | α1-helix; Glu83, Lys86 | *** |
| Tyr50β | >25 | >25 | α1-helix; Glu83, Met87 | *** |
| Asn53β | 1.19 ± 0.12 | 0.73 ± 0.09 | α1-helix; Arg89c | * |
| Glu56βb | 0.72 ± 0.11 | 0.40 ± 0.03 | α1-helix; Lys86 | * |
| CDR3β | ||||
| Tyr103β | 0.82 ± 0.06 | 0.71 ± 0.13 | α2-helix; Gln150 | * |
* represents an effect in which NKT TCR alanine substitution results in a <4-fold reduction in binding to CD1d–α-GalCer compared to the WT value. *** represents an effect in which NKT TCR alanine substitution with a >10-fold reduction in binding to CD1d–α-GalCer when compared to the WT value.
Conserved between the human Jα18 gene segment and the mouse Jα18 gene segment homologue.
CDR2β framework residues.
Denotes contacts that are not maintained in the second NKT TCR–CD1d–α-GalCer complex within the asymmetric unit.
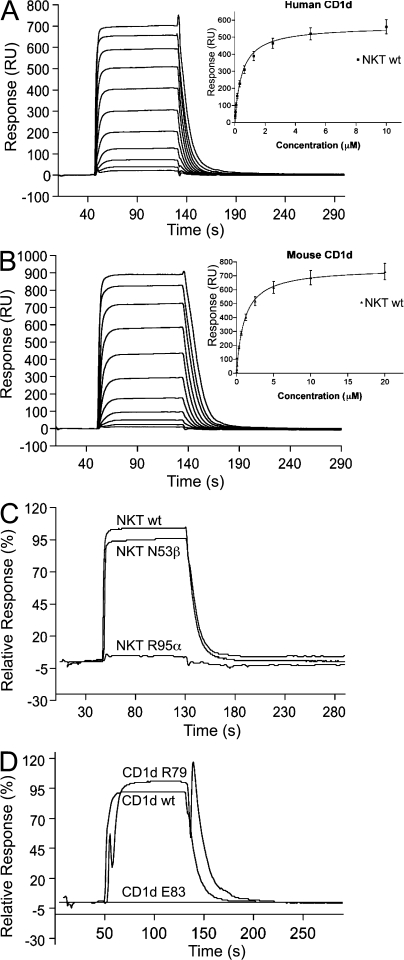
Varying concentrations of the NKT TCR were passed over a research-grade streptavidin sensor chip (GE Healthcare) coupled with biotinylated mouse or human CD1d loaded with α-GalCer and the corresponding unloaded equivalents. The final response was determined by subtracting the unloaded CD1d response from the CD1d–α-GalCer response and the equilibrium binding constant (K deq) was calculated (Fig. 2, A–C). The interaction of WT NKT TCR and human CD1d–α-GalCer had a K deq of ∼0.5 ± 0.05 μM, whereas the K deq for mouse CD1d–α-GalCer was ∼0.9 ± 0.13 μM (Table I) and were broadly consistent with previous measurements (25, 26). Although these WT K deq measurements were reproducible on two separate occasions and using two separate batches of WT NKT TCR, we nevertheless considered it important that all the mutant NKT TCR proteins were analyzed in the same experiment, in duplicate and relative to the same batch of WT NKT TCR. This was to ensure that all the K deq values were comparable and relative to one another (Table I). NKT TCR substitutions that caused less than a fourfold loss in the affinity of the interaction with CD1d–α-GalCer compared with WT NKT TCR were considered to have no major effect. NKT TCR substitutions that caused more than a 10-fold loss in binding affinity were considered crucial to the energetics of the interaction. In each instance, the effect of the NKT TCR substitution on the affinity of the interaction was essentially consistent between mouse and human CD1d–α-GalCer.
Figure 2.
Binding analysis of NKT TCR to human and mouse CD1d–α-GalCer using surface plasmon resonance. A concentration series of the WT NKT TCR was passed over human CD1d–α-GalCer (A) and mouse CD1d–α-GalCer (B). (insets) The equilibrium response versus concentration. (C) Comparison of the relative response of 10 μM of the WT NKT TCR and the Asn53β and Arg95α mutant NKT TCRs to human CD1d–α-GalCer. (D) Comparison of the relative response of 6.25 μM WT NKT TCR to WT human CD1d–α-GalCer and the CD1d mutant proteins carrying Arg79 and Glu83 substitutions.
The CDR1α loop
We mutated Pro28α and Ser30α within the CDR1α loop, both of which interact exclusively with the galactose moiety of α-GalCer (Fig. 1 B). The Ser30αAla substitution did not alter the affinity of the interaction with hCD1d–α-GalCer, even though it forms H-bonds and van der Waals (vdw) contacts with the 3′- and 4′- hydroxyls of the galactose moiety, respectively (Fig. 1 B). This lack of effect was surprising, as the mouse NKT TCR can distinguish between α-GalCer analogues that possess minor structural modifications on the saccharide, such as the positioning of the 2′ and 4′ hydroxyls in α-ManCer (29). Although the Ser at position 30 is not conserved in the mouse NKT TCR homologue, the equivalent substitution in the mouse system (Asn30α→Ala) resulted in a greater than twofold loss of binding of mCD1d tetramers loaded with α-GalCer and the α-GalCer analogues PBS57 and OCH9 (28).
Pro28α contacts the 6′-OH and 5′-O of the galactose moiety of α-GalCer. Although the Pro28αAla substitution had a marked effect on the affinity of the interaction with hCD1d–α-GalCer, our mAb reactivity data suggests the Pro28αAla substitution had an overall effect on the conformational integrity of the Vα24 domain of the NKT TCR, making it difficult to determine whether this substitution was important in the interaction.
The CDR3α loop
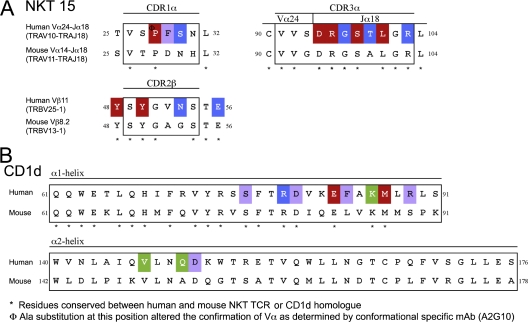
Seven mutations were made in this loop (Table I and Fig. 1, A and C), three of which (Gly96α, Thr98α, and Arg103α) had a negligible effect on the affinity of the interaction. The lack of an effect with the Thr98αAla substitution was not surprising, as the cross-reactive mouse NKT TCR possesses an Ala at that position (Fig. 3 A). Even though Arg103α is conserved in the mouse NKT TCR homologue, its substitution to Ala had no effect on the recognition of hCD1d–α-GalCer. This residue forms an H-bond with the Arg79 on the α1-helix of CD1d in one ternary complex within the asymmetric unit (Table I). The lack of effect of the Gly96αAla substitution was surprising, however, as the main chain of Gly96α H-bonded to the 2′-OH of α-GalCer and also interacted with the 3′-OH of α-GalCer and residues within the α2-helix of CD1d. Nevertheless, four (Asp94α, Arg95α, Ser97α, and Leu99α) of the seven residues substituted within the CDR3α loop of the NKT TCR had a marked effect on the affinity of the interaction with hCD1d–α-GalCer (Table I and Fig. 1 C). The substitutions at these critical positions would result in the loss of salt-bridging interactions with Arg79 and Asp80, and vdw interactions with Ser76 from the α1-helix of CD1d, as well as vdw interactions with residues from the α1- and α2-helices of CD1d. Accordingly, some, but not all of the residues in the strictly conserved Jα18 gene are critically important in the recognition of CD1d.
Figure 3.
Sequence alignment of CD1d and the NKT TCR from both human and mouse. Sequence comparison of the CDR1α, CDR3α, and CDR2β loops of the mouse and human NKT TCR homologues (A) and the CD1d α1- and α2-helices (B). Substituted residues that have a >10-fold reduction in affinity are shown in red; a 4–6-fold reduction in affinity are shown in green; and a <4-fold reduction in affinity are shown in blue; residues that contact either CD1d–α-GalCer or the NKT TCR that were not substituted, purple.
The CDR3β loop
Tyr103β, which is located on the Jβ gene segment of the CDR3β loop of the NKT TCR, is the sole CDR3β residue that contacts CD1d. The Tyr103βAla substitution resulted in the loss of a vdw contact with the α2-helix of hCD1d, but this loss was inconsequential to the affinity of the interaction with hCD1d–α-GalCer, further highlighting the lack of a role of the CDR3β loop of this NKT TCR in the interaction with CD1d–α-GalCer (Fig. 1 D).
The CDR2β loop
The CDR2β loop dominated contacts from the Vβ11 chain of the NKT TCR. Four residues within or surrounding this loop were mutated, but only two of these residues, Tyr48β and Tyr50β, were critical to the energetics of the interaction. These residues are conserved within the mouse NKT TCR homologue and H-bonded to Glu83 and Lys86, as well as forming vdw interactions with residues from the α1-helix (Fig. 1 E). Asn53β and Glu56β were shown to be nonessential residues in the interaction. Accordingly, only two aromatic residues within and next to the CDR2β loop are critical for the interaction with CD1d–α-GalCer.
CD1d substitutions
Based on the crystal structure of the NKT TCR–CD1d–α-GalCer complex, six “alanine-scanning” CD1d mutants were produced, including Arg79, Glu83, Lys86, and Met87 on the CD1d α1-helix and Val147 and Gln150 on the α2-helix. The CD1d mutant proteins were expressed at similar levels to WT CD1d, behaved similarly under reducing and nonreducing SDS-PAGE, and were as reactive in an ELISA probed with anti–human CD1d mAb 51.1 (unpublished data) (30). CD1d proteins were biotinylated, loaded, and coupled to a research-grade streptavidin sensor chip and analyzed for their reactivity against varying concentrations of the WT NKT TCR after the subtraction of the unloaded CD1d response (Fig. 2 D).
The effect of the CD1d substitutions can be grouped into the following three categories: no effect, a moderate effect of 4–6-fold loss of affinity, and a drastic effect of >10-fold loss of affinity (Table II). Arg79, which contacts three residues within the Jα18 region, was the only CD1d residue whose substitution had no effect on the interaction with the NKT TCR (Table II). Two of the substitutions that had the greatest impact on affinity included Glu83 and Met87, which are both located on the α1-helix of CD1d. These substitutions would result in a loss of contacts with Tyr48β and Tyr50β on the CDR2β loop, thereby correlating with the CDR2β mutagenesis (Table I). Consistent with our observation, a mouse CD1d transfectant bearing an Ala substitution at position 83 showed impaired stimulation of semiinvariant mouse NKT cells (31, 32). In addition, Lys86 contacts Tyr48β from the CDR2β loop and also salt bridges to Glu56β, and the Lys86 to Ala substitution had a moderate effect on the interaction with the NKT TCR.
Table II.
The effect of CD1d alanine substitutions on NKT TCR binding
| NKT15 K deq | NKT15 contact | Effect on affinity | ||
|---|---|---|---|---|
| μM | ||||
| WT human CD1d | 0.58 ± 0.13 | - | - | |
| Arg79Aa | 0.35 ± 0.02 | Asp94α, Arg95α, Arg103αb | Jα | * |
| Glu83Aa | >25 | Tyr48β, Tyr50β | 2β | *** |
| Lys86Aa | 2.83 ± 0.13 | Tyr48β | 2β | ** |
| Met87Aa | 7.9 ± 0.32 | Tyr50β | 2β | *** |
| Val147Aa | 3.6 ± 0.17 | Ser97α, Leu99α | Jα | ** |
| Gln150A | 2.5 ± 0.19 | Gly96α, Ser97α, Thr98α, Tyr103β | Jα | ** |
* represents CD1d alanine substitution with <4-fold reduction in binding to NKT TCR compared to the WT value; ** represents CD1d alanine substitution resulting in a 4–6-fold reduction in binding to NKT TCR compared to the WT value; and *** represents CD1d alanine substitution resulting in a >10-fold reduction in binding to NKT TCR compared to the WT value.
Conserved between human and mouse CD1d.
Denotes contacts that are not maintained in the second NKT TCR–CD1d–α-GalCer complex within the asymmetric unit.
Two CD1d residues located on the α2-helix (Val147 and Gln150) were substituted, and each had a moderate effect on the affinity of the interaction with the NKT TCR (Table II). Both contacted residues within the highly selected Jα18 segment of the NKT TCR, with the former making vdw contacts with Ser97α and Leu99α, and the latter with Gly96α, Ser97α, Thr98α, and Tyr103β. The effect of the Val147 to Ala mutant concurs with the NKT TCR mutagenesis data, whereby the reciprocal substitutions of both Ser97α and Leu99α resulted in a marked loss of recognition of CD1d–α-GalCer. On the other hand, not all of the NKT TCR contacts of CD1d residue Gln150 were detrimental to the recognition event when they themselves were substituted. Indeed, only the Ser97α substitution caused a loss, again highlighting the important role of this CDR3α residue.
The energetic hotspots
Next, we mapped the location of NKT TCR residues that were substituted in this study onto the structure of the NKT TCR to ascertain the location and size of the energetic hotspot (Fig. 4 A). The substitutions that had a marked effect on the affinity of the interaction formed a central hotspot over the surface of the antigen-binding domain, whereas noncritical residues were located peripheral to the hotspot (Fig. 4 A). The hotspot included some of the residues from the CDR3α loop (Asp94α, Arg95α, Ser97α, and Leu99α), residues from within or surrounding the CDR2β loop (Tyr48β and Tyr50β).
Figure 4.
Energetic hotspots. (A) The NKT TCR hotspot. Surface representation of the NKT TCR molecule. Yellow, Vα; pale blue, Vβ. The NKT TCR CDR1α, 3α, 2β, and 3β loops and the CD1d α1- and α2-helices are shown as a cartoon representation. (B) The human CD1d hotspot. Surface representation of the human CD1d molecule presenting α-GalCer (pink ball and stick). A cartoon representation of the CD1d α-helices and selected CDR loops are also displayed (C) Critical residues for NKT TCR–CD1d–α-GalCer recognition are biased toward the F′ pocket region. Substituted residues that have >10-fold reduction in affinity are shown in red; 4–6-fold reduction in affinity are shown in green; <4-fold reduction in affinity are shown in blue; residues that were not substituted are shown in purple; α-GalCer is shown in pink; CDR1α loop is shown in yellow; CDR3α is shown in cyan; CDR2β loop is shown in orange; CDR3β loop is shown in blue; and CD1d α-helices are shown in grey.
Similarly, to define CD1d residues that were critical to the NKT TCR interaction, we mapped the residues that were substituted in this study, highlighting residues that formed crucial contacts with the NKT TCR (Fig. 4 B). The CD1d substitutions that caused the greatest loss of NKT TCR recognition included Glu83 and Met87, indicating the disruption of contacts with the α1-helix were the most critical. However, the substitution of other residues on the CD1d α1- and α2-helix, including Lys86, Val147, and Gln150, also caused a moderate loss in the affinity of the interaction. Collectively, these and previous mutagenesis results indicate that the CD1d energetic footprint is localized and centrally located above the F′ pocket of CD1d (Fig. 4, B and C).
Cross-species reactivity
In addition to examining the effect of the 14 NKT TCR mutations on the interaction with hCD1d, we also examined the effect of these mutations on the interaction with mCD1d–α-GalCer (Table I). Although the interaction with mCD1d was consistently approximately twofold weaker when compared with hCD1d, the effect of the NKT TCR mutations on the mCD1d interaction essentially parallelled that observed in our hCD1d–α-GalCer study (Table I). Moreover, of the six residues that were important in the NKT TCR, Asp94α, Arg95α, Ser97α, Leu99α, Tyr48β, and Tyr50β are all conserved in the mouse Vα14Jα18-Vβ8.2 NKT TCR (Table I and Fig. 3 A). Consistently, mouse NKT cells expressing TCRs with these identical individual substitutions (with the exception of Tyr48β, which was not tested) had a marked reduction in mCD1d–α-GalCer tetramer binding (28). Furthermore, of the five CD1d residues that had a moderate-to-marked effect on the binding of the NKT TCR (Glu83, Lys86, Me87, Val147, and Gln150), all but one are conserved in the mouse CD1d homologue (Gln150 to Ala 152; Table II and Fig. 3 B). Collectively, these results indicate that a similar footprint underpins the human–mouse cross-species reactivity, which is reinforced by sequence and structural analyses.
α-GalCer analogues
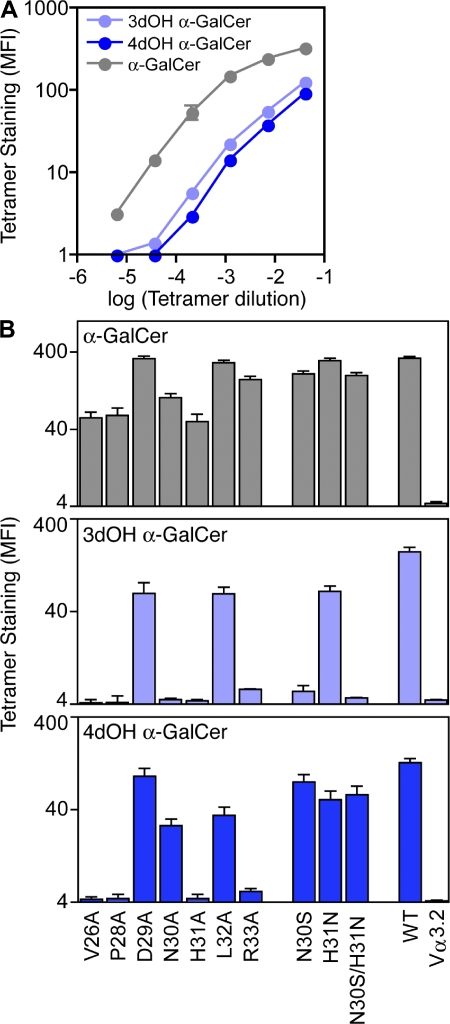
Having shown which NKT TCR residues are required to interact with hCD1d, and that a common structural footprint underpins the reciprocal cross-species reactivity, we then probed the role of the CDR1α loop in interacting with the α-GalCer. Given that the majority of the CDR1α-mediated interactions are with the 3′- and 4′-hydroxyl groups of α-GalCer, we used α-GalCer analogues deficient in either one of these groups. First, we used mouse CD1d tetramers loaded with the α-GalCer analogues and assessed the role these modifications had on the staining of cells expressing the WT mNKT TCR and CDR1α mutants thereof (Fig. 5). The data indicated that WT mNKT TCR could interact with all three analogues of α-GalCer in the following order: α-GalCer>4′-deoxy≈3′-deoxy. Of the alanine-scanning mutations, positions 26 and 28 are likely to affect the conformation and/or mobility of the CDR1α loop, thereby highlighting the overall importance of the conformation of this loop in recognition of α-GalCer. Positions 29, 32, and 33 are, by analogy to the human NKT TCR–CD1d complex, unlikely to participate in contacting α-GalCer, and this was evident from the mutagenesis data. However, the substitutions at positions 30 and 31 of the mNKT TCR are likely to be more specific in the disruption of contacts with CD1d–α-GalCer. Overall, the mutations indicate that substitutions at the 3′-OH are less well tolerated than substitutions at the 4′-OH for α-GalCer.
Figure 5.
α-GalCer analogues. (A) Hybridomas expressing wild-type mouse Vα14i TCRα chain were subjected to staining with the indicated dilutions of mouse CD1d tetramers left unloaded or loaded with α-GalCer, 3′deoxy-α-GalCer, or 4′deoxy-α-GalCer, and mean fluorescence intensity (MFI) of tetramer staining was determined from a thin TCR gate. MFI of background staining with unloaded tetramer was determined for each concentration and subtracted from the MFI of loaded tetramer. Data points represent the mean of MFI ± the range from two independent experiments. (B) Hybridomas expressing WT or indicated mutations of mouse Vα14i TCRα chain were subjected to staining with mouse CD1d tetramers loaded with α-GalCer (top), 3′deoxy-α-GalCer (middle), or 4′deoxy-α-GalCer (bottom). Nonmutated Vα14-Jα18 TCRα chain served as the WT control. The negative control was a Vα14-Jα18 construct in which the Vα14 CDR1 region was swapped for the Vα3.2 CDR1α region (Vα3.2). MFI of tetramer staining for all mutants was determined from a thin TCR gate. Error bars represent the mean of MFI ± the range for two independent experiments.
Furthermore, to account for the sequence variation between the mouse and human CDR1α loops, we reverted single substitutions within the mouse NKT TCR CDR1α loop back to their human counterpart, i.e., Asn30αSer and His31αAsn and also the double Ser-Asn mutant at positions 30 and 31. In comparison to the alanine-scanning mutants, the binding affinity of these mutants was more comparable to the WT mNKT TCR, indicating that the sidechains at positions 30 and 31 contribute some energy, albeit modest, to the interaction (Fig. 5). For example, the hNKT TCR–CD1d–α-GalCer structure reveals that Ser30α H-bonded to the 3′-OH of α-GalCer. This is consistent with the Asn30Ser mutant not being able to rescue binding to the 3′-deoxy derivative of α-GalCer, yet can partially improve the binding affinity of the 4′-deoxy derivative. Interestingly, the Asn30Ala mutant appeared to affect the binding to the 3′- and 4′-deoxy derivatives of α-GalCer (Fig. 5), which, unlike Ser30α of hNKT TCR, suggests that Asn30 of mNKT TCR forms H-bonds to both these moieties, which would be consistent with its longer sidechain.
DISCUSSION
At a structural level, the vast T cell repertoire is manifested in the various ways in which a TCR can interact with the polymorphic pMHC. Despite the repertoire diversity, examples of TCR bias have emerged in antiviral immunity, and such biased TCR usage has been correlated with the recognition of atypical pMHC landscapes (33). The human semiinvariant NKT TCR is an example of biased TCR usage directed against a monomorphic CD1d molecule, in which the NKT TCR α chain is invariant and the β chain is restricted to Vβ11 usage, albeit with variable CDR3β usage. In comparison to the previously determined TCR–pMHC complexes, the NKT TCR adopted an unusual docking strategy to interact with the featureless CD1d–α-GalCer complex (27). We therefore sought to determine the energetic landscape of the NKT TCR–CD1d–α-GalCer interaction.
Our observations demonstrate that although the NKT TCR structural footprint is small, the energetic footprint is smaller still, as not all of the residues of the NKT TCR that mediated contacts with CD1d–α-GalCer contributed to the energetic landscape of the interaction. Namely, we substituted 14 residues of the NKT TCR that contact CD1d–α-GalCer and of these, only 6 contributed to the energetic footprint, with the CDR3α and CDR2β loops of the NKT TCR appearing to be the principal driving force of the interaction. The requirement of the CDR3α loop in the energetics of the interaction argues against the two-step model in TCR-CD1d recognition. The NKT TCR and CD1d substitution results reveal that although only a few contacts are mediated via the Vβ domain, Tyr48β, Tyr50β, and the corresponding binding partners on CD1d are absolutely critical to the interaction. These two Vβ residues have been identified as “recognition codons” in class II–restricted mouse TCRs using the human Vβ11 chain homologue Vβ8.2. The recognition codons are implicated in restricting and defining the binding orientation of the TCR onto the MHC by penetrating between residues on the MHC helices (34). The general use of Tyr48β and Tyr50β highlights a potential evolutionary role of the NKT TCR CDR2β loop for CD1d binding and the use of recognition codons for MHC-like molecules, although it should be noted that the “interaction codon” sites in MHC and CD1d do not reside within structurally equivalent regions (35).
Although some residues from within the CDR3α loop were also deemed critical for the interaction with CD1d, Arg95α was the only energetically important residue that interacts with the Ag. Accordingly, NKT TCR residues interacting with the CD1d seemingly had a greater effect on the energetics of the interaction than those directly contacting the glycolipid ligand, at least in the case of α-GalCer. However, this finding is anomalous given that α-GalCer is a requirement for the staining of NKT cells by CD1d−α-GalCer tetramers and for NKT cell activation. This suggests that the α-GalCer ligand may play a less direct role in the NKT TCR interaction and a more prominent role in influencing the conformation of residues on CD1d, which in turn may influence NKT TCR recognition. Indeed, a comparison of liganded versus unliganded CD1d–α-GalCer (36) shows that four CD1d residues change conformation upon ligation, and all are located on a small stretch of the α1-helix just above the F′ pocket. Furthermore, our CD1d substitution analysis revealed two residues that had a drastic effect on the interaction with the NKT TCR (Glu83 and Met87) and both are located on the α1-helix above the F′ pocket. Consistent with this notion, the conformation of CD1d has been proposed to modulate the affinity of the mouse NKT TCR to two ligands (OCH9 and α-GalCer) that differ only in the length of the predominantly buried sphingosine chain (37).
The apparent lesser contribution of the CDR1α loop was surprising, as this loop solely contacted the α-GalCer, and the loss of binding caused by the Pro28α Ala substitution may be attributable to local changes in the conformation of the loop. However, we cannot exclude the possibility that redundancy exists in the interactions between residues containing α-GalCer that would not have been detected in our single amino acid mutagenesis studies. It also remains possible that substitutions within this loop may have a greater influence on the recognition of other ligands bound to CD1d, and this was analyzed using analogues of α-GalCer. It was clear that the mNKT TCR could interact with the 3′- and 4′-deoxy-galactosyl ceramides, which is consistent with the hNKT TCR CDR1α mutagenesis (Table I). Nevertheless, the alanine-scanning and “humanized” mutagenesis on the CDR1α loop indicated that removal of the 4′-OH moiety was more tolerated than the 3′-OH substitution. The apparent adaptability of the CDR1α–α-GalCer interactions is consistent not only with the relatively low sequence conservation in the CDR1α loop between the mouse and human NKT TCRs (17), but also the presence of some CD1d-restricted, α-GalCer–reactive, Vα24-negative NKT TCRs that differ in the sequence of the CDR1α loop (22, 26).
In contrast to TCR–pMHC interactions, the NKT TCR exhibits reciprocal cross-species reactivity, and the effects of the NKT TCR mutations display an identical pattern when interacting with human CD1d or mouse CD1d, suggesting that the human NKT TCR will dock in a very similar manner on mouse CD1d compared with human CD1d. The similarity of the human NKT TCR footprint on both mouse and human CD1d molecules appears to engender reciprocal cross-reactivity. In summary, the markedly different docking strategies between TCR–pMHC and TCR–CD1d complexes are also mirrored by differences in the energetic footprints between peptide and glycolipid-restricted TCRs.
MATERIALS AND METHODS
Protein expression, refolding, and purification.
The cloning, expression, and purification of the human NKT TCR used in this study (α chain: TRAV10-TRAJ18; β chain TRBV25-1, TRBD1 and TRBJ2-7, with CDR3β sequence 92CASS95 96GLRDRGL102 103Y105EQYFGPGTRLTVT117; and NKT15) has been previously described (25). However, the NKT TCR proteins were further purified using hydrophobic interaction chromatography and a Phenyl HP HiTrap column (GE Healthcare). The NKT TCR was eluted with 20 mM Hepes, pH 7.4, and 0.8 M ammonium sulfate, concentrated, and buffer-exchanged into 10 mM Hepes, pH 7.4, 150 mM NaCl using a 30K Amicon Ultra-4 concentrator (Millipore). The cloning, expression, purification, biotinylation, and loading of human CD1d have been previously described (25). Purified, biotinylated mouse CD1d was supplied by D. Pellicci (University of Melbourne, Parkville, Victoria, Australia) and was generated using a construct originally provided by M. Kronenberg (University of Melbourne).
NKT TCR mutants.
The WT human NKT TCR α- or β chain DNA was used as the template to generate the NKT TCR mutants using site-directed mutagenesis (QuikChange; Stratagene). A total of 14 mutant NKT TCR mutant proteins were made, each coding for a single amino acid substitution to Ala, and each was expressed and purified as per the WT human NKT TCR.
CD1d mutants.
WT CD1d DNA coding for a free C-terminal Cys was used to generate six CD1d mutants using site-directed mutagenesis. The CD1d mutant proteins each contained a single alanine substitution and were expressed in the baculovirus expression system. The mutant proteins were purified, biotinylated, and loaded as per WT CD1d, and then coupled to a SA sensor chip for surface plasmon resonance analysis with WT NKT TCR. Anti-NKT TCR mAbs were provided by N. Crowe, K. Kyparissoudis, and D. Pellicci (University of Melbourne).
ELISA.
The conformational integrity of the CD1d and NKT TCR proteins was determined using an ELISA and relevant conformation-specific mAbs. The integrity of the CD1d proteins was determined using an anti–human CD1d mAb 51.1 (30), whereas the conformation of the NKT TCR proteins was determined using a panel of mAbs including a Vα24-reactive mAb (A2G10), two Vβ11-reactive mAbs (A2G6; 1A6), and a pan-soluble αβ TCR-reactive mAb (12H8). ELISA plates were coated with the CD1d or NKT TCR proteins at 5 or 10 μg/ml, respectively, and tested against serial dilutions of the relevant mAbs. The ELISA was then probed with HRP anti–mouse IgG, followed by OPD substrate, and the ELISA plate read at 492 nm. WT CD1d and NKT TCR proteins were used as positive controls, whereas a recombinant Vα24− Vβ11− TCR, together with its cognate peptide-MHC ligand, was included as a negative control. All experiments were performed at least in duplicate.
Surface plasmon resonance measurements and analysis.
Equilibrium affinity measurements of the NKT TCR–CD1d–α-GalCer interaction were determined by surface plasmon resonance and have been previously described (25). For the NKT TCR mutant analysis, ∼3,000 RU of biotinylated WT human and mouse CD1d were coupled to a SA sensor chip (GE Healthcare) and analyzed against twofold serial dilutions of the WT and mutant NKT TCR. For the CD1d mutant analysis, ∼3,000 RU of human mutant or WT CD1d protein was coupled to a SA sensor chip and analyzed against the WT NKT TCR. For both experiments, the analyte was passed over the sensor chip at 5 μl/min for 80 s at 25°C, and the final response was subtracted from that of unloaded CD1d.
Staining of hybridomas expressing mouse Vα14i TCRs.
Hybridomas expressing WT or mutant mouse Vα14i TCR chains were generated by retroviral transduction of a hybridoma expressing only the DO-11.10 Vβ8.2 TCRβ chain, as previously described (28). For staining with mouse CD1d tetramers, unloaded, biotinylated recombinant CD1d protein was provided by the National Institutes of Health core facility. For multimerization, monomeric biotinylated recombinant CD1d was incubated overnight with α-GalCer, C3′-deoxy-α-GalCer, or C4′deoxy-α-GalCer in PBS and 0.05% Tween 20, followed by addition of streptavidin (SA)-PE. TCR-expressing hybridomas were costained at room temperature for 60 min with indicated tetramer plus anti-TCRβ (H57-597; eBioscience) and data were acquired on a FACSCalibur or FACScan flow cytometer (BD Biosciences). Data files were analyzed using FlowJo software (Tree Star, Inc.) and mean fluorescence intensity was compared for all samples on a narrow TCR gate, as previously described (28).
Acknowledgments
We thank D. Pellicci, N. Crowe, K. Kyparissoudis, and M. Kronenberg for the provision of reagents. We thank Malcolm McConville and Nick Dixon for discussions pertaining to the manuscript.
The Australian Research Council (ARC), the National Health and Medical Research Council of Australia (NHMRC), and the Cancer Council of Victoria supported this research. L. Gapin was supported by a National Institutes of Health (NIH) grant (AI057485), A.R. Howel by NIH grant (AI057519). N.A. Borg and T. Beddoe are supported by a NHMRC Career Development Award, D.I. Godfrey by a NHMRC Principal Research Fellowship, and J. Rossjohn by an ARC Federation Fellowship.
The authors have no conflicting financial interests.
Abbreviations used: α-GalCer, α-galactosylceramide; Ag, antigen; pMHC, peptide-MHC; vdw, van der Waals.
K.S. Wun and N.A. Borg contributed equally to this paper.
References
- 1.Zinkernagel, R.M., and P.C. Doherty. 1997. The discovery of MHC restriction. Immunol. Today. 18:14–17. [DOI] [PubMed] [Google Scholar]
- 2.Brigl, M., and M.B. Brenner. 2004. CD1: antigen presentation and T cell function. Annu. Rev. Immunol. 22:817–890. [DOI] [PubMed] [Google Scholar]
- 3.Ulrichs, T., and S.A. Porcelli. 2000. CD1 proteins: targets of T cell recognition in innate and adaptive immunity. Rev. Immunogenet. 2:416–432. [PubMed] [Google Scholar]
- 4.Moody, D.B., D.M. Zajonc, and I.A. Wilson. 2005. Anatomy of CD1-lipid antigen complexes. Nat. Rev. Immunol. 5:387–399. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi, E., K. Motoki, T. Uchida, H. Fukushima, and Y. Koezuka. 1995. KRN7000, a novel immunomodulator, and its antitumor activities. Oncol. Res. 7:529–534. [PubMed] [Google Scholar]
- 6.Godfrey, D.I., J. McCluskey, and J. Rossjohn. 2005. CD1d antigen presentation: treats for NKT cells. Nat. Immunol. 6:754–756. [DOI] [PubMed] [Google Scholar]
- 7.Stronge, V.S., M. Salio, E.Y. Jones, and V. Cerundolo. 2007. A closer look at CD1d molecules: new horizons in studying NKT cells. Trends Immunol. 28:455–462. [DOI] [PubMed] [Google Scholar]
- 8.Clements, C.S., M.A. Dunstone, W.A. Macdonald, J. McCluskey, and J. Rossjohn. 2006. Specificity on a knife-edge: the alphabeta T cell receptor. Curr. Opin. Struct. Biol. 16:787–795. [DOI] [PubMed] [Google Scholar]
- 9.Rudolph, M.G., R.L. Stanfield, and I.A. Wilson. 2006. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24:419–466. [DOI] [PubMed] [Google Scholar]
- 10.Matsui, K., J.J. Boniface, P. Steffner, P.A. Reay, and M.M. Davis. 1994. Kinetics of T-cell receptor binding to peptide/I-Ek complexes: correlation of the dissociation rate with T-cell responsiveness. Proc. Natl. Acad. Sci. USA. 91:12862–12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsui, K., J.J. Boniface, P.A. Reay, H. Schild, B. Fazekas de St Groth, and M.M. Davis. 1991. Low affinity interaction of peptide-MHC complexes with T cell receptors. Science. 254:1788–1791. [DOI] [PubMed] [Google Scholar]
- 12.Lee, P.U., H.R. Churchill, M. Daniels, S.C. Jameson, and D.M. Kranz. 2000. Role of 2CT cell receptor residues in the binding of self– and allo–major histocompatibility complexes. J. Exp. Med. 191:1355–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manning, T.C., C.J. Schlueter, T.C. Brodnicki, E.A. Parke, J.A. Speir, K.C. Garcia, L. Teyton, I.A. Wilson, and D.M. Kranz. 1998. Alanine scanning mutagenesis of an alphabeta T cell receptor: mapping the energy of antigen recognition. Immunity. 8:413–425. [DOI] [PubMed] [Google Scholar]
- 14.Borg, N.A., L.K. Ely, T. Beddoe, W.A. Macdonald, H.H. Reid, C.S. Clements, A.W. Purcell, L. Kjer-Nielsen, J.J. Miles, S.R. Burrows, et al. 2005. The CDR3 regions of an immunodominant T cell receptor dictate the ‘energetic landscape’ of peptide-MHC recognition. Nat. Immunol. 6:171–180. [DOI] [PubMed] [Google Scholar]
- 15.Wu, L.C., D.S. Tuot, D.S. Lyons, K.C. Garcia, and M.M. Davis. 2002. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature. 418:552–556. [DOI] [PubMed] [Google Scholar]
- 16.Godfrey, D.I., and M. Kronenberg. 2004. Going both ways: immune regulation via CD1d-dependent NKT cells. J. Clin. Invest. 114:1379–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lefranc, M.P. 2001. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 29:207–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gadola, S.D., N. Dulphy, M. Salio, and V. Cerundolo. 2002. Valpha24-JalphaQ-independent, CD1d-restricted recognition of alpha-galactosylceramide by human CD4(+) and CD8alphabeta(+) T lymphocytes. J. Immunol. 168:5514–5520. [DOI] [PubMed] [Google Scholar]
- 19.Dellabona, P., E. Padovan, G. Casorati, M. Brockhaus, and A. Lanzavecchia. 1994. An invariant Vα 24-Jα Q/Vβ 11 T cell receptor is expressed in all individuals by clonally expanded CD4−8− T cells. J. Exp. Med. 180:1171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lantz, O., and A. Bendelac. 1994. An invariant T cell receptor α chain is used by a unique subset of major histocompatibility complex class I–specific CD4+ and CD4−8− T cells in mice and humans. J. Exp. Med. 180:1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porcelli, S., C.E. Yockey, M.B. Brenner, and S.P. Balk. 1993. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4−8− α/β T cells demonstrates preferential use of several Vβ genes and an invariant TCR α chain. J. Exp. Med. 178:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brigl, M., P. van den Elzen, X. Chen, J.H. Meyers, D. Wu, C.H. Wong, F. Reddington, P.A. Illarianov, G.S. Besra, M.B. Brenner, and J.E. Gumperz. 2006. Conserved and heterogeneous lipid antigen specificities of CD1d-restricted NKT cell receptors. J. Immunol. 176:3625–3634. [DOI] [PubMed] [Google Scholar]
- 23.Brossay, L., M. Chioda, N. Burdin, Y. Koezuka, G. Casorati, P. Dellabona, and M. Kronenberg. 1998. CD1d-mediated recognition of an alpha-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J. Exp. Med. 188:1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benlagha, K., A. Weiss, A. Beavis, L. Teyton, and A. Bendelac. 2000. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J. Exp. Med. 191:1895–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kjer-Nielsen, L., N.A. Borg, D.G. Pellicci, T. Beddoe, L. Kostenko, C.S. Clements, N.A. Williamson, M.J. Smyth, G.S. Besra, H.H. Reid, et al. 2006. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J. Exp. Med. 203:661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gadola, S.D., M. Koch, J. Marles-Wright, N.M. Lissin, D. Shepherd, G. Matulis, K. Harlos, P.M. Villiger, D.I. Stuart, B.K. Jakobsen, et al. 2006. Structure and binding kinetics of three different human CD1d-α-galactosylceramide–specific T cell receptors. J. Exp. Med. 203:699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borg, N.A., K.S. Wun, L. Kjer-Nielsen, M.C. Wilce, D.G. Pellicci, R. Koh, G.S. Besra, M. Bharadwaj, D.I. Godfrey, J. McCluskey, and J. Rossjohn. 2007. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 448:44–49. [DOI] [PubMed] [Google Scholar]
- 28.Scott-Browne, J.P., J.L. Matsuda, T. Mallevaey, J. White, N.A. Borg, J. McCluskey, J. Rossjohn, J. Kappler, P. Marrack, and L. Gapin. 2007. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat. Immunol. 8:1105–1113. [DOI] [PubMed] [Google Scholar]
- 29.Sidobre, S., K.J. Hammond, L. Benazet-Sidobre, S.D. Maltsev, S.K. Richardson, R.M. Ndonye, A.R. Howell, T. Sakai, G.S. Besra, S.A. Porcelli, and M. Kronenberg. 2004. The T cell antigen receptor expressed by Valpha14i NKT cells has a unique mode of glycosphingolipid antigen recognition. Proc. Natl. Acad. Sci. USA. 101:12254–12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Exley, M., J. Garcia, S.B. Wilson, F. Spada, D. Gerdes, S.M. Tahir, K.T. Patton, R.S. Blumberg, S. Porcelli, A. Chott, and S.P. Balk. 2000. CD1d structure and regulation on human thymocytes, peripheral blood T cells, B cells and monocytes. Immunology. 100:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burdin, N., L. Brossay, M. Degano, H. Iijima, M. Gui, I.A. Wilson, and M. Kronenberg. 2000. Structural requirements for antigen presentation by mouse CD1. Proc. Natl. Acad. Sci. USA. 97:10156–10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamada, N., H. Iijima, K. Kimura, M. Harada, E. Shimizu, S. Motohashi, T. Kawano, H. Shinkai, T. Nakayama, T. Sakai, et al. 2001. Crucial amino acid residues of mouse CD1d for glycolipid ligand presentation to V(alpha)14 NKT cells. Int. Immunol. 13:853–861. [DOI] [PubMed] [Google Scholar]
- 33.Turner, S.J., P.C. Doherty, J. McCluskey, and J. Rossjohn. 2006. Structural determinants of T-cell receptor bias in immunity. Nat. Rev. Immunol. 6:883–894. [DOI] [PubMed] [Google Scholar]
- 34.Feng, D., C.J. Bond, L.K. Ely, J. Maynard, and K.C. Garcia. 2007. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat. Immunol. 8:975–983. [DOI] [PubMed] [Google Scholar]
- 35.Godfrey, D.I., J. Rossjohn, and J. McCluskey. 2008. The fidelity, occasional promiscuity, and versatility of T cell receptor recognition. Immunity. 28:304–314. [DOI] [PubMed] [Google Scholar]
- 36.Koch, M., V.S. Stronge, D. Shephard, S.D. Gadola, B. Mathew, G. Ritter, A.R. Ferscht, G.S. Besra, R.R. Schmidt, E.Y. Jones, and V. Cerundolo. 2007. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat. Immunol. 8:819–826. [DOI] [PubMed] [Google Scholar]
- 37.McCarthy, C., D. Shepherd, S. Fleire, V.S. Stronge, M. Koch, P.A. Illarionov, G. Bossi, M. Salio, G. Denkberg, F. Reddington, et al. 2007. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J. Exp. Med. 204:1131–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]