Abstract
Eukaryotic DNA replication is regulated to ensure all chromosomes replicate once and only once per cell cycle. Replication begins at many origins scattered along each chromosome. Except for budding yeast, origins are not defined DNA sequences and probably are inherited by epigenetic mechanisms. Initiation at origins occurs throughout the S phase according to a temporal program that is important in regulating gene expression during development. Most replication proteins are conserved in evolution in eukaryotes and archaea, but not in bacteria. However, the mechanism of initiation is conserved and consists of origin recognition, assembly of pre-replication (pre-RC) initiative complexes, helicase activation, and replisome loading. Cell cycle regulation by protein phosphorylation ensures that pre-RC assembly can only occur in G1 phase, whereas helicase activation and loading can only occur in S phase. Checkpoint regulation maintains high fidelity by stabilizing replication forks and preventing cell cycle progression during replication stress or damage.
Keywords: origins, kinase, helicase, checkpoint, replisome, replicons
INTRODUCTION
Le rêve d’une bactérie doit devenir deux bactéries (The dream of a bacterium is to become two bacteria)
François Jacob, 1965
As François Jacob said poetically over 40 years ago, it is a cell’s “dream” to become two cells with identical copies of the genome. Thus the replication of the genome must be an exact process. Errors that result in underreplication or overeplication of the genome in any cell cycle have disastrous consequences and can produce a large array of human genetic diseases, including cancer, birth defects, and many developmental abnormalities (58). Molecular regulatory mechanisms have evolved to ensure that the genome is replicated once and only once and then segregated equally to the resultant daughter cells. This review summarizes recent developments in the field and focuses mainly on cell cycle regulation of DNA replication in eukaryotic cells.
In all cells studied, DNA replication is regulated by recruiting the replication machinery or “replisome” to sites called origins on the chromosome (Figure 1). The replisome is a molecular machine that replicates the DNA bidirectionally from origins in a semiconservative fashion. The recruitment process is called initiation, whereas subsequent replication of the DNA by the replisome is called elongation. It is initiation and hence the recruitment process that is the site of regulation. When a change in replication rate is needed, adjustments are made to initiation. For example, the rate of DNA replication is almost two orders of magnitude faster in embryos than in somatic cells because more origins are used, resulting in more initiations. This review focuses on the important molecules used in the initiation process in the context of the cell cycle. Readers are advised to refer to other excellent reviews (12, 13, 273) for additional information.
Figure 1.
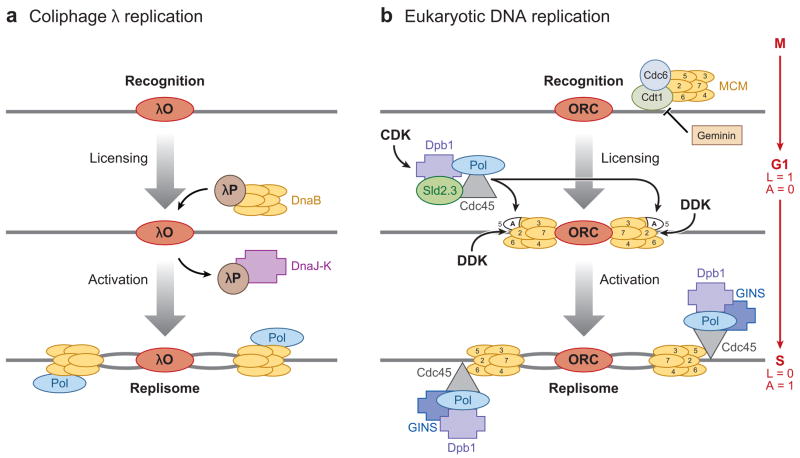
Models of the regulation of DNA replication. (a) In coliphage λ replication, origin is recognized by λO protein, then λP protein loads hexameric DnaB helicase. λP protein is removed by DnaJ-K protein, which activates the helicase and allows replisome to replicate the DNA. (b) In eukaryotic DNA replication, origin is recognized by ORC, then Cdc6 and Cdt1 protein load the hexameric MCM helicase to form the “licensed” (L) pre-RC in G1 phase (L = 1, A = 0). Geminin inhibits Cdt1 and pre-RC formation. CDK and DDK become active in late G1, activate (A) the MCM helicase and load on the replisome that contains the DNA polymerases. In addition, CDK inhibits any further licensing (L = 0, A = 1). Toward this end, CDK phosphorylates Sld2 and Sld3 proteins and DDK phosphorylates MCM proteins, which “pushes out” the “A” domain of Mcm5.
The basic mechanism of regulation is to identify the origin by having an initiator protein bind to it (Figure 1). Protein complexes that load or recruit the replisome to the origin then recognize the initiator protein bound to origin chromatin. The replisome opens the DNA helix, stabilizes the ssDNA that is formed, and allows enzymes (polymerases) to copy the DNA. Coliphage λ replication is an excellent model for the initiation process due to its simplicity (244, 261) because only two viral proteins are required for initiation (Figure 1a). The λO-protein recognizes and binds the single origin on the phage chromosome. λP protein recruits dnaB helicase to the λO-origin complex but inhibits it (7). Replication begins only after heat shock proteins remove λP (6) and the replisome is loaded.
EVOLUTION OF CHROMOSOMAL REPLICATION
One would think that the basic mechanism of regulation has been conserved during evolution as it has adapted to the basic problem of copying a double-stranded antiparallel helix. However, comparative genomic studies of the evolution of the three domains of life (306) have shown that the cellular replication machinery diverged when eubacteria separated from archaea and eukaryotes billions of years ago (83, 158) (Table 1). Many of the replication proteins of eubacteria have no clear orthologues in eukaryotes, yet proteins used for transcription and translation are conserved among the domains (83, 158). Notably, dnaB helicase is used by bacteria whereas archaea and eukaryotes use the MCM (mini-chromosome maintenance) helicase for replication. Similarities in structure and function between the two helicases suggest that they probably resulted from convergent evolution. One explanation is the “replicon takeover” hypothesis, which states that eukaryotic and archaeal replication proteins evolved by nonorthologous gene displacement by viral or plasmid invaders in a common ancestor at the time of the separation (83, 158). Consistent with this hypothesis, a prophage found in the eubacterial Bacillus cereus was found to contain a MCM-homologue (181). The implication is that the MCM helicase evolved from viral invaders and replaced a dnaB-like helicase used by the ancestor of eukaryotes and Archaea. Another possibility is that some replisomal proteins evolved twice in an independent way from the last common ancestor that had a hybrid genome of RNA and DNA (158).
Table 1.
The function of eukaryotic replication proteins
| Protein Namea | Alias/Homologuesb | Function(s)c |
|---|---|---|
| Orc1 | Orp1 Cdc6 | ATPase for DNA binding |
| Orc2 | Orp2 | ATPase |
| Orc3 | Orp3 | ATPase |
| Orc4 | Orp4 | ATPase |
| Orc5 | Orp5 | ATPase |
| Orc6 | Orp6 | ? |
| Initiative Assembly-Helicase loading | ||
| Cdc6 | Cdc18 Orc1 | ATPase helicase clamp loader |
| Cdt1 | Tah11 Dup | Helicase clamp loader |
| Geminin | Inhibits helicase clamp loader | |
| Initiative Assembly-Helicase unwinding | ||
| Mcm2 | Nda1 Cdc19 | ATPase helicase DDK substrate |
| Mcm3 | ATPase helicase | |
| Mcm4 | Cdc54 Cdc21 Dpa | ATPase DDK substrate |
| Mcm5 | Cdc46 Nda4 Bob1 | ATPase helicase regulation |
| Mcm6 | Mis5 | ATPase DDK substrate |
| Mcm7 | Cdc47 Prolifera | ATPase helicase |
| Mcm8 | ATPase helicase | |
| Mcm9 | ATPase helicase | |
| Mcm10 | Dna43 Cdc23 | Helicase Regulation DNA Primase DDK activator |
| Helicase Unwinding/-Elongative Assembly | ||
| CDK-kinase | Cdc28 Cdc2 Cdk2 | Protein kinase for helicase activation and replisome loading. |
| Inhibits helicase loading to block re-replication | ||
| DDK-kinase | Cdc7 Hsk1 | Protein kinase for helicase activation and replisome loading |
| CDK-regulatory | Clb1-6 CycA CycE | Activate Cdk1 and Cdk2 kinases |
| DDK-regulatory | Dbf4 Dfp1 Drf1 Chiffon Ask Him1 Rad35 | Activate Cdc7 Kinase |
| Cdc45 | Sna41 | Helicase activation and replisome loading |
| Sld2 | Drc1 | Cdk substrate; loading of Dpb11 and Cdc45 |
| Sld3 | Cdk substrate; loading of Dpb11 and Cdc45 | |
| Dpb11 | TopBP1 Rad4 Cut5 Mus101 | Helicase activation and replisome loading |
| GINS complex | Helicase activation and replisome loading | |
| Sld5 | Cdc105 | Helicase activation and replisome loading |
| Psf1 | Cdc101 | Helicase activation and replisome loading |
| Psf2 | Cdc102 CG18013 | Helicase activation and replisome loading |
| Psf3 | Cdc103 | Helicase activation and replisome loading |
| Replisome | ||
| Polα primase holoenzyme | Pol1 Cdc17 | Polymerase priming; replication of lagging strands |
| Polδ holoenzyme | Pol3 Cdc2 | Bidirectional replication |
| Polε holoenzyme | Pol2 Cdc20 | Bidirectional replication |
| Rfc1-5 complex | PCNA clamp loading | |
| PCNA | Pol30 | Polymerase clamp |
| RPA1-3 complex | SSB | SSB coats ssDNA |
Refers to the name of the protein most commonly used.
Refers to other names that are used.
Also refer to Figure 1b for description of protein function(s).
Lessons from Archaea
Many archaeal genomes are small and only about 1.5–2.0 Mbp. Investigators have exploited the fact that archaeal replication is similar yet simpler than eukaryotic replication with regard to the number of proteins required (Table 1) [for excellent reviews of archaeal replication, see (12, 133)]. Therefore, Archaea represent a simpler and less complicated model system to analyze the function of the human replication apparatus. For example, the eukaryotic MCM-helicase (see below) is composed of six, paralogous proteins and is inactive either when purified from cells or made using recombinant DNA technology (81), making it difficult to analyze on a molecular level. However, a recombinant form of an archaeal homologue was shown to be an active DNA helicase and both atomic crystal and cryo EM reconstruction structures have been solved (Figure 2). Furthermore, eukaryotic ORC (origin recognition complex) is also composed of six proteins (Orc1–6), and Cdc6 protein is needed to load the MCM helicase at the origin. Cdc6 and Orc1 are homologues and in some Archaea, a single protein Orc1/Cdc6 can act as both ORC and Cdc6.
Figure 2.
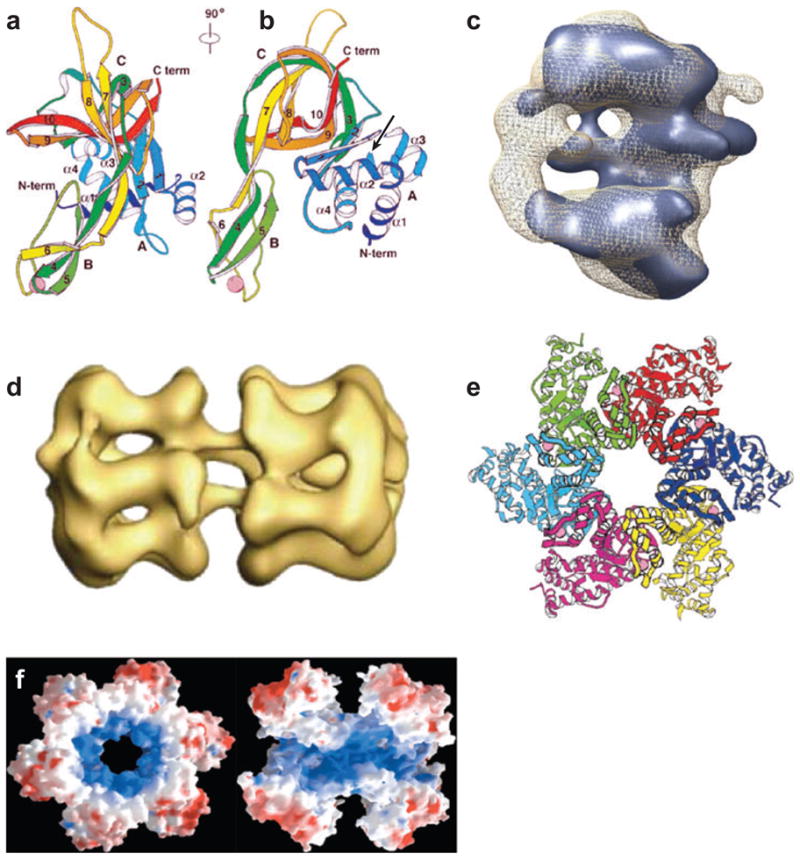
Structures of ORC/Cdc6 and DNA helicases. (a) Ribbon diagram of the atomic structure of the N-terminal fragment of a single archaeal Mth-MCM subunit (b) rotated 90°. A, B, and C domains are indicated. Arrow indicates the position of the P62 residue (76). (c) EM reconstruction of yeast Orc/Cdc6 complex with ORC in blue (258) (d) EM reconstruction of the full-length archaeal double hexameric Mth-MCM complex (96). (e) Ribbon diagram of the atomic structure of a single hexamer of SV40 T antigen (161). (f) Space-filling diagram of the atomic structure of the N-terminal fragment of a single hexamer of the archaeal Mth-MCM complex (left) and a cut side-view (right) with two subunits removed for clarity; blue indicates positively charged amino acids, red indicates negatively charged amino acids (76).
Several groups have utilized the biochemical and structural information gleaned from archaeal proteins to analyze the function of the eukaryotic homologue in vivo using genetic and molecular techniques. For example, the structure of the archaeal Orc1/Cdc6 was used to test the importance of the winged helix domain in binding in eukaryotic cells of Schizosaccharomyces pombe (164). Conversely, the significance of mutations in yeast Mcm5 protein (76) and in mouse Mcm4 (252) was determined by using structural information from the archaeal Mcm protein (36, 76, 77). Thus, the power of each system, Archaea for biochemistry and structure and yeast and mouse for genetics, can be used to determine function both in vivo and in vitro.
ORIGINS AND REPLICONS
Origins of DNA replication are sites in the genome at which replication begins. The DNA replicated from a single origin is called a replicon. Usually, replication begins at an origin and proceeds bidirectionally to complete a single replicon (Figure 3). Eventually, replicons fuse resulting in complete genomic duplication. The chromosome of Escherichia coli has one origin and its entire genome of about 4 Mbp is a single replicon. The replicon hypothesis postulated that origins would be specific DNA sequences (in cis) that would be recognized by DNA-binding proteins (in trans) much like the lacO operator is recognized by the lacI repressor in transcription (119). To a first approximation, the replicon hypothesis is correct with regard to many single-celled prokaryotic organisms (bacteria and Archaea) and to many viruses. In eukaryotes, the situation is more complex in that there are many origins present on a single chromosomal molecule of DNA (Figure 3). To further complicate matters, in many eukaryotes, the replicon hypothesis may not be completely correct in that there seems to be very little sequence-specificity of origins except in the budding yeast Saccharomyces cerevisiae (203, 227). In this review, recent studies about origins in both unicellular and multicellular eukaryotes are compared and contrasted to reveal the important similarities and differences.
Figure 3.
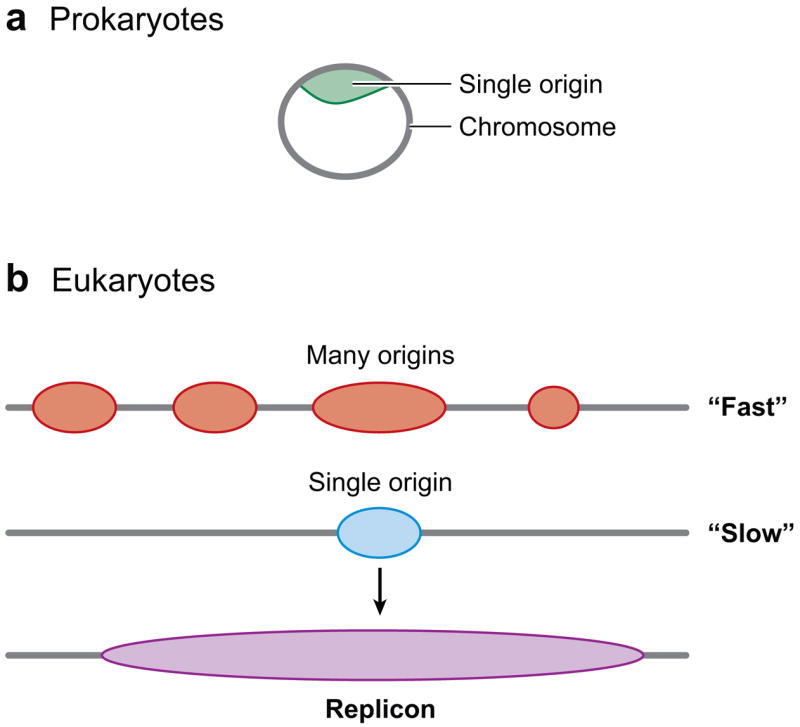
Regulation of DNA replication by origin usage. (a) Prokaryotes have a single origin on a circular chromosome (above). (b) In eukaryotes, multiple origins are found on a single chromosome. When replication is “fast,” many origins are used, whereas only one origin is used in this region when replication is “slow”. Replication proceeds bidirectionally from an origin to form a replicon (below).
In budding yeast S. cerevisiae, origins were isolated using extrachromosomal plasmid-based ARS (autonomously replicating sequence) assays. Simply, the ARS allowed circular bacterial plasmids to replicate as mini-chromosomes and “shuttle” between yeast and bacteria (13). Using 2D-gel analysis, the ARS were shown to be bona fide origins of replication (28). The ARS is about 100 bp and consists of a simple 17-bp consensus A-domain region with an 11-bp ACS (ARS consensus sequence A/TTTTAT/CA/GTTTA/T) that is A-T rich and flanked by poorly conserved B domains (196). The A and B1 domains are binding sites for the ORC, while the other B domains (B2, B3, etc.) act as enhancers of origin efficiency. In some ARS regions, there are degenerate repeats of the ACS instead of the B regions. Thus, the context of the chromatin surrounding the ACS is important for function (13).
Using a combination of classical Meselson-Stahl and high-resolution DNA-DNA microarray technologies, it was shown that budding yeast has about 332 origins among its 16 chromosomes (224, 311), which are spaced apart according to a normal distribution indicating a nearly uniform arrangement (215). More recently, ssDNA genomic mapping was used to identify and confirm these 332 active origins (73). Information about yeast origins can be found in a user-friendly computerized database called OriDB (199) that is based on a phylogenic analysis of sensu stricto Saccharomyces species (200). In this latter study, the ACS in ARS was conserved but not the flanking B2 regions.
Although the efficiency of these origins varied greatly, many were very efficient, used in every cell cycle, and account for most of the replication of the genome. Most origins were found in intergenic regions that were later shown to lack nucleosomes in the region 3′ to the ACS (200, 319). These studies also showed that origins are fired continuously throughout S phase. Most of these origins were also found in combined chromatin immunoprecipitation-microarray (ChIP-chip) studies that determined genome-wide ORC and MCM binding sites (309, 310). Using tiling microarrays and ChIP, 529 ORC-MCM binding sites were found with the ORC-MCM coincidence having the highest predictive value, suggesting that only a subset of origins is used, that is, 332/529 origins (310).
Therefore, budding yeast has specific sequences at origins (ARS), which are recognized and bound by initiator proteins (ORC) according to the replicon hypothesis. The origins are efficient and evenly positioned allowing for complete genome replication in a reasonable amount of time. S. cerevisiae arose from Kluyveromyces waltii by genome duplication, gene selection, and divergence (131). During this evolution, sequences produced at random that contained evenly spaced origins might have been selected to allow optimal chromosomal replication. It is possible that S. cerevisiae uses simple sequences as replication origins because it is “streamlined” from domestication by humans in industrial fermentation. In support, centromeres of S. cerevisiae are also simple sequences of about 100 bp that bind one microtubule but in other eukaryotes they are much larger and bind many microtubules (40).
S. cerevisiae is clearly an exception in that other eukaryotes have many inefficient origins spread randomly throughout their genomes and in many cases, the origins are without any identifiable specific sequences (203, 227). In the distantly related fission yeast S. pombe, combing studies using DNA fibers labeled by DNA analogue precursors and FISH (fluorescence in situ hybridization) showed that origins spacing is distributed in an exponential fashion, indicating that origins fire stochastically and are not inherited in that different origins are used in each generation (215). A low average origin efficiency (<30%) and stochastic origin selection was also seen in other studies using different methods (49). Although the ORC binds a specific sequence in budding yeast and is conserved (see below), there is little sequence conservation among fission yeast origins, which are about 1 kb, A-T rich (about 70%), and are also in non-nucleosomal intergenic regions (49, 246).
Thus, in contrast to budding yeast, fission yeast cells have many inefficient origins that fire randomly and are scattered haphazardly throughout the genome. As pointed out, this could result in large regions being missed by chance, resulting in a “gap” that needs to be replicated passively from very distant origins (227). A similar gap problem also exists in metazoan embryos such as Xenopus and Drosophila (109, 113). One possibility is that origin efficiency may increase as S phase progresses, thereby increasing the chance that an origin will fire in the problematic area (113, 227).
In metazoan cells, the situation is less clear (93, 170, 203). As seen in budding yeast, there are clearly active origins that act as defined, sequence-specific sites and can also function at ectopic sites such as the 1.2-kb human lamin B2 origin (213). In the HBB (human β-globin) locus, there are two 2—3 kb adjacent regions that can act as origins even at ectopic sites (296). Other origins are not sites but broad zones of initiation such as in DHFR (dihydrofolate reductase) with more than 40 sites of initiation in a 55-kb intergenic region (61).
Temporal Program of Initiation
Origins are activated at different times during the S phase according to a temporal program. In budding yeast, origins are activated continually during the S phase with the majority of events in mid-S phase (224). Timing in yeast is regulated after the previous mitosis in early G1 phase (223). Similarly in mammalian cells, the decision to replicate the DHFR locus before β-globin was also made in early G1 phase at the “timing decision point” (162).
However, the function of the temporal program remains unknown. Originally, because transcriptionally active euchromatic regions replicated early and inactive heterochromatic regions late, it was thought that early replication is a prelude to transcription (95). In this hypothesis, transcription “opens up” chromatin to allow easy access to replication factors. Again an analogy to coliphage λ can be made, in which the λcI transcriptional repressor blocks the initiation of DNA replication even if replisomal proteins are supplied in trans (281). Later studies showed transcription is important for the binding of initiator proteins λO and λP (88).
Early evidence in mammals had come from studies in which the active X chromosome replicated early, while the homologous inactive X chromosome in females replicated late (30). Similarly, the heterochromatic inactive B chromosomes of maize also replicate late, while the active A chromosomes replicate early (222). More recent studies using microarray analyses have supported the idea in Drosophila (169) and in human cells (169). The choice of the DHFR for initiation is influenced by transcription (236). In contrast, there is no correlation with transcription and origin usage in the budding yeast (224), and in fission yeast origins fire randomly in each generation (215). However, there is a correlation between origin and transcriptional terminators (200)
The relationship between transcription and replication in regulating the temporal program is unclear in that it has never been established whether transcription of these regions causes replication or vice versa (203). Furthermore, there are a number of exceptions in which transcriptional activation during development correlates with decreased origin usage as in the mouse HoxB locus (100, 203). In fact, the timing of origin activation correlates with a developmental program rather than with transcription per se in chicken cells (54, 204). Large chromatin effects in highly transcribed regions are responsible for increased origin efficiency suggesting a change in global chromatin structure rather than local repressive effects at 5′ ends of specific genes (169, 170).
As described below, origin activation results from the recruitment of the replisome that requires the loading of Cdc45 protein, which is dependent on the action of two protein kinases, CDK (cyclin-dependent kinase) and DDK (Dbf4-dependent kinase). However, because both kinases are active at the beginning of S phase, downstream effects must be responsible for timing (193, 243). One possibility is that the chromatin structure at late origins makes it more difficult for CDK and DDK to act. This is supported by the fact that Cdc45 loading correlates with origin timing as first demonstrated in budding yeast (8, 9, 327). In budding yeast, changing the chromatin context influences timing (85, 260, 320). Posttranslational modification of histones also clearly plays a part in temporal program as deletion of the budding yeast Rpd3 histone deacetylase or targeting the histone acetylase Gcn5 to a late origin, which both increase more acetylated and “open chromatin” at origins, advances replication timing (291).
Another connection between chromatin and replication timing involves the DNA damage and replication checkpoints (see below) (206). Rad53 kinase (known as Chk2 in vertebrates), which is needed for checkpoint control, regulates free histone levels (101) and binds to DNA replication origins (63, 129). Loss of Rad53 protein kinase or its upstream kinase Mec1 (known as ATM/ATR in vertebrates) advances timing (235, 253). When DNA replication is inhibited, Mec1 and Rad53 kinases are activated and prevent replication at late origins in both budding yeast and frog extracts (44, 248). Even during normal replication, the checkpoint pathway is important in regulation of timing in human and frog cells (248, 268).
MODEL IN VITRO SYSTEMS: YEASTS (BUDDING AND FISSION), FROGS, AND MAMMALS
Great progress has been made in dissecting the mechanism of initiation using fully reconstituted in vitro systems with purified proteins and defined DNA substrates in E. coli and in animal viruses such as SV40 (132, 143). However, a eukaryotic in vitro system to measure initiation with all purified components is not yet available. Nonetheless, a number of crude extract in vitro systems have been successful in measuring protein function and assessing biochemical mechanism.
Although budding and fission yeast molecular genetic systems are being used to dissect the mechanism of initiation (79), no yeast in vitro system completely reconstitutes DNA replication. However, partial reconstitution of the pre-RC (prereplication complex) on origin DNA in vitro has been shown (247). In this system, extracts from G1-arrested cells will support assembly of pre-RC containing the ORC (origin recognition complex) and Mcm2–7 DNA helicase onto exogenously added and immobilized ARS DNA. In a more recent version, ORC is depleted from the extracts by ion-exchange chromatography and replaced by recombinant ORC1–6 proteins made in insect cells (26).
In frogs (Xenopus laevis), several embryonic systems are available. The original system consists of injection of DNA into an egg (104). Later a cell-free egg extract was used in which a DNA template is added (21). In both cases, any DNA that is added will replicate once and only once per cell cycle and is packaged into chromatin in a nucleus. Therefore, this system does not require a defined origin sequence, and origin selection is random. A newer, more soluble version does not require the presence of the nuclear membrane (293). This latter system uses two extracts: a concentrated NPE (nucleoplasmic extract) and an egg cytosolic extract. Both of these cell-free systems rely on depletion of the target protein, usually with antibodies (immunodepletion), and then reconstitution with recombinant wild-type or mutant proteins to determine function. As described below, the combination of using budding and fission yeast molecular genetics to identify the protein important for initiation of DNA replication in vivo and the frog extract system to investigate biochemical function in vitro has been very powerful. This is also true of studies of the entire cell cycle (188). Because many of the proteins are conserved in evolution, the conclusions can also be applied and tested using mammalian model systems.
Several mammalian in vitro systems are being used successfully [reviewed in (145)]. Most of the systems use template nuclei isolated from cells in G1 phase and a soluble cytosolic extract made from cells in the S phase. A major caveat is that the system is inefficient and only a small fraction of the nuclei replicate. Furthermore, only early origins are used. As in frog (293), an intact nuclear membrane is not required (144). DNA replication is dependent on the same important proteins (Table 1) (45, 46, 171) found in vivo and in the other systems. These systems have also been used to identify novel regulatory factors such as MCM3AP, a Mcm3 protein acetylase (275). Another use of this type of system is to substitute a frog egg extract to replicate the mammalian nuclei (162), which has been used successfully to analyze the temporal program (236).
CELL CYCLE REGULATION OF THE INITIATION OF DNA REPLICATION
As seen in coliphage λ (Figure 1a), the basic mechanism of initiation occurs in several steps and results in bidirectional replication from the origin:
Recognition: label the origin with the ORC
Initiative assembly or licensing: load the DNA helicase to form the pre-RC
Unwinding: activate the DNA helicase
Elongative assembly: load the replisome including DNA polymerase (POL) holoenyzmes and SSB (single-stranded DNA binding protein).
In the model of eukaryotic replication (Figure 1b), ORC marks the origin and provides a “landing-pad” for other proteins. A simple metaphor for the process is “if you build it, they will come” from the movie “Field of Dreams,” where the building of a baseball field brings about the coming of dead baseball players. In this case, the building of a complex on the origin brings about the coming of important replication proteins and the replisome. Again keeping with the similarity to the centromere, this metaphor also has been used to describe the building of the kinetochore on the centromere (56).
DNA replication is regulated in the cell cycle in the following manner (Figure 1b): ORC recognizes and binds to the origin. In G1 phase, Cdc6 and Cdt1 proteins load the MCM helicase onto the ORC to form the pre-RC. To initiate DNA replication and enter S phase, two protein kinases, CDK (cyclin-dependent kinase) and DDK (Dbf4-dependent kinase), activate the helicase and load the replisome.
“Replication licensing” is a useful term that is used to describe the process in which origins are “licensed” when the MCM helicase is loaded onto them in G1 of the cell cycle (20). Thus, pre-RC formation equates with “licensing.” To avoid confusion, it should be pointed out that the original “licensing” hypothesis is incorrect in that it proposed that the nuclear membrane prevented the “licensing factor’ from entering the nucleus (21), which is not the case [see below; (156)].
Re-replication is prevented by blocking MCM loading during S, G2 and M phases. In other words, pre-RCs can be assembled only in G1 phase, but are activated only during S phase. There never exists a cell cycle phase in which pre-RC formation (licensing) and activation can occur (193). In binary or Boolean terms, if off = 0 and on = 1, then there is no phase in which replication licensing (L) and activation (A) both = 1. In G1 phase, L = 1 and A = 0, while L = 0 and A = 1 in S phase (Figure 1b).
Recognition: ORC Labels the Origin
The ORC is a six-protein complex containing Orc1–6 proteins in equal stoichiometry that was discovered by identification of proteins that bound to the ACS of budding yeast and via genetic screens involving mating-type silencing (15, 262). Although the Orc1–6 proteins are conserved in evolution, the sequence dependence is lost in most other eukaryotes as expected because their origins have no consensus primary DNA sequence. The most striking example of this phenomenon is when recombinant Orc1–6 protein from human replaced the frog Orc1–6 protein in vitro to initiate DNA replication in a sequence-independent manner (94, 290). Human, frog, and S. pombe ORCs clearly prefer an A-T-rich sequence, but there is no consensus (39, 141, 290). The AT content could be important for the “helical instability” of origins that is important for facile unwinding (195, 200).
From these data, it is not clear what DNA or chromatin structure the ORC from most organisms recognizes. In budding yeast, the ACS is necessary but not sufficient for ORC binding in the genome and most ACS do not act as origins (310). Of an estimated 12000 ACS in the S. cerevisiae genome, only about 300 are active, which are conserved among four sensu stricto Saccharomyces species (200). In some cases, two ACS sites may act together in directing ORC binding at origins such as at ARS603 (22). The Orc4 protein in S. pombe has nine AT hook domains that are responsible for binding A-T-rich DNA (89), which is consistent with the A-T-rich origin preference (49, 94). Other Orc4 homologues also prefer A-T-rich sequences but do not have an obvious AT hook domain. Binding of ORC to DNA does not require the Orc6 subunit in budding yeast, whereas all six subunits are needed in Drosophila (13).
ORC may recognize a unique chromatin structure dictated by epigenetic determinants and not primary DNA sequence. In budding yeast, a simple 100-bp sequence is needed (see above), which is not conserved, but the ORC is conserved. Again an analogy can be made to that of the centromere in which a self-replicating chromatin structure assures kinetochore inheritance (40), i.e., the origin is the origin because it has always been the origin. In support, histones at the origins in Drosophila follicle cells are hyperacetylated and changes in the acetylation level affect ORC binding (2). In frog, acetylated histones are preferentially found at active origins (50). As in budding yeast (291), the Rpd3 deacteylase is an important regulator in fly, suggesting that even with the acquisition of specific sequence determinants in evolution, budding yeast still retains this important epigenetic regulation.
What are the roles of the six ORC subunits (Table 1)? Most ORC subunits are in the superfamily of AAA+ ATPases (ATPases Associated with various cellular Activities) with conserved Walker A, B, C, and D motifs (142), except for Orc6 (13). However, only the ATP-binding activity of the Orc1 subunit is required for DNA binding (13). In the frog in vitro system with recombinant human ORC, the ATP binding activity of Orc1, Orc4, and Orc5 subunits is required for replication (94).
The ORC protein ATPase is inhibited when it binds origin DNA. The Orc1 ATPase is activated by the binding of Cdc6 protein (13), which is also an AAA+ ATPase, which then produces a conformational change in the ORC-Cdc6-DNA complex to increase specificity (258, 259). On nonorigin DNA, ATP hydrolysis by Cdc6 causes dissociation from the origin. Origin DNA inhibits ATP hydrolysis by Cdc6 and stabilize the complex, i.e, ORC binding to the origin is not specific unless Cdc6 is also bound.
In the presence of ATP, DNA may be wrapped around the ring-like ORC-Cdc6 molecule (Figure 2) (258) similar to initiator DnaA protein in E. coli and the oriC origin (87). The ORC ring structure was deduced from EM reconstructions at 25 Å resolution with Cdc6 bound in the presence of the nonhydrolyzable analog ATPγS. Addition of Cdc6 to the ORC produces a more ring-like structure that resembles the MCM complex in size and shape (1, 76, 96, 146, 214). This implies that the ORC-Cdc6 ring binds the MCM ring on this surface to facilitate MCM helicase loading onto the origin.
The proposed structure of Cdc6, which was deduced by comparison with the ORC structure, is similar to the atomic structure of the archaeal homologue, Orc1/Cdc6. Orc1 and Cdc6 proteins are homologues, and archaeal species have one protein Orc1/Cdc6 that does both functions, i.e., origin recognition and the loading of the MCM helicase (12). The winged helix domain (WHD) of the archaeal Orc1/Cdc6 protein has similarity to Cdc6 protein and to a number of DNA-binding proteins. This structure was used to show that the WHD is needed for DNA binding by the S. pombe Cdc18 protein, a Cdc6 orthologue (164). Some archaeal species, such as Sulfolobus solfataricus, have multiple Orc1/Cdc6 proteins that bind to two different origins (229). All six ORC subunits have WHD motifs (258).
The role of the Orc6 protein is controversial. ORC6 is required for viability in yeast but is not required for DNA binding in vitro (13). In metazoan cells, complexes with lower amounts of Orc6 than the other Orc1–5 proteins still are active (290) and in Drosophila all six subunits are needed (13).
In yeast, the ORC is bound to origins throughout the cell cycle. However, in other eukaryotes, ORC binding is regulated. In support of the “ORC cycle” hypothesis (57), Orc1 dissociates from the chromatin-bound Orc2–5 complex and is degraded in cells outside of G1 phase. The process is regulated by CDK1-cyclin A phosphorylation (160). In yeast, phosphorylation of Orc2 and Orc6 by CDK1 is also important for preventing rereplication (198).
Initiative Assembly or Licensing-Load of the DNA Helicase to Form the Pre-RC
The next step is to load the DNA helicase onto the origin (Figure 1b). This is accomplished by at least two proteins, Cdc6 and Cdt1 (Table 1). The molecular analogy to this process is “clamp loading” in which the clamp loader loads a ring-shaped molecule onto the DNA by opening of the ring (Figure 4) (52). The model is based on elegant structural studies in E. coli of the pentameric -complex of DNA polymerase III holoenzyme that loads the dimeric β-subunit ring-shaped sliding clamp onto the DNA (52). In eukaryotes, the pentameric Rfc1–5 acts as clamp loader of the trimeric PCNA (proliferating cell nuclear antigen) ring. Clamp loading is an ATP-dependent process and all five RFC subunits are AAA+-ATPases. Different subunits of the clamp loader act as “wrench,” “motor,” and “stator” to unlock the ring. In eukaryotes, Rfc1 is the wrench, Rfc2,3,4 acts as the motor, and Rfc5 is the stator. In this model, binding of ATP by the motor produces a conformational change in the wrench, which binds and opens the ring using the stator as a backboard (52).
Figure 4.
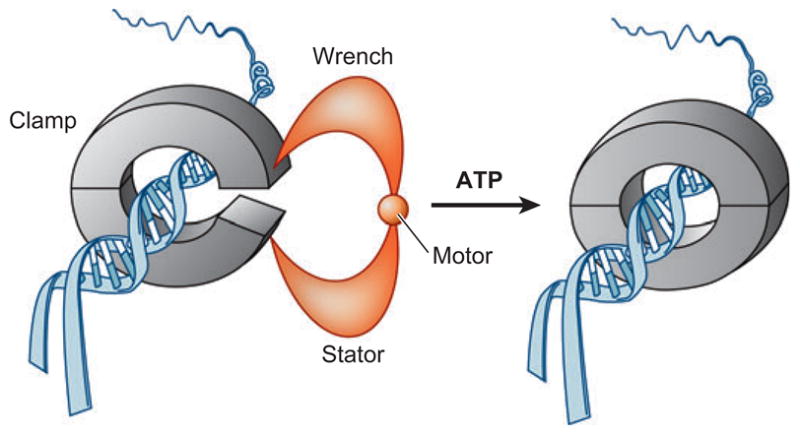
The clamp loading mechanism. Clamp loader (orange) opens clamp using the energy from ATP hydrolysis. Clamp loader is composed of “stator,” “wrench,” and “motor” functions. Clamp loader fixes clamp onto the “stator” while opening the clamp with the “wrench” and “motor.” Open clamp is bound to DNA and then closed. Adapted from (52).
Cdc6 is also an AAA+-ATPase, which is required to load on the MCM helicase in G1 phase as first shown in budding yeast (41) and is proposed to act as a clamp-loader (218, 302). Using the budding yeast in vitro system, Cdc6 ATPase was shown to be required for the subsequent loading of the MCM complex (225). In this process, the Cdc6 and ORC ATPases act sequentially with Cdc6 required initially. Cdc6 and origin chromatin set off a molecular switch in ORC for pre-RC assembly and determine origin specificity as origin mutations can increase Cdc6 ATPase activity, resulting in a less stable Cdc6-DNA complex (258, 259).
Cdt1 protein, like Cdc6 protein, is also required to load the MCM helicase during G1 of the cell cycle of eukaryotes (13). Cdt1 protein (13), which was initially found in fission yeast, was at first missed in budding yeast (201), as it is very divergent (276) but is clearly conserved in eukaryotic evolution. In Drosophila, it is called Dup (double-parked) because mutant cells “park” at two points in the cell cycle (305). As Cdc6 ATPase is needed for Cdt1 binding on the origin in vitro, it has been proposed that a Cdt1-MCM complex is loaded onto the ORC-Cdc6-origin complex during initiation (225). Cdt1 and Cdc6 then dissociate and finally ATP hydrolysis by ORC completes the MCM helicase loading reaction (225, 258, 259).
As stated above, licensing is blocked during S, G2, and M phases of the cell cycle (273). This prevents rereplication. A major level of regulation is catalyzed by CDK, which acts at many redundant levels to block licensing in most eukaryotes (198). These levels include the degradation and localization of several pre-RC components. In fission yeast, it is simpler in that ectopic overexpression of the Cdc6 homologue, Cdc18, is sufficient for rereplication (202). Cdc6 in both yeasts is degraded after CDK phosphorylation (66, 120). In contrast, Cdc6 in mammals is exported from the nucleus after CDK phosphorylation (55, 217).
Another level of regulation to block rereplication occurs through a protein known as Geminin (Table 1; Figure 1b), which was discovered in frog egg extracts (180) and is only found in metazoans, probably because it is important for embryonic development (97, 136). Geminin binds to and inhibits Cdt1 and thus prevents replication licensing by blocking the loading of the MCM helicase (180). Geminin performs a similar role in human somatic cells (308) and in mammalian somatic nuclei incubated in frog egg extracts (267). Geminin forms a negative coiled cylinder (239) that acts in a complex with Cdt1 on origin chromatin (168). Geminin is also important for preventing centrosome rereplication in human cells (269).
The MCM DNA Helicase
The MCM genes were first identified in a genetic screen for mutants that were defective for the maintenance of mini-chromosomes (Mcm phenotype) in budding yeast (Table 1; Figure 1b) (81, 156, 287). The rationale was that mutations that reduce the activity of proteins important for replication would have more drastic effects on mini-chromosomes, which have only one origin or ARS, are not essential, and can be lost from the cells. Conditional cdc (cell division cycle, CDC) mutants of mcm4, mcm7, and mcm5 were also isolated in both budding and fission yeasts. A subset of these mcm mutations is in a family of six paralogous genes numbered MCM2–7, which are conserved in eukaryotes. All six members of the gene family are essential genes in both budding and fission yeast. All are AAA+ ATPases with similarity to DNA helicases. In fission yeast, a complex was identified that contained all six subunits in 1:1:1:1:1:1 stoichiometry and had a ring-like structure (1). However, no ATPase or helicase or DNA-binding activity could be found. This fact still represents a major problem in the field and is discussed further below.
Is the MCM complex the DNA helicase needed for DNA replication (147)? The picture was clouded by the fact that the many mcm-ts mutants had initiation defects at the restrictive temperature, that is, they could not initiate DNA replication, but replication that had already begun was completed. This is in contrast to most dnaB helicase mutants in E. coli, which have elongation defects and stop replication immediately because fork progression terminates (140, 297). The former are called “slow-stop,” and the latter “fast-stop.” This problem was rectified by the construction of “ts-degron” mutants of five of six MCM genes, as the mcm5ts-degron mutant was not viable even at permissive temperature (149). Unlike with conventional ts mutants, the protein is degraded at the restrictive temperature. As expected for a mutant with a defective replicative helicase, all five mutants had a fast-stop phenotype. In contrast, both cdc6ts and degron mutants affect initiation and are slow stop (31, 221). ChIP analyses showed that some Mcm subunits travel with the replication fork (9).
What about DNA helicase activity in vitro? The first breakthrough in biochemical studies was the demonstration that Mcm4/6/7 subcomplexes purified from human cells had ATPase-, ssDNA-, and dsDNA-binding and helicase activity (115, 116). However, the activity was weak and not processive. This is problematic as one would expect a replicative helicase to be processive (216). Later studies by several groups confirmed the results using recombinant Mcm4/6/7 proteins from yeast made in insect cells (153, 241) or in E. coli (127) and also from frog (315) and mouse cells (317). All of these helicase assays are inefficient in that high enzyme:DNA ratios were used, in excess of 5:1, indicating a low turnover number of the enzyme. This fact makes it difficult to perform true processivity assays with low enzyme:DNA ratios.
Although the Mcm4/6/7 complexes had activity, addition of any other subunits inhibited activity, as had been found originally using intact Mcm2–7 complexes (1) or reconstituted fractions purified from cells (115, 116). However, Mcm2–7 complexes from frog and budding yeast had ATPase-but not DNA-binding or helicase activity. A model used to explain these results is that Mcm4/6/7 complexes are catalytic and the Mcm2/3/5 complexes are regulatory (241). However, this model predicts a structure of dimers of trimers, which is at odds with a planar structure found for the Archaeal helicase (76, 96). Furthermore, it was proposed that two trimer complexes dissociate from one another, thereby activating the Mcm4/6/7 for catalysis (317). This idea is inconsistent with the fact that these proposed “regulatory” Mcm2 and Mcm3 subunits are required for fork elongation (149).
The problem with in vitro helicase assays is that artificial substrates are used (216) (Figure 5a). These substrates are composed of a small 40–60-bp primer annealed to a 5-kb ssDNA circle or to a larger primer, which gives a single-stranded tail on which the he-licase translocates in a 3′ to 5′ direction for Mcm4/6/7. In contrast, a large (>100-kb) dsDNA chromosome would have to be unwound at the origin in a eukaryotic cell. In one case, the Mcm4/6/7 complex from fission yeast was more processive (>600 bp) when a substrate containing both 5′ and 3′ tails was used to resemble a true replication fork (Figure 5a) (154). In this model, each trimer of a double trimer of Mcm4/6/7, which exists in solution and on the DNA (154), would bind each one of the leading and lagging strands at the fork. Several other models have been proposed and compared (271) (Figure 5b) (see below).
Figure 5.
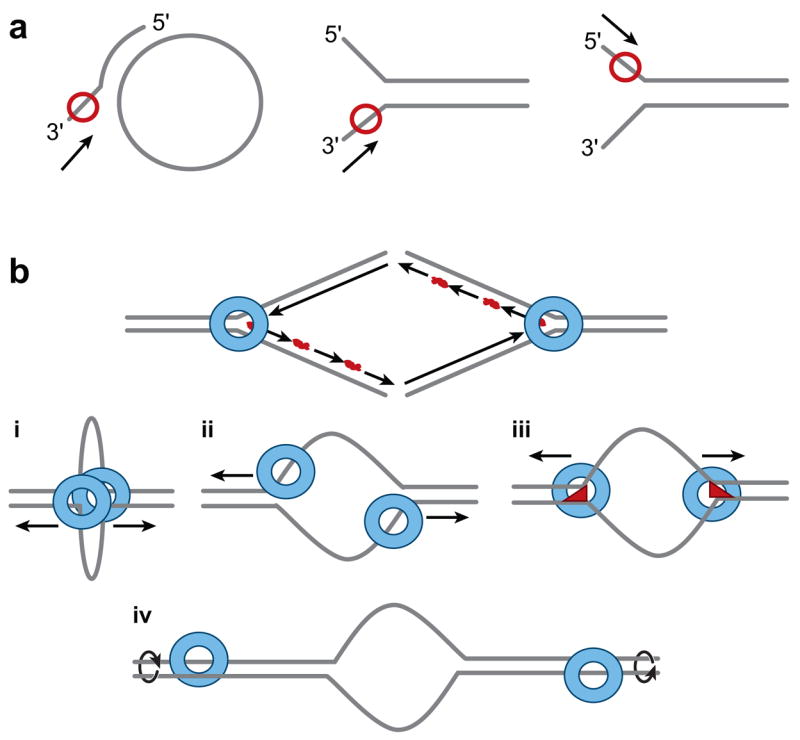
Helicase substrates and models. (a) In vitro helicase substrates that are used frequently have small ssDNA (50 bp) annealed to ssDNA circle (5 kb) with nonhomologous 3′ tail. Helicases (red circle) such as SV40 T antigen or Mcm4/6/7 complex translocate 3′ to 5′ on the tail to unwind DNA and release oligonucleotide from the larger circle. Other substrates used resemble replication forks that are produced by annealing small ssDNA oligonucleotides with nonhomologous ends. Helicases can translocate 3′ to 5′ as above or 5′ to 3′ (DnaB). (b) A single hexameric helicase is depicted as a ring (blue) at the ends of a conventionally drawn replication fork. Lagging strand Okazaki fragments are shown with RNA (red) primers at their 5′ ends. (i) The SV40 T antigen model (161) is made by putting the two rings together forming a loop. In this model, the DNA is pumped into the channel of the double hexamer and then extruded out the holes in the outside C-terminal domains (Figure 2d). (ii) In the “pump-in-ring” model, each single hexamer translocates on a different strand of DNA (127). (iii) In the “ploughshare” model, the ploughshare (red) acts as a wedge and keeps the ssDNA unwound as it emerges from behind each single hexamer (271). (iv) In the “rotary pump” model, different single hexamers twist the DNA at a distance resulting in topological strain and unwinding in the center (151).
The structure of the Mcm2–7 helicase has been deduced by using the archaeal Mth-MCM protein from Methanobacterium thermoautotrophicum and the animal virus SV40 T antigen as models (Figure 2) (38, 76, 78, 96, 161, 214, 244). The Mth-MCM is a true homologue whereas the SV40 T antigen is an analogue that results from convergent evolution. Unlike the eukaryotic Mcm2–7 complex, archaeal MCM homo-oligomeric complexes are fully active and processive, even though all studies used >1:1 ratio of double hexamer proteins to DNA (38, 134, 250). Originally, EM images showed the Mcm2–7 complex from fission yeast to be a ring, and many isomers were seen including planar hexamers, double trimers, and even tetramers plus dimers (1). Both the atomic structure of the N-terminal domain (76) and the cryo-EM reconstruction of the full-length protein showed the Mth-MCM helicase (96, 214) to be a planar, double hexamer in head-to-head conformation (Figure 2).
The N-terminal domain of Mth-MCM is needed for oligomerization and DNA binding through β-fingers or hairpins with positively charged amino acids at the tip that surround the central cavity (76). The C-terminal domain contains the catalytic ATPase and helicase domains (Walker A–D); the structure is yet to be determined. From a comparison with the SV40 T antigen analogue, it has been suggested that dsDNA is pumped bidirectionally into the central opening, unwound, and the ssDNA extruded through the large openings in their C-terminal domains (Figure 5b). The resultant ssDNA would be replicated by the replisome as it exits from these openings. As bidirectional replication proceeds, the loops would grow in size. Again by analogy with SV40 T antigen, ATP binding would produce movement of the second set of β-fingers akin to the closing of an iris diaphragm and release the DNA upon ATP hydrolysis (90, 161). In Mth-MCM, the large opening is big enough (34 Å) to accommodate dsDNA (d = 20 Å), although it is quite small in SV40 T antigen (15–20 Å), which has suggested that each hexamer only binds ssDNA, as seen in the papillomavirus E1 proteinssDNA structure (71). Another possibility is that dsDNA binding changes the size of the hole to accommodate the helix.
Why have six Mcm2–7 paralogues? During evolution, continued gene duplication and divergence produced different MCM genes with different roles in DNA replication. The order of subunits has been inferred to be Mcm5-Mcm3-Mcm7-Mcm4-Mcm6-Mcm2 in a planar ring by reconstitution of subcomplexes using purified recombinant yeast proteins (Figure 1b) (51, 241). This order is consistent with yeast two-hybrid studies of Drosophila, mouse, and human MCM proteins (47, 139, 318). As stated above, Mcm2/3/5 may have evolved to be regulatory subunits. Mcm5 is a unique subunit in that it is the only subunit that is very difficult to tag at either its N or C terminus (149, 197), which is why it has not been studied as extensively as the other subunits. As described below, yeast Mcm2, Mcm4, and Mcm6 proteins may be targets of DDK phosphorylation (175, 251) that results in a conformational change in Mcm5 protein and leads to helicase activation and replisome loading (76, 244, 245).
From all these studies, there are several possibilities for the mechanism of MCM helicase unwinding of the DNA during chromosomal replication (Figure 5b) (244, 271). If Mcm2–7 is a double hexamer, then the pumping model akin to SV40 T antigen might be right. This model assures coordinated bidirectional replication. Yeast two-hybrid studies have clearly shown that two molecules of the same mouse Mcm subunit can bind to each other (139), which is consistent with a double hexameric structure (244). Also consistent with double hexamers at the origin is the fact that frog Mcm2–7 complexes protect about 80 bp of DNA (69). Similar to SV40 T antigen (5, 176, 256), mutant Mth-MCM single hexamers are less active than wild-type double hexamers (78, 244). Even though mutant SV40 single hexamers are about 5% active on artificial helicase substrates (Figure 5a), they are inactive in the SV40 DNA replication system in vitro (11, 303). Furthermore, fission yeast Mcm4/6/7 complexes are dimeric (154). Yet even other archaeal species such as S. solfataricus may use single MCM hexamers in which the DNA enters the helicase C-terminal domain (182).
In the “ploughshare” model (271), a double hexamer is loaded onto the origin, then single hexamers translocate in opposite directions along the dsDNA with a ploughshare protein helping to keep the ssDNA unwound as it emerges from behind the helicase (Figure 5b). In the “pump-in-ring” or steric exclusion model (127), each hexamer also moves bidirectionally on the DNA, but displaces the opposite strand due to the steric hindrance from meeting the dsDNA at the fork. Finally, the “rotary pump” model has different hexamers twisting the DNA at a distance, resulting in topological strain and unwinding in the center (151). It is based on the fact that MCM2–7 proteins are not located at replication foci in the nucleus. These models will be resolved only when atomic structures of full-length archaeal MCM proteins, eukaryotic Mcm4/6/7, or even Mcm2–7 proteins bound to ssDNA and dsDNA are solved.
In some metazoans, there are two other paralogues, Mcm8 and Mcm9, which are found in human and Drosophila, but missing from worm (19, 98, 123, 167, 316). Recombinant frog Mcm8 displays both ATPase and helicase activities in vitro, and in reconstituted egg extracts, Mcm8 is required for fork elongation (172). However, it is not known if Mcm8 forms a multimeric complex. In contrast, human Mcm8 is needed for pre-RC assembly and recruits Cdc6, which then loads the Mcm2–7 complex (292). In Drosophila, Mcm8 is needed for meiotic recombination (19). It is not yet known whether these discrepancies result from the different systems or the methods used. Mcm9 is most similar to Mcm8 (167, 316), but has not been characterized biochemically. Recently, the human MCM-BP protein, a distant homologue that lacks important MCM motifs, has been shown to regulate the Mcm2–7 complex by replacing Mcm2 in the hexamer (231).
Unwinding: Activate the DNA Helicase
In G1 phase, pre-RCs with the Mcm2–7 helicase bound are present on all origins. Nearly 90% of all origins that are bound by ORC also have MCM complex bound (309, 310). The next step is to activate the MCM helicase and load on the replisome (Figure 1b), with helicase activation being dependent on replisome loading, which both require phosphorylation by CDK and DDK enzymes. In this manner, the cooperation between helicase activation and replisome loading assures coordinated replication. A lack of coordination could allow the helicase to produce unreplicated ssDNA at the fork, which occurs when DNA replication is blocked (32).
The large multiprotein complex that is formed at this step has been referred to as the pre-IC (pre-initiation complex) (326). The original idea was that the pre-IC contains all the proteins needed for DNA replication and then a final activation step is required, which is catalyzed by DDK. The nature of the activation step is unclear as DDK was later shown to be required in the formation of the pre-IC (327). It is possible that the pre-IC does not exist for very long and that replication begins as soon as the pre-IC is formed. Resolution must await an in vitro system that can follow all the events in time.
As described above, intact Mcm2–7 complexes are inactive in vitro. Furthermore, excess MCM complexes are loaded on origins of about 40 MCM hexamers per origin in frog (69) and about 10 in budding yeast (65). Only a subset of these hexamers are selected to become active replication origins by binding a protein called Cdc45. Cdc45 moves with the replication fork (8) and was shown to be needed for both initiation and fork elongation by using a cdc45ts-degron mutant (279). Although Cdc45 protein is conserved from yeast to humans, no insight can be gleaned about its biochemical function from primary sequence.
Yet addition of these inactive recombinant Mcm2–7 complexes to frog extracts previously depleted of it restores DNA replication (315). This implies that the complex must be activated in some manner when it is added to the extracts. Mcm2–7 complexes isolated from frog egg extracts displayed helicase activity if bound to Cdc45 protein (177). Cdc45 protein is needed for loading of the replisome, including DNA polymerases and RPA, the eukaryotic SSB (8). The interaction of Cdc45 protein with the MCM complex was initially inferred from extragenic suppressor studies in which mcm5ts mutations were suppressed by cdc45cs mutations (108). Both neutralizing antibodies to Cdc45 or a fragment of the Rb (retinoblastoma) protein that binds Cdc45 inhibit MCM helicase activity and further unwinding of the template even after replication has begun in the frog NPE system, which supports the idea that Cdc45 is a helicase cofactor (211). A complex of Mcm2–7, Cdc45, and GINS (see below) purified from Drosophila embryos has helicase activity in vitro. From these data, it appears that the Mcm2–7 complex may be activated by the binding of other initiation proteins (189).
Elongative Assembly: Load the Replisome Including DNA Polymerase Holoenyzmes and SSB
CDK and DDK are the two conserved protein kinases required for helicase activation and replisome loading. In budding yeast, there is only Cdk1 or Cdc28 enzyme, but there are six B-type cyclins (Clb1–6) needed for S and M phases (188). Most likely, Cdk1-Clb5 complexes are the most active in regulating DNA replication and Cdk1-Clb2 for regulating mitosis (194). There is little overlap in function in that Cdk1-Clb5 phosphorylates different substrates than Cdk1-Clb2 (48, 165, 183). Some replication proteins (Table 1) are Cdk1-Clb5 substrates because Clb5 (but not Clb2) has a unique hydrophobic patch that binds the RXL or Cy motif in the substrate (165). This supports the idea that it is substrate specificity by different Cdk-cyclin complexes that drives the cell cycle. However, in other eukaryotes this may not be the case (183). For example, frog mitotic Cdk1-cyclin B1 complexes can promote DNA replication when targeted to the nucleus (187).
In mammals, there are many different CDKs and cyclins with at least four CDKs (Cdk1–4) and four classes of cyclins (A, B, D, and E) required for cell cycle progression (188). The Cdk2 homologue is probably used in DNA replication, although Cdk1 (also known as Cdc2) can substitute (4, 266), accounting for the viability of Cdk2 knockout mice (208). By analogy, Cdk2-cyclin E and Cdk2-cyclin A act as yeast Cdk1-Clb5 for DNA replication, whereas Cdk1-cyclin B act as yeast Cdk1-Clb2 for mitosis.
CDK and DDK enzymes are regulated independently of each other, but by similar mechanisms (174, 188, 243). Both kinase subunits are inactive in monomeric form and are activated by the binding of an unstable activating subunit, Cyclin and Dbf4/Drf1 protein for CDK and DDK, respectively. Thus, CDK is Cyclin-dependent kinase and DDK is Dbf4-dependent kinase. Cell cycle regulation of the unstable subunit assures cell cycle regulation of activity. With CDKs, other levels of regulation occur including protein inhibitor binding, phosphorylation by other kinases, and cyclin subcellular localization (188). With DDK, it is simpler in that Dbf4 protein is absent in G1 phase because it is targeted for proteosomal degradation by the APC (anaphase promotion complex) (174, 243). As cells enter S phase, the APC is inactivated by CDK phosphorylation and Dbf4 is stabilized. In vertebrates, a Dbf4 paralogue has been identified called Drf1 (185, 270, 278, 314). In frog, Cdc7-Drf1 is mainly the embryonic form found in egg extracts, while Cdc7-Dbf4 functions in somatic cells (270).
How do the two protein kinases activate the MCM helicase and load on the replisome (Figure 1b)? CDK might have evolved later to coordinate the cell cycle with DNA replication, while DDK is simpler in having only a very specific role in DNA replication (191). Evidence indicates that the Mcm2–7 complex is a target of phosphorylation by DDK, which is needed to load on the Cdc45 protein (245, 327). Initially, Mcm2 was shown to be a preferred substrate of DDK in vitro and in vivo in budding yeast (157), in fission yeast (29), and in human (237, 285). Genetic studies also demonstrated an interaction between DDK and the MCM complex. The mcm5-bob1 allele (P83L) in budding yeast bypasses the role of DDK in replication (103) and a dbf4 mutation suppresses the defect in a mcm2ts mutant (157). Later studies confirmed that all Mcm2–7 subunits except for Mcm5 protein are DDK substrates in vitro in several eukaryotes [reviewed in (13)]. Recently, phosphorylation sites in the N terminus of budding yeast Mcm4 were mapped and shown to be important for formation of the pre-IC and for DNA replication by using nonphosphorylatable serine/threonine-to-alanine substitution mutants (251). However, phosphorylation of these sites is important but not essential. In similar studies in fission yeast, it was proposed that the N terminus of Mcm4 or Mcm2 or Mcm6 could be phosphorylated by DDK to allow for replication (175). The hypothesis is that there is considerable redundancy in the system with any of 3 different subunits of the MCM complex acting as targets for DDK. Support for this hypothesis would show that the phenotype of any of these phosphorylation mutants is suppressible by mcm5-bob1, which can bypass DDK function (103).
Phosphorylation of the MCM complex by DDK leads to the loading of Cdc45 protein in frog egg extracts (121, 294), budding yeast (245, 327), and in fission yeast (312). In budding yeast, there is a low amount of Cdc45 bound to the origin detectable by ChIP studies in G1 phase when both CDK and DDK are inactive (8, 245, 327). In another ChIP study, Cdc45 protein binding to early origins was appreciable even in G1 phase cells (124). Similarly, Cdc45 protein binding to bulk chromatin was found to be independent of DDK (326). It is possible that Cdc45 protein may be weakly bound to origin chromatin in G1 phase that is later stabilized by combined CDK and DDK action. In the mcm5-bob1 mutant, Cdc45 loading at early origins occurs in G1 phase and mcm5-bob1 bypass is dependent on Cdk1-Clb5 activity (245). Although DDK is bypassed, it was hypothesized that the cells are alive because DNA replication is still regulated in the cell cycle by CDK (245). Recently, the role of CDK in replication was also bypassed, which led to synthetic lethality with DDK bypass by the mcm5-bob1 mutation and to replication occurring in G1 arrested cells (321). Therefore, CDK and DDK regulate similar events independently, i.e., helicase activation and loading of the replisome (24). This explains why both kinases are needed for replication in all eukaryotes examined.
However, the order of action by the two kinases is different in different systems. In budding yeast, CDK acts before DDK (205). In frog egg extracts, DDK acts first (121, 294). This discrepancy may be due to the different experimental systems used or it may be that there is really a difference between replication in the two organisms. Recently, the order of action in fission yeast was found to be similar to that in frog (312), indicating that these differences may indeed be organism specific. Budding yeast, like most somatic cells, coordinates cell size and division by CDK in G1 phase, whereas fission yeast coordinates size and division mainly in G2 phase and embryos lack the coordination completely (188). Thus, budding yeast CDK is already being used in G1 phase and would act before DDK.
DDK may have a relaxed specificity of phosphorylation in that there is a weak consensus phosphorylation site that consists of serine or threonine residues with nearby acidic residues or serine and threonine residues that are phosphorylated by CDK and are also acidic (37). In fact, the seven DDK phosphorylation sites in the N terminus of yeast Mcm4 could be replaced by a synthetic sequence with serines and acidic amino acids or the N terminus of Mcm2 (251). However, efficient phosphorylation by DDK in vivo and in vitro requires a second region that docks the Mcm4 substrate to the DDK.
How would phosphorylation of Mcm2, Mcm4, or Mcm6 by DDK activate the helicase and load the replisome? One hypothesis is that DDK phosphoryation results in a conformational change in the Mcm5 protein that activates the helicase and is a signal for the binding of Cdc45 protein (Figure 1b) (76, 244, 245). This hypothesis is based on structural modeling of the budding yeast mcm5-bob1 P83L mutant that bypasses DDK function, using the structure of the archaeal MtMCM N terminus with a similar mutation, P62L (76). The N terminus of MthMCM has three domains called A, B, and C (Figure 2a,b). Amino acids with large side chains such as leucine at the P83 residue of Mcm5 would “push-out” the A domain, which is an alpha-helical domain at the very N terminus. The assumption is that this structure would normally be caused by phosphorylation by the other subunits in the hexameric complex. Using high-resolution genomic footprinting, the structure of ARS1 origin chromatin in S phase was shown to be more accessible to permanganate cleavage than in G1 phase and was dependent on DDK function. Similar origin accessibility was also seen in the mcm5-bob1 mutant arrested in G1 phase (92). Thus, structural changes of origin chromatin in the mcm5-bob1 mutant mimic the effect of DDK phosphorylation. These changes correlate with the binding of Cdc45 protein that occurs in G1 phase in the mcm5-bob1 mutant (245).
Recently, CDK’s role in promoting origin activation has been determined (277, 312, 322). In order to understand this role, a number of important replication proteins must be described. The GINS complex, which is based on the numbers 5, 1, 2, and 3 in Japanese (Go, Ichi, Nii, San), is composed of the Sld5, Psf1, Psf2, and Psf3 proteins, and is required for replication (Table 1). The budding yeast GINS complex was identified using a combination of yeast two-hybrid approaches and extragenic suppressor analyses, coupled with biochemical purification studies (272). The approach centered on Dpb11 (DNA Polymerase B possible subunit), a subunit of DNA polymerase ε holoenzyme, which is also called Pol2 or PolB (179). DPB11 was isolated as a high-copy suppressor of pol2ts mutants (10). Sld mutants are Synthetic Lethal with Dpb11, while Psf proteins interact or Partner with Sld5 protein in two-hybrid analyses. The Sld2 and Sld3 proteins were also identified in this way in that a cdc45ts mutant is suppressed by high-copy SLD3, and SLD4 is allelic with CDC45 (124). The GINS complex has also been found in frogs (146) and in a large-scale degron screen for yeast DNA replication mutants (125). In both yeast cells and frog egg extracts, GINS functions interdependently with Cdc45 protein in the loading of the replisome (146, 272) and has a ring-like structure in the EM (146). Sld2 and Sld3 proteins are also important for replisome loading with Cdc45 protein in both budding and fission yeasts, but Sld3 is absent in metazoans (124, 192). Thus, a large number of proteins are needed in addition to Cdc45 to load on the replisome. These proteins may also help to activate the MCM helicase in that Mcm2–7 complex can be activated by the binding of Cdc45 and GINS (189), thereby coupling helicase activation and replisome loading (24).
The role of CDK in this process is to phosphorylate Sld2 (178) and Sld3 (277, 322), causing them to bind to the BRCT repeats in Dpb11, which then binds origin chromatin and recruits the replisome with Cdc45. The role of CDK in replication can be bypassed by using phosphomimetic mutation of the phosphorylation site sld2-T84D and fusion of Sld3 to Dpb11. Similar to DDK bypass, bypass of CDK is not sufficient for DNA replication in cells arrested in G1 phase because DDK is inactive as Dbf4 protein is absent due to proteosomal degradation (243). However, overexpression of Dbf4 protein, which can overcome the degradation and produce active DDK (243), together with CDK bypass, allow for DNA replication in G1 arrested cells (277, 322). Similarly, this CDK bypass and DDK bypass by mcm5-bob1 are synthetically lethal (322). In both cases, lethality occurs because the cell cycle regulation of DNA replication has been completely ablated. Because CDK is inhibited in these cells by the overexpression of a nondegradable Sic1 mutant, CDK cannot prevent Mcm2–7 loading and rereplication occurs. Thus, both papers demonstrate positive and negative roles for CDK in DNA replication. In fission yeast, Sld3 loading is not dependent on CDK and GINS but only on DDK (312, 313). In this case, Sld3 may load first followed by GINS, Cdc45, and finally the replisome, perhaps explaining why DDK acts before CDK in fission yeast. In frog, the Sld2 (RecQ14) protein acts very late, does not need CDK, and is required only for RPA SSB protein loading (234).
Also identified in the original mcm screen was the MCM10 gene, which is not a Mcm2–7 homologue (81, 156, 287). Mcm10 is needed for the loading of the Cdc45 protein after pre-RC formation and for stabilizing the replisome as shown in human cells (118), frog egg extracts (307), budding (228, 238) and fission yeast (228). Mcm10 protein may act by stimulating DDK (155) and DNA polymerase α activities at the fork. Fission yeast Mcm10 protein also has primase activity in vitro, which is required in vivo (75).
In summary (Figure 1b), activation of the helicase and loading of the replisome are regulated by the combined action of DDK and CDK. In this proposed model, DDK phosphorylates any one of Mcm2/4/6 proteins, resulting in the “pushing out” of the A domain of Mcm5 in the hexamer. The Mcm5 protein structural change together with CDK phosphorylation of Sld2 and Sld3, Mcm10 and the binding of Cdc45 and Dpb11 proteins load the replisome, which then activates the Mcm2–7 helicase. The DNA is unwound by the helicase and the DNA is replicated by the replisome. It is not clear what the exact role of Mcm10 is in this model but it is known to be required for Cdc45 loading and replisome stability (see above). Perhaps, the loading complex falls apart without it.
PREMEIOTIC DNA REPLICATION
During meiosis, a diploid cell undergoes one round of DNA replication followed by two successive nuclear divisions to yield four haploid products. After DNA replication, homologous chromosomes pair, recombine, and synapse. The first division (MI) is reductional; the newly recombined homologous chromosomes segregate from one another. The second meiotic division (MII) is a mitotic, equational division, during which sister chromatids segregate [reviewed in (230)]. As in the mitotic cell cycle, meiotic cells exhibit controls that ensure DNA replication occurs once and only once throughout meiosis. Unlike in the mitotic cell cycle, however, additional mechanisms must function during meiosis for chromosomes to segregate properly into four haploid progeny. First, meiosis-specific features are included within chromatin during meiotic S phase to promote meiotic recombination. Meiotic recombination creates a physical connection and tension between homologues prior to MI, thus promoting proper segregation of homologues during MI. Additionally, meiotic cells must also inhibit an additional round of DNA replication between MI and MII.
In every model organism studied, the duration of the meiotic S phase is longer than that of the mitotic S phase (17, 33, 112). In budding yeast, meiotic S phase is two to three times longer than S phase in mitotic cells (35). The explanation for a prolonged meiotic S phase is unclear, because origin usage and the rate of replication fork progression appear to be similar in both mitotic and meiotic S phases. For example, in S. cerevisiae DNA replication initiates at the same ARS elements during mitosis and meiosis (42, 111), and replication fork progression from these ARS elements occurs at similar rates during both S phases (42). In fission yeast, meiotic S phase requires the same genes for progression of DNA replication as in the mitotic S phase (80). It has been proposed that a prolonged meiotic S phase allows for the establishment of meiosis-specific features of chromatin to promote homologous recombination (35, 112). Two meiosis-specific proteins, the chromatid cohesin Rec8 (138) and the transesterase Spo11 that catalyzes formation of DNA double-strand breaks (DSBs) to initiate recombination, are thought to play a role in controlling meiotic S phase progression (35, 130). This model is supported by the fact that at least in S. cerevisae, DNA replication and homologous recombination are directly related to one another. Chromosomal regions where DNA replication is blocked or delayed are also blocked or delayed for formation of DSBs. Furthermore, the time between replication and DSB formation is constant (1.5 to 2 h) for all regions of the genome studied (23).
Regulation of the initiation of meiotic S phase utilizes much of the same cellular machinery (Table 1) as in the mitotic S phase [reviewed in (80, 263)]. It has been difficult to determine genetically if mitotic replication factors are also required for meiotic S phase because meiosis is an inherently temperature-sensitive process. Because the restrictive temperature is often lower for meiosis than for mitosis, temperature-sensitive alleles that display a strong S-phase defect in mitosis may display a leaky intermediate phenotype when incubated at a lower restrictive temperature for meiosis. Furthermore, meiotic cells appear to be able to function with lower levels of some replication factors than mitotic cells (82). For instance, in S. pombe temperature-sensitive cdc18/cdc6, mcm2, and mcm4 mutants were proficient in meiosis and meiotic S phase in conditions that caused arrest in the mitotic cell cycle, suggesting that these replication initiation proteins were not required for meiotic replication (82). However, this conclusion was disputed in later studies. In one study, the same mutants of S. pombe displayed a meiotic replication defect when sporulated at higher, more restrictive temperatures (190). Additional work in fission yeast has shown that MCM proteins are chromatin-associated during meiotic S phase, suggesting a possible role in DNA replication, and a double temperature-sensitive degron mcm4 mutant does not replicate its DNA in meiosis (163). In this case, both the degron cassette at the N terminus of mcm4 and a hypomorphic mcm4ts mutation were required to completely block DNA replication. Taken together, licensing in the meiotic S phase requires Cdc18 (or Cdc6 in S. cerevisiae) and the Mcm proteins as found in mitotic cells.
As in the mitotic cell cycle, CDK and DDK also play vital roles during meiotic S phase for helicase activation and replisome loading. Earlier works in S. cerevisiae with temperature-sensitive alleles of Cdc28/Cdk1 suggested this protein was dispensable for initiation of meiotic S phase (254). However, an analogue-sensitive mutant, cdc28-as1, is blocked for DNA replication during meiotic S phase in the presence of the analogue inhibitor (16). Cdc28/Cdk1 may also play additional roles in meiosis downstream of DNA replication such as regulating DSB formation (107). In S. cerevisiae, clb5Δ clb6Δ mutants deleted for both S-phase Clb5 and Clb6 cyclins that are used for initiation in the mitotic S phase (242) do not replicate their DNA during meiosis (264). This is because in mitotic cells the remaining four Clb cyclins in the cell are functionally redundant with Clb5 and Clb6 (242).
The precise role of DDK during meiosis is unclear, but studies involving temperature-sensitive and analog-sensitive DDK mutants suggest that this complex plays a role in meiosis beyond DNA replication. In S. cerevisiae, cdc7 temperature-sensitive mutants arrest in meiosis after DNA has been replicated but before DSB formation and recombination (111, 240), suggesting that CDC7 plays a unique role in meiosis beyond its mitotic replication functions. An analogue-sensitive mutant, cdc7-as3, exhibits a similar meiotic defect in the presence of inhibitor, though DNA replication takes about 4 h longer than wild-type cells in the presence of inhibitor (295). Temperature-sensitive cdc7 (hsk1) mutants of S. pombe also display prolonged meiotic S phase and defects in DSB formation (207). Dbf4, the regulatory subunit of DDK, may also have multiple meiotic functions. In budding yeast, depletion of Dbf4 before meiosis greatly delays meiotic S phase, and cells in which Dbf4 has been depleted after initiation of S phase arrest before anaphase I, and this arrest is independent of defects in DNA replication or recombination (289). Thus, DDK regulates both DNA replication during meiotic S phase and DSB formation after meiotic S phase.
Despite overlap between proteins that regulate DNA replication in mitosis and meiosis, one gene is specifically required for meiotic S phase. Mutants of S. cerevisiae lacking MUM2 fail to completely replicate their DNA and arrest prior to MI. Though MUM2 is expressed throughout the mitotic yeast cell cycle, mum2Δ mutants do not appear to have any defects in vegetative growth (72). MUM2 genetically interacts with ORC2 and with POL1 (Table 1), which encodes the catalytic subunit of the DNA polymerase α-primase complex, suggesting that Mum2p affects the DNA replication machinery (53), though its precise function is unknown.
Another unique aspect of meiotic DNA replication is that a second round of DNA replication must be inhibited between the two nuclear divisions. In S. cerevisiae, three CDK-dependent and nonredundant mechanisms inhibit rereplication of DNA during the cell cycle: CDK inhibits Cdc6 activity and expression, promotes nuclear exclusion of Mcm2–7, and downregulates ORC ac through inhibitory phosphorylation (99, 198). Work with Xenopus oocytes and S. pombe indicates that similar mechanisms prevent rereplication of DNA in meiosis. Degradation of Cdc6 protein and repression of its synthesis prevents rereplication of DNA in immature Xenopus oocytes (159, 304). Exclusion of Orc proteins from the nucleus of immature Xenopus oocytes also prevents pre-RC formation, thus providing a second mechanism for inhibiting DNA replication (304). In fission yeast, Mcm4p is not associated with chromatin during the time between MI and MII (163), even though Mcm4p levels are constant throughout meiosis (82). Although the precise mechanisms inhibiting DNA rereplication during meiosis may not be identical to inhibition of mitotic DNA rereplication, both processes involve modification, degradation, and/or relocation of components of the pre-RC.
CHECKPOINT REGULATION OF DNA REPLICATION
DNA checkpoint mechanisms evolved to monitor the successful completion of cell cycle events involving DNA replication and mitosis. If DNA replication is blocked or the DNA is damaged, a signal transduction pathway (Figure 6) is activated, resulting in a block to further initiation events in S phase and to entry into mitosis (206). This pathway consists of sensors (RFC, RPA, PCNA, 9-1-1, Polε), amplifying mediators (Rad9, Mrc1/Claspin), transducers or transmitters of the signal (Mec1, Tel1, ATM, ATR, Chk2/Rad53, and Chk1 protein kinases) and effector target proteins (Dun1 and Cdk1 protein kinases, Cdc45, p53 transcription factor, MCM helicase, Polζ) that produce the response (cell cycle arrest, transcription, DNA repair, translesion synthesis, etc.). The pathway may not be linear in that replication forks may be both sensors and effectors (280). These checkpoint responses are important medically as defects in human Chk2, ATM, and p53 lead to human cancer by increasing genomic instability (128). [For reviews on checkpoint response see (27, 84, 128, 206, 265).]
Figure 6.
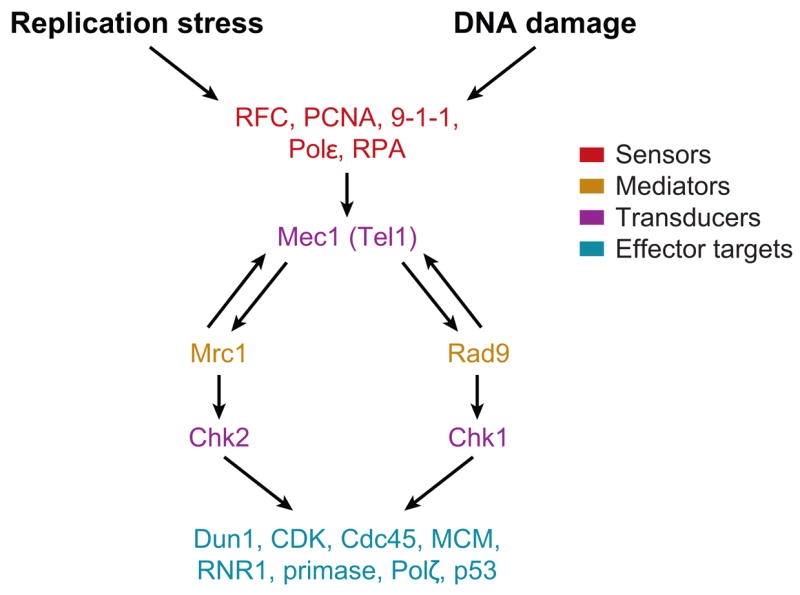
DNA replication and damage checkpoint regulation. Replication stress or blockage or DNA damage induces activation of a signal transduction pathway of many different proteins. The proteins are in different classes indicated as sensors, mediators, transducers, and effector targets. For example, if DNA replication is blocked, ssDNA coated by RPA sends a signal to activate Mec1 protein kinase. Mec1 binds to Mrc1, which amplifies the signal by binding to and activating Chk2 (Rad53) protein kinase. Chk2, in turn, inhibits Cdc45 helicase activation and loading of the replisome.
The checkpoint response was initially discovered in budding yeast using the following rationale: If replication is blocked, cells are prevented from entering mitosis by an active process. Checkpoint mutants are defective in the process, do not sense that replication has stopped, enter mitosis with unreplicated chromosomes, and die (106, 299, 301). For example, if wild-type cells are treated with HU (hydroxyurea), an inhibitor of RNR (ribonucleotide reductase), they will arrest in S phase, but retain viability and are alive after HU is removed. In contrast, mec1 or rad53 checkpoint mutants are more sensitive to HU because they die rather than arrest in its presence (301). Another example is with DNA polymerase α mutants (cdc17ts), which arrest with a G1-content of unreplicated DNA at 38°, but with a nearly complete G2-content of DNA at the partially restrictive temperature of 34°(300). Only the arrest at 34° was dependent on known checkpoint genes because the cells have incompletely replicated DNA.
Why do the cdc17ts mutants arrest in G1 phase at 38°? Another checkpoint must have prevented the cells from entering mitosis with completely unreplicated DNA. Many other cdc replication mutants from the Hartwell collection, such as cdc7 and cdc6, also do not make DNA at the restrictive temperature and arrest at the G1/S boundary (31, 105, 243). It has been suggested that cdc mutants of this type arrest because they are “leaky” and produce some replicated or damaged DNA that activates the known DNA checkpoint (221, 280). However, in precise measurements of DNA replication by heavy isotopic labeling of cdc7ts mutants at the restrictive temperature, most cells make very little DNA, with a small percentage of the cells (<30%) that replicate only chromosome III (226), which is only about 2% of the genome. It is unlikely that there is enough “leakiness” to account for the arrest that occurs in the 70% of cells that do not initiate DNA replication.
Another explanation is that some strains of budding yeast may lack a novel checkpoint mechanism that detects unreplicated DNA. These strains enter mitosis with un-replicated DNA (148, 221, 279, 280, 284) and undergo a “reductional anaphase” (221). This novel checkpoint, the “G1/M-phase checkpoint” (284), is based on the fact that cdc7ts and dbf4ts mutants arrest in G1 phase only in some genetic backgrounds and independent of allelic “leakiness.” Strains without this checkpoint still retain the known DNA replication, damage, and mitotic checkpoint pathways. This checkpoint is independent of known checkpoint genes such as MEC1 and RAD53, but it has not been genetically defined as it is dependent on too many unknown genes (284, 301). Fission yeast replication mutants, e.g., cdc18, also lack this undefined checkpoint and will actually septate or cut the unreplicated DNA after mitosis to yield a “cut” phenotype (132).
In budding yeast, late origins did not fire when cells were released from G1 into S phase in the presence of HU (235). In mec1 or rad53 checkpoint mutants, replication occurs at the late origins. The rationale is that HU lowers the level of dNTPs to slow replication, which activates the checkpoint. Obviously, there still must be enough dNTPs in HU for replication as it does occur at the late origins if the checkpoint is mutated. In fact, a considerable amount of replication occurs in HU, producing ssDNA birectionally from origins, which has been used to map origins in budding and fission yeast using microarray technology (73). With rad53 checkpoint mutants in HU, replication forks stall at the origins (73) because the replisome dissociates and forks are reversed, producing a “chicken-foot” morphology (166). Therefore, the checkpoint response is important for stabilization and restarting of stalled replication forks (27).
How does the checkpoint response prevent firing at late origins? In yeast, frog, and human systems, ATM/ATR (Mec1/Tel1), Chk1, and Chk2 (Rad53) kinases are important (44, 235, 248, 268). In fact, all origins in these studies fire earlier even under normal conditions. The hypothesis is that ssDNA generated during normal replication activates ATR/ATM pathway to inhibit initiation at late origins. The inhibition occurs by preventing CDK- and DDK-dependent Cdc45 protein loading, thus blocking helicase activation and replisome recruitment (8, 248, 268).
The role of DDK in the checkpoint response is controversial in that different results have been obtained even in the same experimental system (13, 63, 68, 122, 220, 243). In frog egg extracts, DNA damage by etoposide, which produces ssDNA breaks, inhibits DDK by dissociating the Cdc7-Dbf4 complex (44). Similar results were obtained in human BCR-ABL transformed cells (60). Dbf4 protein also becomes phosphorylated by Rad53 (Chk2) kinase in response to replication arrest in both yeast species (257, 302). In this scenario, DDK would be inhibited when the checkpoint is activated.
However, the major form of DDK in frog egg extracts is Cdc7-Drf1 and not Cdc7-Dbf4 (270), and Cdc7-Drf1 is not down-regulated in response to dsDNA breaks or replication inhibition with aphidicolin, a polymerase inhibitor (220, 314). In a number of human cell lines including the same BCR-ABL transformed cells used above, both Cdc7-Dbf4 and Cdc7-Drf1 DDK complexes were unaffected by either replication blocks or DNA-damaging agents (278). Assays of DDK were increased in HU-arrested budding yeast cells (210), but reduced in another study (302). It is difficult to compare these two studies as the former used Mcm2 and the latter used Mcm7 as a DDK substrate. Yet phosphorylation of known DDK physiological substrates Mcm2 and Mcm4 is unaffected by replication inhibitors in human (186, 278) and in budding yeast cells in vivo (157, 251).
DDK may also be required for the checkpoint, which is difficult to reconcile with inhibiting it during the checkpoint response, as discussed above. In fission yeast, DDK (Hsk1) is required for the checkpoint response (68, 274). However, in cdc7Δ mcm5-bob1 budding yeast cells in which DDK is deleted and bypassed by the mcm5-bob1 mutation, both DNA damage (219) and replication (302) checkpoints are intact and Rad53 (Chk2) kinase becomes activated (63, 219). In fact, cdc7Δ mcm5-bob1 budding yeast cells are sensitive to DNA-damaging agents because they have a defect in the Rev3-Rev7 (DNA polymerase)-dependent translesion synthesis subpath-way (219) of the Rad6 epistasis group that also affects mutagenesis (84). As expected, cdc7 mutants are also defective in both UV-induced and chemical mutagenesis (243). In addition, CDK but not DDK is needed to activate the checkpoint during dsDNA break repair by homologous recombination in budding yeast (114). Thus, it is possible that DDK may be required for some aspects of the checkpoint in some organisms, but clearly not in budding yeast. DDK is also not required for maintaining the checkpoint (280).
A possible explanation for some of these conflicting results is that DDK may be needed for replication restart after the checkpoint response has decayed (68). This is based on finding that dbf4 (dfp1) mutants of fission yeast are sensitive to damage by MMS, yet retain the intra-S phase checkpoint (86). This is consistent with the fact that cdc7Δ mcm5-bob1 mutants are sensitive to HU because they may not be able to adapt or recover from HU and restart replication (302). In checkpoint adaptation, cells continue in the cell cycle even in the presence of DNA damage (233). Adaptation is regulated by the budding yeast Cdc5 protein, a polo-like protein kinase (91), which also has a role in the initiation of DNA replication and interacts with DDK (102). This effect on adaptation may also explain the Rev3-Rev7 epistatic relationship (219) if one assumes that DDK is needed to restart replication with DNA polymeraseζ. Furthermore, inhibition of late replication to slow S phase during replication stress may not be as important for viability as replication restart, thus obviating the need to inhibit DDK (68). In support of this idea, the mec1–100 hypomorphic mutant of budding yeast is defective in late origin firing but is still resistant to DNA damage by stabilizing replication forks (212, 280).
The Mcm2–7 helicase may be a target of the checkpoint response (249). Budding yeast Mcm2–7 complex is important to establish but not maintain the checkpoint, but pre-RCs do not play a direct role in checkpoint control during chromosome replication (148). Replication forks are also important for checkpoint response (280). In human cells, Mcm2 and Mcm3 are phosphorylated by the ATM/ATR kinases during checkpoint activation, and lowered Mcm7 levels inhibit the intra-S-phase checkpoint (43). In frog egg extracts, if DNA polymerase progression is inhibited by aphidicolin or by impassible DNA lesions in the template, the Mcm2–7 helicase still unwinds and produces ssDNA, which is coated with the RPA and activates the checkpoint as a sensor (Figure 6) (32). This may explain why reduced Mcm7 inhibits the checkpoint (43).
Other replication proteins important for the checkpoint include Mrc1 (Claspin), Dbp11, PCNA, RFC, and DNA polymeraseε. Mrc1 acts as a mediator for amplifying the Rad53 and Chk1 signal when replication is blocked in both yeast species (3) and in frog it is called Claspin (152). Mrc1 is part of the replisome and monitors the progress of replication fork (152, 209) and interacts with checkpoint proteins Tof1and Csm3 when forks are arrested (129). Dpb11 and its loader Sld2 (Drc1) are also important for the replication checkpoint and activating Rad53 (Chk2). (152). In frog, the N terminus of xRTS protein is similar to Sld2 and also loads the replisome (234). However, the remaining nonconserved C-terminal domain of xRTS is a RecQ helicase family member (234), which is an orthologue of the gene mutated in human Rothmund-Thomson syndrome, a familial degenerative skin disorder with increased susceptibility for bone cancer (150). DNA polymerase ε has an unknown function in the checkpoint and in DNA replication because its polymerase activity is not required for DNA replication, but the C terminus is needed for the checkpoint and for viability (67, 74, 137).
PCNA and RFC are clamps and clamp loader, respectively (52) (see above). PCNA is a trimer of three identical subunits, whereas RFC is a pentamer with five different subunits, Rfc1–5. In replication, PCNA is the processivity factor that clamps the replicative DNA polymerases δ and ε on the DNA. When DNA replication is blocked, alternative clamps or clamp loaders are used. The hypothesis is that alternative clamps are loaded during DNA damage to recruit DNA repair proteins or to activate the checkpoint or for other responses such as chromosome cohesion (173). With the clamp loader, the Rfc1 subunit is replaced by another protein such as Rad17 to form a Rad17-Rfc2–5 complex during DNA repair (18, 70). This alternative clamp would load on an alternative clamp such as the 9-1-1 complex instead of PCNA. 9-1-1 is composed of three different subunits called Rad9, Rad1, and Hus1 from fission yeast and is conserved in other eukaryotes. There are at least four different clamp loaders and two different clamps. In some cases, PCNA is used as the clamp but is modified by mono-ubiquitination (110, 288) to load on DNA polymerase η for translesion synthesis during polymerase switching (126). Ubiquitination is catalyzed by the Rad6-Rad18 E2:E3 conjugating:ligase complex (110), which explains why translesion synthesis by polymerase η (RAD30) in yeast is in the RAD6 epistasis group (see above) (84). The combination of different clamps and clamp loaders could produce a variety of molecules on the DNA to elicit a number of different cellular responses (173). Indeed, the clamp and clamp loader analogy can explain many of the interactions of proteins with long molecules such as DNA.
Some checkpoint proteins also have positive roles in DNA replication such as Mrc1, Dpb11, PCNA, and Drc1 (discussed above). Yeast mrc1Δ mutants have a reduced rate of replication (209). The replication and checkpoint functions of Mrc1 are separable in that only the checkpoint response is affected if the Mec1 phosphorylation sites in Mrc1 are mutated.
Similar results are found with Rad53 protein kinase (63, 101, 323). In budding yeast, RAD53 or MEC1 are essential genes that can be rescued by overexpression of RNR1, or by down-regulation of the Sml1 protein, which is an inhibitor of Rnr1 (59, 232, 323–325). However, these mutants are still checkpoint defective. Yet rad53Δ sml1Δ strains grow slower than mec1Δ sml1Δ strains (101, 323). Similarly, only the growth defect of the rad53Δ sml1Δ strains can be suppressed by deletion of the HHT2 HHF2 genes, the major histone H3/H4 genes (101). Therefore, Rad53 has a role in DNA replication that does not involve the checkpoint function and may involve regulation of histone levels during the S phase (63, 101). In support of this idea, Rad53 protein binds to origins (63, 129) and interacts with two important replication genes, CDC7 and MCM5 (Table 1) (62, 63). Again, the same is not true of mec1 mutants or mutations of any other checkpoint genes. Rad53 protein kinase is likely involved in the initiation of DNA replication as rad53 mutants display reduced origin firing using 2D-gel analysis (166) or the Mcm assay (63). Rad53 kinase also may regulate origin firing based upon growth conditions to optimize the rate of DNA replication (255). SLD6 was found to be allelic with RAD53 (124), indicating a possible interaction with DPB11, which is important in replisome loading (Figure 1b) (see above).
Regions of the genome such as the yeast rDNA repeats that are difficult to replicate because of compact chromatin structure and high levels of transcription in the opposite direction to the replication fork require Rrm3, a DNA helicase (117, 282). The Tof1 and Csm3 checkpoint proteins regulate the process by inhibiting Rrm3 and produce a pause site at the Ter termination site in the rDNA (184). Surprisingly, Mrc1 protein is not needed in this case. Similar results are seen at replication “slow zones” in mec1 and rad53 mutants (34). Thus, the checkpoint proteins can help regulate replication through difficult genomic regions.
SUMMARY POINTS.
The mechanism of DNA replication has been conserved in evolution. Because replication proteins for initiation have been conserved between eukaryotic and archaeal organisms, archaeal proteins are being used as simpler, structural models.
Most eukaryotes except for budding yeast have ill-defined origins of replication that rely on epigenetic mechanisms for molecular recognition by initiator proteins. In this regard, budding yeast may have become streamlined during evolution in both origin and centromere recognition.
Replication is initiated at multiple origins along the DNA using a conserved mechanism that consists of four steps: origin recognition, assembly of a prereplicative initiation complex, followed by activation of the helicase and loading of the replisome.
Cell cycle regulation of DNA replication occurs through protein phosphorylation by two protein kinases, CDK and DDK. Both kinases are needed for helicase activation and for loading of the replisome in S phase. CDK also inhibits initiation by preventing pre-RC assembly during S phase. Thus, pre-RC formation can only occur in G1 phase and helicase activation can only occur in S phase. In Boolean terms, there is never a phase in which both events are true as this would result in rereplication due to reloading and subsequent activation.
The same proteins are used in premeiotic replication that occurs during gamete production, but in reduced activity. DDK has an additional function after DNA replication but before DSB formation during the meiotic cell cycle.
FUTURE ISSUES.
Most of the progress in this field has been made by combining the knowledge gained from yeast genetic in vivo studies with in vitro studies in the frog. These studies have identified the important regulatory molecules and their functions. Important contributions have also been made from genetic studies in Drosophila and mice and from human cultured cells using siRNA approaches. These studies have revealed more complex regulation in metazoan cells. Structural insights have been made by solving atomic structures of simpler, archaeal proteins that are easier to produce and are more stable at ambient temperatures (Figure 2). A working model of DNA replication under cell cycle control has resulted (Figure 1b). However, a fully reconstituted in vitro replication system in which both initiation and elongation occur with all purified components is still lacking. Given all the tools of modern, recombinant DNA technology, this issue should be resolved in the future.
In most eukaryotes, origin recognition is still a thorny issue. We do not know the type of chromatin or DNA structure the ORC recognizes. The fact that it is not sequence dependent makes this hard to analyze. Resolution will require higher-order chromatin structure analysis. Another issue is the importance of the temporal program. What is the physiological significance of the fact that origins are activated at different times during the S phase? The connection with gene expression is unclear (203). In fission yeast, it is random every generation (227). Perhaps, late origins exist to be a “fail-safe” mechanism ensuring complete DNA replication before mitosis. In this model, late origins would only be used if that part of the replicon was still unreplicated at the end of S phase. Even this model may be doubtful as a recent report suggests that yeast cells do not know if normal replication is actually completed before entering mitosis (283, 298).
The role of protein phosphorylation is becoming clear (Figure 1b), even though there is a fair amount of redundancy in the process. Both CDK and DDK are activated in late G1 phase using different regulatory subunits. Yet they regulate the same process. This ensures that replication cannot occur in a haphazard manner, as it requires two independent events. Before the coordination of growth and division was optimized during evolution, only DDK may have been used (191). Then CDK evolved to ensure coordination and to prevent rereplication. DDK is rate limiting for initiation in both yeasts (25, 64; N. Rhind, personal communication). Future studies will focus on defining how phosphorylation of important serine and threonine residues in substrates by the two kinases produces the biochemical and structural changes needed to ensure function.
Checkpoint regulation is very important in cancer progression by regulating genome stability (128). It is clear that DNA replication is at the heart of checkpoint regulation (Figure 6). Replication proteins are important regulators of the checkpoint and replication itself is needed to generate the signals needed. Tumor suppressors such as p53, BRCA1/2 (Breast Cancer), BLM (Bloom’s syndrome), and FANC (Fanconi’s anemia) genes are all important players in the response (135). Knowledge of DNA replication is important for cancer therapy also in that most modalities either block or damage DNA replication. Future direction will be on designing drugs that intercept the checkpoint. For example, resveratrol, which is a natural compound found in red wine and used in chemoprevention, activates the checkpoint only in cancer cells and not in normal cells (286).
Although phosphorylation by CDK and DDK is also needed for DNA replication in the meiotic cell cycle, their exact roles are unknown. In addition, both kinases have roles after DNA replication. Only identification of the physiological substrates that are phosphorylated by these kinases during meiosis will reveal the molecular nature of these roles.
Acknowledgments
Supported by PHS grant to R. A. Sclafani (GM35078). Thanks to Drs. Xiaojiang Chen, Rui Zhao, and Bruce Stillman for permission to use published figures, and to Dr. Nicholas Rhind for communication of his unpublished work.
- MCM
mini-chromosome maintenance
- pre-RC
prereplication complex
- CDK
cyclin-dependent kinase
- DDK
Dbf4-dependent kinase
- Licensing
process by which pre-RC has been assembled on origins
- Replicon
DNA replicated from a single origin
- RF
replication factor (A or C)
- PCNA
proliferating cell nuclear antigen
- ARS
autonomously replicating sequence
- ACS
ARS consensus sequence
- Checkpoint
process in which cells monitor the completion of important events such as DNA replication and mitosis
- POL
DNA polymerase
- AAA+
ATPases associated with various cellular activities
- CDC
cell division cycle
- Replisome
replication complex that polymerizes or copies the DNA
- Origin
site on the chromosome at which DNA replication begins
- Helicase
enzyme that converts dsDNA into ssDNA
- ORC
origin recognition complex
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Adachi Y, Usukura J, Yanagida M. A globular complex formation by Nda1 and the other five members of the MCM protein family in fission yeast. Genes Cells. 1997;2:467–79. doi: 10.1046/j.1365-2443.1997.1350333.x. [DOI] [PubMed] [Google Scholar]
- 2.Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–76. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- 3.Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, et al. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol. 2001;3:958–3. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- 4.Aleem E, Kiyokawa H, Kaldis P. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat Cell Biol. 2005;7:831–7. doi: 10.1038/ncb1284. [DOI] [PubMed] [Google Scholar]
- 5.Alexandrov AI, Botchan MR, Cozzarelli NR. Characterization of simian virus 40 T-antigen double hexamers bound to a replication fork. The active form of the helicase. J Biol Chem. 2002;277:44886–277. doi: 10.1074/jbc.M207022200. [DOI] [PubMed] [Google Scholar]
- 6.Alfano C, McMacken R. Heat shock protein-mediated disassembly of nucleoprotein structures is required for the initiation of bacteriophage λ DNA replication. J Biol Chem. 1989;264:10709–264. [PubMed] [Google Scholar]
- 7.Alfano C, McMacken R. Ordered assembly of nucleoprotein structures at the bacteriophage lambda replication origin during the initiation o DNA replication. J Biol Chem. 1989;264:10699–264. [PubMed] [Google Scholar]
- 8.Aparicio OM, Stout AM, Bell SP. Differential assembly of cdc45p and DNA polymerases at early and late origins of DNA replication. Proc Natl Acad Sci USA. 1999;96:9130–35. doi: 10.1073/pnas.96.16.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during the S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 10.Araki H, Leem SH, Phongdara A, Sugino A. Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci USA. 1995;92:11791–95. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbaro BA, Sreekumar KR, Winters DR, Prack AE, Bullock PA. Phosphorylation of simian virus 40 T antigen on Thr 124 selectively promotes double-hexamer formation on subfragments of the viral core origin. J Virol. 2000;74:8601–74. doi: 10.1128/jvi.74.18.8601-8613.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barry ER, Bell SD. DNA replication in the archaea. Microbiol Mol Biol Rev. 2006;70:876–70. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–71. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 14.Deleted in proof
- 15.Bell SP, Stillman B. Nucleotide dependent recognition of chromosomal origins by a multi-protein complex. Nature. 1992;357:128–34. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 16.Benjamin KR, Zhang C, Shokat KM, Herskowitz I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 2003;17:1524–17. doi: 10.1101/gad.1101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett MD, Smith JB. The effects of polyploidy on the meiotic duration and pollen development in cereal anthers. Proc R Soc London Ser B. 1972;181:81–107. [Google Scholar]
- 18.Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, et al. Loading of the human 9–1–1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc Natl Acad Sci USA. 2003;100:1633–38. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanton HL, Radford SJ, McMahan S, Kearney HM, Ibrahim JG, Sekelsky J. REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS Genet. 2005;1:e40. doi: 10.1371/journal.pgen.0010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blow JJ, Hodgson B. Replication licensing–defining the proliferative state? Trends Cell Biol. 2002;12:72–78. doi: 10.1016/s0962-8924(01)02203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blow JJ, Laskey RA. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988;332:546–48. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- 22.Bolon YT, Bielinsky AK. The spatial arrangement of ORC binding modules determines the functionality of replication origins in budding yeast. Nucleic Acids Res. 2006;34:5069–34. doi: 10.1093/nar/gkl661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borde V, Goldman AS, Lichten M. Direct coupling between meiotic DNA replication and recombination initiation. Science. 2000;290:806–9. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- 24.Botchan M. Cell biology: a switch for S phase. Nature. 2007;445:272–74. doi: 10.1038/445272a. [DOI] [PubMed] [Google Scholar]
- 25.Bousset K, Diffley JFX. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12:480–12. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol Cell. 2004;16:967–78. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 27.Branzei D, Foiani M. The Rad53 signal transduction pathway: replication fork stabilization, DNA repair, and adaptation. Exp Cell Res. 2006;312:2654–312. doi: 10.1016/j.yexcr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 28.Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–71. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 29.Brown GW, Kelly TJ. Purification of Hsk1, a minichromosome maintenance protein kinase from fission yeast. J Biol Chem. 1998;273:22083–273. doi: 10.1074/jbc.273.34.22083. [DOI] [PubMed] [Google Scholar]
- 30.Brown SW. Heterochromatin. Science. 1966;151:417–25. doi: 10.1126/science.151.3709.417. [DOI] [PubMed] [Google Scholar]
- 31.Bueno A, Russell P. Dual functions of CDC6: a yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 1992;11:2167–11. doi: 10.1002/j.1460-2075.1992.tb05276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–19. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Callan HG. DNA replication in the chromosomes of eukaryotes. Cold Spring Harbor Symp Quant Biol. 1974;38:195–38. doi: 10.1101/sqb.1974.038.01.023. [DOI] [PubMed] [Google Scholar]
- 34.Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–6. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- 35.Cha RS, Weiner BM, Keeney S, Dekker J, Kleckner N. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 2000;14:493–14. [PMC free article] [PubMed] [Google Scholar]
- 36.Chen YJ, Yu X, Kasiviswanathan R, Shin JH, Kelman Z, Egelman EH. Structural polymorphism of Methanothermobacter thermautotrophicus MCM. J Mol Biol. 2005;346:389–94. doi: 10.1016/j.jmb.2004.11.076. [DOI] [PubMed] [Google Scholar]
- 37.Cho WH, Lee YJ, Kong SI, Hurwitz J, Lee JK. CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proc Natl Acad Sci USA. 2006;103:11521–26. doi: 10.1073/pnas.0604990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chong JP, Hayashi MK, Simon MN, Xu RM, Stillman B. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc Natl Acad Sci USA. 2000;97:1530–35. doi: 10.1073/pnas.030539597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chuang RY, Kelly TJ. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci USA. 1999;96:2656–61. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–21. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 41.Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JFX. An essential role of the Cdc6 protein in forming the prereplicative complexes of budding yeast. Nature. 1995;379:180–82. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 42.Collins I, Newlon CS. Chromosomal DNA replication initiates at the same origins in meiosis and mitosis. Mol Cell Biol. 1994;14:3524–14. doi: 10.1128/mcb.14.5.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cortez D, Glick G, Elledge SJ. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc Natl Acad Sci USA. 2004;101:10078–83. doi: 10.1073/pnas.0403410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell. 2003;11:203–13. doi: 10.1016/s1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 45.Coverley D, Laman H, Laskey RA. Distinct roles for cyclins E and A during DNA replication complex assembly and activation. Nat Cell Biol. 2002;4:523–4. doi: 10.1038/ncb813. [DOI] [PubMed] [Google Scholar]
- 46.Coverley D, Wilkinson HR, Madine MA, Mills AD, Laskey RA. Protein kinase inhibition in G2 causes mammalian Mcm proteins to reassociate with chromatin and restores ability to replicate. Exp Cell Res. 1998;238:63–238. doi: 10.1006/excr.1997.3829. [DOI] [PubMed] [Google Scholar]
- 47.Crevel G, Ivetic A, Ohno K, Yamaguchi M, Cotterill S. Nearest neighbour analysis of MCM protein complexes in Drosophila melanogaster. Nucleic Acids Res. 2001;29:4834–42. doi: 10.1093/nar/29.23.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cross FR, Maria Yuste-Rojas M, Gray S, Jacobson MD. Specialization and targeting of B-type cyclins. Mol Cell. 1999;4:11–19. doi: 10.1016/s1097-2765(00)80183-5. [DOI] [PubMed] [Google Scholar]
- 49.Dai J, Chuang RY, Kelly TJ. DNA replication origins in the Schizosaccharomyces pombe genome. Proc Natl Acad Sci USA. 2005;102:337–42. doi: 10.1073/pnas.0408811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Danis E, Brodolin K, Menut S, Maiorano D, Girard-Reydet C, Mechali M. Specification of a DNA replication origin by a transcription complex. Nat Cell Biol. 2004;6:721–6. doi: 10.1038/ncb1149. [DOI] [PubMed] [Google Scholar]
- 51.Davey MJ, Indiani C, O’Donnell M. Reconstitution of the mcm2-7p heterohexamer, subunit arrangement, and ATP site architecture. J Biol Chem. 2003;278:4491–278. doi: 10.1074/jbc.M210511200. [DOI] [PubMed] [Google Scholar]
- 52.Davey MJ, Jeruzalmi D, Kuriyan J, O’Donnell M. Motors and switches: AAA+ machines within the replisome. Nat Rev Mol Cell Biol. 2002;3:826–35. doi: 10.1038/nrm949. [DOI] [PubMed] [Google Scholar]
- 53.Davis L, Barbera M, McDonnell A, McIntyre K, Sternglanz R, et al. The Sac-charomyces cerevisiae MUM2 gene interacts with the DNA replication machinery and is required for meiotic levels of double strand breaks. Genetics. 2001;157:1179–89. doi: 10.1093/genetics/157.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dazy S, Gandrillon O, Hyrien O, Prioleau MN. Broadening of DNA replication origin usage during metazoan cell differentiation. EMBO Rep. 2006;7:806–7. doi: 10.1038/sj.embor.7400736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delmolino LM, Saha P, Dutta A. Multiple mechanisms regulate subcellular localization of human CDC6. J Biol Chem. 2001;276:26947–276. doi: 10.1074/jbc.M101870200. [DOI] [PubMed] [Google Scholar]
- 56.DeLuca JG, Salmon ED. Kinetochores: If you build it, they will come. Curr Biol. 2004;14:R921–23. doi: 10.1016/j.cub.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 57.DePamphilis ML. The ‘ORC cycle’: a novel pathway for regulating eukaryotic DNA replication. Gene. 2003;310:1–15. doi: 10.1016/s0378-1119(03)00546-8. [DOI] [PubMed] [Google Scholar]
- 58.DePamphilis ML. DNA Replication and Human Disease. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 2006. p. 814. [Google Scholar]
- 59.Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–12. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dierov J, Dierova R, Carroll M. BCR/ABL translocates to the nucleus and disrupts an ATR-dependent intra-S phase checkpoint. Cancer Cell. 2004;5:275–85. doi: 10.1016/s1535-6108(04)00056-x. [DOI] [PubMed] [Google Scholar]
- 61.Dijkwel PA, Wang S, Hamlin JL. Initiation sites are distributed at frequent intervals in the Chinese hamster dihydrofolate reductase origin of replication but are used with very different efficiencies. Mol Cell Biol. 2002;22:3053–22. doi: 10.1128/MCB.22.9.3053-3065.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dohrmann PR, Oshiro G, Tecklenburg M, Sclafani RA. RAD53 regulates DBF4 independently of checkpoint function in Saccharomyces cerevisiae. Genetics. 1999;151:965–77. doi: 10.1093/genetics/151.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dohrmann PR, Sclafani RA. Novel role for checkpoint Rad53 protein kinase in the initiation of chromosomal DNA replication in Saccharomyces cerevisiae. Genetics. 2006;174:87–99. doi: 10.1534/genetics.106.060236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Donaldson AD, Fangman WL, Brewer B. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev. 1998;12:491–12. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Donovan S, Harwood J, Drury LS, Diffley JFX. Cdc6p-dependent loading of Mcm proteins onto prereplicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–16. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drury LS, Perkins G, Diffley JF. The cyclin-dependent kinase cdc28p regulates distinct modes of cdc6p proteolysis during the budding yeast cell cycle. Curr Biol. 2000;10:231–10. doi: 10.1016/s0960-9822(00)00355-9. [DOI] [PubMed] [Google Scholar]
- 67.Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274:22283–88. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- 68.Duncker BP, Brown GW. Cdc7 kinases (DDKs) and checkpoint responses: lessons from two yeasts. Mutat Res. 2003;532:21–532. doi: 10.1016/j.mrfmmm.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 69.Edwards MC, Tutter AV, Cvetic C, Gilbert CH, Prokhorova TA, Walter JC. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J Biol Chem. 2002;277:33049–277. doi: 10.1074/jbc.M204438200. [DOI] [PubMed] [Google Scholar]
- 70.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–75. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 72.Engebrecht J, Masse S, Davis L, Rose K, Kessel T. Yeast meiotic mutants proficient for the induction of ectopic recombination. Genetics. 1998;148:581–98. doi: 10.1093/genetics/148.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Feng W, Collingwood D, Boeck ME, Fox LA, Alvino GM, et al. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat Cell Biol. 2006;8:148–8. doi: 10.1038/ncb1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng W, D’Urso G. Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control. Mol Cell Biol. 2001;21:4495–504. doi: 10.1128/MCB.21.14.4495-4504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fien K, Hurwitz J. Fission yeast Mcm10p contains primase activity. J Biol Chem. 2006;281:22248–281. doi: 10.1074/jbc.M512997200. [DOI] [PubMed] [Google Scholar]
- 76.Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS. The structure and function of MCM from archaeal M. thermoautotrophicum. Nat Struct Biol. 2003;10:160–67. doi: 10.1038/nsb893. The MCM structure is based on the atomic archaeal Mth-MCM structure. [DOI] [PubMed] [Google Scholar]
- 77.Fletcher RJ, Chen XS. Biochemical activities of the BOB1 mutant in Methanobacterium thermoautotrophicum MCM. Biochemistry. 2006;45:462–67. doi: 10.1021/bi051754z. [DOI] [PubMed] [Google Scholar]
- 78.Fletcher RJ, Shen J, Gomez-Llorente Y, Martin CS, Carazo JM, Chen XS. Double hexamer disruption and biochemical activities of Methanobacterium thermoautotrophicum MCM. J Biol Chem. 2005;280:42405–10. doi: 10.1074/jbc.M509773200. [DOI] [PubMed] [Google Scholar]
- 79.Forsburg SL. The art and design of genetic screens: yeast. Nat Rev Genet. 2001;2:659–2. doi: 10.1038/35088500. [DOI] [PubMed] [Google Scholar]
- 80.Forsburg SL. Only connect: linking meiotic DNA replication to chromosome dynamics. Mol Cell. 2002;9:703–11. doi: 10.1016/s1097-2765(02)00508-7. [DOI] [PubMed] [Google Scholar]
- 81.Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev. 2004;68:109–68. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Forsburg SL, Hodson JA. Mitotic replication initiation proteins are not required for premeiotic S phase. Nat Genet. 2000;25:263–25. doi: 10.1038/77015. [DOI] [PubMed] [Google Scholar]
- 83.Forterre P. Displacement of cellular proteins by functional analogues from plasmids or viruses could explain puzzling phylogenies of many DNA informational proteins. Mol Microbiol. 1999;33:457–33. doi: 10.1046/j.1365-2958.1999.01497.x. [DOI] [PubMed] [Google Scholar]
- 84.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006. p. 1118. [Google Scholar]
- 85.Friedman KL, Diller JD, Ferguson BM, Nyland SV, Brewer BJ, Fangman WL. Multiple determinants controlling activation of yeast replication origins late in S phase. Genes Dev. 1996;10:1595–10. doi: 10.1101/gad.10.13.1595. [DOI] [PubMed] [Google Scholar]
- 86.Fung AD, Ou J, Bueler S, Brown GW. A conserved domain of Schizosaccharomyces pombe dfp1+ is uniquely required for chromosome stability following alkylation damage during S phase. Mol Cell Biol. 2002;22:4477–90. doi: 10.1128/MCB.22.13.4477-4490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Funnell BE, Baker TA, Kornberg A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J Biol Chem. 1987;262:10327–34. [PubMed] [Google Scholar]
- 88.Furth ME, McLeester C, Dove WF. Specificity determinants for bacteriophage lambda DNA replication. I. A chain of interactions that controls the initiation of replication. J Mol Biol. 1978;126:195–126. doi: 10.1016/0022-2836(78)90359-5. [DOI] [PubMed] [Google Scholar]
- 89.Gaczynska M, Osmulski PA, Jiang Y, Lee JK, Bermudez V, Hurwitz J. Atomic force microscopic analysis of the binding of the Schizosaccharomyces pombe origin recognition complex and the spOrc4 protein with origin DNA. Proc Natl Acad Sci USA. 2004;101:17952–57. doi: 10.1073/pnas.0408369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell. 2004;119:47–60. doi: 10.1016/j.cell.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 91.Galgoczy DJ, Toczyski DP. Checkpoint adaptation precedes spontaneous and damage-induced genomic instability in yeast. Mol Cell Biol. 2001;21:1710–21. doi: 10.1128/MCB.21.5.1710-1718.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Geraghty DS, Ding M, Heintz NH, Pederson DS. Premature structural changes at replication origins in a yeast minichromosome maintenance (MCM) mutant. J Biol Chem. 2000;275:18011–275. doi: 10.1074/jbc.M909787199. [DOI] [PubMed] [Google Scholar]
- 93.Gilbert DM. In search of the holy replicator. Nat Rev Mol Cell Biol. 2004;5:848–5. doi: 10.1038/nrm1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Giordano-Coltart J, Ying CY, Gautier J, Hurwitz J. Studies of the properties of human origin recognition complex and its Walker A motif mutants. Proc Natl Acad Sci USA. 2005;102:69–74. doi: 10.1073/pnas.0408690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Goldman MA, Holmquist GP, Gray MC, Caston LA, Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984;224:686–92. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- 96.Gomez-Llorente Y, Fletcher RJ, Chen XS, Carazo JM, San Martin C. Polymorphism and double hexamer structure in the archaeal minichromosome maintenance (MCM) helicase from Methanobacterium thermoautotrophicum. J Biol Chem. 2005;280:40909–15. doi: 10.1074/jbc.M509760200. [DOI] [PubMed] [Google Scholar]
- 97.Gonzalez MA, Tachibana KE, Adams DJ, vander Weyden L, Hemberger M, et al. Geminin is essential to prevent endoreduplication and to form pluripotent cells during mammalian development. Genes Dev. 2006;20:1880–20. doi: 10.1101/gad.379706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gozuacik D, Chami M, Lagorce D, Faivre J, Murakami Y, et al. Identification and functional characterization of a new member of the human Mcm protein family: hMcm8. Nucleic Acids Res. 2003;31:570–31. doi: 10.1093/nar/gkg136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Green BM, Morreale RJ, Ozaydin B, Derisi JL, Li JJ. Genome-wide mapping of DNA synthesis in Saccharomyces cerevisiae reveals that mechanisms preventing reinitiation of DNA replication are not redundant. Mol Biol Cell. 2006;17:2401–14. doi: 10.1091/mbc.E05-11-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gregoire D, Brodolin K, Mechali M. HoxB domain induction silences DNA replication origins in the locus and specifies a single origin at its boundary. EMBO Rep. 2006;7:812–7. doi: 10.1038/sj.embor.7400758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–49. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 102.Hardy CF, Pautz A. A novel role for Cdc5p in DNA replication. Mol Cell Biol. 1996;16:6775–16. doi: 10.1128/mcb.16.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hardy CFJ, Dryga O, Seematter S, Pahl PMB, Sclafani RA. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci USA. 1997;94:3151–55. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Harland RM, Laskey RA. Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell. 1980;21:761–71. doi: 10.1016/0092-8674(80)90439-0. [DOI] [PubMed] [Google Scholar]
- 105.Hartwell LH. Genetic control of the cell division cycle in yeast II. Genes controlling DNA replication and its initiation. J Mol Biol. 1971;59:183–59. doi: 10.1016/0022-2836(71)90420-7. [DOI] [PubMed] [Google Scholar]
- 106.Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–34. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 107.Henderson KA, Kee K, Maleki S, Santini PA, Keeney S. Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell. 2006;125:1321–32. doi: 10.1016/j.cell.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hennessy KM, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991;5:958–5. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- 109.Herrick J, Jun S, Bechhoefer J, Bensimon A. Kinetic model of DNA replication in eukaryotic organisms. J Mol Biol. 2002;320:741–320. doi: 10.1016/s0022-2836(02)00522-3. [DOI] [PubMed] [Google Scholar]
- 110.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–41. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 111.Hollingsworth REJ, Sclafani RA. Yeast premeiotic DNA replication utilizes mitotic origin ARS1 independent of CDC7 function. Chromosoma. 1993;102:415–20. doi: 10.1007/BF00360406. [DOI] [PubMed] [Google Scholar]
- 112.Holm PB. The premeotic DNA replication of euchromatin and heterochromatin in Lilium longiflorum (Thunb) Carlsberg Res Commun. 1977;42:249–81. [Google Scholar]
- 113.Hyrien O, Marheineke K, Goldar A. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays. 2003;25:116–25. doi: 10.1002/bies.10208. [DOI] [PubMed] [Google Scholar]
- 114.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–17. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ishimi Y. A DNA helicase is associated with an Mcm4, -6, -7 protein complex. J Biol Chem. 1997;272:24508–272. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 116.Ishimi Y, Ichinose S, Omori A, Sato K, Kimura H. Binding of human minichromosome maintenance proteins with histone H3. J Biol Chem. 1996;271:24115–271. doi: 10.1074/jbc.271.39.24115. [DOI] [PubMed] [Google Scholar]
- 117.Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell. 2003;12:1525–36. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 118.Izumi M, Yanagi K, Mizuno T, Yokoi M, Kawasaki Y, et al. The human homolog of Saccharomyces cerevisiae Mcm10 interacts with replication factors and dissociates from nuclease-resistant nuclear structures in G(2) phase. Nucleic Acids Res. 2000;28:4769–77. doi: 10.1093/nar/28.23.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jacob F, Brenner S. On the regulation of DNA synthesis in bacteria: the hypothesis of the replicon. C R Seances Acad Sci Ser D. 1963;256:298–300. [PubMed] [Google Scholar]
- 120.Jallapelli PV, Brown GW, Muzi-Falconi M, Tien D, Kelly TJ. Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev. 1997;11:2767–79. doi: 10.1101/gad.11.21.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jares P, Blow JJ. Xenopus Cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev. 2000;14:1528–14. [PMC free article] [PubMed] [Google Scholar]
- 122.Jares P, Donaldson A, Blow JJ. The Cdc7/Dbf4 protein kinase: target of the S phase checkpoint? EMBO Rep. 2000;1:319–22. doi: 10.1093/embo-reports/kvd076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Johnson EM, Kinoshita Y, Daniel DC. A new member of the MCM protein family encoded by the human MCM8 gene, located contrapodal to GCD10 at chromosome band 20p12.3-13. Nucleic Acids Res. 2003;31:2915–31. doi: 10.1093/nar/gkg395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kamimura Y, Tak YS, Sugino A, Araki H. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 2001;20:2097–107. doi: 10.1093/emboj/20.8.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature. 2003;423:720–24. doi: 10.1038/nature01692. [DOI] [PubMed] [Google Scholar]
- 126.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 127.Kaplan DL, Davey MJ, O’Donnell M. Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J Biol Chem. 2003;278:49171–278. doi: 10.1074/jbc.M308074200. [DOI] [PubMed] [Google Scholar]
- 128.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–23. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 129.Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–83. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 130.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–84. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 131.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–24. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 132.Kelly TJ, Brown GW. Regulation of chromosome replication. Annu Rev Biochem. 2000;69:829–69. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- 133.Kelman Z, Hurwitz J. Structural lessons in DNA replication from the third domain of life. Nat Struct Biol. 2003;10:148–10. doi: 10.1038/nsb0303-148. [DOI] [PubMed] [Google Scholar]
- 134.Kelman Z, Lee JK, Hurwitz J. The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum DeltaH contains DNA helicase activity. Proc Natl Acad Sci USA. 1999;96:14783–88. doi: 10.1073/pnas.96.26.14783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kennedy RD, D’Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19:2925–19. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 136.Kerns SL, Torke SJ, Benjamin JM, McGarry TJ. Geminin prevents rereplication during Xenopus development. J Biol Chem. 2007;282:5514–282. doi: 10.1074/jbc.M609289200. [DOI] [PubMed] [Google Scholar]
- 137.Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3:679–85. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- 138.Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, et al. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- 139.Kneissl M, Putter V, Szalay AA, Grummt F. Interaction and assembly of murine prereplicative complex proteins in yeast and mouse cells. J Mol Biol. 2003;327:111–327. doi: 10.1016/s0022-2836(03)00079-2. [DOI] [PubMed] [Google Scholar]
- 140.Kohiyama M, Cousin D, Ryter A, Jacob F. Thermosensitive mutants of Escherichia coli K 12. I. Isolation and rapid characterization. Ann Inst Pasteur. 1966;110:465–86. [PubMed] [Google Scholar]
- 141.Kong D, DePamphilis ML. Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit. Mol Cell Biol. 2001;21:8095–103. doi: 10.1128/MCB.21.23.8095-8103.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Koonin EV. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21:2541–21. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kornberg A, Baker T. DNA Replication. New York: Freeman; 1992. [Google Scholar]
- 144.Krude T. Initiation of human DNA replication in vitro using nuclei from cells arrested at an initiation-competent state. J Biol Chem. 2000;275:13699–275. doi: 10.1074/jbc.275.18.13699. [DOI] [PubMed] [Google Scholar]
- 145.Krude T. Initiation of chromosomal DNA replication in mammalian cell-free systems. Cell Cycle. 2006;5:2115–22. doi: 10.4161/cc.5.18.3248. [DOI] [PubMed] [Google Scholar]
- 146.Kubota Y, Takase Y, Komori Y, Hashimoto Y, Arata T, et al. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev. 2003;17:1141–17. doi: 10.1101/gad.1070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Labib K, Diffley JF. Is the MCM2-7 complex the eukaryotic DNA replication fork helicase? Curr Opin Genet Dev. 2001;11:64–70. doi: 10.1016/s0959-437x(00)00158-1. [DOI] [PubMed] [Google Scholar]
- 148.Labib K, Kearsey SE, Diffley JF. MCM2-7 proteins are essential components of prereplicative complexes that accumulate cooperatively in the nucleus during G1-phase and are required to establish, but not maintain, the S-phase checkpoint. Mol Biol Cell. 2001;12:3658–67. doi: 10.1091/mbc.12.11.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Labib K, Tercero JA, Diffley JF. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–47. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 150.Larizza L, Magnani I, Roversi G. Rothmund-Thomson syndrome and RECQL4 defect: splitting and lumping. Cancer Lett. 2006;232:107–232. doi: 10.1016/j.canlet.2005.07.042. [DOI] [PubMed] [Google Scholar]
- 151.Laskey RA, Madine MA. A rotary pumping model for helicase function of MCM proteins at a distance from replication forks. EMBO Rep. 2003;4:26–4. doi: 10.1038/sj.embor.embor706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lee J, Kumagai A, Dunphy WG. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol Cell. 2003;11:329–40. doi: 10.1016/s1097-2765(03)00045-5. [DOI] [PubMed] [Google Scholar]
- 153.Lee JK, Hurwitz J. Isolation and characterization of various complexes of the minichromosome maintenance proteins of Schizosaccharomyces pombe. J Biol Chem. 2000;275:18871–78. doi: 10.1074/jbc.M001118200. [DOI] [PubMed] [Google Scholar]
- 154.Lee JK, Hurwitz J. Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc Natl Acad Sci USA. 2001;98:54–59. doi: 10.1073/pnas.98.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lee JK, Seo YS, Hurwitz J. The Cdc23 (Mcm10) protein is required for the phosphorylation of minichromosome maintenance complex by the Dfp1-Hsk1 kinase. Proc Natl Acad Sci USA. 2003;100:2334–39. doi: 10.1073/pnas.0237384100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lei M. The MCM complex: its role in DNA replication and implications for cancer therapy. Curr Cancer Drug Targets. 2005;5:365–80. doi: 10.2174/1568009054629654. [DOI] [PubMed] [Google Scholar]
- 157.Lei M, Kawasaki Y, Young MR, Kihara M, Sugino A, Tye BK. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–11. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Leipe DD, Aravind L, Koonin EV. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3389–401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lemaitre JM, Bocquet S, Mechali M. Competence to replicate in the unfertilized egg is conferred by Cdc6 during meiotic maturation. Nature. 2002;419:718–22. doi: 10.1038/nature01046. [DOI] [PubMed] [Google Scholar]
- 160.Li CJ, Vassilev A, DePamphilis ML. Role for Cdk1 (Cdc2)/cyclin A in preventing the mammalian origin recognition complex’s largest subunit (Orc1) from binding to chromatin during mitosis. Mol Cell Biol. 2004;24:5875–24. doi: 10.1128/MCB.24.13.5875-5886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li D, Zhao R, Lilyestrom W, Gai D, Zhang R, et al. Structure of the replicative helicase of oncoprotein SV40 large tumour antigen. Nature. 2003;423:512–18. doi: 10.1038/nature01691. [DOI] [PubMed] [Google Scholar]
- 162.Li F, Chen J, Izumi M, Butler MC, Keezer SM, Gilbert DM. The replication timing program of the Chinese hamster beta-globin locus is established coincident with its repositioning near peripheral heterochromatin in early G1 phase. J Cell Biol. 2001;154:283–154. doi: 10.1083/jcb.200104043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lindner K, Gregan J, Montgomery S, Kearsey SE. Essential role of MCM proteins in premeiotic DNA replication. Mol Biol Cell. 2002;13:435–44. doi: 10.1091/mbc.01-11-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Liu J, Smith CL, DeRyckere D, DeAngelis K, Martin GS, Berger JM. Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol Cell. 2000;6:637–48. doi: 10.1016/s1097-2765(00)00062-9. [DOI] [PubMed] [Google Scholar]
- 165.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–8. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 166.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–61. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 167.Lutzmann M, Maiorano D, Mechali M. Identification of full genes and proteins of MCM9, a novel, vertebrate-specific member of the MCM2-8 protein family. Gene. 2005;362:51–56. doi: 10.1016/j.gene.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 168.Lutzmann M, Maiorano D, Mechali M. A Cdt1-geminin complex licenses chromatin for DNA replication and prevents rereplication during S phase in Xenopus. EMBO J. 2006;25:5764–25. doi: 10.1038/sj.emboj.7601436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.MacAlpine DM, Rodriguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18:3094–105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 171.Madine MA, Swietlik M, Pelizon C, Romanowski P, Mills AD, Laskey RA. The roles of the MCM, ORC, and Cdc6 proteins in determining the replication competence of chromatin in quiescent cells. J Struct Biol. 2000;129:198–129. doi: 10.1006/jsbi.2000.4218. [DOI] [PubMed] [Google Scholar]
- 172.Maiorano D, Cuvier O, Danis E, Mechali M. MCM8 is an MCM2-7-related protein that functions as a DNA helicase during replication elongation and not initiation. Cell. 2005;120:315–28. doi: 10.1016/j.cell.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 173.Majka J, Burgers PM. The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol. 2004;78:227–78. doi: 10.1016/S0079-6603(04)78006-X. [DOI] [PubMed] [Google Scholar]
- 174.Masai H, Arai K. Cdc7 kinase complex: a key regulator in the initiation of DNA replication. J Cell Physiol. 2002;190:287–190. doi: 10.1002/jcp.10070. [DOI] [PubMed] [Google Scholar]
- 175.Masai H, Taniyama C, Ogino K, Matsui E, Kakusho N, et al. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J Biol Chem. 2006;281:39249–281. doi: 10.1074/jbc.M608935200. [DOI] [PubMed] [Google Scholar]
- 176.Mastrangelo IA, Hough PV, Wall JS, Dodson M, Dean FB, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature. 1989;338:658–62. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 177.Masuda T, Mimura S, Takisawa H. CDK- and Cdc45-dependent priming of the MCM complex on chromatin during S-phase in Xenopus egg extracts: possible activation of MCM helicase by association with Cdc45. Genes Cells. 2003;8:145–61. doi: 10.1046/j.1365-2443.2003.00621.x. [DOI] [PubMed] [Google Scholar]
- 178.Masumoto H, Muramatsu S, Kamimura Y, Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–55. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- 179.Masumoto H, Sugino A, Araki H. Dpb11 controls the association between DNA polymerases alpha and epsilon and the autonomously replicating sequence region of budding yeast. Mol Cell Biol. 2000;20:2809–20. doi: 10.1128/mcb.20.8.2809-2817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–53. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 181.McGeoch AT, Bell SD. Eukaryotic/archaeal primase and MCM proteins encoded in a bacteriophage genome. Cell. 2005;120:167–68. doi: 10.1016/j.cell.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 182.McGeoch AT, Trakselis MA, Laskey RA, Bell SD. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat Struct Mol Biol. 2005;12:756–12. doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- 183.Miller ME, Cross FR. Cyclin specificity: How many wheels do you need on a unicycle? J Cell Sci. 2001;114:1811–20. doi: 10.1242/jcs.114.10.1811. [DOI] [PubMed] [Google Scholar]
- 184.Mohanty BK, Bairwa NK, Bastia D. The Tof1p-Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2006;103:897–902. doi: 10.1073/pnas.0506540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Montagnoli A, Bosotti R, Villa F, Rialland M, Brotherton D, et al. Drf1, a novel regulatory subunit for human Cdc7 kinase. EMBO J. 2002;21:3171–21. doi: 10.1093/emboj/cdf290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Montagnoli A, Tenca P, Sola F, Carpani D, Brotherton D, et al. Cdc7 inhibition reveals a p53-dependent replication checkpoint that is defective in cancer cells. Cancer Res. 2004;64:7110–64. doi: 10.1158/0008-5472.CAN-04-1547. [DOI] [PubMed] [Google Scholar]
- 187.Moore JD, Kirk JA, Hunt T. Unmasking the S-phase-promoting potential of cyclin B1. Science. 2003;300:987–90. doi: 10.1126/science.1081418. [DOI] [PubMed] [Google Scholar]
- 188.Morgan DO. The Cell Cycle. London: New Science; 2007. p. 297. [Google Scholar]
- 189.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103:10236–41. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Murakami H, Nurse P. Meiotic DNA replication checkpoint control in fission yeast. Genes Dev. 1999;13:2581–13. doi: 10.1101/gad.13.19.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Murray AW. Recycling the cell cycle. Cyclins revisited. Cell. 2004;116:221–34. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- 192.Nakajima R, Masukata H. SpSld3 is required for loading and maintenance of SpCdc45 on chromatin in DNA replication in fission yeast. Mol Biol Cell. 2002;13:1462–72. doi: 10.1091/mbc.02-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Nasmyth K. 1996. Control of S phase. See Ref. 58, pp. 331–86
- 194.Nasmyth K. Viewpoint: putting the cell cycle in order. Science. 1996;274:1643–45. doi: 10.1126/science.274.5293.1643. [DOI] [PubMed] [Google Scholar]
- 195.Natale DA, Umek RM, Kowalski D. Ease of DNA unwinding is a conserved property of yeast replication origins. Nucleic Acids Res. 1993;21:555–21. doi: 10.1093/nar/21.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Newlon CS. 1996 . DNA replication in yeast. See Ref. 58, pp. 873–914
- 197.Nguyen VQ, Co C, Irie K, Li JJ. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr Biol. 2000;10:195–10. doi: 10.1016/s0960-9822(00)00337-7. [DOI] [PubMed] [Google Scholar]
- 198.Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA rereplication through multiple mechanisms. Nature. 2001;411:1068–73. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- 199.Nieduszynski CA, Hiraga S, Ak P, Benham CJ, Donaldson AD. OriDB: a DNA replication origin database. Nucleic Acids Res. 2007;35:D40–46. doi: 10.1093/nar/gkl758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Nieduszynski CA, Knox Y, Donaldson AD. Genome-wide identification of replication origins in yeast by comparative genomics. Genes Dev. 2006;20:1874–20. doi: 10.1101/gad.385306. A comparative approach in different yeast species to demonstrate the evolution and importance of origin sequences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Nishitani H, Lygerou Z, Nishimoto T, Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–28. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- 202.Nishitani H, Nurse P. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- 203.Norio P. DNA replication: the unbearable lightness of origins. EMBO Rep. 2006;7:779–7. doi: 10.1038/sj.embor.7400766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, et al. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell. 2005;20:575–87. doi: 10.1016/j.molcel.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 205.Nougarede R, Della Seta F, Zarzov P, Schwob E. Hierarchy of S-phase-promoting factors: Yeast Dbf4-cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol Cell Biol. 2000;20:3795–20. doi: 10.1128/mcb.20.11.3795-3806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet. 2002;36:617–36. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 207.Ogino K, Hirota K, Matsumoto S, Takeda T, Ohta K, et al. Hsk1 kinase is required for induction of meiotic dsDNA breaks without involving checkpoint kinases in fission yeast. Proc Natl Acad Sci USA. 2006;103:8131–36. doi: 10.1073/pnas.0602498103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ortega S, Prieto I, Odajima J, Martin A, Dubus P, et al. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet. 2003;35:25–35. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- 209.Osborn AJ, Elledge SJ. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 2003;17:1755–17. doi: 10.1101/gad.1098303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Oshiro G, Owens JC, Shellman Y, Sclafani RA, Li JJ. Cell cycle control of cdc7p kinase activity through regulation of dbf4p stability. Mol Cell Biol. 1999;19:4888–19. doi: 10.1128/mcb.19.7.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Pacek M, Walter JC. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 2004;23:3667–23. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Paciotti V, Clerici M, Scotti M, Lucchini G, Longhese MP. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol Cell Biol. 2001;21:3913–21. doi: 10.1128/MCB.21.12.3913-3925.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Paixao S, Colaluca IN, Cubells M, Peverali FA, Destro A, et al. Modular structure of the human lamin B2 replicator. Mol Cell Biol. 2004;24:2958–24. doi: 10.1128/MCB.24.7.2958-2967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Pape T, Meka H, Chen S, Vicentini G, van Heel M, Onesti S. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 2003;4:1079–4. doi: 10.1038/sj.embor.7400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Patel PK, Arcangioli B, Baker SP, Bensimon A, Rhind N. DNA replication origins fire stochastically in fission yeast. Mol Biol Cell. 2006;17:308–16. doi: 10.1091/mbc.E05-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu Rev Biochem. 2000;69:651–69. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 217.Pelizon C, Madine MA, Romanowski P, Laskey RA. Unphosphorylatable mutants of Cdc6 disrupt its nuclear export but still support DNA replication once per cell cycle. Genes Dev. 2000;14:2526–14. doi: 10.1101/gad.176300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Perkins G, Diffley JF. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- 219.Pessoa-Brandao L, Sclafani RA. CDC7/DBF4 functions in the translesion synthesis branch of the RAD6 epistasis group in Saccharomyces cerevisiae. Genetics. 2004;167:1597–610. doi: 10.1534/genetics.103.021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Petersen P, Chou DM, You Z, Hunter T, Walter JC, Walter G. Protein phosphatase 2A antagonizes ATM and ATR in a Cdk2- and Cdc7-independent DNA damage checkpoint. Mol Cell Biol. 2006;26:1997–26. doi: 10.1128/MCB.26.5.1997-2011.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a “reductional” anaphase in the budding yeast Saccharomyces cerevisiae. EMBO. 1995;14:3788–99. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Pryor A, Faulkner K, Rhoades MM, Peacock WJ. Asynchronous replication of heterochromatin in maize. Proc Natl Acad Sci USA. 1980;77:6705–9. doi: 10.1073/pnas.77.11.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Raghuraman MK, Brewer BJ, Fangman WL. Cell cycle-dependent establishment of a late replication program. Science. 1997;276:806–8. doi: 10.1126/science.276.5313.806. [DOI] [PubMed] [Google Scholar]
- 224.Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, et al. Replication dynamics of the yeast genome. Science. 2001;294:115–21. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- 225.Randell JC, Bowers JL, Rodriguez HK, Bell SP. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol Cell. 2006;21:29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 226.Reynolds AE, McCarroll RM, Newlon CS, Fangman WL. Time of replication of ARS elements along yeast chromosome III. Mol Cell Biol. 1989;9:4488–9. doi: 10.1128/mcb.9.10.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Rhind N. DNA replication timing: random thoughts about origin firing. Nat Cell Biol. 2006;8:1313–8. doi: 10.1038/ncb1206-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ricke RM, Bielinsky AK. Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol Cell. 2004;16:173–85. doi: 10.1016/j.molcel.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 229.Robinson NP, Dionne I, Lundgren M, Marsh VL, Bernander R, Bell SD. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell. 2004;116:25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- 230.Roeder GS. Meiotic chromosomes: It takes two to tango. Genes Dev. 1997;11:2600–11. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- 231.Sakwe AM, Nguyen T, Athanasopoulos V, Shire K, Frappier L. Identification and characterization of a novel component of the human minichromosome maintenance complex. Mol Cell Biol. 2007;27:3044–27. doi: 10.1128/MCB.02384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–60. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 233.Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–39. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 234.Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, et al. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–98. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 235.Santocanale C, Diffley JF. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–18. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 236.Sasaki T, Ramanathan S, Okuno Y, Kumagai C, Shaikh SS, Gilbert DM. The Chinese hamster dihydrofolate reductase replication origin decision point follows activation of transcription and suppresses initiation of replication within transcription units. Mol Cell Biol. 2006;26:1051–26. doi: 10.1128/MCB.26.3.1051-1062.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Sato N, Arai K-I, Masai H. Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: in vitro phosphorylation of MCM subunits by a putative human homolgue of Cdc7. EMBO J. 1997;16:4340–51. doi: 10.1093/emboj/16.14.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Sawyer SL, Cheng IH, Chai W, Tye BK. Mcm10 and Cdc45 cooperate in origin activation in Saccharomyces cerevisiae. J Mol Biol. 2004;340:195–202. doi: 10.1016/j.jmb.2004.04.066. [DOI] [PubMed] [Google Scholar]
- 239.Saxena S, Yuan P, Dhar SK, Senga T, Takeda D, et al. A dimerized coiled-coil domain and an adjoining part of geminin interact with two sites on Cdt1 for replication inhibition. Mol Cell. 2004;15:245–58. doi: 10.1016/j.molcel.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 240.Schild D, Byers B. Meiotic effects of DNA-defective cell division cycle mutations of Saccharomyces cerevisiae. Chromosoma. 1978;70:109–30. doi: 10.1007/BF00292220. [DOI] [PubMed] [Google Scholar]
- 241.Schwacha A, Bell SP. Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol Cell. 2001;8:1093–104. doi: 10.1016/s1097-2765(01)00389-6. [DOI] [PubMed] [Google Scholar]
- 242.Schwob E, Bohm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S phase transition in Saccharomyces cerevisiae. Mol Cell Biol. 1994;79:233–44. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 243.Sclafani R. Cdc7p-Dbf4p becomes famous in the cell cycle. J Cell Sci. 2000;113:2111–113. doi: 10.1242/jcs.113.12.2111. [DOI] [PubMed] [Google Scholar]
- 244.Sclafani RA, Fletcher RJ, Chen XS. Two heads are better than one: regulation of DNA replication by hexameric helicases. Genes Dev. 2004;18:2039–18. doi: 10.1101/gad.1240604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Sclafani RA, Tecklenburg M, Pierce A. The mcm5-bob1 bypass of Cdc7p/Dbf4p in DNA replication depends on both Cdk1-independent and Cdk1-dependent steps in S. cerevisiae. Genetics. 2002;161:47–57. doi: 10.1093/genetics/161.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Segurado M, de Luis A, Antequera F. Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep. 2003;4:1048–53. doi: 10.1038/sj.embor.7400008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Seki T, Diffley JF. Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc Natl Acad Sci USA. 2000;97:14115–20. doi: 10.1073/pnas.97.26.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol. 2004;6:648–6. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- 249.Shechter D, Gautier J. MCM proteins and checkpoint kinases get together at the fork. Proc Natl Acad Sci USA. 2004;101:10845–46. doi: 10.1073/pnas.0404143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Shechter DF, Ying CY, Gautier J. The intrinsic DNA helicase activity of Methanobacterium thermoautotrophicum delta H minichromosome maintenance protein. J Biol Chem. 2000;275:15049–59. doi: 10.1074/jbc.M000398200. [DOI] [PubMed] [Google Scholar]
- 251.Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24:101–13. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, et al. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007;39:93–39. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- 253.Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, et al. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–21. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 254.Shuster EO, Byers B. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics. 1989;123:129–43. doi: 10.1093/genetics/123.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Sidorova JM, Breeden LL. Precocious s-phase entry in budding yeast prolongs replicative state and increases dependence upon rad53 for viability. Genetics. 2002;160:123–36. doi: 10.1093/genetics/160.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Smelkova NV, Borowiec JA. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J Virol. 1997;71:8766–71. doi: 10.1128/jvi.71.11.8766-8773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Snaith HA, Brown GW, Forsburg SL. Schizosaccharomyces pombe hsk1p is a potential cds1p target required for genome integrity. Mol Cell Biol. 2000;20:7922–32. doi: 10.1128/mcb.20.21.7922-7932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol. 2005;12:965–12. doi: 10.1038/nsmb1002. Contains an elegant movie that can be downloaded to show the ORC-Cdc6 structure in 3D and how the MCM hexamer may bind to its surface during loading. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Speck C, Stillman B. Cdc6 ATPase activity regulates ORC-CDC6 stability and the selection of specific DNA sequences as origins of DNA replication. J Biol Chem. 2007;282:11705–282. doi: 10.1074/jbc.M700399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Stevenson JB, Gottschling DE. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev. 1999;13:146–13. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Stillman B. Initiation of chromosomal DNA replication in eukaryotes. J Biol Chem. 1994;269:7047–269. [PubMed] [Google Scholar]
- 262.Stillman B. Origin recognition and the chromosome cycle. FEBS Lett. 2005;579:877–579. doi: 10.1016/j.febslet.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 263.Strich R. Meiotic DNA replication. Curr Top Dev Biol. 2004;61:29–61. doi: 10.1016/S0070-2153(04)61002-7. [DOI] [PubMed] [Google Scholar]
- 264.Stuart D, Wittenberg C. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev. 1998;12:2698–12. doi: 10.1101/gad.12.17.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Su TT. Cellular responses to DNA damage: one signal, multiple choices. Annu Rev Genet. 2006;40:187–40. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- 266.Su TT, Stumpff J. Promiscuity rules? The dispensability of cyclin E and Cdk2. Sci STKE. 2004;2004:pe11. doi: 10.1126/stke.2242004pe11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Sun WH, Coleman TR, DePamphilis ML. Cell cycle-dependent regulation of the association between origin recognition proteins and somatic cell chromatin. EMBO J. 2002;21:1437–21. doi: 10.1093/emboj/21.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–25. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Tachibana KE, Nigg EA. Geminin regulates multiple steps of the chromosome inheritance cycle. Cell Cycle. 2006;5:151–54. doi: 10.4161/cc.5.2.2363. [DOI] [PubMed] [Google Scholar]
- 270.Takahashi TS, Walter JC. Cdc7-Drf1 is a developmentally regulated protein kinase required for the initiation of vertebrate DNA replication. Genes Dev. 2005;19:2295–19. doi: 10.1101/gad.1339805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Takahashi TS, Wigley DB, Walter JC. Pumps, paradoxes and ploughshares: mechanism of the MCM2-7 DNA helicase. Trends Biochem Sci. 2005;30:437–30. doi: 10.1016/j.tibs.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 272.Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003;17:1153–17. doi: 10.1101/gad.1065903. All aspects of yeast genetics (suppressors: synthetic lethality: two-hybrid): molecular biology (ChIP) and biochemistry (affinity chromatography: mass spectrometry) used to identify the novel GINS complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Takeda DY, Dutta A. DNA replication and progression through S phase. Oncogene. 2005;24:2827–43. doi: 10.1038/sj.onc.1208616. [DOI] [PubMed] [Google Scholar]
- 274.Takeda T, Ogino K, Tatebayashi K, Ikeda H, Arai K, Masai H. Regulation of initiation of S phase, replication checkpoint signaling, and maintenance of mitotic chromosome structures during S phase by Hsk1 kinase in the fission yeast. Mol Biol Cell. 2001;12:1257–74. doi: 10.1091/mbc.12.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Takei Y, Assenberg M, Tsujimoto G, Laskey R. The MCM3 acetylase MCM3AP inhibits initiation, but not elongation, of DNA replication via interaction with MCM3. J Biol Chem. 2002;277:43121–277. doi: 10.1074/jbc.C200442200. [DOI] [PubMed] [Google Scholar]
- 276.Tanaka S, Diffley JF. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat Cell Biol. 2002;4:198–4. doi: 10.1038/ncb757. [DOI] [PubMed] [Google Scholar]
- 277.Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–32. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 278.Tenca P, Brotherton D, Montagnoli A, Albanese C, Santocanale C. Cdc7 is an active kinase in human cancer cells undergoing replication stress. J Biol Chem. 2007;282:208–282. doi: 10.1074/jbc.M604457200. The only comprehensive study in which the effects of the DNA damage and replication checkpoints on human DDK function are rigorously examined using the same reagents and procedures; provides substantial evidence against the hypothesis that DDK is a target of the DNA checkpoint. [DOI] [PubMed] [Google Scholar]
- 279.Tercero JA, Labib K, Diffley JFX. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J. 2000;19:2082–19. doi: 10.1093/emboj/19.9.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Tercero JA, Longhese MP, Diffley JF. A central role for DNA replication forks in checkpoint activation and response. Mol Cell. 2003;11:1323–36. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 281.Thomas R, Bertani LE. On the control of the replication of temperate bacteriophages superinfecting immune hosts. Virology. 1964;24:241–53. doi: 10.1016/0042-6822(64)90163-1. [DOI] [PubMed] [Google Scholar]
- 282.Torres JZ, Schnakenberg SL, Zakian VA. Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol Cell Biol. 2004;24:3198–212. doi: 10.1128/MCB.24.8.3198-3212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Torres-Rosell J, De Piccoli G, Cordon-Preciado V, Farmer S, Jarmuz A, et al. Anaphase onset before complete DNA replication with intact checkpoint responses. Science. 2007;315:1411–15. doi: 10.1126/science.1134025. [DOI] [PubMed] [Google Scholar]
- 284.Toyn JH, Johnson AL, Johnston LH. Segregation of unreplicated chromosomes in Saccharomyces cerevisiae reveals a novel G1/M-phase checkpoint. Mol Cell Biol. 1995;15:5312–21. doi: 10.1128/mcb.15.10.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Tsuji T, Ficarro SB, Jiang W. Essential role of phosphorylation of MCM2 by Cdc7/Dbf4 in the initiation of DNA replication in mammalian cells. Mol Biol Cell. 2006;17:4459–72. doi: 10.1091/mbc.E06-03-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Tyagi A, Singh RP, Agarwal C, Siriwardana S, Sclafani RA, Agarwal R. Resveratrol causes Cdc2-tyr15 phosphorylation via ATM/ATR-Chk1/2-Cdc25C pathway as a central mechanism for S phase arrest in human ovarian carcinoma Ovcar-3 cells. Carcinogenesis. 2005;26:1978–87. doi: 10.1093/carcin/bgi165. [DOI] [PubMed] [Google Scholar]
- 287.Tye B-K. MCM proteins in DNA replication. Annu Rev Biochem. 1999;68:649–68. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 288.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–19. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Valentin G, Schwob E, Della Seta F. Dual role of the Cdc7-regulatory protein Dbf4 during yeast meiosis. J Biol Chem. 2006;281:2828–281. doi: 10.1074/jbc.M510626200. [DOI] [PubMed] [Google Scholar]
- 290.Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–17. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol Cell. 2002;10:1223–33. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- 292.Volkening M, Hoffmann I. Involvement of human MCM8 in prereplication complex assembly by recruiting hcdc6 to chromatin. Mol Cell Biol. 2005;25:1560–25. doi: 10.1128/MCB.25.4.1560-1568.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Walter J, Sun L, Newport JW. Regulated chromosomal DNA replication in the absence of a nucleus. Mol Cell. 1998;1:519–1. doi: 10.1016/s1097-2765(00)80052-0. A soluble in vitro system (frog egg crude extracts) was used to measure all steps of DNA replication from initiation to elongation. [DOI] [PubMed] [Google Scholar]
- 294.Walter JC. Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J Biol Chem. 2000;275:39773–275. doi: 10.1074/jbc.M008107200. [DOI] [PubMed] [Google Scholar]
- 295.Wan L, Zhang C, Shokat KM, Hollingsworth NM. Chemical inactivation of cdc7 kinase in budding yeast results in a reversible arrest that allows efficient cell synchronization prior to meiotic recombination. Genetics. 2006;174:1767–74. doi: 10.1534/genetics.106.064303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Wang L, Lin CM, Brooks S, Cimbora D, Groudine M, Aladjem MI. The human beta-globin replication initiation region consists of two modular independent replicators. Mol Cell Biol. 2004;24:3373–24. doi: 10.1128/MCB.24.8.3373-3386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Wechsler JA, Gross JD. Escherichia coli mutants temperature-sensitive for DNA synthesis. Mol Gen Genet. 1971;113:273–84. doi: 10.1007/BF00339547. [DOI] [PubMed] [Google Scholar]
- 298.Weinert T. Cell biology. What a cell should know (but may not) Science. 2007;315:1374–75. doi: 10.1126/science.1140759. [DOI] [PubMed] [Google Scholar]
- 299.Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–22. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 300.Weinert TA, Hartwell LH. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–8. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 302.Weinreich M, Stillman B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 1999;18:5334–18. doi: 10.1093/emboj/18.19.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Weisshart K, Taneja P, Jenne A, Herbig U, Simmons DT, Fanning E. Two regions of simian virus 40 T antigen determine cooperativity of double-hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. J Virol. 1999;73:2201–73. doi: 10.1128/jvi.73.3.2201-2211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Whitmire E, Khan B, Coue M. Cdc6 synthesis regulates replication competence in Xenopus oocytes. Nature. 2002;419:722–25. doi: 10.1038/nature01032. [DOI] [PubMed] [Google Scholar]
- 305.Whittaker AJ, Royzman I, Orr-Weaver TL. Drosophila double parked: a conserved, essential replication protein that colocalizes with the origin recognition complex and links DNA replication with mitosis and the down-regulation of S phase transcripts. Genes Dev. 2000;14:1765–76. [PMC free article] [PubMed] [Google Scholar]
- 306.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–90. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Wohlschlegel JA, Dhar SK, Prokhorova TA, Dutta A, Walter JC. Xenopus Mcm10 binds to origins of DNA replication after Mcm2-7 and stimulates origin binding of Cdc45. Mol Cell. 2002;9:1–20. doi: 10.1016/s1097-2765(02)00456-2. [DOI] [PubMed] [Google Scholar]
- 308.Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–12. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 309.Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, et al. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science. 2001;294:2357–60. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- 310.Xu W, Aparicio JG, Aparicio OM, Tavare S. Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics. 2006;7:276. doi: 10.1186/1471-2164-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Yabuki N, Terashima H, Kitada K. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells. 2002;7:781–89. doi: 10.1046/j.1365-2443.2002.00559.x. [DOI] [PubMed] [Google Scholar]
- 312.Yabuuchi H, Yamada Y, Uchida T, Sunathvanichkul T, Nakagawa T, Masukata H. Ordered assembly of Sld3, GINS and Cdc45 is distinctly regulated by DDK and CDK for activation of replication origins. EMBO J. 2006;25:4663–25. doi: 10.1038/sj.emboj.7601347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Yamada Y, Nakagawa T, Masukata H. A novel intermediate in initiation complex assembly for fission yeast DNA replication. Mol Biol Cell. 2004;15:3740–50. doi: 10.1091/mbc.E04-04-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Yanow SK, Gold DA, Yoo HY, Dunphy WG. Xenopus Drf1, a regulator of Cdc7, displays checkpoint-dependent accumulation on chromatin during an S-phase arrest. J Biol Chem. 2003;278:41083–278. doi: 10.1074/jbc.M307144200. [DOI] [PubMed] [Google Scholar]
- 315.Ying CY, Gautier J. The ATPase activity of MCM2-7 is dispensable for pre-RC assembly but is required for DNA unwinding. EMBO J. 2005;24:4334–24. doi: 10.1038/sj.emboj.7600892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Yoshida K. Identification of a novel cell-cycle-induced MCM family protein MCM9. Biochem Biophys Res Commun. 2005;331:669–331. doi: 10.1016/j.bbrc.2005.03.222. [DOI] [PubMed] [Google Scholar]
- 317.You Z, Ishimi Y, Masai H, Hanaoka F. Roles of Mcm7 and Mcm4 subunits in the DNA helicase activity of the mouse Mcm4/6/7 complex. J Biol Chem. 2002;277:42471–277. doi: 10.1074/jbc.M205769200. [DOI] [PubMed] [Google Scholar]
- 318.Yu Z, Feng D, Liang C. Pairwise interactions of the six human MCM protein subunits. J Mol Biol. 2004;340:1197–340. doi: 10.1016/j.jmb.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 319.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–30. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 320.Zappulla DC, Sternglanz R, Leatherwood J. Control of replication timing by a transcriptional silencer. Curr Biol. 2002;12:869–12. doi: 10.1016/s0960-9822(02)00871-0. [DOI] [PubMed] [Google Scholar]
- 321.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:272–445. doi: 10.1038/nature05432. Yeast genetics and molecular biology used to determine the previously unknown positive role of CDK in DNA replication and link it to the role of DDK. [DOI] [PubMed] [Google Scholar]
- 322.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–85. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 323.Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 2001;20:3544–20. doi: 10.1093/emboj/20.13.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–40. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 325.Zhao X, Rothstein R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad Sci USA. 2002;99:3746–51. doi: 10.1073/pnas.062502299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280:593–96. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
- 327.Zou L, Stillman B. Assembly of a complex containing cdc45p, replication protein A, and mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol Cell Biol. 2000;20:3086–20. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]