Ammonium and nitrate are primary nitrogen sources for many organisms. Though the energy cost required for ammonium assimilation is lower than that of nitrate (2) and many species use ammonium preferentially, nitrate is the nitrogen source most preferred because of its major abundance (about 10 to 1,000 times more available) in natural soils, except in some ecosystems such as coniferous forests (59). This preference occurs even though nitrate concentrations are very heterogeneous, fluctuating by several orders of magnitude from 0 to >10 mM (10). Thus, organisms have evolved to have an adequate supply of genes that are specific for ensuring efficient nitrate assimilation. Even plants, especially crop herbaceous plants, utilize nitrate as the preferred nitrogen form for growth in combination with ammonium (3, 26), since in addition, nitrate provides an efficient signal for modulating many cell processes such as root development, root-shoot balance, or stomatal opening (24, 55, 67).
Nitrate assimilation is a fundamental pathway for incorporating the macroelement N, used by many living beings including the crops responsible for supporting human food, like wheat, barley, or corn. Due to the economic importance of nitrogen for crop productivity and the ecological side effects derived from the massive use of N fertilizers (26), molecular developments in this area have made it a fast-moving field. The initial molecular approaches focused on the main enzymes of the pathway, nitrate reductase (NR) and nitrite reductase (NiR) (46, 65), which subsequently changed to the transport step, providing nitrate to the cells and constituting a versatile point of nutrient control. Recently, attention has started to point to the regulatory control of the pathway, the integration of N assimilation with that of other nutrients, and the improvement of nitrate assimilation efficiency.
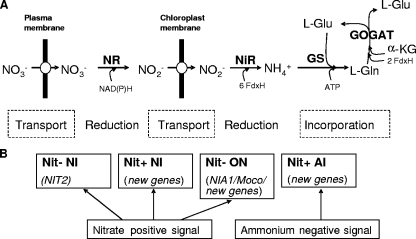
Nitrate assimilation is an apparently simple process in photosynthetic eukaryotes. The process involves two transport and two reduction steps to produce ammonium in the chloroplast, the main site of ammonium incorporation into carbon skeletons, and takes place by the glutamine synthetase/glutamate synthase cycle (Fig. 1A) (8, 17). The transport steps consist of the entry of nitrate into the cell and nitrite into the chloroplast. The first reduction step from nitrate to nitrite occurs in the cytosol and is catalyzed by the NR enzyme, and the second reduction step from nitrite to ammonium in the chloroplast is catalyzed by NiR.
FIG. 1.
Nitrate assimilation pathway in photosynthetic eukaryotes (A) and genes for its regulation (B). (A) Two transport and two reduction steps are required to produce ammonium in the chloroplast, where it is subsequently incorporated. FdxH, reduced ferredoxin. (B) Phenotypes of mutants defective in positive and negative signaling and the possible genes affected (21). NI, nitrate insensitive; ON, overexpression in nitrate; AI, ammonium insensitive.
Functional genomics strategies with the green alga Chlamydomonas sp. have shown the complexity of the process and have helped to discover the fundamental regulatory steps (Fig. 1B). This haploid unicellular organism lacks the structural complexity present in plants, though it has a remarkable flexibility to adapt to extreme environmental conditions (25). This review focuses mostly on the current progress attained in understanding the nitrate assimilation of Chlamydomonas and how the generated data are providing clues to understanding this process in other photosynthetic organisms, especially in the more complex systems of plants.
The advantages of using Chlamydomonas in molecular studies are clear nowadays, since, in addition to its easy microbiological handling, its haploid genetics, and the transformation of its three genomes (25, 52), the sequencing of its genome reveals the evolution of key animal and plant functions (38) that mark highlights on its future perspectives. In fact, Chlamydomonas has become an excellent system with which to study fundamental biological processes such as photosynthesis, chloroplast inheritance and biology, mitochondrial genetics, carbon and nitrogen metabolism, and nutrient deficiency, among others (52).
KEY POINTS OF NITRATE ASSIMILATION IN CHLAMYDOMONAS
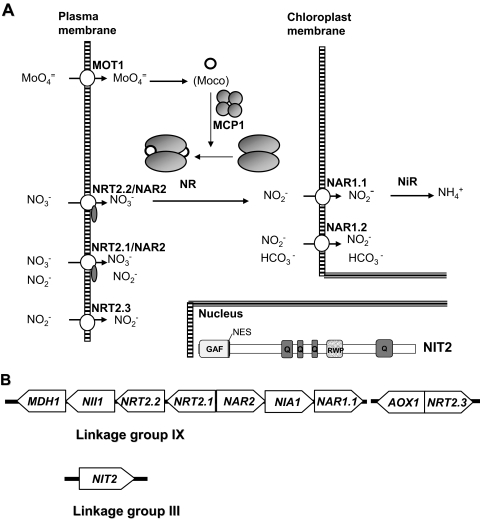
NR and NiR are highly conserved at the sequence level in both algae and plants (5, 9, 66). The NR enzyme has a homodimeric structure with three prosthetic groups: FAD, heme b557, and molybdenum cofactor (Moco). Thus, molybdenum is a micronutrient essential for nitrate assimilation. In the Chlamydomonas system, two proteins for molybdate metabolism have been identified: MOT1, a high-affinity molybdate transporter that defines a new family of transporters distantly related to those of sulfate and is present in eukaryotes and prokaryotes (60), and MCP1, a Moco carrier protein involved in Moco transfer to apo-NR (1, 18) (Fig. 2A). MOT1 from Arabidopsis sp. has also been described recently (61). The NR enzyme was the center of attention for a long time as the critical step in the pathway, since it is highly regulated (9, 23). However, nitrate and nitrite transporters at the plasma membrane and at the chloroplast envelope membrane (Fig. 2A) are important players in the control of the overall efficiency of the nitrate pathway (20).
FIG. 2.
Nitrate assimilation in Chlamydomonas. (A) Scheme of transporters, reductases, and the regulatory NIT2 protein for nitrate assimilation. The main structural motifs in NIT2 are indicated: the GAF domain, nuclear export sequence (NES), poly-glutamine (Q), and the RWP-RK (RWP) motif. (B) Nitrate gene clusters. Two gene clusters in linkage group IX contain nitrate assimilation genes in Chlamydomonas. The regulatory gene NIT2 is in linkage group III. Genes and some of their encoded products are indicated in panel A (25, 48, 49). Other details are given in the text.
In fact, three different protein families, NRT1 (one member), NRT2 (six members), and NAR1 (six members), related to nitrate and nitrite transport are deduced from the Chlamydomonas genome (14). The NRT2 genes that encode high-affinity nitrate/nitrite transporters (HANNiT) have also been cloned from mosses, fungi, algae, yeast, and bacteria, and in almost all plant species, they appear as a multigene family (19, 20, 40, 64). Some NRT2 proteins require a second component named NAR2. The requirement for the NAR2 protein for the functionality of high-affinity nitrate transporters (HANT) has been corroborated not only with Chlamydomonas (69) but also with other systems such as the moss Physcomitrella patens (64) and plants (39, 41, 62). The functionality of NRT2 proteins has been shown only for some of them, as follows: NRT2.1/NAR2 as a bispecific HANNiT, NRT2.2/NAR2 as a HANT, and NRT2.3 as a high-affinity nitrite transporter (14, 47-49) (Fig. 2A).
The NAR1 gene family is mostly present only in bacteria and in eukaryotic organisms such as fungi, yeast, algae, and protozoan parasites but not in plants. In Chlamydomonas, some of the NAR1 genes are clearly regulated by carbon or nitrogen (36), and functionality studies of NAR1.1 have provided the first molecular evidence of a specific nitrite transporter in the chloroplast, showing that this is a regulated process (50). NAR1.1 mediates a nitrite transport with a Km of about 5 μM and improves nitrate use efficiency for cell growth under light/dark cycles and low CO2 environments (35, 50). A second member of the family, NAR1.2, was shown to be a bispecific nitrite/bicarbonate transporter (36).
Interestingly, NIA1, the gene encoding NR, is clustered with other genes that were initially named nitrate assimilation related (NAR) because of their clear relationship to the nitrate pathway. Like NIA1, all of them show a coordinated regulation by the nitrogen source, nitrate induction, and ammonium repression (15). A second cluster of nitrate-regulated genes of about 10 kb was also found in linkage group IX (Fig. 2B) and contains NRT2.3 (encoding a nitrite transporter) and AOX1 (encoding a mitochondrial alternative oxidase) (48, 51). The clustering of nitrate assimilation genes also occurs in Aspergillus nidulans (27) and Hansenula polymorpha (43). This fact might represent a cell strategy to optimize the regulation of this pathway.
REGULATORY GENES FOR NITRATE ASSIMILATION
Notwithstanding the importance of the regulated transport of nutrients for nitrate use efficiency, another key level of control exists. This corresponds to the regulatory genes mediating positive and negative signaling of nitrate and reduced N compounds, respectively. These regulatory genes have been well characterized molecularly in fungi and yeast (37, 57). The NIRA, NIT4, and YNA1 genes from A. nidulans, Neurospora crassa, and H. polymorpha, respectively, are pathway-specific genes involved in nitrate induction and encode GAL4-like CYS6/Zn2-type binuclear zinc cluster proteins (37, 57). AREA and NIT2 from A. nidulans and N. crassa are major regulatory proteins which mediate nitrogen repression from rich nitrogen sources such as ammonium and glutamine and encode GATA-binding transcription factors (37). However, attempts to extrapolate these data and clone specific regulatory genes for nitrate assimilation in photosynthetic eukaryotes have failed (11, 63), most probably because of significant structural differences among regulatory proteins from fungi/yeast and photosynthetic eukaryotes.
Valuable information about regulatory genes might be provided from photosynthetic eukaryotes, by systems such as Chlamydomonas. In fact, a positive regulatory locus, NIT2, has been known for a long time in this alga genus, and a lot of research of different topics, aside from nitrate assimilation, has been performed using strain 137c (lacking functional NIA1 and NIT2) as a wild-type strain (13, 25). Chlamydomonas NIT2-defective mutant strains have a direct effect on nitrate and nitrite assimilation (12), and the expression of a number of genes (NII1, NRT2.1, NRT2.2, NRT2.3, NAR2, NIA1, NAR1.1, and NAR1.6) encoding key elements of the pathway is prevented (36, 47-49). This NIT2 gene was cloned by transposon tagging and shown to be repressed in ammonium and expressed in nitrogen-free medium, and thus, it might mediate metabolite repression of the nitrate assimilation pathway in Chlamydomonas (56).
NIT2 cDNA was isolated, and the structural characteristics of the NIT2 protein were analyzed together with the mechanism of nitrate sensing (6). That NIT2 is induced in a nitrogen-free medium (56) does not indicate that nitrate is not needed for this induction, since it is well known that nitrate contaminates all N-free media and is able to accumulate intracellularly by the HANT at concentrations high enough to mimic nitrate effects for positive signaling of NIA1 expression (31). By using HANT-deficient mutants, it was shown that intracellular nitrate is not essential for NIT2 expression that takes place in the absence of reduced N sources; however, nitrate causes a stabilization of NIT2 transcripts (6).
NIT2 appears to be a signaling transcription factor with important structural and sequence differences between the nitrate regulatory proteins of fungi and those of yeast, which might explain the lack of success with previous attempts to isolate nitrate regulatory genes from plants (11, 63). NIT2 has an RWP-RK motif within a leucine zipper and shows conservation with putative transcription coactivators from plants as nodule inception (NIN) proteins and the minus dominance (MID) protein of Chlamydomonas (6, 16, 29, 54). In addition, NIT2 shows a GAF domain at the N-terminal fragment of the protein, which is unable to bind specifically to DNA (Fig. 2A). Glutamine-rich domains essential for NIT2 functionality are known to be involved in protein-protein interactions (6, 42). The GAF domain is known to bind small molecules such as oxoglutarate, nitric oxide, or cGMP (4, 30, 53); however, nitrate was found not to bind to this domain (6). A nuclear export sequence (Fig. 2A) was found in NIT2 that binds specifically to the NIA1 promoter regions previously identified as essential for a regulated expression of this gene (6, 33).
In an effort to identify regulatory genes for nitrate assimilation, a functional genomics approach was used with Chlamydomonas. To this purpose, an ordered mutant library of about 22,000 insertional mutant strains covering most of the genome was obtained (21). It is well known that foreign DNA insertions cause a deletion at the insertion position, ranging from a few kilobases to more than 50 kb (7, 28). The rationale for that approach was to interfere with the function of regulatory genes for nitrate assimilation in the insertional mutants, obtained with the mutagenic paromomycin marker gene (58) in strain 704, having the chimeric reporter construction of the Chlamydomonas arylsulfatase (ARS) gene under the control of the NIA1 gene promoter as a sensor for regulatory signals (34). Mutant selection was undertaken from the different screenings of phenotypes (Fig. 1B). First, essential genes mediating the positive effect of nitrate were identified from NIT− ARS− mutants. Only five such mutants were isolated, and identification of the affected region by restriction enzyme site-directed amplification-PCR (22) showed that all of them were affected at the NIT2 gene location (6). Second, another screening was based on strains insensitive to nitrate (low ARS) and able to grow on nitrate, NIT+ (21). These mutants are probably affected at genes not essential for the expression of nitrate assimilation genes but which are required for the efficient modulation of nitrate-positive effects (Fig. 1B). Forty such mutants were isolated, and their characterization is in progress in our laboratory. Third, those mutants overexpressing the ARS marker activity in the presence of nitrate were unable to grow on nitrate, NIT−. These mutants are affected at NIA1, Moco biosynthesis genes, and new genes currently under study (21, 32). Finally, NIT+ mutants expressing ARS in ammonium-containing medium, proposed to be affected by the negative signaling of the pathway by ammonium or ammonium derivatives, were isolated.
These ammonium-insensitive mutants are particularly interesting, since this kind of mutant has been reported previously only in Chlamydomonas (NRG1-4, FAR1), though affected genes are unknown (44, 45, 68). Mutants from the Chlamydomonas mutant collection were found to be defective at genomic regions bearing putative genes that could be related to nitrate assimilation regulation. These are related to regulatory functions such as peptidyl-prolyl isomerase, DNA binding, guanylate cyclase, or protein kinase (21). Generated data are consistent with a complex network mediating negative signaling of nitrate assimilation.
From the data discussed above, it can be concluded that Chlamydomonas is an attractive system for unraveling fundamental points of nitrate assimilation such as genes and functions for transporters at the plasma and plastid membranes and for positive and negative regulatory networks of the pathway in photosynthetic eukaryotes. Though valuable new information has recently been generated from this pathway, much more data are needed to get a better understanding of the regulatory and metabolic circuits involved.
Acknowledgments
This work was funded by the Ministerio de Educación y Ciencia, Spain (grant BFU2005-07521), the Junta de Andalucía, Spain (PAI, CVI-0128 and grant no. P06-CVI-1609), and the University of Córdoba, Spain.
Footnotes
Published ahead of print on 29 February 2008.
REFERENCES
- 1.Ataya, F. S., C. P. Witte, A. Galvan, M. I. Igeno, and E. Fernandez. 2003. Mcp1 encodes the molybdenum cofactor carrier protein in Chlamydomonas reinhardtii and participates in protection, binding, and storage functions of the cofactor. J. Biol. Chem. 27810885-10890. [DOI] [PubMed] [Google Scholar]
- 2.Bloom, A. J., S. S. Sukrapanna, and R. L. Warner. 1992. Root respiration associated with ammonium and nitrate absorption and assimilation by barley. Plant Physiol. 991294-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Britto, D. T., M. Y. Siddiqi, A. D. M. Glass, and H. J. Kronzucker. 2001. Futile transmembrane NH4+ cycling: a cellular hypothesis to explain ammonium toxicity in plants. Proc. Natl. Acad. Sci. USA 984255-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Busch, A., K. Strube, B. Friedrich, and R. Cramm. 2005. Transcriptional regulation of nitric oxide reduction in Ralstonia eutropha H16. Biochem. Soc. Trans. 33193-194. [DOI] [PubMed] [Google Scholar]
- 5.Caboche, M., and P. Rouze. 1990. Nitrate reductase a target for molecular and cellular studies in higher plants. Trends Genet. 6187-192. [DOI] [PubMed] [Google Scholar]
- 6.Camargo, A., A. Llamas, R. A. Schnell, J. J. Higuera, D. Gonzalez-Ballester, P. A. Lefebvre, E. Fernandez, and A. Galvan. 2007. Nitrate signaling by the regulatory gene NIT2 in Chlamydomonas. Plant Cell 193491-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cenkci, B., J. L. Petersen, and G. D. Small. 2003. REX1, a novel gene required for DNA repair. J. Biol. Chem. 27822574-22577. [DOI] [PubMed] [Google Scholar]
- 8.Crawford, N. M. 1995. Nitrate: nutrient and signal for plant growth. Plant Cell 7859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford, N. M., and B. G. Forde. March 2002, posting date. Molecular and developmental biology of inorganic nitrogen nutrition, p. 1-25. In C. Somerville, E. Meyerowitz (ed.), The Arabidopsis book. American Society of Plant Biologists, Rockville, MD. doi: 10.1199/tab.0011. http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- 10.Crawford, N. M., and A. D. M. Glass. 1998. Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3389-395. [Google Scholar]
- 11.Daniel-Vedele, F., and M. Caboche. 1993. A tobacco cDNA clone encoding a GATA-1 zinc finger protein homologous to regulators of nitrogen metabolism in fungi. Mol. Gen. Genet. 240365-373. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez, E., and J. Cardenas. 1982. Regulation of the nitrate-reducing system enzymes in wild type and mutant strains of Chlamydomonas reinhardtii. Mol. Gen. Genet. 186164-169. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez, E., and R. F. Matagne. 1984. Genetic analysis of nitrate reductase-deficient mutants in Chlamydomonas reinhardtii. Curr. Genet. 8635-640. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez, E., and A. Galvan. 2007. Inorganic nitrogen assimilation in Chlamydomonas. J. Exp. Bot. 582279-2287. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez, E., R. A. Schnell, L. P. W. Ranum, S. C. Hussey, C. D. Silflow, and P. Lefebvre. 1989. Isolation and characterization of the nitrate reductase structural gene in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 866449-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferris, P. J., E. V. Armbrust, and U. W. Goodenough. 2002. Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics 160181-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer, P., and U. Klein. 1988. Localization of nitrogen-assimilating enzymes in the chloroplast of Chlamydomonas reinhardtii. Plant Physiol. 88947-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer, K., A. Llamas, M. Tejada-Jimenez, N. Schrader, J. Kuper, F. S. Ataya, A. Galvan, R. R. Mendel, E. Fernandez, and G. Schwarz. 2006. Function and structure of the molybdenum cofactor carrier protein from Chlamydomonas reinhardtii. J. Biol. Chem. 28130186-30194. [DOI] [PubMed] [Google Scholar]
- 19.Forde, B. G. 2000. Nitrate transporters in plants: structure, function and regulation. Biochim. Biophys. Acta 1465219-235. [DOI] [PubMed] [Google Scholar]
- 20.Galvan, A., and E. Fernandez. 2001. Eukaryotic nitrate and nitrite transporters. Cell Mol. Life Sci. 58225-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Ballester, D., A. de Montaigu, J. J. Higuera, A. Galvan, and E. Fernandez. 2005. Functional genomics of the regulation of the nitrate assimilation pathway in Chlamydomonas. Plant Physiol. 137522-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez-Ballester, D., A. de Montaigu, A. Galvan, and E. Fernandez. 2005. Restriction enzyme site-directed amplification PCR: a tool to identify regions flanking a marker DNA. Anal. Biochem. 340330-335. [DOI] [PubMed] [Google Scholar]
- 23.Guerrero, M. G., J. M. Vega, and M. Losada. 1981. The assimilatory nitrate-reducing system and its regulation. Annu. Rev. Plant Physiol. 32169-204. [Google Scholar]
- 24.Guo, F. Q., J. Young, and N. M. Crawford. 2003. The nitrate transporter AtNRT1.1 (CHL1) functions in stomatal opening and contributes to drought susceptibility in Arabidopsis. Plant Cell 15107-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris, E. 1989. The Chlamydomonas sourcebook. Academic Press, New York, NY.
- 26.Hirsch, R. E., and M. R. Sussman. 1999. Improving nutrient capture from soil by the genetic manipulation of crop plants. Trends Biotechnol. 17356-361. [DOI] [PubMed] [Google Scholar]
- 27.Johnstone, I. L., P. C. McCabe, P. Greaves, S. J. Gurr, G. E. Cole, M. A. D. Brow, S. E. Unkless, A. J. Clutterbuck, J. R. Kinghorn, and M. A. Inn. 1990. Isolation and characterisation of the crna-niiA-niaD gene cluster for nitrate assimilation in Aspergillus nidulans. Gene 90181-192. [DOI] [PubMed] [Google Scholar]
- 28.Kindle, K. L. 1998. Nuclear transformation: technology and applications, p. 41-61. In J. D. Rochaix, M. Goldmichdt-Clermont, and S. Merchant (ed.), The molecular biology of chloroplasts and mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 29.Lin, H., and U. W. Goodenough. 2007. Gametogenesis in the Chlamydomonas reinhardtii mating type is controlled by two genes, NID and MTD1. Genetics 176913-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Little, R., and R. Dixon. 2003. The amino-terminal GAF domain of Azotobacter vinelandii NifA binds 2-oxoglutarate to resist inhibition by NifL under nitrogen-limiting conditions. J. Biol. Chem. 27828711-28718. [DOI] [PubMed] [Google Scholar]
- 31.Llamas, A., M. I. Igeño, A. Galvan, and E. Fernandez. 2002. Nitrate signaling on the nitrate reductase gene promoter depends directly on the activity of the nitrate transport systems in Chlamydomonas. Plant J. 30261-271. [DOI] [PubMed] [Google Scholar]
- 32.Llamas, A., M. Tejada-Jimenez, D. González-Ballester, J. J. Higuera, G. Schwarz, A. Galván, and E. Fernández. 2007. Chlamydomonas reinhardtii CNX1E reconstitutes molybdenum cofactor biosynthesis in Escherichia coli mutants. Eukaryot. Cell 61063-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loppes, R., and M. Radoux. 2002. Two short regions of the promoter are essential for activation and repression of the nitrate reductase gene in Chlamydomonas reinhardtii. Mol. Genet. Genomics 26842-48. [DOI] [PubMed] [Google Scholar]
- 34.Loppes, R., M. Ohresser, M. Radoux, and R. F. Matagne. 1999. Transcriptional regulation of the Nit1 gene encoding nitrate reductase in Chlamydomonas reinhardtii: effect of various environmental factors on the expression of a reporter gene under the control of the Nit1 promoter. Plant Mol. Biol. 41701-711. [DOI] [PubMed] [Google Scholar]
- 35.Mariscal, V., J. Rexach, E. Fernandez, and A. Galvan. 2004. The plastidic nitrite transporter NAR1.1 improves nitrate use efficiency for growth in Chlamydomonas. Plant Cell Environ. 271321-1328. [Google Scholar]
- 36.Mariscal, V., P. Moulin, M. Orsel, A. J. Miller, E. Fernandez, and A. Galvan. 2006. Differential regulation of the Chlamydomonas Nar1 gene family by carbon and nitrogen. Protist 157421-433. [DOI] [PubMed] [Google Scholar]
- 37.Marzluf, G. A. 1997. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 6117-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merchant, S. S., et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318245-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okamoto, M., A. Kumar, W. Li, Y. Wang, M. Y. Siddiqi, N. M. Crawford, and A. D. Glass. 2006. High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol. 1401036-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orsel, M., A. Krapp, and F. Daniel-Vedele. 2002. Analysis of the NRT2 nitrate transporter family in Arabidopsis. Structure and gene expression. Plant Physiol. 129886-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orsel, M., F. Chopin, O. Leleu, S. J. Smith, A. Krapp, F. Daniel-Vedele, and A. J. Miller. 2006. Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein-protein interaction. Plant Physiol. 1421304-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Owens, B. M., Y.-X. Zhu, T.-C. Suen, P.-X. Wang, J. F. Greenblatt, P. E. Goss, and R. G. Hawley. 2003. Specific homeodomain-DNA interactions are required for HOX11-mediated transformation. Blood 1014966-4974. [DOI] [PubMed] [Google Scholar]
- 43.Perez, M. D., C. Gonzalez, J. Avila, N. Brito, and J. M. Siverio. 1997. The YNT1 gene encoding the nitrate transporter in the yeast Hansenula polymorpha is clustered with genes YNI1 and YNR1 encoding nitrite reductase and nitrate reductase, and its disruption causes inability to grow in nitrate. Biochem. J. 321397-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez-Alegre, M., A. Dubus, and E. Fernandez. 2005. REM1, a new type of long terminal repeat retrotransposon in Chlamydomonas reinhardtii. Mol. Cell. Biol. 2510628-10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prieto, R., A. Dubus, A. Galvan, and E. Fernandez. 1996. Isolation and characterization of two regulatory mutants for nitrate assimilation in Chlamydomonas reinhardtii. Mol. Gen. Genet. 251461-471. [DOI] [PubMed] [Google Scholar]
- 46.Privalle, L. S., K. N. Lahners, M. A. Mullins, and S. Rothstein. 1989. Nitrate effects on nitrate reductase activity and nitrite reductase mRNA levels in maize suspension cultures. Plant Physiol. 90962-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quesada, A., A. Galvan, and E. Fernandez. 1994. Identification of nitrate transporters in Chlamydomonas reinhardtii. Plant J. 5407-419. [DOI] [PubMed] [Google Scholar]
- 48.Quesada, A., J. Hidalgo, and E. Fernandez. 1998. Three Nrt2 genes are differentially regulated in Chlamydomonas reinhardtii. Mol. Gen. Genet. 258373-377. [DOI] [PubMed] [Google Scholar]
- 49.Quesada, A., A. Galvan, R. A. Schnell, P. A. Lefebvre, and E. Fernandez. 1993. Five nitrate assimilation related loci are clustered in Chlamydomonas reinhardtii. Mol. Gen. Genet. 240387-394. [DOI] [PubMed] [Google Scholar]
- 50.Rexach, J., E. Fernandez, and A. Galvan. 2000. The Chlamydomonas reinhardtii Nar1 gene encodes a chloroplast membrane protein involved in nitrite transport. Plant Cell 121441-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rexach, J., B. Montero, E. Fernandez, and A. Galvan. 1999. Differential regulation of the high affinity nitrite transport systems III and IV in Chlamydomonas reinhardtii. J. Biol. Chem. 2742781-2786. [DOI] [PubMed] [Google Scholar]
- 52.Rochaix, J.-D., M. Goldschmidt-Clermont, and S. Merchant (ed.). 1998. The molecular biology of chloroplasts and mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 53.Rybalkin, S. D., I. G. Rybalkina, M. Shimizu-Albergine, X. B. Tang, and J. A. Beavo. 2003. PDE5 is converted to an activated state upon cGMP binding to the GAF A domain. EMBO J. 22469-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schauser, L., W. Wieloch, and J. Stougaard. 2005. Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J. Mol. Evol. 60229-237. [DOI] [PubMed] [Google Scholar]
- 55.Scheible, W. R., M. Lauerer, E. D. Schulze, M. Caboche, and M. Stitt. 1997. Accumulation of nitrate in the shoot acts as a signal to regulate shoot-root allocation in tobacco. Plant J. 11671-691. [Google Scholar]
- 56.Schnell, R. A., and P. A. Lefebvre. 1993. Isolation of the Chlamydomonas regulatory gene nit2 by transposon tagging. Genetics 1394737-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siverio, J. M. 2002. Assimilation of nitrate by yeasts. FEMS Microbiol. Rev. 26277-284. [DOI] [PubMed] [Google Scholar]
- 58.Sizova, I., M. Fuhrmann, and P. Hegemann. 2001. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277221-229. [DOI] [PubMed] [Google Scholar]
- 59.Stark, J. M., and S. C. Hart. 1977. High rates of nitrification and nitrate turnover in undisturbed coniferous forests. Nature 38561-64. [Google Scholar]
- 60.Tejada-Jimenez, M., A. Llamas, E. Sanz-Luque, A. Galvan, and E. Fernandez. 2007. A high affinity molybdate transporter in eukaryotes. Proc. Natl. Acad. Sci. USA 10420126-20130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tomatsu, H., J. Takano, H. Takahashi, A. Watanabe-Takahashi, N. Shibagaki, and T. Fujiwara. 2007. An Arabidopsis thaliana high-affinity molybdate transporter required for efficient uptake of molybdate from soil. Proc. Natl. Acad. Sci. USA 10418807-18812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tong, Y., J. J. Zhou, Z. Li, and A. J. Miller. 2005. A two-component high-affinity nitrate uptake system in barley. Plant J. 41442-450. [DOI] [PubMed] [Google Scholar]
- 63.Truong, H. N., M. Caboche, and F. Daniel-Vedele. 1997. Sequence and characterization of two Arabidopsis cDNAs isolated by functional complementation of a yeast gln3 gdh1 mutant. FEBS Lett. 410213-218. [DOI] [PubMed] [Google Scholar]
- 64.Tsujimoto, R., H. Yamazaki, S. Maeda, and T. Omata. 2007. Distinct roles of nitrate and nitrite in regulation of expression of the nitrate transport genes in the moss Physcomitrella patens. Plant Cell Physiol. 48484-497. [DOI] [PubMed] [Google Scholar]
- 65.Vincentz, M., and M. Caboche. 1991. Constitutive expression of nitrate reductase allows normal growth and development of Nicotiana plumbaginifolia plants. EMBO J. 101027-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wray, J. L., and J. R. Kinghorn. 1989. Molecular and genetic aspects of nitrate assimilation. Oxford Science Publications, Oxford, United Kingdom.
- 67.Zhang, H., and B. G. Forde. 1998. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279407-409. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, D., and P. A. Lefebvre. 1997. FAR1, a new negative regulatory locus required for the repression of the nitrate reductase gene in Chlamydomonas reinhardtii. Genetics 146121-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou, J. J., E. Fernandez, A. Galvan, and A. J. Miller. 2000. A high affinity nitrate transport system from Chlamydomonas requires two gene products. FEBS Lett. 466225-227. [DOI] [PubMed] [Google Scholar]