Abstract
In this report, we present features of the neuronal SNARE complex determined by atomistic molecular dynamics simulations. The results are robust for three models, varying force fields (AMBER and GROMOS) and solvent environment (explicit and implicit). An excellent agreement with experimental findings is observed. The SNARE core complex behaves like a stiff rod, with limited conformational dynamics. An accurate picture of the interactions within the complex emerges with a characteristic pattern of atomic contacts, hydrogen bonds, and salt bridges reinforcing the underlying layer structure. This supports the metaphor of a molecular Velcro strip that has been used by others to describe the neuronal fusion complex. No evidence for directionality in the formation of these interactions was found. Electrostatics largely dominates all interactions, with an acidic surface patch structuring the hydration layers surrounding the complex. The interactions within the four-helix bundle are asymmetric, with the synaptobrevin R-SNARE notably exhibiting an increased rigidity with respect to the three Q-SNARE helices. The interaction patterns we observe provide a new tool for interpreting the impact of mutations on the complex.
INTRODUCTION
Membrane fusion is essential for a manifold of biological processes such as fertilization, cellular transport, and viral infection. Transport processes including intracellular traffic or the secretion of hormones and neurotransmitters require an exchange of material from inside the transport vesicle to another cellular compartment or to the surrounding medium. Such exchange is prevented by the membranes protecting both the vesicle and the target organelle and thus requires their fusion. Vesicle fusion is achieved in a multistep process starting with priming and docking followed by the actual fusion event, where both membranes are first brought into close contact and a fusion pore is then formed (1). Membranes do not fuse spontaneously as several forces including electrostatic and hydration repulsion oppose their approach. It is thought that in all eukaryotes, fusion is achieved via specialized protein complexes involving the soluble n-ethylmaleimide-sensitive factor attachment protein receptors (SNARE) family of proteins (2,3). The SNARE proteins form a tight complex that is central to membrane fusion. This complex is thought to provide the necessary force for overcoming intermembrane repulsion and thus catalyze the fusion event. Part of the complex also contributes to the formation of the fusion pore. After fusion, dedicated molecular machinery carries out the disassembly of the SNARE complex. The molecular details of both assembly and disassembly of the SNARE complex are poorly understood. In this work, we focus on a description of intrinsic structural and dynamic properties of the neuronal SNARE complex to shed some light upon the probable driving forces for assembly and disassembly.
The mechanism of SNARE assembly is presently unknown. A zipping mechanism, by which the complex forms sequentially starting from the membrane-distal region, has been suggested (4). Data from a recent mutational study, however, does not support such a vectorial process (5). Once assembled, the fusion complex is extraordinarily stable. It supports temperatures up to 80°C (6) and is resistant to SDS unless boiled (7). The structure of the assembled neuronal SNARE complex consists of a parallel four-helical bundle forming a ternary assembly of the synaptobrevin (Sb), syntaxin (Sx), and SNAP-25 (Sn) proteins (see Fig. 1 A). Two C-terminal transmembrane domains are present in Sb and Sx, respectively anchored in the vesicular and plasma membranes. SNAP-25, which contributes two helices to the complex, is attached to the plasma membrane via its palmitoylated flexible linker domain. The crystal structures of the synaptic and endosomal SNARE complexes have revealed a characteristic layer structure with 15 hydrophobic layers and one central ionic layer (8–10). The ionic layer is conserved and the four helices forming the bundle contribute one arginine and three glutamines, respectively, leading to their classification as R-, Qa-, Qb-, and Qc-SNAREs (11).
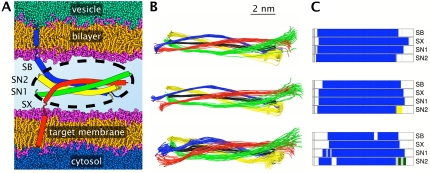
FIGURE 1.
Schematic view of the SNARE complex embedded in the vesicular and target membranes. (A) Inside of the vesicle represented in green and the cytosol in blue. Lipid headgroups are shown in pink, aliphatic tails in orange. The four helices of the complex are synaptobrevin (Sb, blue), syntaxin (Sx, red), and SNAP-25 (Sn1 and Sn2, green and yellow). The palmitoylated linker of SNAP-25 is not shown. The solvent in the space separating both membranes is omitted for clarity. The black, dotted ellipse represents the cytosolic core of the complex that was simulated using molecular dynamics. (B) Cumulated snapshots of the helical axes determined using P-Curves. The overall helical axis is shown in black. Results for the last 2 ns of the three simulation systems Q1 (top), Q2 (center), and Q2GB (bottom) are presented at a 100-ps interval. (C) Dominant secondary structure present for >90% of the total simulation time for Q1, Q2, and Q2GB from top to bottom. The color code is α-helix, blue; bend, olive; turn, yellow; and coil, gray.
The extraordinary stability of the synaptic fusion complex requires an adapted mechanism for its disassembly. Dedicated ATP-powered molecular machinery is necessary to recycle the fusion proteins. Disassembly is mediated by the n-ethylmaleimide-sensitive factor (NSF), an ATPase, and by its adaptor protein, α-soluble NSF attachment protein (α-SNAP). Three α-SNAPs bind the SNARE complex in an antiparallel orientation. This, in turn, leads to ATP hydrolysis by NSF and ultimately to the disassembly of the SNARE complex. Electrostatic interactions between α-SNAP and the acidic surface of the fusion complex seem to play an important role in this process (12).
Previous combined experimental and computational studies of the synaptic fusion complex have focused on mutations in the central ionic (13) and membrane proximal (14,15) layers, as well as on N-terminal mutations in SNAP-25 (16). Here, we describe a detailed global view of the interactions within the core complex.
METHODS
System preparation
The initial configuration of the simulation system was constructed from the crystal structure of the neuronal SNARE complex at 2.4 Å resolution (10). We combined parts from the three distinct copies of the SNARE complex in the crystal unit cell. The backbone root mean-square deviation (RMSD) between the copies is <1 Å. The constructed model of the four-helical SNARE bundle comprises 299 amino acids and consists of synaptobrevin-II (Sb) N25 to M96, syntaxin-1a (Sx) G180 to R262, and SNAP-25B (Sn) M7-K83 and V120-G206. The N-terminal HABC domain of Sx and the palmitoylated linker in Sn were not included. Our model corresponds to the soluble part of the complex highlighted in Fig. 1 A. Simulated annealing and energy minimization of this structure were carried out with the Yasara software (17).
pKa calculations
We have used pKa calculations as implemented in the Yasara program to determine the most probable protonation state of the hydrated complex (18). Sx:H199 was found to be neutral (δ-protonated), whereas all other histidines are protonated. Other amino acids are in their standard protonation state.
Simulation setup and equilibration
The GROMACS 3 and AMBER 8 program suites were used to carry out molecular dynamics (MD) simulations (19,20). With GROMACS, we used the ffG43a2 force field (21), and with AMBER, the ff99 force field (22) for explicit solvent simulations. The generalized Born (GB) approach was used to represent solvent electrostatic damping in an additional implicit solvent simulation (23,24). In analogy to the explicit solvent simulations with minimal salt concentration, the GB simulations were run with 0 M salt. After energy minimization in vacuum, the explicit solvent systems were neutralized with 10 K+ counterions (25) and hydrated in rectangular boxes of TIP3P (AMBER (26)) or SPC (GROMACS (27)) water molecules, respectively; see Table 1 for details. Simulations were carried out with periodic boundary conditions, and electrostatic interactions were treated with the particle mesh Ewald (PME) algorithm (28). The direct space cutoff was 10 Å in the GROMACS run (denoted as Q1) and 9 Å in the AMBER run (denoted as Q2). An integration time step of 2 fs was used. With AMBER, all bonds involving hydrogen atoms were constrained using SHAKE (29). With GROMACS, all bond lengths were constrained using the LINCS algorithm (30). The production runs were carried out in the NPT ensemble using a Berendsen barostat (31) with a pressure (P) of 1 bar, and a Berendsen thermostat (31) at a reference temperature (T) of 300 K. In the Q1 simulation, the protein complex was first relaxed in water by a short minimization, then the counterions were added. A 200-ps molecular dynamics equilibration in the NPT ensemble followed with 24 kcal mol−1 Å−2 position restraints on the heavy atoms. During the subsequent production run, center of mass translations were removed every step. In simulation Q2, the water molecules and ions were first relaxed by an energy minimization using 25 kcal mol−1 Å−2 restraints on all solute atoms. The temperature was then increased during 10 ps from 100 K to 300 K and held at this temperature during a further 40 ps of constant volume simulation. The restraints on the solute were then gradually relaxed from 5 to 0.5 kcal mol−1 Å−2 during six successive minimizations and 25-ps MD equilibrations at constant pressure. Fifty picoseconds of unrestrained simulation were performed before the production phase. The production runs with AMBER were carried out with an optimized calculation module (PMEMD) (32). For the implicit solvent simulations, denoted Q2GB, the system was first relaxed by an energy minimization using a 4r distance-dependent dielectric function, then by energy minimization with the GB model. The cutoff for the nonbonded interactions was set to 15 Å. The cutoff for the GB pairwise summation was set to 12 Å. During the production phase, an integration time step of 2 fs was used. All bonds involving hydrogen atoms were constrained using SHAKE (29). A Langevin thermostat was used at a temperature of 300 K (33), with a collision frequency of 1 ps−1. The translational and rotational center-of-mass motion were removed every 1000 steps.
TABLE 1.
Characteristics of the simulation systems
Analysis and graphics programs used
Secondary structure elements were identified using two approaches, the DSSP method by Kabsch and Sander (34) and the STRIDE method by Heinig and Frishman (35). Graphical representations were prepared with the VMD (36), Yasara (37), and VTK (38) programs and toolkits. Standard conformational analysis was carried out using tools from the GROMACS package as well as the Ptraj module of AMBER. Helicoidal parameters were analyzed via P-Curves (39). Energy components were analyzed using the MDS software (unpublished). Further analysis was carried out with local code. Statistical and data analysis was performed using the R statistical software package (41).
RESULTS
Structural drift and secondary structure
We analyzed the structural drift within the SNARE complex by calculating the time series of the root mean-square deviation (RMSD) for the central part of the complex including its 16 layers. In explicit solvent, the structure of the four-helical bundle stays close to the crystal structure determined by Sutton et al. (10) (0.8–1.4 Å RMSD per helix for the Cα-atoms; see Table 2), which will be used as reference throughout this work. An increased RMSD of 1.9 to 2.4 Å is observed for simulation Q2GB with implicit solvent. For comparison, the variation within all available crystal structures reaches 0.6–1.1 Å. In all cases, the RMSD shows little time evolution and levels off before the end of the simulation, although in Q2GB the RMSD increases to ∼3 Å for Sb and Sx toward the very end of the simulation. The cumulated views in Fig. 1 B show that the differences in RMSD arise from differences in flexibility, rather than from differences in the overall structure of the complex.
TABLE 2.
Root mean-square deviations
| Helix | Q1 | Q2 | Q2GB | PX |
|---|---|---|---|---|
| Sb | 0.8 | 1.2 | 2.4 | 0.9 |
| Sx | 0.8 | 0.8 | 2.1 | 0.8 |
| Sn1 | 0.8 | 1.2 | 1.9 | 1.1 |
| Sn2 | 0.8 | 1.4 | 2.2 | 0.6 |
Values are indicated in ångströms. The simulation values are average RMSDs calculated for the Cα atoms of the 53 central residues of each helix over the last 2 ns after fitting on the same set of Cα atoms. The PX column shows the largest deviations observed between the asymmetric units of the 2.4 Å protein crystal by Sutton et al. (10) and the 1.4 Å structure by Ernst and Brunger (9).
A secondary structure analysis confirms the very high structural integrity of the synaptic fusion complex (Fig. 1 C). The amount of secondary structure observed in the simulations follows the order Q1 ∼ Q2 > Q2GB with an overall percentage of α-helical content of 89%, 89%, and 82%, respectively. The Sb and Sx helices maintain their α-helicity between layers −7 and +8 (see Fig. 2 C for layer numbering) throughout the explicit solvent simulations Q1 and Q2. The same is true for layers −7 to +6 in synaptosome-associated protein of 25 kDa (SNAP-25) helix 1 (Sn1) and −7 to +5 in SNAP-25 helix 2 (Sn2). Loss of secondary structure occurs mainly at the termini. With implicit solvent (Q2GB), a decrease in α-helicity of ∼15% is observed locally, in particular for Sb (layers −3 to −1, +4/+5, +7/+8) and Sx (layers −2/−1 and +8).
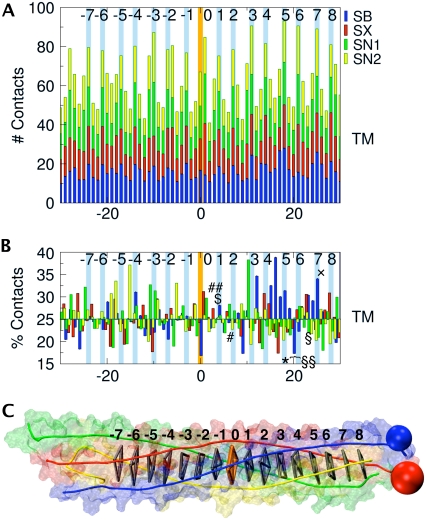
FIGURE 2.
(A) Number of contacts as a function of residue position. The central ionic layer is numbered 0 on the abscissa. The numbering of the hydrophobic and central layers is shown top right. The transmembrane (TM) part of the complex extends to the right of the plot. The color code is the same as in Fig. 1. At each residue position, the contacts for the four helices are cumulated. (B) Percentage contribution of each helix to the overall contacts at a given position. Tetanus (TeNT) and botulinum (BoNT) neurotoxin cleavage sites are indicated as follows: TeNT (*), BoNT/A (§), BoNT/B (§§), BoNT/C (♦), BoNT/D ($), BoNT/E (#), BoNT/F (##), and BoNT/G (×). The structure of the SNARE complex shown in panel C is aligned with plots A and B. The hydrophobic layers (gray) and the central ionic layer (orange) are indicated by virtual bonds between the corresponding Cα positions, as well as the connection to the TM helices (blue and red spheres).
Contact analysis and layer structure of the SNARE complex
Fig. 2 A shows a plot of the atomic contacts between residues versus residue position in the four-helical SNARE bundle. For each position, the contacts of equivalent residues in all four helices are summed. The total number of contacts shows a periodic pattern of maxima corresponding to the 15 hydrophobic layers and the central ionic layer separated by stretches of 2–3 residues with reduced contacts. This illustrates the close packing of the core of the complex and emphasizes the structural role of these layers. Residues next to layer-residues, notably in positions −2 and −3, can show a number of contacts quite close to the maximum, or even superior in the case of the central ionic layer. It is important to note that contacts were normalized by the number of atoms in each amino-acid residue. Since the central ionic layer involves large amino-acid side chains, its total number of contacts, which is indeed a maximum, is lowered more than the neighboring residues by the normalization.
On average, all four helices contribute equally to each layer. For the core zone, between layers −7 and +8 all contributions lie between 16% and 45%. Notable deviations from the average occur for Sb with a contribution of up to 40% for layers +3 to +6, for Sn1 with a contribution of 40% adjacent to layer +3, and for Sn2 with up to 40% in layers −6 to −4 (Fig. 2 B). These overall trends are consistent for normalized and raw contact data and throughout all simulations, with only minor variations at the precise locations. The contact pattern we observe is expected to result from stabilizing interactions such as hydrogen bonds, salt bridges, and hydrophobic contacts, which will now be analyzed.
Hydrogen bonds and salt bridges form a stabilizing pattern of interactions
Based on our simulations and on available structural data, we analyzed the percentage of existence of all possible hydrogen bonds and salt bridges in the SNARE complex. The intrahelical n/n+3, n/n+4, and n/n+5 backbone hydrogen bonds were not included in this analysis. The crystal structures of the neuronal SNARE complex at 2.4 and 1.4 Å resolution will be used as in the literature (9,10), providing four distinct coordinate sets, and thus, a first rough classification of the probability for each interaction. Fig. 3, A and C, respectively summarize intra- and interhelical hydrogen bonds and salt bridges comparing results averaged over the Q1, Q2, and Q2GB simulations (plain upward curve) with the crystals (hatched downward curve). The agreement between experiment and simulation is very good, with correlation coefficients of 0.7 and 0.9 for hydrogen bonds and salt bridges, respectively. The simulation results are more precise, given the larger number of structures that could be used for analysis compared to only four available crystal structures. Hydrogen bonds missing from the crystal structure analysis are predominantly intrahelical. None of the experimentally detected interactions are missing in the simulations, although they may exist only for a fraction of the simulation time in a small number of cases. Among the three simulations, some differences due to the force field and treatment of the solvent are observed, although the overall patterns are very robust. One important difference occurs at the C-terminal Sn1/Sn2 interface, where several hydrogen bonds are missing in the simulations with the GROMACS force field. General trends indicate increased hydrogen bonding for the implicit solvent simulation Q2GB and lower percentages for the Q1 simulation with the GROMACS force field. In all cases, the hydrogen-bond and salt-bridge interaction patterns for each helix are distinct and highly variable. Interactions between nonadjacent helices are, however, negligible.
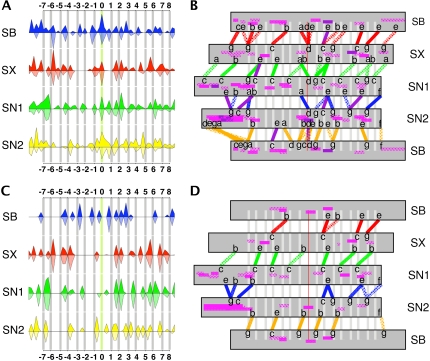
FIGURE 3.
Percentage occurrence of hydrogen bonds (top; A and B) and salt bridges (bottom; C and D) in the SNARE complex. The same color code as in Fig. 1 is used for A and C, where results averaged over simulations Q1, Q2, and Q2GB are shown by the full upward-facing curve. The hatched downward-facing curve is derived from the available crystal structures as described in the text. (B and D) Detailed view of the interactions between adjacent helices averaged over simulations Q1, Q2, and Q2GB. Three levels of interactions are shown as different filling patterns of the connecting lines: diamonds >25%, small dots 50–75%, and full lines 75% to 100%. Dark purple lines are interactions beyond 100% due to several cumulated contacts. Intrahelical interactions are shown in pink with similar fill patterns. The positions of the heptad-repeat scheme are indicated with letters a–g, where a and d correspond to hydrophobic layers.
Detailed interactions between adjacent pairs of helices averaged over simulation Q1, Q2, and Q2GB are shown in Fig. 3, B and D. Hydrogen bonds very often bridge across one or two hydrophobic layers acting like a reinforcing clip strengthening these positions by holding them firmly in place. The locations of these reinforcements correspond to strategic places within the SNARE complex, notably, layers −7/−6 at the N-terminus, layers +6/+7/+8 at the C-terminus, and layers −1/+1/+2 around the central ionic belt. For each pair of helices, a different distribution of strong and fragile regions is observed. A good example is the Sn1/Sn2 pair where a prominent gap appears between layers −6 and −2, both for hydrogen bonds and for salt bridges. Other weak spots occur around layers +3 to +7 for Sb/Sx, Sx/Sn1, and Sb/Sn2. Among all pairs, Sb and Sn2 have a tight network of hydrogen bonds, in particular toward the N-terminus. Layers that are particularly poor in hydrogen bonds are −5, −3, −2, +4, and +5. Different hydrogen-bonding scenarios between arginine 56 and the three glutamines are observed for the central ionic layer. A hydrogen bond between Sb:R56 and Sx:N226 is present for >80% of the simulation time in Q1 and Q2GB, made possible by a rearrangement of the arginine side chain early on in the simulation. This hydrogen bond is absent in simulation Q2.
Salt bridges are roughly half as numerous as hydrogen bonds, but are less labile as a function of simulation time. The heptad repeat leads to preferential positions for the formation of salt bridges, which overwhelmingly adopt an orientation aligned in one direction, spanning one or two hydrophobic layers. Table 3 shows the distribution of salt-bridge types, where each heptad is labeled with letters a to g, a and d being hydrophobic layers. Results for the three simulations and for the crystal structures are very similar. The most frequent salt bridges are of the g-b and c-e type. The core of the Sn1/Sn2 pair shows an infrequent salt-bridge configuration of the g-e(−1) type between Sn1:E61 and Sn2:K184, which is oriented differently than the others. Salt bridges are concentrated on the C-terminal half of the bundle, except for the Sb/Sn2 pair. Only Sb/Sn2 and Sx/Sn1 have strong salt bridges toward the N-terminus. In contrast to the hydrogen-bonding pattern, salt bridges generally do not reinforce the central ionic layer. This layer is, however, surrounded by two salt bridges between Sb and Sn2, spanning layers −1 and +1, as well as intrahelical salt bridges within these two helices. While Sb and Sn2 strengthen the bundle in the N-terminal half and around the ionic layer, the other helix pairs reinforce layers in the C-terminal half. Three salt bridges notably stabilize layer +2. Layers +5 and +7 are also well protected by a dense network of salt bridges. On the whole, hydrogen-bonding and salt-bridge interactions are synergistic, reinforcing specific parts of the complex.
TABLE 3.
Salt-bridge distribution
| Positions | Clip | Q1 | Q2 | Q2GB | PX |
|---|---|---|---|---|---|
| g-b | 1 | 5 | 5 | 5 | 5 |
| c-e | 1 | 7 | 7 | 7 | 8 |
| c-b | 2 | 3 | 5 | 9 | 8 |
| g-e | 2 | 1 | 2 | 2 | 2 |
| g-e(−1) | 0 | 3 | 2 | 2 | 4 |
| g-f(−1) | 0 | 1 | 1 | 0 | 1 |
| g-f | 2 | 0 | 1 | 1 | 0 |
| c-f | 1 | 0 | 0 | 1 | 0 |
Statistics of the residue positions forming salt bridges. The seven residues of a helical heptad repeat are labeled a–g, with a and d being hydrophobic layers. The Clip column indicates the number of hydrophobic layers that are spanned by the salt bridge. Only salt bridges with at least 25% occurrence were counted.
Electrostatic interactions dominate
To add to the structural analysis presented in the previous paragraphs, we examined the energy components of each residue's interactions with its environment. We considered electrostatic, Lennard-Jones, and internal energy components. Supplementary Material, Fig. S1 illustrates the cumulated interaction energies for the Q1 simulation. The total interaction energy is negative and dominated by the interlayer residues, which interact electrostatically and with surrounding water molecules. Minima are observed for the hydrophobic layers. The central ionic layer confirms its special role and does show important interactions with its environment. An above average stabilization is observed for layers +5 to +8. The electrostatic profile has precisely the same characteristics as the total interaction energy, suggesting that it dominates the interaction profile. Protein-protein interactions are mainly stabilizing, except in the central −1 to +4 layer region, where they are largely counterbalanced by attractive interactions with water. Lennard-Jones interactions, which are an order-of-magnitude smaller than electrostatic interactions, reflect the layer structure very well.
Considering the individual helix contributions, local variations are observed. Compared to expected contributions of +25%, helices may actually contribute between +40% and −20% (destabilizing) outside the hydrophobic layers, but no clear pattern emerges.
Helical parameters and flexibility distinguish synaptobrevin from the other helices
The characteristic conformational dynamics of protein helices are best described using helical parameters and can be analyzed using the P-Curves algorithm (39). Table 4 lists such parameters calculated for the Sb helix. A first comparison of the average helical parameters with the values for ideal secondary structure motifs shows that the simulations with the GROMACS force field are closest to the parameters of an ideal right-handed α-helix. The values obtained for the AMBER force field are shifted toward a 3-10 helix and this shift is still more marked for the GB implicit solvent simulation. In all cases, the helices in the four-helix bundle simulations show parameters that are much closer to an ideal helix than for a single free helix of the same length in solution.
TABLE 4.
Helicoidal parameters for standard secondary structure motifs, for synaptobrevin in the SNARE complex, and free in solution
| Structure | X displacement | Y displacement | Inclination | Tip | Rise | Twist |
|---|---|---|---|---|---|---|
| α(r) | 0.1 | 1.5 | −6.1 | −20.6 | 1.5 | 100.2 |
| Q1 | 0.14 | 1.54 | −8 | −18.8 | 1.5 | 100 |
| Q2 | 0.15 | 1.54 | −12 | −18 | 1.5 | 101 |
| Q2GB | 0.22 | 1.49 | −15 | −15 | 1.6 | 100 |
| 3-10 | 0.3 | 1.1 | −20.1 | −13.3 | 1.8 | 111.9 |
| Sol. | 0.5 | 1.4 | −26 | −7 | 1.8 | 97 |
Helicoidal parameters calculated via the P-Curves algorithm for the SNARE simulations Q1, Q2, and Q2GB. The ideal values for a right-handed α-helix and a 3-10 helix (in boldface) are taken from Sklenar et al. (39). The values for a single helix in solution determined from unpublished results are given at the bottom (Sol.). Translational parameters are in ångströms and rotational parameters are in degrees.
The residue-dependent profiles of the helical parameters do not reveal characteristic patterns. Fig. 4 shows the average Y-displacement parameter for helix Sb in simulation Q1. The standard deviations of the helical parameters (see Table 5), and Y-displacement in particular (which is related to the diameter of the helix), reveal that Sb behaves differently from the other three helices. The standard deviations—and thus the helix fluctuations—are much reduced, indicating that Sb is more rigid than Sx, Sn1, and Sn2.
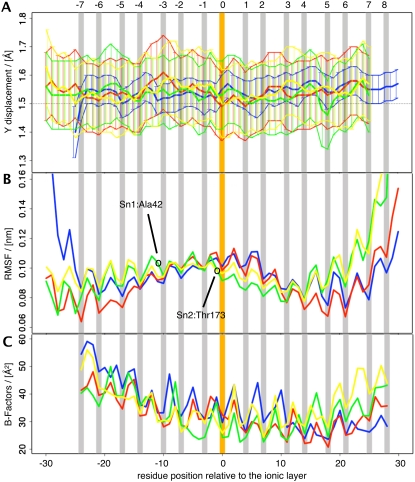
FIGURE 4.
Flexibility analysis using P-Curves (top, A); root mean-square fluctuations (RMSF; middle, B); and experimental B-factors (bottom, C). (A) The Y-displacement parameter as a function of residue position with respect to the central ionic layer at 0 averaged over the last 8 ns of simulation Q1. The same color code as in Fig. 1 is used. Error bars are shown along with upper and lower boundary plots to facilitate the visual comparison of the fluctuations. (B) RMSF of simulation Q1 after least-squares fitting of the whole trajectory with respect to the Cα atoms of the 16 layers. (C) B-factors averaged for all available crystal structures.
TABLE 5.
Mean standard deviation of the helicoidal parameters for simulation Q1
| Helix | X displacement | Y displacement | Inclination | Tip | Rise | Twist |
|---|---|---|---|---|---|---|
| Sb (R) | 0.06 | 0.06 | 3.6 | 2.1 | 0.18 | 8.8 |
| Sx (Qa) | 0.06 | 0.12 | 3.6 | 2.3 | 0.21 | 9.4 |
| Sn1 (Qb) | 0.08 | 0.13 | 3.8 | 2.9 | 0.22 | 10 |
| Sn2 (Qc) | 0.07 | 0.14 | 4.2 | 2.9 | 0.23 | 12 |
Values were calculated for residues −30 to +30 of each helix of the SNARE complex in simulation Q1. Translational parameters are in ångströms and rotational parameters are in degrees.
An alternative way to analyze the flexibility of the SNARE complex is using root mean-square fluctuations (RMSFs). RMSF plots shown in Fig. 4 B indicate that layer residues are less flexible than off-layer residues. This trend can also be seen in the experimental B-factors (Fig. 4 C). A more detailed analysis of the RMSFs was not carried out, as it would be biased by the dependence on the choice of the fitting procedure for least-squares superposition. Such artifacts are amplified in the SNARE complex due to its elongated form. Small changes at the extremities can thus result in artifactually large fluctuations.
Water inserts into the complex and is structured by acidic surface residues
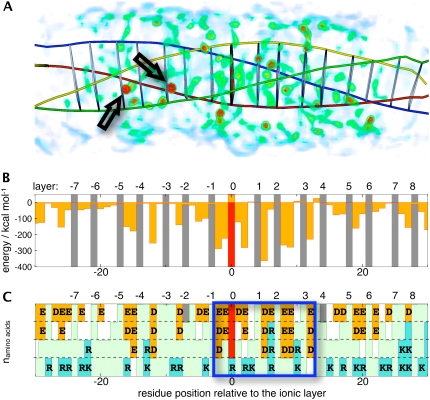
Water molecules can insert into hydrophobic layers of the SNARE complex, as documented by the 1.4 Å crystal structure 1N7S, where four inserted water molecules are observed: between layers −5/−4, −3/−2, in layer 0, and in layer +8. Similar water insertion was observed early on in our explicit solvent simulations Q1 and Q2. Fig. 5 A shows the average water occupancy in the Q1 simulation, highlighting two inserted water molecules, the first between layers −5/−4 and the second between −3/−2. In none of our simulations, however, did a water molecule insert into the ionic layer. The volume rendering of Fig. 5 A illustrates the structuring of the water by the complex, showing an anisotropic distribution of regions of high water occupancy, with isolated hot spots, and less occupied regions. Water occupancy is particularly high near the central ionic layer. Fig. 5 B shows the per-residue interaction energy of the SNARE complex with the surrounding water molecules. A distinct pattern is apparent with a prominent region of attractive interactions between layers −1 and +4. These interaction energies correlate very well with the distribution of acidic amino-acid residues shown in Fig. 5 C. Basic amino acids seem to play a minor role.
FIGURE 5.
(A) Volume-rendered water occupancy map averaged over simulation Q1. Two predominant positions of inserted water molecules are highlighted with arrows. The helical axes and layers positions (using Cα virtual bonds) are also indicated. The color codes for water occupancy are green, 53%; yellow, 59%; and red, >65%. Occupancies <42% are excluded for clarity. Panels B and C are aligned with A. (B) Interaction energy between protein residues at a given residue position and all water molecules for simulation Q1. (C) Residue distribution for each residue position. Acidic residues are shown in orange, basic residues in blue, and hydrophobic residues in light green. A central region with a high number of acidic residues described in the text is highlighted.
DISCUSSION
Validity of our model and comparison to experiment
We have presented simulation results on the cytosolic core of the four-helix SNARE bundle. Our model does not include the Sb and Sx transmembrane domains, the N-terminal domain of Sx, or the palmitoylated linker in the SNAP-25 protein. Our study targets the properties of the central soluble portion of the SNARE complex and the features that are likely to be preserved throughout the SNARE family (42). The validity of studying this core region as a minimal model for the synaptic fusion complex is supported by in vitro experiments on deletion mutants and protein fragments (7,12,43,44).
The conformational dynamics of the core complex, as observed in our MD simulations, indicate a stable and compact structure. The RMSD is low and little overall motion is observed except for fluctuations around the average structure. The straight overall helical axes shown in Fig.1 B corroborate the high structural integrity of the complex. The most striking feature is the robustness of these properties for two different force fields, AMBER ff99 and GROMACS ffG43a2, and for an explicit versus implicit solvent representation. The dynamic properties of the SNARE complex therefore appear to be strongly constrained by its sequence and construction, leaving little freedom for variations due to the details of the molecular simulation. Nevertheless, by averaging over three different simulations, we obtain a consensus picture, less affected by the particularities of each approach. Here, as in other fields (45,46), averaging over simulations with different force fields improves agreement with experiment.
The comparison with available experimental data, in particular with the crystallographic structures of the complex, shows good agreement with the simulation results, notably as concerns a detailed analysis of the hydrogen-bonding and salt-bridge patterns. Here the simulations do actually provide a more detailed, time-resolved picture (47), which is particularly interesting in the case of the intrahelical hydrogen bonds. Previous interpretations of these interactions, as in Sutton et al. (10), may need to be revised. The predominantly helical structure observed in the simulations is in qualitative agreement with circular dichroism results (48). A quantitative comparison of simulated and experimental circular dichroism data was not carried out, as it has been shown for other systems that such comparisons are intrinsically unreliable (49). Other data come from electron paramagnetic resonance (EPR) experiments that have provided structural information on the SNARE complex (50), its possible intermediates (51–53) and adjacent domains (54). For the core complex, an excellent agreement with the crystallographic structures is found. The line shapes measured in EPR experiments provide information about conformational flexibility of individual protein residues. Interestingly Poirier et al. (50) suggested an increased local flexibility for the Sn1:A42 residue, which is clearly visible in Fig. 4 B, whereas such a trend is not present in the experimental B-factors. The same is true for the unusually immobile Sn2:T173 residue observed by Margittai et al. (53), which shows up in our simulations, but not in the crystallographic data. The syntaxin residues E224, S225, E238, and H239 as well as the synaptobrevin residues S61 and T79 do not show a low mobility in our simulations, supporting the interpretation provided by the authors of the experimental EPR study that surface contacts between several SNARE complexes are at the origin of their reduced flexibility.
Biological and functional implications
How can our results be interpreted with respect to possible fusion mechanisms? The contact analysis carried out has confirmed the clear layer structuring of the complex. Hydrogen bonds and salt bridges reinforce these layers locally. There is, however, no evidence that these interactions should form in any particular sequence. Regarding currently proposed mechanisms, directional N- to C-terminal zippering of the complex (2,56), concerted formation of the contacts (57) and C-terminal initiation (58) are all compatible with these findings. The main driving force for SNARE association seems to be the formation of the large number of stabilizing interactions between the helices. One should, however, point out the somewhat unexpected and underestimated role for electrostatics, which completely dominate the energetics of the SNARE helix interactions. Previously, it was suggested that a membrane-proximal patch of basic residues could be of biological importance by attracting negatively charged phospholipids (59,60). We show here that acidic surface residues play a key role in structuring the surrounding water shell. This observed water structure, which is most clearly defined and extensive in the center of the complex, could contribute actively to the fusion event by geometrically preorganizing the adjacent vesicular and target membranes due to its wedgelike shape. By introducing repulsion with the negatively charged membrane surfaces, the acidic residues could function in synergy with the basic patch. However, it remains unclear how such interactions would be modified by available mono- and divalent cations and whether an overall repulsion or attraction would be observed. The biological importance of the acidic residues is emphasized by the fact that many of these residues are conserved (11).
Disassembly of the SNARE complex may also rely on solvent-exposed acidic residues involved in binding to α-SNAP (12). Marz et al. (12) identified 14 acidic residues forming three potential binding sites. In our simulations, seven of these residues actively participate in the salt-bridge and hydrogen-bond network shown in Fig. 3. These residues are Sb:E55, Sn2:E183 (Site1), Sn1:E38, Sn1:D41 (Site 2), and Sx:E238, Sn1:E55, Sn2:D166 (Site 3). By forming alternative interactions with basic surface residues of α-SNAP, the established interaction network will be disrupted at several places, in turn destabilizing the SNARE complex and facilitating disassembly. Residues not directly involved in interactions could also be important for attracting and docking α-SNAP to the SNARE complex. Hydrophobic, acidic, and basic surface patches are all potential interaction sites with different effector proteins (11). Furthermore, binding of α-SNAP to the SNARE complex likely involves dehydration of the fusion complex. The loss of the stabilizing water interactions of Fig. S1 D will, in turn, lead to a destabilization of the central −1 to +4 layer region, revealing unfavorable interprotein interactions (Fig. S1 C).
Interactions in the hydrophobic and central ionic layers
Despite the overall structural integrity of the SNARE helix-bundle, it is not a symmetric complex. A comparison of the four constitutive helices makes the vesicular synaptobrevin stand out. The analysis of the helicoidal parameters has revealed that its flexibility is significantly reduced compared to the other three helices. This goes hand in hand with the fact that Sb is the only helix contributing an arginine to the central ionic layer. The particularity of the Sb helix could suggest a role as central element for the complex formation, around which the synaptic plasma membrane protein helices adopt their structure. Locally, further asymmetries are found within the complex, as shown by the contact analysis plot of Fig. 2 B. The above average number of contacts of Sb in layers +3 to +5 is the single most marked feature. Beyond the +6 layer another peak of Sb contributions is observed. These layers overlap with region Sb77-94 that has been implicated in calmodulin binding (61). Some peaks observed in Fig. 2 B do coincide with toxin cleavage sites as indicated. There is, however, no clear pattern. Some peaks correspond to an above-average number of contacts, others to below-average ones. Sutton et al. (10) have suggested that there are significantly fewer surface interactions originating from Sb compared to those originating from Sx, Sn1, or Sn2. This is only partially confirmed by our detailed analysis.
The comparison of the individual hydrophobic layers indicates that the section from layers +5 to +8 exhibits particular stabilization. Fig. S1 A, the analysis of the total interaction energy per residue, shows this quite clearly. Interestingly, these interactions are dominated by the interlayer residues and not by the residues forming the characteristic hydrophobic layer structure. The hydrogen-bond and salt-bridge analysis (Fig. 3, A and C) corroborates this finding and the number of contacts in this region is also above average (Fig. 2 A). Mutational studies confirm the importance of this C-terminal part of the synaptic fusion complex, in particular of the +5 layer (5,14,15,62). These layers may play a key role in the biological function of the SNARE complex. They are positioned right next to the membrane-anchored domains of Sb and Sx. Providing a rigid scaffold in the vicinity of the vesicular and target bilayers could also be important for the formation of a fusion pore (5,63). It is interesting to note that the interaction patterns show helix- and layer-dependent specificity and are not at all homogeneously distributed.
Our study also confirms the special role of the central ionic layer in several respects. It has a particularly favorable hydrogen-bonding pattern, reinforced by adjacent salt bridges. This is somewhat in contrast with the previous suggestion that the layers flanking the ionic “0” layer are primarily maintained by hydrophobic interactions (10) and that the central ionic layer might be a weak spot in the complex (12,64). As mentioned above, dehydration could, however, significantly weaken this region in particular. Energetically, the interactions with water and with the rest of the protein are maximal in and around this polar layer. As suggested by Sutton et al. (10), sealing the ionic “0” layer itself from water seems to enhance its electrostatic interactions, given that interactions with water are repulsive (Fig. S1 D). We did not observe any direct interaction of a water molecule within the layer, which is in agreement with the ionic layer structure discussed in Sutton et al. (10), but unlike the high resolution crystal structure by Ernst and Brunger (9) where a water molecule was found interacting with Sb:R56. The precise role of this layer, however, remains unknown. It has been suggested that it could serve as register to position the four helices in a precise way (11). This idea is compatible with the hydrogen-bonding patterns, where the marked central layer motif provides a visible reference point. It would be interesting to carry out additional simulations where one or several helices are shifted along the overall axis to test this hypothesis, but this is beyond the scope of this study.
Simulation methodology
The 10-ns MD simulations presented in this work are clearly too short to allow for exhaustive exploration of conformational space. Previous work has shown that at least an order-of-magnitude increase of simulation time would be necessary to improve sampling (65). This is currently beyond our available computing power. However, in the particular case of the soluble SNARE core complex, sampling does not appear to be a major issue. The conformational dynamics of this complex are characterized by an exceptional stability. Given the absence of loops and other very mobile domains, no undersampled regions are apparent.
Concerning the details of the simulations carried out, the main features that were presented in this work are robust with respect to two different force fields, two explicit water models, and an implicit solvent representation. There are, however, some detailed differences, in particular for the implicit solvent approach (Q2GB) which shows increased flexibility, higher RMSD values, and lower secondary structure content. The first two of these features are expected and have been reported earlier (66). They are related to the fact that the implicit solvent representation removes friction and hydrostatic pressure. The localized loss of secondary structure for Sb and Sx near the central layer and near the C-terminus is, however, in contrast with recent work indicating a bias toward α-helical structures with GB models (67). In the case of the SNARE complex, this may be related to the absence of the highly structured surrounding water shell in an implicit model. Globally, the α-helical structure is clearly shifted toward a 3-10 helix as revealed by the P-Curves analysis of the implicit solvent simulation. We did observe some minor artifacts related to the GB approach. Fragile regions in the complex had lower hydrogen-bond occurrence than in the explicit solvent simulations, whereas the overall number of hydrogen-bonding interactions tended to increase, thus compensating the loss of direct solvent interactions. A large amount of motion was observed at the termini. Salt-bridge interactions were stabilized with respect to the explicit solvent simulations as has been described previously in the literature (68).
CONCLUSIONS AND OUTLOOK
The observed similarity between our three simulations and the excellent agreement with experiment provide a strong support for the validity of the established interaction patterns and features of the SNARE complex we have described. We confirm the image of a stiff, rodlike complex being held together by interactions between the helices, acting like molecular Velcro strips. A new finding is that the hydrophobic layers at the core of the complex contribute little to the total energy balance, which is dominated by electrostatics. The asymmetry in the complex, particularly between the R- and Q-SNARE helices, is also somewhat unexpected. These simulation results are complementary to existing experimental studies and provide a more detailed, time-resolved picture of molecular properties that are difficult to characterize experimentally.
Based on our results, two general suggestions can be made. Concerning the interpretation of mutations in the synaptic fusion complex, previous work has focused on the importance of hydrophobic layer mutants, whereas interlayer residues have received little attention. In the light of the interaction network we observe and the corresponding energy contributions per residue, it would be interesting to analyze mutational data in terms of potential disruption of the complex hydrogen-bond and salt-bridge network, although such disruptions can of course be induced by the steric effect of hydrophobic layer mutations. In addition, the role of the surface residues of the complex seems to be underestimated. In particular, the importance of the charged acidic surface residues and electrostatic interactions in general could be tested by experiments analyzing the influence of varying electrolyte concentration on the properties of the SNARE complex, as has been done in the case of α-SNAP binding (12).
An obvious extension of the current work involves studying a membrane-embedded SNARE complex. This is a challenging task due to the complexity of its environment (69). Taking into account the transmembrane parts of the complex and the lipid bilayers to which they are anchored requires important computing resources and long simulation times, but we are now attempting such simulations. Further studies of the impact of mutations within the hydrophobic layers on the interaction network of the synaptic fusion complex could also provide new insights.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
We thank the French supercomputer centers IDRIS and CINES for providing computer resources (grants No 041714, 051714, and LBT2411). We thank Etienne Engler and Georges Wipff for access to the MDS analysis software. Tsjerk Wassenaar has contributed a modified version of the trjconv analysis program. We thank Fabien Cailliez for contributing to the analysis of the helical axes. Joséphine Abi Ghanem, François Darchen, Brigitte Hartmann, Jean-Pierre Henry, and Brahim Heddi are thanked for stimulating discussions.
M.-P.D. acknowledges support from the French Ministry of Research.
Editor: Klaus Schulten.
References
- 1.Harbury, P. A. 1998. Springs and zippers: coiled coils in SNARE-mediated membrane fusion. Structure. 6:1487–1491. [DOI] [PubMed] [Google Scholar]
- 2.Jahn, R., T. Lang, and T. C. Sudhof. 2003. Membrane fusion. Cell. 112:519–533. [DOI] [PubMed] [Google Scholar]
- 3.Chen, Y. A., and R. H. Scheller. 2001. SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2:98–106. [DOI] [PubMed] [Google Scholar]
- 4.Fiebig, K., L. Rice, E. Pollock, and A. Brunger. 1999. Folding intermediates of SNARE complex assembly. Nat. Struct. Biol. 6:117–123. [DOI] [PubMed] [Google Scholar]
- 5.Han, X., and M. Jackson. 2006. Structural transitions in the synaptic SNARE complex during Ca2+-triggered exocytosis. J. Cell Biol. 172:281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fasshauer, D., H. Otto, W. Eliason, R. Jahn, and A. Brünger. 1997. Structural changes are associated with soluble n-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J. Biol. Chem. 272:28036–28041. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi, T., H. McMahon, S. Yamasaki, T. Binz, Y. Hata, T. C. Sudhof, and H. Niemann. 1994. Synaptic vesicle membrane fusion complex: action of clostridial neurotoxins on assembly. EMBO J. 13:5051–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonin, W., D. Fasshauer, S. Becker, R. Jahn, and T. R. Schneider. 2002. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat. Struct. Biol. 9:107–111. [DOI] [PubMed] [Google Scholar]
- 9.Ernst, J. A., and A. T. Brunger. 2003. High resolution structure, stability, and synaptotagmin binding of a truncated neuronal SNARE complex. J. Biol. Chem. 278:8630–8636. [DOI] [PubMed] [Google Scholar]
- 10.Sutton, R. B., D. Fasshauer, R. Jahn, and A. T. Brunger. 1998. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature. 395:347–353. [DOI] [PubMed] [Google Scholar]
- 11.Fasshauer, D., R. B. Sutton, A. T. Brunger, and R. Jahn. 1998. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA. 95:15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marz, K. E., J. M. Lauer, and P. I. Hanson. 2003. Defining the SNARE complex binding surface of α-SNAP: implications for SNARE complex disassembly. J. Biol. Chem. 278:27000–27008. [DOI] [PubMed] [Google Scholar]
- 13.Ossig, R., H. Schmitt, B. de Groot, D. Riedel, S. Keränen, H. Ronne, H. Grubmüller, and R. Jahn. 2000. Exocytosis requires asymmetry in the central layer of the SNARE complex. EMBO J. 19:6000–6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sørensen, J., K. Wiederhold, E. Müller, I. Milosevic, G. Nagy, B. de Groot, H. Grubmüller, and D. Fasshauer. 2006. Sequential N- to C-terminal SNARE complex assembly drives priming and fusion of secretory vesicles. EMBO J. 25:955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagow, R., H. Bao, E. Cohen, R. Daniels, A. Zuzek, W. Williams, G. Macleod, R. Sutton, and B. Zhang. 2007. Modification of a hydrophobic layer by a point mutation in syntaxin 1A regulates the rate of synaptic vesicle fusion. PLoS Biol. 5:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagy, G., I. Milosevic, D. Fasshauer, E. Müller, B. de Groot, T. Lang, M. Wilson, and J. Sørensen. 2005. Alternative splicing of SNAP-25 regulates secretion through nonconservative substitutions in the SNARE domain. Mol. Biol. Cell. 16:5675–5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krieger, E., T. Darden, S. B. Nabuurs, A. Finkelstein, and G. Vriend. 2004. Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins. 57:678–683. [DOI] [PubMed] [Google Scholar]
- 18.Krieger, E., J. E. Nielsen, C. A. Spronk, and G. Vriend. 2006. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model. 25:481–486. [DOI] [PubMed] [Google Scholar]
- 19.Case, D. A., T. E. Cheatham 3rd, T. Darden, H. Gohlke, R. Luo, K. M. Merz, Jr., A. Onufriev, C. Simmerling, B. Wang, and R. J. Woods. 2005. The AMBER biomolecular simulation programs. J. Comput. Chem. 26:1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindahl, E., B. Hess, and D. van der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 7:306–317. [Google Scholar]
- 21.Van Gunsteren, W. F., X. Daura, and A. Mark. 1998. GROMOS force field. In Encyclopedia of Computational Chemistry, Vol. 2. P. von Ragué Schleyer, editor. Wiley-VCH, Chichester, UK.
- 22.Wang, J., P. Cieplak, and P. A. Kollman. 2000. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 21:1049–1074. [Google Scholar]
- 23.Tsui, V., and D. A. Case. 2000. Theory and applications of the generalized Born solvation model in macromolecular simulations. Biopolymers. 56:275–291. [DOI] [PubMed] [Google Scholar]
- 24.Onufriev, A., D. Bashford, and D. A. Case. 2004. Exploring protein native states and large-scale conformational changes with a modified generalized Born model. Proteins. 55:383–394. [DOI] [PubMed] [Google Scholar]
- 25.Aqvist, J. 1990. Ion-water interaction potentials derived from free energy perturbation simulations. J. Phys. Chem. 94:8021–8024. [Google Scholar]
- 26.Jorgensen, W. L., J. Chandrasekhar, and J. D. Madura. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–936. [Google Scholar]
- 27.Berendsen, H. J. C., and J. P. M. Postma. W. F. v. Gunsteren, and J. Hermans. 1981. Interaction models for water in relation to protein hydration. In Intermolecular Forces. B. Pullman, editor. Reidel Publishing Company, Dordrecht, The Netherlands.
- 28.Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald—an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- 29.Ryckaert, J.-P., G. Ciccotti, and H. Berendsen. 1977. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23:327–341. [Google Scholar]
- 30.Hess, B., H. Bekker, H. J. C. Berendsen, and J. Fraaije. 1997. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [Google Scholar]
- 31.Berendsen, H. J. C., J. P. M. Postma, W. F. Vangunsteren, A. Dinola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 32.Duke, R. E., and L. G. Pedersen. 2003. PMEMD 3. University of North Carolina-Chapel Hill, Chapel Hill, NC.
- 33.Loncharich, R., B. Brooks, and R. Pastor. 1992. Langevin dynamics of peptides: the frictional dependence of isomerization rates of n-acetylalanyl-n′-methylamide. Biopolymers. 32:523–535. [DOI] [PubMed] [Google Scholar]
- 34.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 22:2577–2637. [DOI] [PubMed] [Google Scholar]
- 35.Heinig, M., and D. Frishman. 2004. STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 32:W500–W502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:27–38. [DOI] [PubMed] [Google Scholar]
- 37.Krieger, E. 1993–2007. YASARA, a molecular graphics, modeling and simulation program for Linux, Windows and Mac OS X. Yasara Biosciences, Graz, Austria.
- 38.Will, S., M. M. Kenneth, and E. L. William. 1998. An Object-Oriented Approach to the Visualization Toolkit, 2nd Ed. Prentice-Hall, Englewood Cliffs, NJ.
- 39.Sklenar, H., C. Etchebest, and R. Lavery. 1989. Describing protein structure: a general algorithm yielding complete helicoidal parameters and a unique overall axis. Proteins. 6:46–60. [DOI] [PubMed] [Google Scholar]
- 40.Reference deleted in proof.
- 41.R Development Core Team. 2007. R: A Language and Environment for Statistical Computing. Vienna, Austria.
- 42.Antonin, W., C. Holroyd, D. Fasshauer, S. Pabst, G. F. Von Mollard, and R. Jahn. 2000. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 19:6453–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parlati, F., T. Weber, J. A. McNew, B. Westermann, T. H. Sollner, and J. E. Rothman. 1999. Rapid and efficient fusion of phospholipid vesicles by the α-helical core of a SNARE complex in the absence of an N-terminal regulatory domain. Proc. Natl. Acad. Sci. USA. 96:12565–12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fasshauer, D., W. Eliason, A. Brünger, and R. Jahn. 1998. Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly. Biochemistry. 37:10354–10362. [DOI] [PubMed] [Google Scholar]
- 45.Martin, M., and M. Biddy. 2005. Monte Carlo molecular simulation predictions for the heat of vaporization of acetone and butyramide. Fluid Phase Equil. 236:53–57. [Google Scholar]
- 46.Martin, M., and A. Thompson. 2004. Industrial property prediction using Towhee and LAMMPS. Fluid Phase Equil. 217:105–110. [Google Scholar]
- 47.van Gunsteren, W. F., D. Bakowies, R. Baron, I. Chandrasekhar, M. Christen, X. Daura, P. Gee, D. P. Geerke, A. Glattli, P. H. Hunenberger, M. A. Kastenholz, C. Oostenbrink, M. Schenk, D. Trzesniak, N. F. van der Vegt, and H. B. Yu. 2006. Biomolecular modeling: goals, problems, perspectives. Angew. Chem. Int. Ed. Engl. 45:4064–4092. [DOI] [PubMed] [Google Scholar]
- 48.Fasshauer, D., D. Bruns, B. Shen, R. Jahn, and A. Brünger. 1997. A structural change occurs upon binding of syntaxin to SNAP-25. J. Biol. Chem. 272:4582–4590. [DOI] [PubMed] [Google Scholar]
- 49.Daura, X., D. Bakowies, D. Seebach, J. Fleischhauer, W. F. van Gunsteren, and P. Kruger. 2003. Circular dichroism spectra of β-peptides: sensitivity to molecular structure and effects of motional averaging. Eur. Biophys. J. 32:661–670. [DOI] [PubMed] [Google Scholar]
- 50.Poirier, M., W. Xiao, J. Macosko, C. Chan, Y. Shin, and M. Bennett. 1998. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat. Struct. Biol. 5:765–769. [DOI] [PubMed] [Google Scholar]
- 51.Xiao, W., M. Poirier, M. Bennett, and Y. Shin. 2001. The neuronal t-SNARE complex is a parallel four-helix bundle. Nat. Struct. Biol. 8:308–311. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, F., Y. Chen, D. Kweon, C. Kim, and Y. Shin. 2002. The four-helix bundle of the neuronal target membrane SNARE complex is neither disordered in the middle nor uncoiled at the C-terminal region. J. Biol. Chem. 277:24294–24298. [DOI] [PubMed] [Google Scholar]
- 53.Margittai, M., D. Fasshauer, S. Pabst, R. Jahn, and R. Langen. 2001. Homo- and heterooligomeric SNARE complexes studied by site-directed spin labeling. J. Biol. Chem. 276:13169–13177. [DOI] [PubMed] [Google Scholar]
- 54.Margittai, M., D. Fasshauer, R. Jahn, and R. Langen. 2003. The HABC domain and the SNARE core complex are connected by a highly flexible linker. Biochemistry. 42:4009–4014. [DOI] [PubMed] [Google Scholar]
- 55.Reference deleted in proof.
- 56.Matos, M., K. Mukherjee, X. Chen, J. Rizo, and T. Südhof. 2003. Evidence for SNARE zippering during Ca2+-triggered exocytosis in PC12 cells. Neuropharmacology. 45:777–786. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, F., Y. Chen, Z. Su, and Y. Shin. 2004. SNARE assembly and membrane fusion, a kinetic analysis. J. Biol. Chem. 279:38668–38672. [DOI] [PubMed] [Google Scholar]
- 58.Kweon, D., C. Kim, and Y. Shin. 2003. Regulation of neuronal SNARE assembly by the membrane. Nat. Struct. Biol. 10:440–447. [DOI] [PubMed] [Google Scholar]
- 59.Weimbs, T., K. Mostov, S. H. Low, and K. Hofmann. 1998. A model for structural similarity between different SNARE complexes based on sequence relationships. Trends Cell Biol. 8:260–262. [DOI] [PubMed] [Google Scholar]
- 60.Montal, M. 1999. Electrostatic attraction at the core of membrane fusion. FEBS Lett. 447:129–130. [DOI] [PubMed] [Google Scholar]
- 61.Quetglas, S., C. Leveque, R. Miquelis, K. Sato, and M. Seagar. 2000. Ca2+-dependent regulation of synaptic SNARE complex assembly via a calmodulin- and phospholipid-binding domain of synaptobrevin. Proc. Natl. Acad. Sci. USA. 97:9695–9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fergestad, T., M. Wu, K. Schulze, T. Lloyd, H. Bellen, and K. Broadie. 2001. Targeted mutations in the syntaxin H3 domain specifically disrupt SNARE complex function in synaptic transmission. J. Neurosci. 21:9142–9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cho, S., M. Kelly, K. Rognlien, J. Cho, J. Hörber, and B. Jena. 2002. SNAREs in opposing bilayers interact in a circular array to form conducting pores. Biophys. J. 83:2522–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen, X., D. Tomchick, E. Kovrigin, D. Araç, M. Machius, T. Südhof, and J. Rizo. 2002. Three-dimensional structure of the complexin/SNARE complex. Neuron. 33:397–409. [DOI] [PubMed] [Google Scholar]
- 65.Faraldo-Gomez, J. D., L. R. Forrest, M. Baaden, P. J. Bond, C. Domene, G. Patargias, J. Cuthbertson, and M. S. Sansom. 2004. Conformational sampling and dynamics of membrane proteins from 10-nanosecond computer simulations. Proteins. 57:783–791. [DOI] [PubMed] [Google Scholar]
- 66.Cornell, W., R. Abseher, M. Nilges, and D. A. Case. 2001. Continuum solvent molecular dynamics study of flexibility in interleukin-8. J. Mol. Graph. Model. 19:136–145. [DOI] [PubMed] [Google Scholar]
- 67.Roe, D. R., A. Okur, L. Wickstrom, V. Hornak, and C. Simmerling. 2007. Secondary structure bias in generalized Born solvent models: comparison of conformational ensembles and free energy of solvent polarization from explicit and implicit solvation. J. Phys. Chem. B. 111:1846–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Geney, R., M. Layten, R. Gomperts, V. Hornak, and C. Simmerling. 2006. Investigation of salt bridge stability in a generalized Born solvent model. J. Chem. Theory Comput. 2:115–127. [DOI] [PubMed] [Google Scholar]
- 69.Krieger, E., L. Leger, M.-P. Durrieu, N. Taib, P. Bond, M. Laguerre, R. Lavery, M. S. Sansom, and M. Baaden. 2007. Atomistic modeling of the membrane-embedded synaptic fusion complex: a grand challenge project on the DEISA HPC infrastructure. In ParCo 2007, Parallel Computing: Architectures, Algorithms and Applications. C. B. G. R. Joubert, F. Peters, T. Lippert, M. Bücker, P. Gibbon, and B. Mohr, editors. John von Neumann Institute for Computing, Juelich, Germany.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.