Abstract
The transcriptional regulator p53 plays an essential role in tumor suppression. Accordingly, it is found mutated, and its activity is reduced, in many human cancers. Recent reports show that some cancers are characterized by a loss of function of wild-type p53, which, in several cases, accumulates in intracellular aggregates. Although the nature of such aggregates is still unclear, recent evidence indicates that the p53 C-terminal and core domains can undergo amyloid aggregation in vitro under mild denaturing conditions, although no information is available on the largely unstructured N-terminal transactivation domain. We therefore decided to investigate the amyloid propensity of the acidic unfolded 1–63 fragment of the transactivation domain, cloned, expressed, and purified from a bacterial strain. Here we show that, when exposed to acidic pH, the 1–63 fragment forms thioflavine T-positive aggregates whose amyloid nature was confirmed by Fourier transform infrared spectroscopy analysis, atomic force microscopy, and x-ray diffraction. These aggregates were shown to be cytotoxic to human SH-SY5Y cells by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide reduction, lactate dehydrogenase release, and caspase-3 activity assays. These results add new significant details to the picture describing the propensity of single domains of p53 to aggregate, further suggesting that, under suitable destabilizing conditions, the whole protein may aggregate into amyloid assemblies in vivo.
INTRODUCTION
The tumor suppressor protein p53 is a 393-amino acid phosphoprotein containing a single zinc ion whose homotetramer plays a critical role as a transcriptional enhancer in cell cycle control and apoptotic activation in response to DNA damage (1,2). The key antitumoral role of p53 is supported by the fact that it is mutated in around 50% of human cancers (3); indeed, p53 inactivation by specific mutations or interaction with cellular proteins appears to be a critical step in the origin of many cancers, thus making this protein a major target for anticancer therapy.
The three-dimensional structure of p53 is organized into three domains. The unstructured N-terminal domain (residues 1–93) harbors an acidic transactivation domain (residues 1–62), where the binding site for mdm2 (an E3 ubiquitin ligase (4)) is located, and a proline-rich domain (residues 63–93). The DNA-binding core (residues 102–292) is the main domain responsible for p53-DNA interaction. Finally, the C-terminal domain (residues 293–393) contains a tetramerization domain and a regulatory domain. p53 displays a limited structural stability and contains large regions substantially devoid of ordered structure (5) with high conformational flexibility. Although the solution and x-ray structures of the DNA-binding domain and the tetramerization domain have been determined (6,7), there is a substantial lack of structural and biophysical information on the full-length protein as well as on N-terminal domain. In particular, the latter appears completely disordered with the typical features of the natively unfolded proteins (8).
Tumors involving p53 impairment can also be independent of the presence of specific p53 mutations. In fact, cells expressing wild-type p53 can contain an inactive form of the protein, allowing malignancy to arise. In these cells p53 inactivation can result, at least in part, from the loss of the bound zinc, whose deleterious effect on protein stability and activity is amplified by subsequent aggregation (9). The zinc-free p53 folding variant displays dominant negative effects that are able to drive the active, wild-type protein into an inactive mutant conformation (10) through a process with some similarity to prion amplification. Actually, accumulation of wild-type inactive p53 has been described in various cancers including neuroblastoma, retinoblastoma, and breast and colon cancers (11,12). In these cells, p53 is aggregated (13) in large cytoplasmic and/or nuclear deposits (11) possibly arising from a conformational change of the protein (14). It is not known whether the p53 found in these aggregates displays amyloid-like structure, but this possibility should be considered.
Amyloidoses are ∼25 protein-misfolding diseases of the degenerative type affecting either the central nervous system or peripheral tissues and organs; they include Alzheimer's, Parkinson's, and prion diseases, type 2 diabetes, and several systemic amyloidoses (15). Each of these conditions is characterized by the intracellular and/or extracellular presence, in the affected tissues, of fibrillar deposits of polymers of 1 of 25 peptides or proteins, each characteristic of a specific disease (16). The presence of amyloid fibrils is a key feature of amyloid diseases and is considered a main cause of the clinical signs (17). Amyloid fibrils arise from a nucleation-polymerization process on a destabilized peptide/protein; destabilization can follow mutations, chemical modifications, overexpression, or any impairment of peptide/protein degradation by the ubiquitin proteasome pathway (17,18). Despite the structural heterogeneity of their monomeric constituents, amyloid fibrils share surprisingly similar structural features. In fact, their key signature is the presence of an ordered β-structure-rich core in which each monomer contributes two or more β-strands that result in a continuous double parallel or antiparallel β-sheet propagating along the main axis of the fibril and in which each β-strand runs perpendicular to the fibril (16,19).
Presently, amyloid diseases are considered substantially gain-of-function conditions because cell damage is thought to arise from the acquired ability of the destabilized monomers to polymerize into toxic assemblies rather than from the loss of their biological function (19,20). Currently, the generally accepted idea is that the true cytotoxic species are not mature amyloid fibrils but their oligomeric precursors, often known as prefibrillar aggregates or protofibrils. An increasingly growing number of data show that prefibrillar aggregates are unstable species that interact with cell membranes and other cell components impairing their integrity and/or function. In particular, structural alteration of the cell membranes with disruption of their selective permeability, resulting in free calcium balance and redox homeostasis derangement (21), is considered a key biochemical modification in most amyloid diseases (16).
Because of the lack of data on either the aggregation potential of the p53 N-terminal domain, particularly the 1–62 transactivation domain, or the mechanisms of cytotoxicity, if any, of its aggregates, we sought to provide information on these topics by cloning, expressing, and purifying the p53 1–63 N-terminal domain (p53(1–63)). The aggregation propensity of the latter was investigated together with the morphological features and cytotoxicity of the differing aggregates arising during the fibrillization path. Our data show that, similarly to the core domain and the tetramerization domain (both containing unstructured regions), the p53 unstructured N-terminal domain can aggregate in amyloid assemblies exhibiting cytotoxic features. These data complete those previously reported on the aggregation potential of the p53 domains supporting a possible role of the N-terminal domain as a trigger of p53 aggregation in the highly crowded intracellular environment.
MATERIALS AND METHODS
Materials
The primers for PCR amplification were synthesized by Genenco (Florence, Italy). HotStartTaq mix was from Qiagen GmbH (Hilden, Germany). Restriction enzymes were from Invitrogen (Carlsbad, CA). isopropyl-β-d-thiogalactopyranoside was from Inalco (Milan, Italy). Sepharose-GSH affinity resin was from Pharmacia (Piscataway, NJ). The CytoTox-ONE Homogeneous Membrane Integrity Assay was from Promega (Madison, WI). The Ac-DEVD-AMC caspase-3 fluorometric substrate and Ac-DEVD-CHO caspase-3 inhibitor were from Biomol (Plymouth Meeting, PA). Rabbit polyclonal anti-p53 (FL-393) antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Peroxidase-conjugated antirabbit antibody was from Chemicon (Temecula, CA). SuperSignal West Pico Chemiluminescent Substrate was from Pierce (Rockford, IL). All other reagents, media, enzymes and inhibitors were from Sigma Chemical (St. Louis, MO).
Cloning and purification of the (1–63) p53 N-terminal fragment
The pUC18p53 plasmid containing the gene coding for whole-length human p53 was kindly provided by Prof. Nobua Tsuchida, Dept. of Molecular Cellular Oncology and Microbiology, Tokyo Medical and Dental University. PCR amplification of the DNA fragment corresponding to residues 1–63 of the whole p53 gene product was carried out using the direct primer 5′-AAAGGGGGATCCATGGAGGAGCCGCAGTCAGATCCT-3′ containing the restriction site for BamHI and the reverse primer 5′-AAAAGGGAATTCTCAAGCTTCATCTGGACCTGGGTCT-3′ containing the restriction site for EcoRI; 10 pmol of each primer was added to 20 ng of template DNA and 12.5 μL of 2× PCR HotStartTaq mix in a final volume of 25 μl.
The fragment resulting from PCR amplification (95°C for 15 min for hot start, followed by 25 cycles consisting of 95°C for 1.0 min, 64°C for 1.0 min, 72°C for 1.0 min) was digested with BamHI and EcoRI and ligated into pGEX-2T vector downstream and in frame with glutathione S-transferase. Sequence determination performed by BMR Genomics (Padova, Italy) confirmed the integrity of the cloned fragment.
Fusion protein expression was performed in E. coli BL21 cells transformed with the pGEX-2T(1–63) plasmid. Cells were grown overnight at 37°C in LB medium, diluted 1:10 in fresh medium, and treated with 0.1 mM isopropyl-β-d-thiogalactopyranoside for 4.0 h for the induction of fusion protein expression when they reached an OD of 0.6 at 600 nm. Bacterial cells were centrifuged at 4000 rpm for 15 min at 4.0°C, and the pellet was resuspended in a volume of PBS (20 mM Na2HPO4, pH 7.3, 250 mM NaCl, 1.0 mM EDTA, 1.0 mM β-mercaptoethanol, 0.1 mM phenylmethylsulphonyl fluoride (PMSF)) corresponding to 1% of the cell culture volume. Cells were lysed by incubation for 30 min in ice in the presence of 44 U/ml of lysozyme and 0.1% Triton X-100, followed by sonication (50 kHz and 60 W for 30 s, six pulses), and debris was removed by centrifugation at 16,000 rpm for 40 min at 4.0°C. The supernatant was loaded onto a Sepharose-GSH affinity column and, after extensive washing with PBS and TBS (50 mM Tris-HCl buffer, pH 8.0, 150 mM NaCl), an in-column digestion of the fusion protein was performed by incubating the resin overnight in the presence of 50 U of human thrombin. p53(1–63) was then eluted from the column and concentrated by ultrafiltration to 3.0 mg/ml. Protein concentration was determined by UV absorption at 280 nm using an extinction coefficient of 4.17 (1.0 mg/ml, 1.0-cm optical path). Protein purity and quality were checked by SDS-PAGE (silver staining), electrospray mass spectrometry, and amino acid analysis, confirming the expected amino acid sequence GSMEEPQSDPSVEPPLSQETFSDLWKLLPENNVLSPLPSQAMDDLMLSPDDIEQWFTEDPGPDEA, corresponding to a Mr = 7.2 kDa and an isoelectric pH of 3.28. The Gly-Ser dipeptide present at the N-terminus results from thrombin cleavage of the fusion protein cloned in pGEX-2T.
Th-T assay
The p53(1–63) bulk solution (3.0 mg/ml) was diluted to 0.4 mg/ml in 20 mM citrate buffer at different pH values (3.0, 3.3, 3.6, 4.0) (assay buffer) and incubated at 25°C or 4.0°C. Then, 60 μl of this solution was withdrawn at different incubation times and mixed with 440 μl of 25 μM ThT in 25 mM phosphate buffer, pH 6.0. In some experiments the aggregation mixture was centrifuged at 18,000 rpm for 5.0 min, and the supernatant was assayed for ThT binding. Sample fluorescence was measured at 25°C (440Ex/485Em) (22) in a 2 × 10 mm path-length cell using a Perkin-Elmer LS 55 spectrofluorimeter (Wellesley, MA) equipped with a thermostated cell holder connected to a Haake F8 water-bath (Karlsruhe, Germany).
FT-IR spectroscopy
The p53(1–63) solution was incubated at 0.4 mg/ml in 20 mM citrate buffer, pH 3.0, at 25°C for increasing time periods. Peptide aggregates were lyophilized and resuspended in D2O (minimum 99.96 atom % D) at 6.7 mg/ml, and 15-μl aliquots of the suspension were placed between CaF2 windows using a 6.0-μm spacer and analyzed by a Jasco 4200 FT-IR spectrometer. Each spectrum was obtained by coadding 100 interferograms at a spectrum resolution of 4.0 cm−1. The spectrum of the native p53(1–63) was obtained by lyophilizing a solution of 100 μg of the protein in 50 mM Tris-HCl buffer, pH 8.0, containing 150 mM NaCl followed by dissolution of the lyophilized peptide in 15 μl D2O for FT-IR analysis. Blank analysis was carried out by lyophilizing and redissolving the buffers in D2O before FT-IR.
ANS fluorescence
Aliquots of the p53(1–63) aggregation mixture (0.4 mg/ml in 20 mM citrate buffer, pH 3.0, at 25°C) were withdrawn at various time points and mixed into a cuvette containing 450 μl of 55 μM ANS in citrate buffer (final protein concentration: 0.07 mg/ml). Samples were excited at 380 nm, and emission was registered between 410 and 630 nm. At any time point, aliquots of the reaction mixture were also centrifuged at 18,000 rpm for 5.0 min, and supernatant and pellet were separately assayed for their ability to bind ANS. p53(1–63) native peptide was similarly assayed for its ability to bind ANS in 50 mM Tris-HCl, pH 8.0, containing 150 mM NaCl.
Dot-blot analysis
p53(1–63) was incubated in 20 mM citrate buffer, pH 3.0, for 30 min at 25°C; then an aliquot of the mixture was centrifuged at 18,000 rpm for 5.0 min, and different amounts of the supernatant were spotted onto a nitrocellulose membrane together with identical quantities of total p53(1–63). As a positive control, corresponding amounts of whole-length p53 were similarly applied to the membrane, and 1.0 μg of an unrelated peptide was used as a negative control. After incubation with anti-p53, peroxidase-conjugated antirabbit antibodies and a chemiluminescent substrate, the image was acquired with a ChemiDoc system (Bio-Rad, Hercules, CA).
AFM
A 0.4 mg/ml p53(1–63) solution was incubated at 4.0°C in 20 mM citrate buffer, pH 3.0. Aliquots of this solution were withdrawn at fixed time intervals and diluted 100 times with bidistilled water; 20-μl aliquots of the diluted sample were deposited on a freshly cleaved mica substrate and dried under a gentle nitrogen flux. Dilution of the sample was required to avoid salt crystallization. Tapping-mode AFM images were acquired in air using a Dimension 3000 microscope (Digital Instruments-Veeco, Santa Barbara, CA) equipped with a “G” scanning head (maximum scan size 100 μm) and driven by a Nanoscope IIIa controller and a Multimode Scanning Probe Microscope (Digital Instruments-Veeco), equipped with an “E” scanning head (maximum scan size 10 μm) and driven by a Nanoscope IV controller. Single-beam uncoated silicon cantilevers (type RTESPA, Veeco, and type OMCL-AC160TS, Olympus, Tokyo, Japan) were used. The drive frequency was around 300 kHz; the scan rate was between 0.3 and 0.8 Hz.
Aggregate size was measured from the height in cross section of topographic AFM images. Because sample drying results in a reduction of aggregate size with respect to fully hydrated conditions, the values reported in the Results section should be corrected by taking this effect into account. The correction factor was evaluated to be 1.4 from the comparison of globular proteins imaged in air and under liquid. Other factors that may affect the measured size are the presence of amorphous material partially embedding the aggregate and aggregate flattening on the mica substrate. The aggregate width was measured at the full aggregate height; because width values are affected by the tip size, they should be considered only as a rough estimate.
X-ray fibril diffraction
p53(1–63) fibrils obtained after 1 month of incubation at 25°C in 20 mM sodium citrate buffer, pH 3.0, were centrifuged at 3500 rpm and 21°C for 10 min. The resulting pellet was collected and inserted into a 1.0-mm capillary with an open end. The capillary was placed into a support of expanded polystyrene to avoid its rupture and gently centrifuged; finally, the supernatant was removed. The pellet in the capillary was exposed to air to achieve solvent evaporation for ∼1 week. Then the capillary was exposed to x rays at room temperature using a MAR Research (Norderstedt, Germany) 345 imaging-plate detector coupled to a Rigaku (Tokyo, Japan) RU-H3R rotating-anode generator, using monochromatic Cu Kα radiation. Fibril diffraction image analysis and spacing estimates were achieved using the Mosflm program (23).
Cytotoxicity assay
Human SH-SY5Y neuroblastoma cells were obtained from the American Type Culture Collection (Manassas, VA) and cultured in 1:1 Ham's F-10:DMEM medium supplemented with 10% fetal calf serum, 3.0 mM glutamine, 100 μg/ml streptomycin, and 100 units/ml penicillin, in a 5.0% CO2 humidified atmosphere at 37°C. Aggregate cytotoxicity was assessed by the MTT reduction inhibition assay (24). Cells were plated on 96-well plates at a density of 6000 cells/well in 200 μl of fresh medium. After 72 h, the cells were exposed for 24 h to different concentrations (ranging from 0.4 μM to 12 μM) of p53(1–63) aggregates precipitated by centrifugation (18,000 rpm for 5.0 min) and resuspended in 50 μl of fresh medium. At the end of the incubation, the cell culture medium was removed, and the cells were incubated for 2.0 h with 100 μl of a 0.5 mg/ml MTT solution in DMEM without phenol red. After incubation, cell lysis solution (100 μl/well: 20% SDS, 50% N,N-dimethylformamide) was added, and the samples were incubated overnight at 37°C in a humidified incubator. Blue formazan absorbance was measured at 570 nm using an automatic plate reader (Bio-Rad). Control experiments were performed by exposing cells to the nonaggregated monomeric protein at a final concentration of 12 μM for the same lengths of time.
Necrotic and apoptotic markers
Membrane integrity in cells exposed to p53(1–63) aggregates was evaluated by measuring LDH release into the culture medium. LDH activity was measured with the CytoTox-ONE homogeneous membrane integrity assay, according to instructions provided by the manufacturer. The assay couples the reduction of resazurin to resofurin, catalyzed by diaphorase, to the oxidation of lactate to pyruvate, catalyzed by LDH. The intensity of the resulting fluorescence (560Ex/590Em) is a measure of LDH activity in the sample. Briefly, the cells, plated and cultured as described for the MTT reduction assay, were treated for 24 h with 12 μM p53(1–63) aggregates aged for 100 min in 20 mM citrate buffer, pH 3.0, at 25°C or 4.0°C, resuspended in DMEM without phenol red and sodium pyruvate and supplemented with 5.0% fetal calf serum. The reagent was added to the culture medium, and the increase in fluorescence emission was measured after 2.0 h using a Fluoroskan Ascent FL fluorescence plate reader (Thermo Scientific, Madison, WI). For each sample, the fluorescence value was normalized with respect to the “total LDH activity” determined after cell lysis.
The caspase-3 activity assay was carried out as follows. Cells were exposed for 24 h to 12 μM p53(1–63) aggregates obtained as for the LDH assay and resuspended in complete medium. Control cells were treated with the same concentration of soluble p53(1–63). After 24 h, the cells were lysed with 20 mM Tris-HCl buffer, pH 7.4, containing 250 mM NaCl, 2.0 mM EDTA, 0.1% Triton X-100, 5.0 μg/ml leupeptin, 5.0 μg/ml aprotinin, 0.5 mM PMSF, 4.0 mM vanadate, and 1.0 mM DTT for 20 min in ice. Lysis was completed by sonication, and total protein content was determined in the clarified lysate with the Bradford reagent. Then, 50 μg of total protein was diluted in 0.5 ml of 50 mM HEPES-KOH buffer, pH 7.0, containing 10% glycerol, 0.1% 3-[(3-cholamidopropyl)-dimethylammonium]-1-propane sulfonate, 2.0 mM EDTA, 10 mM DTT, in the presence of 50 μM Ac-DEVD-AMC (caspase-3 fluorometric substrate). Incubation was carried out for 2.0 h at 37°C, and fluorescence values were recorded (380Ex/460Em). To determine nonspecific substrate degradation, the same assay was performed by preincubating samples for 15 min at 37°C in the presence of 100 nM Ac-DEVD-CHO (caspase-3 inhibitor) before substrate addition.
Statistical analysis
Dunnett multiple-comparisons test was performed on data obtained from MTT, LDH, and caspase-3 assays.
RESULTS
p53(1–63) forms ThT-positive aggregates in vitro
The p53(1–63) transactivation domain displays typical features of the intrinsically unstructured proteins including a low content of hydrophobic residues that are flanked, in sequence, by numerous acidic residues. These features inhibit the formation of a hydrophobic core, disfavor the organization of rigid secondary structure elements, and result in an extended conformation caused by electrostatic repulsion (25).
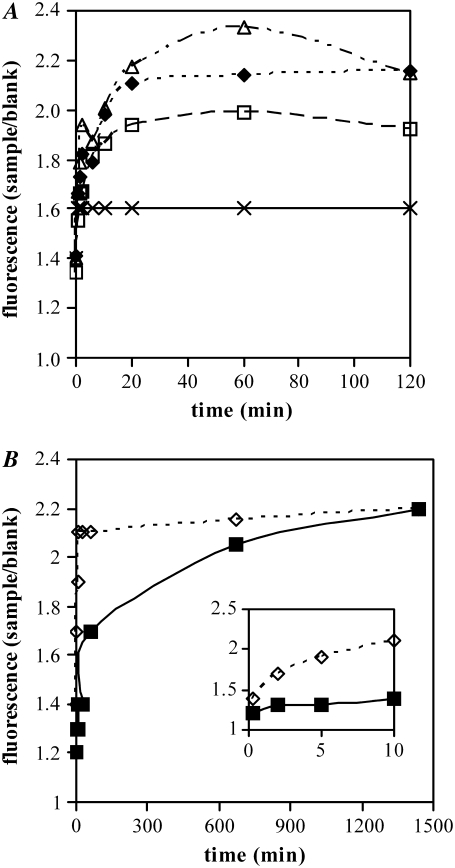
Initially, we determined the propensity of p53(1–63) to form amyloid aggregates at pH values approaching its isoelectric point (3.28), a condition known to favor protein-protein interaction. The peptide (3.0 mg/ml in 50 mM Tris-HCl buffer, pH 8.0, 150 mM NaCl) was diluted to 0.4 mg/ml in 20 mM citrate buffer at differing pH values (3.0, 3.3, 3.6, 4.0) and analyzed at 25°C for its ability to bind ThT by monitoring the time-course of the increase of fluorescence at 485 nm (Fig. 1 A). ThT positivity is considered a strong indicator of the presence of ordered aggregated material of amyloid type (22). We controlled the citrate buffer so that the pH did not change on addition of the peptide solution by mixing the corresponding volumes of citrate and Tris-HCl buffers. We found that p53(1–63) rapidly forms ThT-positive aggregates in the 3.0–3.6 pH range; however, within the time scales of our experiments, no significant aggregation was observed at pH 4.0. The ThT fluorescence increase reached a plateau after 30 min of incubation. As expected, reducing the incubation temperature to 4.0°C resulted in a slower increase of ThT fluorescence (Fig. 1 B), although the plateau value, reached at these conditions in the 12- to 24-h interval, was substantially unchanged, indicating that p53(1–63) aggregation is driven mainly by entropic contributions as reported for the aggregation of other proteins/peptides (26).
FIGURE 1.
ThT assay on p53(1–63). The peptide was incubated in 20 mM citrate buffered at pH 3.0 (black diamonds), 3.3 (white squares), 3.6 (white triangles), and 4.0 (crosses) at 25°C for up to 120 min (A) or in 20 mM citrate buffer pH 3.0 at 4.0°C (black squares) or 25°C (white diamonds) for up to 24 h (B). At regular time intervals aliquots were withdrawn from the reaction mixtures and assayed for their ability to bind ThT. Fluorescence of samples was normalized with respect to the fluorescence of a blank. These are representative experiments of four giving qualitatively identical results.
p53(1–63) forms ANS-positive aggregates
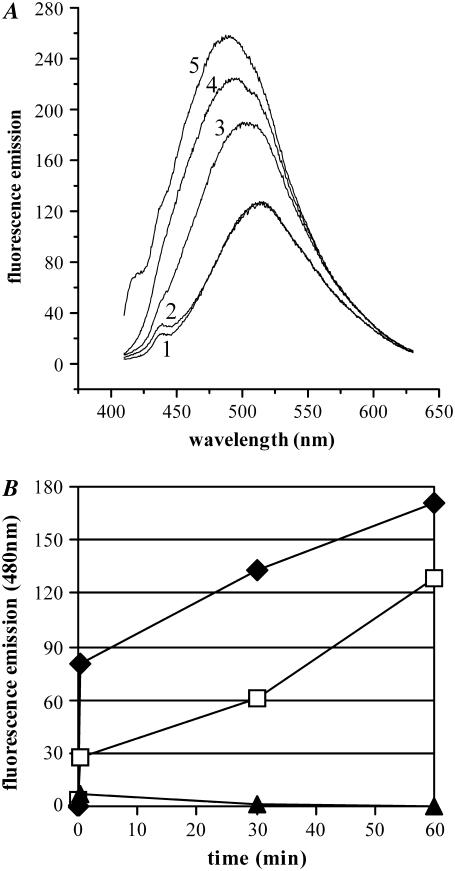
The hydrophobic dye ANS is a dye whose fluorescence is greatly enhanced on binding to hydrophobic surfaces, displaying a characteristic blue shift in its fluorescence maximum from 515 nm to 480 nm (27,28). ANS is largely used as a probe to reveal the increased exposure of hydrophobic surfaces that is characteristic of partially structured folding intermediates and amyloid aggregates (27,28). As early as after 30 s of incubation of p53(1–63) at pH 3.0, we observed an increase in ANS binding (Fig. 2 A) characterized by a pronounced blue shift of the emission spectrum which increased with aggregation. The supernatant obtained by centrifugation of the aggregation mixture displayed no ANS-binding while the pellet was strongly positive (Fig. 2 B), suggesting that the early precipitation of p53(1–63) is associated to a progressive exposure of hydrophobic clusters resulting from the structural organization of the polypeptide chain.
FIGURE 2.
ANS test on p53(1–63). (A) The emission spectrum of ANS without protein was acquired (1). The ANS spectrum was then acquired in the presence of native p53(1–63) (2) or of p53(1–63) incubated in 20 mM citrate buffer, pH 3.0, and assayed after 30 s (3), 30 min (4), or 1 h (5). (B) The pellet and the supernatant obtained by centrifuging aliquots of the reaction mixture were separately assayed for their ability to bind ANS by measuring their emission at 480 nm (total, black diamonds; pellet, white squares; supernatant, black triangles). The data are representative of three experiments carried out under identical conditions, giving qualitatively identical results.
Structural analysis of p53(1–63) aggregates
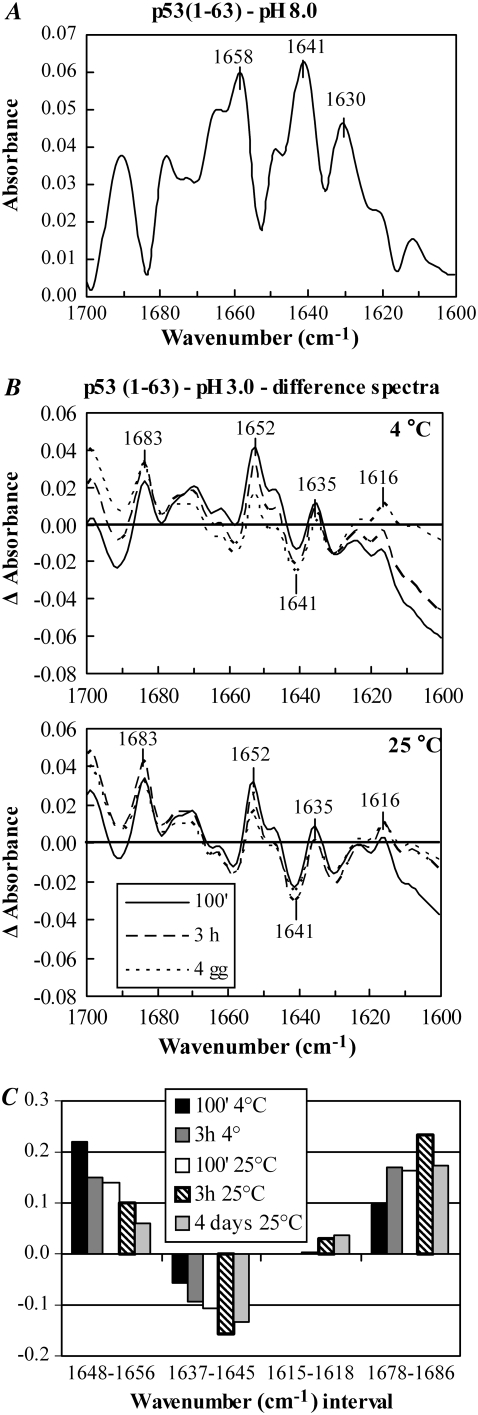
To gain more information on the structural features of the aggregating peptide in the precipitate, we performed an FT-IR analysis of the soluble and aggregated peptide. Aliquots of p53(1–63) dissolved in 50 mM Tris-HCl buffer, pH 8.0, 150 mM NaCl, lyophilized, and dissolved in D2O, were analyzed by FT-IR. The spectrum of this soluble form of the peptide confirmed its partially unfolded nature, characterized by a peak around 1640 cm−1, although the presence of a peak at 1659 cm−1 and of a minor one at 1631 cm−1 indicated some α-helix and β-sheet secondary structure (Fig. 3 A). p53(1–63) was then aged in the aggregation medium for various lengths of time at 25°C or 4.0°C, lyophilized, and resuspended in D2O. To better monitor the variations in the absorbance at characteristic wavenumbers during the time course of aggregation, the FT-IR spectra were first scaled to the same area, and difference spectra were then calculated by subtracting the spectrum of the native peptide from the spectrum of the aggregated peptide (Fig. 3 B). Then the area of the resulting Δ-absorbance peaks (taken above the baseline for positive peaks and under the baseline for negative peaks) was calculated (Fig. 3 C). The results of the FT-IR analysis revealed a progressive increase of the absorbance around 1615 cm−1 and 1683 cm−1 that is typical of the intermolecular β-sheet found in amyloid assemblies (29), paralleled by a decrease in the absorbance around 1640 cm−1, suggesting a transition from disordered to ordered aggregates in the precipitate. The latter appeared to be slower for aggregates grown at 4.0°C than at 25°C; nevertheless, after 4 days the spectra of the peptide incubated at these different temperatures are similar, indicating that the samples reached comparable states regardless of the temperature (Fig. 3 B).
FIGURE 3.
(A) FT-IR spectrum of p53(1–63) dissolved in 50 mM Tris-HCl buffer, pH 8.0, containing 150 mM NaCl, lyophilized, and resuspended in D2O. (B) Structural changes occurring during p53(1–63) aggregation were determined by incubating the peptide in 20 mM citrate buffer, pH 3.0, for increasing lengths of time at 4.0°C or 25°C. The FT-IR analysis was carried out on the peptide lyophilized and resuspended in D2O. The spectra were scaled to the same area, and difference spectra were calculated by subtracting the spectrum of the native peptide from the spectrum of the aggregated peptide at any time point. (C) For every Δ-absorbance spectrum, the areas comprised into distinct wavenumber intervals, taken above the zero line for positive Δ-absorbance peaks and below the zero line for negative Δ-absorbance peaks, were calculated. The analysis was repeated three times, giving closely similar results.
Detailed peak analysis showed that after 100 min of incubation in the aggregation medium, p53(1–63) displayed a decrease of the unfolded structure with respect to the native peptide, as shown by the negative peak in the 1637–1645 cm−1 range in the difference spectrum. The absolute value of this difference peak was higher for the aggregates obtained at 25°C than for those obtained at 4.0°C, in agreement with the faster aggregation kinetics of the former (Fig. 3 C). At this time, transitory increases in α-helix (as suggested by the positive difference peaks in the 1648–1656 cm−1 interval) and in β-sheet nonamyloid and amyloid structures (as shown by the positive difference peaks around 1635 and 1683 cm−1, respectively) are evident. After 3.0 h of incubation, the contribution of unfolded, α-helix, and β-sheet nonamyloid structures was reduced, whereas an increase in amyloid β-structure (1615–1618 cm−1 and 1678–1686 cm−1) was apparent. The slower kinetics of these changes at 4.0°C than at 25°C is evident. In fact, at 25°C, these peaks reach their maxima at 3.0 h (Fig. 3 C), although after 4 days at either 4.0°C or 25°C, the difference spectra were similar and displayed a prevalence of amyloid structure over all other secondary structures.
Taken together, these data suggest that p53(1–63) aggregation proceeds through the early precipitation of the peptide that subsequently undergoes a progressive transition leading it to acquire some amyloid structural features, a process mainly driven by entropic contributions that apparently needs a transient increase in the content of nonamyloid secondary structure, as previously shown for other systems (26,30).
AFM used after p53(1–63) aggregation
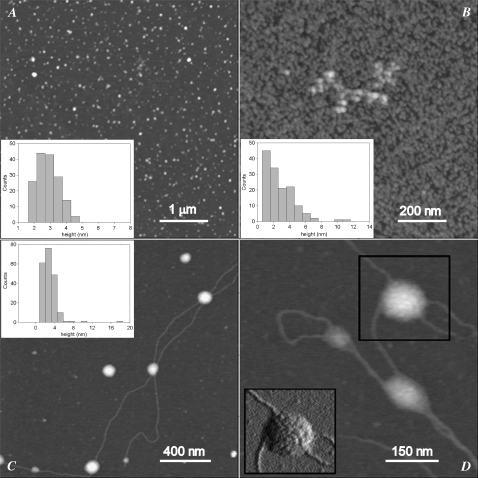
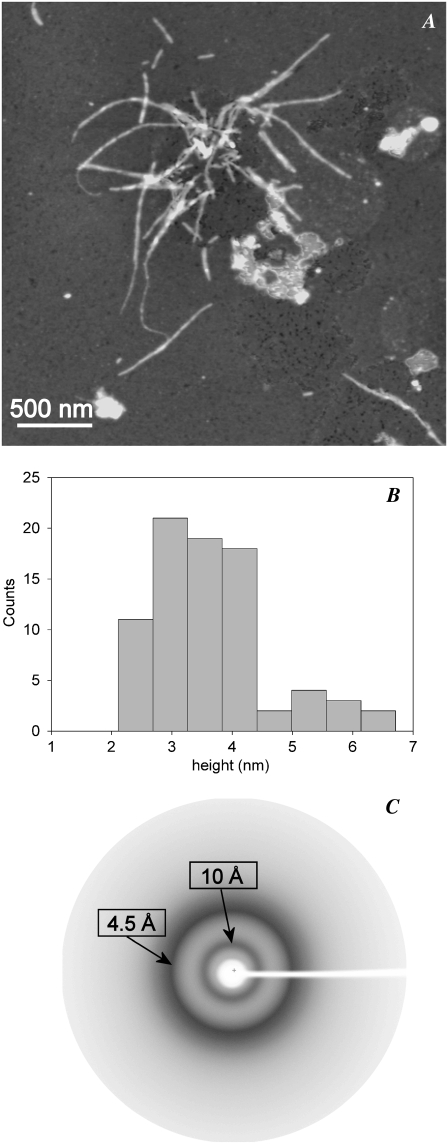
Tapping-mode AFM was used to investigate the morphological features of the aggregates resulting from the incubation of p53(1–63) in citrate buffer at pH 3.0. Initially, globular particles randomly present on the whole sample surface were observed with height distribution peaking at 1.3 nm (not shown). When the correction factor for dehydration effects (see Materials and Methods) is considered, this size may correspond to the monomeric protein. At early aggregation times (20 min), globular particles (Fig. 4 A) with an average size of 3.1 ± 0.1 nm (Fig. 4 A, inset) were present. These globules displayed the tendency to assemble into short beaded chains composed of several units at later (3.0 h) times (Fig. 4 B) with a broader size distribution (Fig. 4 B, inset). After 24 h, very thin filaments 0.5 nm high coexisted with lenticular structures 4.0 nm high and 60–240 nm wide (Fig. 4, C and D). The latter consisted of entangled filaments, as shown in the amplitude image of Fig. 4 D, and these were probably spheroidal structures deformed by the adhesion to the mica substrate. Comparably similar structures were observed in the path of aggregation of other amyloid-forming proteins (31,32). Fibrillar structures 2.0 nm high were observed after 10 days (not shown), whereas at longer incubation times (∼1 month), thicker fibrils, often supercoiled, were found (Fig. 5 A). Accordingly, the size distribution of these mature fibrils was bimodal, with peaks at 3.3 and 5.5 nm (Fig. 5 B), further indicating that they were formed by different numbers of protofilaments.
FIGURE 4.
Tapping-mode AFM images (height data) of p53(1–63) N-terminal domain aggregated at 4.0°C and pH 3.0 for (A) 20 min, (B) 3.0 h, or (C and D) 24 h. Scan size: (A) 5.0 μm; (B) 1.0 μm; (C) 1.9 μm; (D) 0.68 μm. Z range 10 nm. The corresponding aggregate height distributions are shown in the insets. (D) Inset is a 250 nm × 240 nm portion of the corresponding amplitude data image (scan size 0.68 μm) showing the spheroidal structure indicated by the black square.
FIGURE 5.
(A) Tapping-mode AFM image (height data) of mature p53(1–63) fibrils. Scan size 3.1 μm, Z range 10 nm. (B) Height distribution of mature p53 fibrils showing the coexistence of fibril populations of different sizes. (C) X-ray diffraction diagram of p53(1–63) fibrils collected on sedimented fibrillar aggregates showing the equatorial and meridional reflections at 4.5 and 10.0 Å, respectively, characteristic of the cross-β-structure.
p53(1–63) fibrils display the shared cross-β-amyloid structure under x-ray diffraction
It is known that in some cases protein aggregation results in fibrillar assemblies in which the monomers substantially maintain their native structure and are arranged differently than in amyloid fibrils (33,34) even though their aggregates display spectroscopic features of the amyloids such as ThT and Congo red positivity. X-ray analysis reveals with full reliability structural features, such as the cross-β-structure, that are key signatures of amyloid fibrils. Therefore, p53(1–63) fibrillar aggregates, obtained by incubating the protein in 20 mM sodium citrate buffer, pH 3.0, were sedimented in a capillary and analyzed by x-ray diffraction. The x-ray diffraction pattern (Fig. 5 C) is consistent with the presence of a cross-β-conformation typical of amyloid fibrils. In particular, the diffraction pattern shows a strong reflection around 4.5 Å, corresponding to the hydrogen bonding distance between consecutive β-strands in β-sheets, and a strong reflection around 10 Å, which indicates the sheet distance of the two β-sheets propagating along each protofilament. These findings definitely indicate that our fibrillar material is amyloid in nature.
p53(1–63) aggregates are cytotoxic
p53(1–63) was aggregated at 25°C or 4.0°C for increasing time periods. At the end of each incubation time, the aggregates were recovered by centrifugation, suspended in complete medium, and added to SH-SY5Y neuroblastoma cell cultures at 12.0 μM final protein concentration for 24 h. The stability of the aggregates during the time course of cell exposure was confirmed by the ThT assay (data not shown). The quantity of aggregates recovered after centrifugation was evaluated by determining the amount of protein present in the supernatant by Dot-blot analysis with anti-p53 antibodies, confirming that no p53(1–63) remained in the supernatant after aggregate centrifugation (not shown). Furthermore, a ThT assay performed on the aggregate mixture and on the supernatant (after aggregate precipitation) revealed that, although the whole mixture was strongly ThT positive, no positivity was detected in the supernatant (data not shown).
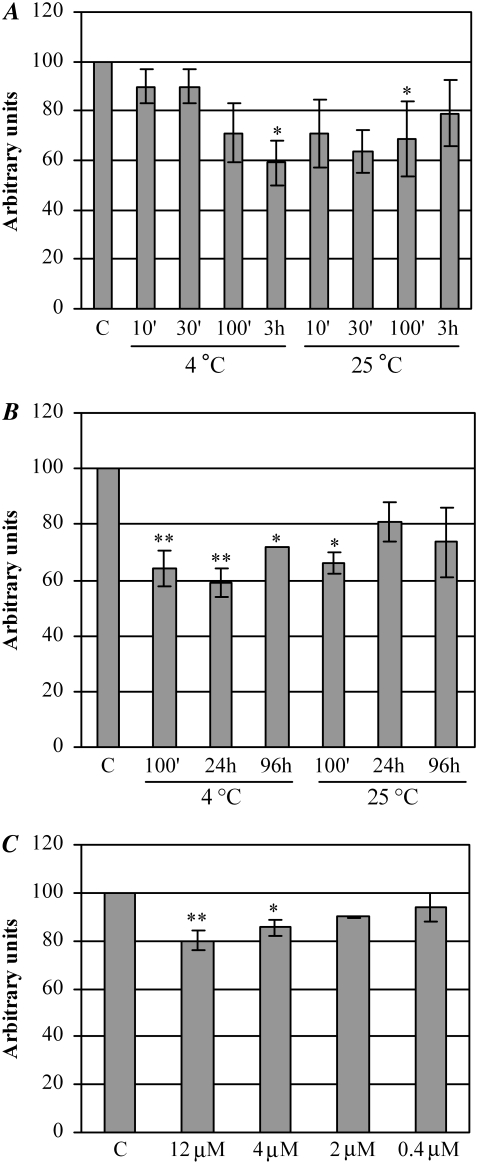
p53(1–63) aggregate cytotoxicity was determined by exposing cultured human SH-SY5Y neuroblastoma cells to protein aggregates aged for differing lengths of time. Control experiments were conducted by exposing cells to 12.0 μM p53(1–63) in its soluble form. The viability of the cells so treated was assayed by the MTT reduction assay. Fig. 6 A shows that the aggregates obtained after 100 min at 25°C displayed significant cytotoxicity, whereas the peptide incubated in the aggregation solution at 4.0°C started to be significantly cytotoxic only after 3 h of aging, in agreement with its slower aggregation kinetics. Moreover, aggregate toxicity progressively decreased with aggregation time, the reduction being more evident for aggregates aged at 25°C than for aggregates aged at 4.0°C (Fig. 6 B), concomitant with the increasing presence of fibrillar material. A dose-dependence analysis of aggregate toxicity was also performed by incubating the cells for 24 h with different amounts (12.0, 4.0, 2.0, and 0.4 μM, monomeric protein concentration) of p53(1–63) aggregates aged in 20 mM citrate buffer, pH 3.0, at 25°C for 100 min. In these conditions, the toxicity was progressively reduced with the decrease of aggregate concentration, becoming irrelevant at 2 μM, which can be taken as the lower limit of toxicity under our conditions (Fig. 6 C).
FIGURE 6.
Evaluation of p53(1–63) cytotoxicity by the MTT reduction assay. (A and B) Human SH-SY5Y cells were exposed for 24 h to 12.0 μM p53(1–63) aggregates obtained by incubating the peptide in 20 mM citrate buffer, pH 3.0, at 4.0°C or 25°C for increasing lengths of time. (C) The cells were exposed to different concentrations of p53(1–63) aggregates obtained by incubating the peptide at 0.4 mg/ml in 20 mM citrate buffer, pH 3.0, for 100 min at 25°C. Control cells were treated with 12.0 μM native p53(1–63). Cytotoxicity was evaluated by the MTT reduction assay (see Materials and Methods). Data, obtained from three different experiments carried out under identical conditions, were normalized with respect to the absorbance of the controls, which was taken as 100% for each experiment; SEM is reported. Statistical analysis was performed comparing the value of every treatment with respect to control by means of the Dunnett multiple-comparisons test. *p < 0.05; **p < 0.01.
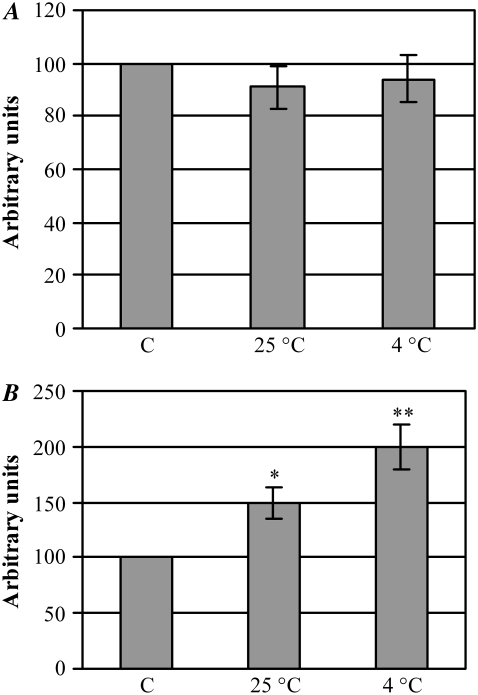
We then sought to assess whether cell stress given by the aggregates resulted in membrane disruption and/or apoptotic activation. First, we performed a membrane integrity test in exposed cells by assaying the LDH release in the culture medium during cell exposure to p53(1–63) aggregates obtained by incubating the domain at pH 3.0 for 100 min at 25°C or at 4.0°C, respectively. Fig. 7 A shows no LDH release in the medium from cells exposed to aggregates aged at both temperatures as compared with control cells, indicating that aggregate toxicity does not result in cell disruption. To check whether cell impairment after exposure to the aggregates matched the activation of the apoptotic pathway, we assayed the activity of caspase-3, a major executioner caspase, in cells exposed for 24 h to the aggregates. Both 100% and 50% increases in caspase-3 activity were observed in cells treated with aggregates aged for 100 min at 4.0°C or at 25°C, respectively (Fig. 7 B), in agreement with previously reported data for these and other cell cultures exposed to HypF-N prefibrillar aggregates (35–37). At these times the extent of apoptotic activation did not match the MTT test values, being higher in cells exposed to aggregates aged at 4.0°C. However, it must be taken into account that the MTT test is a measure of cell viability in terms of mitochondrial functionality, whereas the activation of caspase-3 can also result from the extrinsic pathway starting from cell membrane signals; under our conditions, it is likely that the extrinsic apoptotic pathway is triggered by aggregates added to the cell culture that are likely to interact with the cell membrane, as has been repeatedly reported (36,37). This agrees with the slower kinetics of the aggregation process at 4.0°C than at 25°C, suggesting that the aggregates arising at 4.0°C after 100 min of incubation display less ordered structural features, favoring their interaction with the cell membrane.
FIGURE 7.
(A) Evaluation of SH-SY5Y necrosis after cell exposure to p53(1–63) aggregates. Cells were treated for 24 h with 12.0 μM p53(1–63) aggregates obtained by incubating the peptide in 20 mM citrate buffer, pH 3.0, for 100 min at 25°C or 4.0°C. LDH release in the culture medium after necrotic rupture of the cell membrane was determined by an assay coupled to the oxidation of the fluorescent dye Resazurin. For each sample, LDH activity in the medium was normalized with respect to total LDH activity determined after cell lysis. (B) Evaluation of SH-SY5Y apoptosis after cell exposure to p53(1–63) aggregates. Cells were treated as above, and their caspase-3 activity was determined using the fluorescent substrate Ac-DEVD-AMC. Data obtained from three different experiments carried out under identical conditions were normalized with respect to the value of controls (cells treated with the same concentration of soluble p53(1–63)), which was taken as 100%. SEM is reported. Statistical analysis was performed comparing the value of every treatment with respect to control by means of the Dunnett multiple-comparisons test. *p < 0.05; **p < 0.01.
DISCUSSION
The N-terminal domain of p53 displays the canonical features of an intrinsically disordered protein with important implications for the functional mechanism of the whole p53 protein and is involved in p53 turnover by binding mdm2. p53 mediates transcription signals by forming complexes with several proteins before transcription initiation. In this study, we analyzed the structural properties of the purified p53 N-terminal transactivation domain (residues 1–63) using different approaches. The results from FT-IR, ThT, and ANS spectroscopy, AFM, and x-ray diffraction show that, when it is incubated at low pH, this domain acquires an ordered secondary structure aggregating reproducibly into amyloid-like fibrils, in agreement with other unstructured proteins rich in acidic residues (38,39).
FT-IR analysis showed that, at low pH, the natively unfolded p53(1–63) acquires a transient partially folded conformation as shown by either the decrease in absorbance at 1641 cm−1 (typical of the unfolded structure) and the increases at 1652 and 1635 cm−1, indicative of α-helix and intramolecular β-sheet, respectively. A transitory increase in α-helix is not unusual during amyloid aggregation; for instance, human amylin aggregation in the presence of lipid membranes (a condition resembling that of its aggregation in vivo) proceeds through a transitory shift from unfolded to mainly α-helical before conversion into β-amyloid structure (40). Similarly, the partial helical structure initially found in the amyloid-β peptides in the presence of critical concentrations of organic solvents increases the rate of amyloid fibril formation, possibly by favoring the intermolecular packing within the oligomers (41). Subsequently, a decrease of the α-helix component accompanied by a striking increase in the signal around 1615 and 1683 cm−1 indicative of intermolecular β-structure was seen in p53(1–63). This transition could be favored by a decrease of the intramolecular electrostatic repulsions at pH values approaching the isoelectric point, with the increased availability of the peptide backbone, and by an increased strength of the hydrophobic interactions resulting from the exposure of contiguous hydrophobic surfaces, as confirmed by the positivity of the ANS test. These structural modifications of the unfolded p53(1–63) are consistent with the fact that, in general, amyloid fibril formation requires both secondary and tertiary interactions to be established among monomers. Actually, in vitro amyloidogenesis from folded proteins is frequently observed to occur under mild destabilizing conditions still favoring secondary interactions established by the peptide backbone (42) but only partially weakening the tertiary ones.
Under our experimental conditions, oligomers and fibrils were predominantly populated in the aggregation medium after a few hours or from day 2 of incubation, respectively, as shown by AFM, ThT, and ANS binding. The oligomers converted into fibrils during aging in the aggregation medium in agreement with the current view on the mechanisms of amyloid formation (42,43). Subsequent fibril maturation resulting in fibril thickening, as measured by AFM, was possibly related to an increase of the number of protofilaments.
Amyloid oligomers obtained at different times of incubation of p53(1–63) in acidic conditions impair SH-SY5Y cell viability, a cell model previously shown to be highly vulnerable to amyloid toxicity (37). When cells were treated for 24 h with amyloid oligomers obtained at different times of incubation in acidic conditions, cell stress was revealed by the inhibition of MTT reduction and by caspase-3 activation. Under these conditions, we did not observe any LDH release from the exposed cells, suggesting that apoptosis was most likely triggered through the activation of signal transduction pathways at the cell surface without membrane disruption (35,44). However, our preliminary observations suggest that samples of p53(1–63) aged in acidic conditions for longer time periods can induce a necrosis-like cell death as indicated by the increase in LDH release in the culture medium (data not shown), suggesting that mature amyloid fibrils could be cytotoxic even through a less specific mechanism. Cell necrotic death triggered by amyloid fibrils has previously been reported in some cases (45,46).
The formation of off-pathway, nonfunctional partially structured protein folding variants is referred to as kinetic misfolding (47); in the case of several wild-type proteins found aggregated in human diseases such as senile systemic amyloidosis and some types of cancer, the latter often results in the aggregation of the off-pathway intermediates (48). This mechanism is attractive in the case of p53 because it describes how a population of natively folded, biologically active molecules can acquire a nonfunctional, aggregation-prone state over time. It is known that aggregates of p53, either wild type or, more frequently, containing specific mutations are found in many tumor cells (49). Recently, the possible origin of these aggregates from any perturbation of the folded/unfolded transition equilibrium of the p53 DNA binding domain has been highlighted. The latter equilibrium seems to involve a partially folded intermediate with increased exposure of the hydrophobic core, which undergoes reversible aggregation into a 10- to 20-mer. This finding can explain the tendency of p53 to depart from the native state to an inactive one undergoing aggregation and making the cell susceptible to malignant transformation (49). Under mild denaturing conditions, the DNA binding domain and a peptide from a mutant of the tetramerization domain are also able to form β-sheet-rich fibrillar aggregates very similar to those found in amyloid diseases (50–52) and endowed with similar cytotoxicity (50,53).
These and our data support the idea that p53 may be involved in tumor genesis by losing its biological function in two ways: a loss of the antitumor function of the wild-type or mutated protein and/or a possible gain of function increasing the aggregation potential of the misfolded mutant or wild-type p53; once present, p53 aggregates can propagate the structural alteration to other functional, correctly folded p53 molecules in a prion-like way, as already suggested (9,49). This parallelism is further strengthened by a computational analysis revealing a high similarity of p53 and PrP hydrophobicity patterns in terms of marked departure from randomness, at odds with the majority of proteins (54). Such highly deterministic hydrophobicity patterns can easily be perturbed by single amino acid substitutions, and this is the reason why both proteins present an unusually low ratio of polymorphisms (i.e., silent mutations/total mutations).
The lack of functional p53 by these or other mechanisms can be considered one of the fundamental events favoring malignant transformation of a cell. In this perspective, our results on an isolated domain under in vitro conditions are of interest, showing for the first time that even the p53 N-terminal domain can potentially aggregate into cytotoxic amyloid assemblies. It could be argued that our in vitro observations lack physiological relevance because p53(1–63) was made to fibrillize in vitro at low pH. However, some cellular compartments, e.g., endosomes and lysosomes, associated with protein translocation and degradation, respectively, are characterized by a low internal pH. In addition, the anionic phospholipids found in the inner leaflet of the cell membrane have been proposed to be potent inductors of β-sheet structures by acting as conformational catalysts for amyloids (55). The local low pH found near negatively charged lipid membranes favors partial unfolding, and hence formation of aggregation-prone intermediates, of adsorbed protein molecules (56). Moreover, it cannot be excluded that the aggregation of the whole p53 can be favored even at physiological pH values by the excluded volume effects associated with the macromolecular crowding of the intracellular environment (57). Under these conditions, aggregation could possibly start from the unstructured N-terminal domain in some functional state where it is not associated with mdm2, in agreement with other examples of unfolded domain-driven aggregation (58–61). Finally, because of the aggregation propensity of the tetramerization and the core domain (51,52), some cooperativity among p53 domains in favoring aggregation under in vivo conditions and/or in the presence of suitable mutations could be hypothesized.
Actually, the majority of cancer-associated p53 mutations are located within the DNA-binding domain and the tetramerization domain. However, the IARC TP53 Mutation Database (http://www-p53.iarc.fr) reports that some mutations within the N-terminal domain of p53 are also associated with human cancer. Many of these mutations, which are not implicated in the transcriptional activity of the N-terminal domain, could potentially perturb the p53 aggregation rate according to a previously reported algorithm (62). Such a formula evaluates the intrinsic contribution of any amino acid substitution to the aggregation kinetics of an unfolded polypeptide chain (Table 1), considering the resulting quantitative modification of hydrophobicity, net charge, and propensity to form a β-sheet. The possibility that these mutations can favor the aggregation of p53(1–63) or even of the whole protein, under conditions closer to the physiological ones, is currently under investigation. In conclusion, the propensity of the N-terminal domain of p53 to form amyloid aggregates could help to explain the susceptibility of p53 to undergo in vivo structural modification from the native to an inactive aggregated state, making the cell highly vulnerable to malignant transformation.
TABLE 1.
Cancer-associated p53(1–63) mutations are supposed to affect the aggregation rate of the domain
| Mutations | ln(Vmut/Vwt) |
|---|---|
| D7H | 3.46 |
| E11Q | 0.555 |
| E17D | −1.4398 |
| K24N | 1.139 |
| L35F | 0.79785 |
| L43S | −1.14 |
| S46F | 2.681 |
| P47S/L | 0 |
| D49H | 3.924 |
| A63V | 1.494 |
For every reported mutation the parameter ln(Vmut/Vwt) was calculated according to Chiti et al. (62). Positive or negative values respectively indicate increased or reduced aggregation propensity of the mutated peptide relative to the wild type. Vmut, aggregation rate of the mutated peptide; Vwt, aggregation rate of the wild-type peptide.
Acknowledgments
We thank Prof. Nobua Tsuchida for providing the pUC18p53 plasmid and Silvia Torrassa for help in the AFM measurements. This work was supported by grants from the Italian MURST (PRIN Project n. 2005053998_001 and _005), from the Ente Cassa di Risparmio di Firenze (grant n. 2005.07.01), and Fondazione CARIGE.
Stefania Rigacci and Monica Bucciantini contributed equally to this work.
Abbreviations used: PCR, polymerase chain reaction; FT-IR, Fourier transform infrared spectroscopy; AFM, atomic force microscopy; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; LDH, lactic dehydrogenase; ThT, thioflavine T; ANS, 1-anilinonaphthalene-8-sulfonic acid.
Editor: Heinrich Roder.
References
- 1.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell. 88:323–331. [DOI] [PubMed] [Google Scholar]
- 2.Vousden, K. H., and X. Lu. 2002. Live or let die: the cell's response to p53. Nat. Rev. Cancer. 2:594–604. [DOI] [PubMed] [Google Scholar]
- 3.Hollstein, M., D. Sidransky, B. Vogelstein, and C. C. Harris. 1991. p53 mutations in human cancers. Science. 253:49–53. [DOI] [PubMed] [Google Scholar]
- 4.Li, M., C. L. Brooks, F. Wu-Baer, D. Chen, R. Baer, and W. Gu. 2003. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 302:1972–1975. [DOI] [PubMed] [Google Scholar]
- 5.Bell, S., C. Klein, L. Müller, S. Hansen, and J. Buchner. 2002. p53 contains large unstructured regions in its native state. J. Mol. Biol. 322:917–927. [DOI] [PubMed] [Google Scholar]
- 6.Cho, Y., S. Gorina, P. D. Jeffrey, and N. P. Pavletich. 1994. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 265:346–355. [DOI] [PubMed] [Google Scholar]
- 7.Jeffrey, P. D., S. Gorina, and N. P. Pavletich. 1995. Crystal structure of the tetramerization domain of the p53 tumour suppressor at 1.7 angstroms. Science. 267:1498–1502. [DOI] [PubMed] [Google Scholar]
- 8.Dawson, R., L. Müller, A. Dehner, C. Klein, H. Kessler, and J. Buchner. 2003. The N-terminal domain of p53 is natively unfolded. J. Mol. Biol. 332:1131–1141. [DOI] [PubMed] [Google Scholar]
- 9.Butler, J. S., and S. N. Loh. 2003. Structure and aggregation of the zinc-free form of the p53 DNA binding domain. Biochemistry. 42:2396–2403. [DOI] [PubMed] [Google Scholar]
- 10.Milner, J., and E. A. Medcalf. 1991. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell. 65:765–774. [DOI] [PubMed] [Google Scholar]
- 11.Moll, U. M., A. G. Ostermeyer, R. Haladay, B. Winkfield, M. Frazier, and G. Zambetti. 1996. Cytoplasmic sequestration of wild-type p53 protein impairs the G1 checkpoint after DNA damage. Mol. Cell. Biol. 16:1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottifredi, V., and C. Prives. 2001. Getting p53 out of the nucleus. Science. 292:1851–1852. [DOI] [PubMed] [Google Scholar]
- 13.Ostermeyer, A. G., E. Runko, B. Winkfield, B. Ahn, and U. M. Moll. 1996. Cytoplasmically sequestered wild-type p53 protein in neuroblastoma is relocated to the nucleus by a C-terminal peptide. Proc. Natl. Acad. Sci. USA. 93:15190–15194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolff, A., A. Technau, C. Ihling, K. Technau-Ihling, R. Erber, F. X. Bosch, and G. Brandner. 2001. Evidence that wild-type p53 in neuroblastoma cells is in a conformation refractory to integration into the transcriptional complex. Oncogene. 20:1307–1317. [DOI] [PubMed] [Google Scholar]
- 15.Stefani, M. 2004. Protein misfolding and aggregation: new examples in medicine and biology of the dark side of the protein world. Biochim. Biophys. Acta. 1739:5–25. [DOI] [PubMed] [Google Scholar]
- 16.Stefani, M., and C. M. Dobson. 2003. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J. Mol. Med. 81:678–699. [DOI] [PubMed] [Google Scholar]
- 17.Selkoe, D. J. 2003. Folding proteins in fatal ways. Nature. 426:900–904. [DOI] [PubMed] [Google Scholar]
- 18.Sherman, M. Y., and A. L. Goldberg. 2001. Cellular defences against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 29:15–32. [DOI] [PubMed] [Google Scholar]
- 19.Serpell, L. S. 2000. Alzheimer's amyloid fibrils: structure and assembly. Biochim. Biophys. Acta. 1502:16–30. [DOI] [PubMed] [Google Scholar]
- 20.Hegde, R. S., P. Tremblay, D. Groth, S. J. DeArmond, S. B. Prusiner, and V. R. Lingappa. 1999. Transmissible and genetic prion diseases share a common pathway of neurodegeneration. Nature. 402:822–826. [DOI] [PubMed] [Google Scholar]
- 21.Lashuel, H. A., and P. T. Lansbury. 2006. Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q. Rev. Biophys. 39:167–201. [DOI] [PubMed] [Google Scholar]
- 22.LeVine, H. 3rd. 1999. Quantification of β-sheet amyloid fibril structures with Thioflavin T. Methods Enzymol. 309:274–284. [DOI] [PubMed] [Google Scholar]
- 23.Leslie, A. G. W. 1992. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, No. 26. SERC Daresbury Laboratory, Warrington, UK.
- 24.Abe, K., and H. Kimura. 1996. Amyloid β toxicity consists of a Ca2+-independent early phase and a Ca(2+)-dependent late phase. J. Neurochem. 67:2074–2078. [PubMed] [Google Scholar]
- 25.Uversky, V. N. 2002. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 11:739–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding, F., J. M. Borreguero, S. V. Buldyrey, H. E. Stanley, and N. V. Dokholyan. 2003. Mechanism for the α-helix to β-hairpin transition. Proteins. 53:220–228. [DOI] [PubMed] [Google Scholar]
- 27.Engelhard, M., and P. A. Evans. 1995. Kinetics of interaction of partially folded proteins with a hydrophobic dye: evidence that molten globule character is maximal in early folding intermediates. Protein Sci. 4:1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semisotnov, G. V., N. A. Rodionova, O. I. Razgulyaev, V. N. Uversky, A. F. Gripas, and R. I. Gilmanshin. 1991. Study of the molten globule intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers. 31:119–128. [DOI] [PubMed] [Google Scholar]
- 29.Zandomeneghi, G., M. R. Krebs, M. G. McCammon, and M. Fandrich. 2004. FTIR reveals structural differences between native β-sheet proteins and amyloid fibrils. Protein Sci. 13:3314–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerdà-Costa, N., A. Esteras-Chopo, F. X. Aviles, L. Serrano, and V. Villegas. 2007. Early kinetics of amyloid fibril formation reveals conformational reorganisation of initial aggregates. J. Mol. Biol. 366:1351–1363. [DOI] [PubMed] [Google Scholar]
- 31.Relini, A., R. Rolandi, M. Bolognesi, A. Gliozzi, M. Aboudan, G. Merlini, and V. Bellotti. 2004. Ultrastructural organization of ex-vivo amyloid fibrils formed by the apolipoprotein A-I Leu174Ser variant: an atomic force microscopy study. Biochim. Biophys. Acta. 1690:33–41. [DOI] [PubMed] [Google Scholar]
- 32.Calamai, M., C. Canale, A. Relini, M. Stefani, F. Chiti, and C. M. Dobson. 2005. Reversal of protein aggregation provides evidence for multiple aggregated states. J. Mol. Biol. 346:603–616. [DOI] [PubMed] [Google Scholar]
- 33.Lomas, D. A., and R. W. Carrell. 2002. Serpinopathies and the conformational dementias. Nat. Rev. Genet. 3:759–768. [DOI] [PubMed] [Google Scholar]
- 34.Fay, N., Y. Inoue, L. Bousset, H. Taguchi, and R. Melki. 2003. Assembly of the yeast prion Ure2p into protein fibrils. Thermodynamic and kinetic characterization. J. Biol. Chem. 278:30199–30205. [DOI] [PubMed] [Google Scholar]
- 35.Bucciantini, M., E. Giannoni, F. Chiti, F. Baroni, L. Formigli, J. Zurdo, N. Taddei, G. Ramponi, C. M. Dobson, and M. Stefani. 2002. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 416:507–511. [DOI] [PubMed] [Google Scholar]
- 36.Bucciantini, M., S. Rigacci, A. Berti, L. Pieri, C. Cecchi, D. Nosi, L. Formigli, F. Chiti, and M. Stefani. 2005. Patterns of cell death triggered in two different cell lines by HypF-N prefibrillar aggregates. FASEB J. 19:437–439. [DOI] [PubMed] [Google Scholar]
- 37.Cecchi, C., S. Baglioni, C. Fiorillo, A. Pensalfini, G. Liguri, S. Rigacci, M. Bucciantini, and M. Stefani. 2005. Insights into the molecular basis of the differing susceptibility of varying cell types to the toxicity of amyloid aggregates. J. Cell Sci. 18:3459–3470. [DOI] [PubMed] [Google Scholar]
- 38.Uversky, V. N., J. Li, and A. L. Fink. 2001. Evidence for a partially folded intermediate in α-synuclein fibril formation. J. Biol. Chem. 276:10737–10744. [DOI] [PubMed] [Google Scholar]
- 39.Fändrich, M., and C. M. Dobson. 2002. The behaviour of polyamino acids reveals an inverse side chain effect in amyloid structure formation. EMBO J. 21:5682–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jayasinghe, A. A., and R. Langen. 2005. Lipid membranes modulate the structure of islet amyloid polypeptide. Biochemistry. 44:12113–12119. [DOI] [PubMed] [Google Scholar]
- 41.Fezoui, Y., and D. B. Teplow. 2002. Kinetic studies of amyloid β-protein fibril assembly. Differential effects of α-helix stabilization. J. Biol. Chem. 277:36948–36954. [DOI] [PubMed] [Google Scholar]
- 42.Kelly, J. W. 2000. Mechanisms of amyloidogenesis. Nat. Struct. Biol. 7:824–826. [DOI] [PubMed] [Google Scholar]
- 43.Harper, J. D., S. S. Wong, C. M. Lieber, and P. T. Jr Lansbury. 1999. Assembly of Aβ amyloid protofibrils: an in vitro model for a possible early event in Alzheimer's disease. Biochemistry. 38:8972–8990. [DOI] [PubMed] [Google Scholar]
- 44.Bucciantini, M., G. Calloni, F. Chiti, L. Formigli, D. Nosi, C. M. Dobson, and M. Stefani. 2004. Prefibrillar amyloid protein aggregates share common features of cytotoxicity. J. Biol. Chem. 279:31374–31382. [DOI] [PubMed] [Google Scholar]
- 45.Gharibyan, A. L., V. Zamotin, K. Yanamandra, O. S. Moskaleva, B. A. Margulis, I. A. Kostanyan, and L. A. Morozova-Roche. 2007. Lysozyme amyloid oligomers and fibrils induce cellular death via different apoptotic/necrotic pathways. J. Mol. Biol. 365:1337–1349. [DOI] [PubMed] [Google Scholar]
- 46.Novitskaya, V., O. V. Bocharova, I. Bronstein, and I. V. Baskakov. 2006. Amyloid fibrils of mammalian prion proteins are highly toxic to cultured cells and primary neurons. J. Biol. Chem. 281:13828–13836. [DOI] [PubMed] [Google Scholar]
- 47.Silva, J. L., D. Foguel, and C. A. Royer. 2001. Pressure provides new insight into protein folding, dynamics and structure. Trends Biochem. Sci. 26:612–618. [DOI] [PubMed] [Google Scholar]
- 48.Kim, Y. S., T. W. Randolph, F. J. Stevens, and J. F. Carpenter. 2002. Kinetics and energetics of assembly, nucleation and growth of aggregates and fibrils for an amyloidogenic protein. Insight into transition states from pressure, temperature and co-solute studies. J. Biol. Chem. 277:27240–27246. [DOI] [PubMed] [Google Scholar]
- 49.Ishimaru, D., L. M. T. R. Lima, L. F. Maia, P. M. Lopez, A. P. A. Bom, A. P. Valente, and J. L. Silva. 2004. Reversible aggregation plays a crucial role on the folding landscape of p53 core domain. Biophys. J. 87:2691–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishimaru, D., L. R. Andrade, L. S. P. Teixeira, P. A. Quesado, L. M. Maiolino, P. M. Lopez, Y. Cordeiro, L. T. Costa, W. M. Heckl, G. Weissmuller, D. Foguel, and J. L. Silva. 2003. Fibrillar aggregates of the tumor suppressor p53 core domain. Biochemistry. 42:9022–9027. [DOI] [PubMed] [Google Scholar]
- 51.Lee, A. S., C. Galea, E. L. DiGiammarino, B. Jun, G. Murti, R. C. Ribeiro, G. Zambetti, C. P. Schultz, and R. W. Kriwacki. 2003. Reversible amyloid formation by the p53 tetramerization domain and a cancer-associated mutant. J. Mol. Biol. 327:699–709. [DOI] [PubMed] [Google Scholar]
- 52.Higashimoto, Y., Y. Asanomi, S. Takakusagi, M. S. Lewis, K. Uosaki, S. R. Durell, C. W. Anderson, E. Appella, and K. Sakaguchi. 2006. Unfolding, aggregation and amyloid formation by the tetramerization domain from the mutant p53 associated with lung cancer. Biochemistry. 45:1608–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosal, R., M. R. Pincus, P. W. Brandt-Rauf, R. L. Fine, J. Michi, and H. Wang. 2004. NMR solution structure of a peptide from the mdm-2 binding domain of the p53 protein that is selectively cytotoxic to cancer cells. Biochemistry. 43:1854–1861. [DOI] [PubMed] [Google Scholar]
- 54.Porrello, A., S. Soddu, J. P. Sbilut, M. Crescenzi, and A. Giuliani. 2004. Discrimination of single amino acid mutations of the p53 protein by means of deterministic singularities of recurrence quantification analysis. Proteins. 55:743–755. [DOI] [PubMed] [Google Scholar]
- 55.Zhao, H., E. K. J. Tuominen, and P. K. J. Kinnunen. 2004. Formation of amyloid fibers triggered by phosphatidylserine-containing membranes. Biochemistry. 43:10302–10307. [DOI] [PubMed] [Google Scholar]
- 56.Grudzielanek, S., V. Smirnovas, and R. Winter. 2007. The effects of various membrane physical-chemical properties on the aggregation kinetics of insulin. Chem. Phys. Lipids. 149:28–39. [DOI] [PubMed] [Google Scholar]
- 57.van den Berg, B., J. Ellis, and C. M. Dobson. 1999. Effects of macromolecular crowding on protein folding and aggregation. EMBO J. 18:6927–6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baxa, U., K. L. Taylor, J. S. Wall, M. N. Simon, N. Q. Cheng, R. B. Wickner, and A. C. Steven. 2003. Architecture of Ure2p prion filaments: the N-terminal domains form a central core fiber. J. Biol. Chem. 278:43717–43727. [DOI] [PubMed] [Google Scholar]
- 59.Paushkin, S. V., V. V. Kushnirov, V. N. Smirnov, and M. D. Ter-Avanesyan. 1996. Propagation of the yeast prion-like [ψ+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 12:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 60.Balguerie, A., S. Dos Reis, C. Ritter, S. Chaignepain, B. Coulary-Salin, V. Forge, K. Bathany, I. Lascu, J. M. Schmitter, R. Riek, and J. Saupe. 2003. Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. J. Mol. Biol. 22:2071–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fandrich, M., V. Forge, K. Buder, M. Kittler, C. M. Dobson, and S. Diekmann. 2003. Myoglobin forms amyloid fibrils by association of unfolded polypeptide segments. Proc. Natl. Acad. Sci. USA. 26:15463–15468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chiti, F., M. Stefani, N. Taddei, G. Ramponi, and C. M. Dobson. 2003. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 14:6950–6953. [DOI] [PubMed] [Google Scholar]