Abstract
Members of the RGS7 (R7) family and Gβ5 form obligate heterodimers that are expressed predominantly in the nervous system. R7-Gβ5 heterodimers are GTPase-activating proteins (GAPs) specific for Gi/o-class Gα subunits, which mediate phototransduction in retina and the action of many modulatory G protein-coupled receptors (GPCRs) in brain. Here we have focused on R7BP, a recently identified palmitoylated protein that can bind R7-Gβ5 complexes and is hypothesized to control the intracellular localization and function of the resultant heterotrimeric complexes. We show that: 1) R7-Gβ5 complexes are obligate binding partners for R7BP in brain because they co-immunoprecipitate and exhibit similar expression patterns. Furthermore, R7BP and R7 protein accumulation in vivo requires Gβ5. 2) Expression of R7BP in Neuro2A cells at levels approximating those in brain recruits endogenous RGS7-Gβ5 complexes to the plasma membrane. 3) R7BP immunoreactivity in brain concentrates in neuronal soma, dendrites, spines or unmyelinated axons, and is absent or low in glia, myelinated axons, or axon terminals. 4) RGS7-Gβ5-R7BP complexes in brain extracts associate inefficiently with detergent-resistant lipid raft fractions with or without G protein activation. 5) R7BP and Gβ5 protein levels are upregulated strikingly during the first 2-3 weeks of postnatal brain development. Accordingly, we suggest that R7-Gβ5-R7BP complexes could regulate signaling by modulatory Gi/o-coupled GPCRs in the developing and adult nervous systems.
Keywords: postnatal brain development, signal transduction, palmitoylation, lipid raft, G protein-coupled receptors, immunohistochemistry
INTRODUCTION
G protein coupled receptors (GPCRs) regulate neuronal structure and function by controlling ion channel activity, second messenger production and protein kinase activation (Gainetdinov et al., 2004, Schubert et al., 2006). Modulatory GPCR signaling pathways are tightly controlled by mechanisms such as GPCR phosphorylation and arrestin binding (Lefkowitz and Shenoy, 2005, Dhami and Ferguson, 2006).
GPCR signaling regulation also involves the action of RGS proteins (regulators of G protein signaling), which accelerate GTP hydrolysis by Gα subunits (i.e. RGS proteins are GTPase-activating proteins or GAPs (Berman et al., 1996, Hunt et al., 1996, Watson et al., 1996)). The RGS7 (R7) family (RGS6, RGS7, RGS9-1, RGS9-2 and RGS11) is thought to be particularly important for regulating GPCR function in the nervous system. R7 family members form obligate heterodimers with Gβ5 (Cabrera et al., 1998, Snow et al., 1998, Makino et al., 1999, He et al., 2000, Kovoor et al., 2000, Witherow et al., 2000, Zhang and Simonds, 2000, Chen et al., 2003), which are expressed significantly only in retina and the nervous system (Witherow et al., 2000, Jones et al., 2004). R7-Gβ5 heterodimers have GAP activity specific for Gi/o-class α subunits (Hooks et al., 2003), which mediate phototransduction and modulatory GPCR signaling. RGS9 is the best understood R7 family member. Genetic inactivation of RGS9 slows the termination kinetics of phototransduction in mice (Chen et al., 2000, Krispel et al., 2003), and causes light adaptation and contrast detection defects in humans (Nishiguchi et al., 2004). RGS9 knockout mice also exhibit augmented response to opioids and cocaine (Rahman et al., 2003, Zachariou et al., 2003).
The R7-family binding protein, R7BP, was identified recently as a protein that can interact with R7-Gβ5 heterodimers to form heterotrimeric complexes (Drenan et al., 2005, Martemyanov et al., 2005). When palmitoylated, overexpressed R7BP directs R7-Gβ5-R7BP heterotrimers to the plasma membrane, whereas when depalmitoylated it shuttles them into the nucleus of transiently transfected cells (Drenan et al., 2005, Song et al., 2006). R7BP palmitoylation and plasma membrane targeting augment the ability of RGS7-Gβ5-R7BP complexes to accelerate the kinetics of Gi/o-mediated signaling (Drenan et al., 2005, Drenan et al., 2006).
Despite such information, little is known about the function of R7BP in the nervous system. For example, it is unknown whether R7-Gβ5 complexes are obligate binding partners for R7BP in vivo, whether R7BP is expressed in neurons and/or glia, or whether expression of R7BP at normal levels in neuronal cells is sufficient to recruit endogenous R7-Gβ5 complexes to the plasma membrane. Likewise, little is known about the regional, subcellular or developmental expression of R7BP in brain, which would suggest when or where R7-Gβ5-R7BP complexes could regulate signaling by modulatory GPCRs. Here we have addressed these questions in order to better understand the function of R7-Gβ5-R7BP complexes in brain.
EXPERIMENTAL PROCEDURES
Vertebrate animals
All procedures involving vertebrate animals were performed in strict accord with the NIH Guide for the Care and Use of Laboratory Animals and approved by the Animal Study Committees of Washington University School of Medicine, Emory University School of Medicine and the University of Miami School of Medicine. Mice were housed in animal facilities with free access to water and food, and with a 12-h light/dark cycle. A breeding pair of Gβ5 -/+ mice in a mixed genetic background was generously provided to one of us (V.S.) by Dr. Jason Chen (Virginia Commonwealth University). These mice were backcrossed with C57BL/6J mice until the F3 generation of Gβ5-/+ mice was obtained. Gβ5-/+ mice were interbred to obtain Gβ5-/-, Gβ5-/+ and wild type mice. Neonatal rat pups were obtained by purchasing timed pregnant Sprague Dawley rats (Charles River Laboratories, Inc.). Except for studies involving Gβ5-/- mice, all wild type mice used were of the pure C57BL/6 (Taconic Labs) strain.
Generation of affinity purified chicken and rabbit anti-R7BP antibodies
MBP and GST fusion proteins bearing full-length mouse R7BP at their C-termini were expressed and purified from derivatives of pMAL-C2H10T and pGEX-2T in E. coli strain BL2DE3-pLysS. MBP-R7BP (>90% purity) was used to immunize chickens (Aves Laboratories) and rabbits (Zymed Laboratories). The resultant antibodies were affinity purified by the commercial antibody providers on columns coupled to GST-R7BP. The concentrations of affinity-purified R7BP antibody stocks were 0.7 mg/ml (chicken antibody) and 0.21 mg/ml, (rabbit antibody); these stocks were diluted as described below for use in various experiments.
Confocal fluorescence immunohistochemistry
Mice were anesthetized deeply (87 mg/kg ketamine HCl, 13.4 mg/kg xylazine HCl i.p.) and perfused intracardially with PBS followed by 4% paraformaldehyde in PBS. Brain was removed and kept overnight in 4% paraformaldehyde at 4°C. For immunohistochemistry experiments using free floating sections (40 μm-thick), brains were cryoprotected using 30% sucrose in PBS at 4°C and sections were cut using a sliding microtome. Samples then were processed for immunohistochemistry according to instructions provide by the manufacturer of the Vectastain kit (Vector Labs). Briefly, tissue sections were rinsed in PBS and endogenous peroxidase was inactivated by treating with 0.04%Triton X-100 in PBS containing 0.5% H2O2 for 1 h. To block non-specific antibody binding, sections were incubated in blocking buffer (5% non-fat milk, 1% normal goat serum in PBS) for 1h. Sections then were incubated at 4°C overnight with affinity purified anti-R7BP antibodies (1: 2000, chicken antibody; 1:1000 rabbit antibody, each in blocking buffer). After several rinses with PBS, sections were incubated 1 h at room temperature with biotinylated secondary anti-chicken or anti-rabbit antibody (1:1000; Vector Labs), rinsed in PBS and incubated for 1 h with avidin-biotin-HRP complex (ABC Vectastain kit, Vector Labs). After several rinses in TBS, sections were incubated for 10 min in 0.05% diaminobenzidine (Sigma) and 0.015% hydrogen peroxide, rinsed in PBS, mounted, dried, dehydrated and embedded in Cytoseal 60 medium under coverslips. Blocked anti-R7BP antibodies were prepared by incubating affinity purified chicken or rabbit antibodies (1μl antibody stock diluted in 1-200μl of blocking buffer) with MBP-R7BP fusion protein (16.5μg) for 2h at 37°C. Regional distribution of R7BP was analyzed with the aid of the rodent brain atlas (Paxinos and Watson, 1982). For experiments using thin (10 μm) sections of brain, we embedded tissue in paraffin before sectioning. For immunofluorescence labeling, paraffin-embedded sections were rehydrated and permeabilized for 25 min with trypsin (1mg/ml trypsin tablet for immunohistochemistry; Sigma) in PBS. Sections were washed with PBS and incubated at 4°C overnight with chicken or rabbit anti-R7BP antibody (1:100 in blocking buffer) and co-stained overnight with monoclonal mouse anti-neuronal nuclear antigen (NeuN, Chemicon International Inc.; 1:100) or with polyclonal rabbit anti-glial fibrillary acidic protein (GFAP) antibody ( 1:150). After several rinses in PBS, sections were incubated 1 h at room temperature with biotinylated secondary anti-chicken, anti-rabbit and/or anti-mouse antibodies (Vector Labs) diluted 1:1000 in PBS, rinsed in PBS and incubated for 1h in avidin-biotin-HRP complex (ABC Vectastain kit, Vector Labs). Sections were visualized by using the TSA Plus amplification system (NEN Life Science Products), which converts an HRP substrate to a fluorescent product. Sections were mounted on slides for confocal microscopy (model LSM-510; Carl Zeiss MicroImaging, Inc.). In each experiment we used sections from brain obtained from at least three animals.
Electron microscopic immunocytochemistry
Five adult rats were perfusion-fixed with a mixture of 4% paraformaldehyde/0.1% glutaraldehyde and used for electron microscopic localization of R7BP protein in the striatum and the thalamus, two brain regions significantly enriched in R7BP immunoreactivity. Following perfusions, brains were removed from the skull, post-fixed in 4% paraformaldehyde for 2 - 24 h, cut into 60-μm-thick sections using a vibrating microtome and stored in PBS at 4°C until processed for immunocytochemistry. Prior to immunocytochemical processing, all sections were put into a 1% sodium borohydride solution for 20 minutes and then washed with PBS. Following sodium borohydride treatment, sections were placed in a cryoprotectant solution (0.05M Na-phosphate, pH 7.4, 25% sucrose, and 10% glycerol) for 20 min, frozen at -80°C for 20 min, returned to a decreasing gradient of cryoprotectant solutions, and rinsed in PBS. Sections were then incubated in primary and secondary antibody solutions, identical to those used for light microscopy, with two exceptions: 1) the omission of Triton X-100 and 2) incubation in primary antibody for 48 hours at 4°C. After the DAB reaction, the tissue was rinsed in PB (0.1M, pH 7.4) and treated with 1% OsO4 for 20 min. It was then returned to PB and dehydrated with increasing concentrations of ethanol. When exposed to 70% ETOH, 1% uranyl acetate was added to the solution for 35 min to increase the contrast of the tissue in the electron microscope. Following dehydration, sections were treated with propylene oxide and embedded in epoxy resin for 12 h (Durcupan ACM, Fluka, Buchs, Switzerland), mounted onto slides and placed in a 60°C oven for 48 h. Separate samples of the dorsal striatum and the ventrobasal thalamic nuclei were cut out of the larger sections, mounted onto resin blocks and cut into 60-nm sections using an ultramicrotome (Leica Ultracut T2). The 60-nm sections were collected on Pioloform-coated copper grids, stained with lead citrate for 5 min to enhance tissue contrast and examined on the Zeiss EM-10C electron microscope. Electron micrographs were taken with a CCD camera (DualView 300W; Gatan, Inc., Pleasanton, CA) controlled by DigitalMicrograph software (Gatan, Inc.). Although the overall quality of immunostaining was very similar across these five rats, three animals (DT9, DT12 and DN19) were chosen for quantitative analysis because of their optimal ultrastructural preservation.
From each of these rats, one block of tissue was taken at the level of the dorsolateral striatum and the ventrolateral thalamus. Ultrathin sections taken from the first 5-7 μm of each block were examined and 150 electron micrographs (50 micrographs/block) of randomly selected immunoreactive elements were digitized at 25,000X. Labeled elements were categorized as dendrites, spines, unmyelinated axons, myelinated axons, and axon terminals on the basis of established ultrastructural features (Peters et al., 1991). The relative percentages of labeled elements were calculated by dividing the number of immunoreactive elements of a specific category by the total number of labeled elements in the striatum and thalamus. The same material was used to quantify the relative percentage of immunoreactive and non-immunoreactive spines in the striatum. From each micrograph, spine heads were categorized as labeled or unlabeled based on the presence or not of the DAB amorphous deposit. The respective values for each group were then divided by the total number of spines counted in the striatal tissue and expressed as the percentage of labeled spines in the striatum.
Tissue fractionation, immunoblotting and immunoprecipitation
Two sets of procedures were used. The first set was employed for analysis of wild type and Gβ5-/-littermates. Adult wild type and Gβ5-/- littermates were sacrificed by inhalation of carbon dioxide vapor. Brains were removed and homogenized in 4 ml of TEBS buffer (20 mM Tris-HCl, pH 7.5, 1 mM EDTA, 50 mM NaCl, 2 mM β-mercaptoethanol, and protease inhibitor cocktail). The total brain homogenate was centrifuged at 150,000g for 1.5 h at 4°C. The supernatant fraction (cytosolic extract) was collected, and the pellet fraction containing membrane proteins was washed twice in ice cold TEBS buffer. The pellet was suspended in 4ml TEBS buffer containing 1% sodium cholate and left on ice for 15 min. The detergent extract was centrifuged at 150,000g for 45 min and the supernatant was retained as the membrane detergent extract. A second set of methods was used for all other fractionation and immunoprecipitation experiments. For subcellular fractionation of whole brain tissue, we used a Dounce homogenizer to disrupt tissue in homogenization buffer (25 mM Tris pH 7.4, 150 mM NaCl, 5 mM EDTA supplemented with protease inhibitors (Roche) and 1mM PMSF). The homogenate was passed through a 22 gauge needle at least 5 times. Brain lysates were centrifuged at 1000 g for 15 min to remove cell debris and then at 200,000g for 30 min to pellet membranes. The membrane pellet was washed twice in homogenization buffer, and suspended in detergent solubilization buffer (50 mM sodium phosphate, 150 mM NaCl, 1% Triton X-100, protease inhibitor tablets (Roche) and 1 mM PMSF) and incubated for 1 h at 4°C. The detergent extract was centrifuged at 200,000g for 30 min to remove insoluble matter. The detergent extract supernatant was used for immunoprecipitation. Immunoprecipitation of cytosolic and membrane fractions was performed using chicken or rabbit anti-R7BP antibodies, as follows. Briefly, tissues were homogenized in Triton buffer (50mM sodium phosphate, 150 mM NaCl, 1% Triton X-100, protease inhibitor tablets (Roche) and 1mM PMSF). Detergent insoluble material was removed by centrifugation (10,000g) for 20 min at 4°C. The resultant tissue extracts or membrane detergent extracts were pre-cleared by incubating 1h with PrecipHen agarose (Aves Labs) or Protein G Sepharose (Amersham Biosciences) beads according to the manufacturer’s instructions and then centrifuged at 1000g for 10 min. We then added 5 μl of affinity purified chicken R7BP antibody or 7 μl of anti- Gβ5 antibody to 1 mg of cleared protein extract. After a 2h incubation at 4°C, we added 50 μl of PrecipHen agarose or 70 μl of Protein G Sepharose (50% slurry) and incubated samples with gentle agitation overnight at 4°C. Beads were harvested by centrifugation at 1000g for 2 min and washed three times with 500 μl of Triton buffer. Proteins were eluted by adding SDS-PAGE sample buffer and boiling, and then resolved and detected by western blotting. For immunoblotting whole tissues, we homogenized intact tissue in RIPA buffer supplemented with protease inhibitor tablets (Roche) and 1mM PMSF. Cell debris and unbroken tissues were pelleted by centrifugation (15,000g) for 10 min at 4°C. Supernatant fractions were used directly for western blotting. For all immunoblotting procedures, blots were blocked for 1 h in 5% milk/TBST followed by overnight incubation with primary antibodies (anti-R7BP diluted 1:7000, anti-actin diluted 1:1000, affinity purified anti Gβ5 antibodies, generously provided by Dr. William Simonds (Zhang and Simonds, 2000), diluted 1:1000). Blots were washed and incubated for 1 h with HRP-conjugated secondary antibodies. Signals were detected with an ECL reagent kit (Amersham Biosciences).
Isolation of detergent resistant membrane fractions
We adapted published methods of isolating detergent resistant membranes (Shogomori et al., 2005) for use with mouse brain. For each experiment, two mouse brains were homogenized in TNE buffer (25 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, pH 7.4 containing 1 mM PMSF and protease inhibitor tablets (Roche)) using a Dounce homogenizer and several passes through a 22-gauge needle. To prepare detergent resistant membrane fractions, we added brain (2 mg of protein in 500 μl) to an equal volume of 2% Triton X-100 and mixed the samples by end-over-end agitation for 1 h at 4°C. We then added 2 ml of TNE buffer plus 80% sucrose (all sucrose solutions contained protease inhibitors) in an ultracentrifuge tube, overlaid this solution with 6 ml of TNE containing 38% sucrose and then 3 ml of TNE containing 5% sucrose. Gradients were subjected to centrifugation for 14 h at 200,000g at 4°C using an SW41Ti rotor. Beginning from the top, 10 fractions (1.2 ml each) were harvested. Aliquots of equal volume (15 μl) from each fraction were analyzed by SDS-PAGE and Western blotting using antibodies against the following proteins: caveolin (BD Biosciences; diluted 1:1000 in TBST plus 5% non-fat milk), flotillin (BD Biosciences; 1:2000), transferrin receptor (Zymed; 1:1000), PSD95 (Affinity Bioreagents; 1:2500), Giα (B087 (Linder et al., 1993); 1:5000); Gβ5 (ATDG antibody (Zhang and Simonds, 2000); 1: 2500); RGS7 ((Levay et al., 1999); 1:1000); R7BP (chicken 1: 7500); rabbit anti-chicken HRP-conjugate (Pierce, 1:5000); goat anti-mouse HRP-conjugate (Pierce, 1:5000) and goat anti-rabbit HRP-conjugate (Pierce, 1:5000).
Culturing primary neurons and generating N2a cells stably expressing FLAG-R7BP
Primary neurons were cultured essentially according to published protocols (Mao and Wang, 2001). Sprague Dawley rat pups (postnatal day 1-15) were sacrificed by decapitation. Brains were excised and placed separately in Petri dishes containing sterile PBS. Striatum, hippocampus and thalamus were isolated according to published procedures (Miller and Vogt, 1984, Misgeld and Dietzel, 1989), and placed in a dish containing ice-cold PBS for removal blood vessels and meninges. Tissue was cut in approximately 1 mm square pieces, incubated in sterile PBS containing 0.25% trypsin for 30 min at 37°C, and centrifuged for 2 min at 200g. Cells were dissociated mechanically by gentle trituration through fire-polished Pasteur pipettes (bore diameter: ∼0.4 mm) in sterile PBS containing bovine serum albumin (1 mg/ml), DNase I (10 μg/ml), and soybean trypsin inhibitor (Sigma; 0.5 mg/ml). After centrifugation for 5 min at 1000g, cells were suspended in DMEM/F12 medium containing 10% fetal bovine serum, B27 supplement (Invitrogen), glucose (10 g/l), gentamicin (10 mg/l), and penicillin/streptomycin (10 mg/l), stained with Trypan Blue and counted on a hemocytometer. Cell death, as assessed by Trypan Blue exclusion, was less than 5%, and the average yield was 0.9-1.2×106 cells per neonatal striatum. Cells were diluted to a final concentration of 3×105 cells/ml and plated in 0.01% poly-D-lysine-coated 6 well plates containing glass cover slides and incubated at 37°C in a 5% CO2 and humidified atmosphere. After 4 d, the medium was replaced by adding one-half volume of Neurobasal media (Invitrogen) containing B27 supplement and 5 μM cytosine arabinoside (Ara-c). The medium was changed every 3-5 d with Neurobasal media containing B27 supplement but lacking Ara-c.
Mouse Neuro2a (N2a) neuroblastoma cells were obtained from Dr. David Harris (Washington University, St. Louis, MO) and cultured in Dulbecco’s minimal essential media supplemented with 10% fetal bovine serum. N2a cell lines stably expressing mouse FLAG-R7BP were generated as described previously (Gorodinsky and Harris, 1995), with minor modifications. N2a cells, grown to 70% confluence in 6-well plates, were transfected using 2 μg of plasmid DNA expressing FLAG-tagged wild type R7BP (Drenan et al., 2005) and Effectene (QIAGEN) according to the manufacturer’s protocol. Colonies surviving selection (21 d) in Geneticin (500 μg/ml) were expanded, purified by limited dilution (10-20 cells/well in a 24-well plate), and analyzed for FLAG-R7BP expression by western blotting using chicken anti-R7BP antibodies and an anti-FLAG-M2 HRP conjugate (Sigma-Aldrich; 1:5000). Stable clones expressing FLAG-R7BP at levels similar to endogenous R7BP in mouse brain were maintained in Dulbecco’s minimal essential media supplemented with 10% fetal bovine serum and Geneticin. N2a cells were differentiated into neuron-like cells by serum starvation for 48 h. For confocal immunofluorescence microscopy, N2a cells were plated on cover slips, differentiated or not as described above and prepared for microscopy as described previously (Drenan et al., 2005). Confocal fluorescence microscopy was performed with a laser scanning confocal microsope (model LSM-510; Carl Zeiss Microimaging, Inc.), and the following antibodies: chicken anti-R7BP (1:1000), rabbit anti-R7BP (1:250), rabbit anti-Gβ5 (1:200 (Zhang and Simonds, 2000)), anti-FLAG-M2 FITC conjugate (Sigma-Aldrich; 1:500), and Alexa 488-conjugated goat anti-chicken and goat anti-rabbit (Molecular probes; 1:100). Nuclei were stained with DAPI (0.5 μg/ml).
RESULTS
Generation and characterization of R7BP antibodies
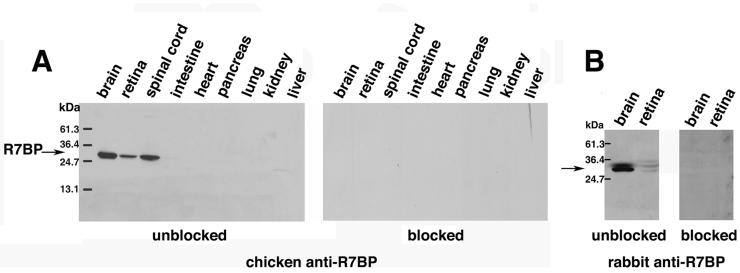
To analyze the expression and localization of endogenous R7BP and its ability to interact with R7-Gβ5 complexes in brain, we generated affinity-purified polyclonal anti-R7BP antibodies in chickens and rabbits. Previously described R7BP antibodies were unsuitable because they could not detect endogenous R7BP in Western blots of tissue extracts (Song et al., 2007). In contrast, our affinity-purified chicken R7BP antibody recognized a single species of the MW expected for R7BP (∼30kDa), which was detected readily in Western blots of total brain, retina and spinal cord extracts from mice (Figure 1A) or rats (see below), consistent with previous Northern blotting studies (Drenan et al., 2005). The 30kDa band was not detected when chicken antibodies were blocked with excess MBP-R7BP fusion protein. The 30kDa band in brain extracts was also the major species recognized by the rabbit antibody (Figure 1B). However, in both brain and retina extracts the rabbit antibody also recognized a minor species of slightly slower mobility that appeared to be a cross-reacting molecule, because it was not recognized when blots were stripped and reprobed with the chicken antibody (data not shown). No bands were detected when the rabbit antibody was blocked by preincubation with an excess of MBP-R7BP (Figure 1B). Despite some limited potential for cross-reactivity, both the chicken and rabbit antibodies detected very similar patterns of immunoreactivity in brain at both the light and electron microscopic levels (see below). In subsequent experiments we therefore used, as indicated, affinity-purified chicken and/or rabbit R7BP antibodies.
Figure 1.
Characterization of anti-R7BP antibodies. Immunoblots of mouse tissue extracts (25μg/lane) that were probed with affinity purified anti-R7BP antibodies from chicken (A) and rabbit (B) that were unblocked or blocked by preincubation with an excess of GST-R7BP fusion protein. Results shown are representative of four independent experiments.
R7-Gβ5 heterodimers are obligate binding partners of R7BP
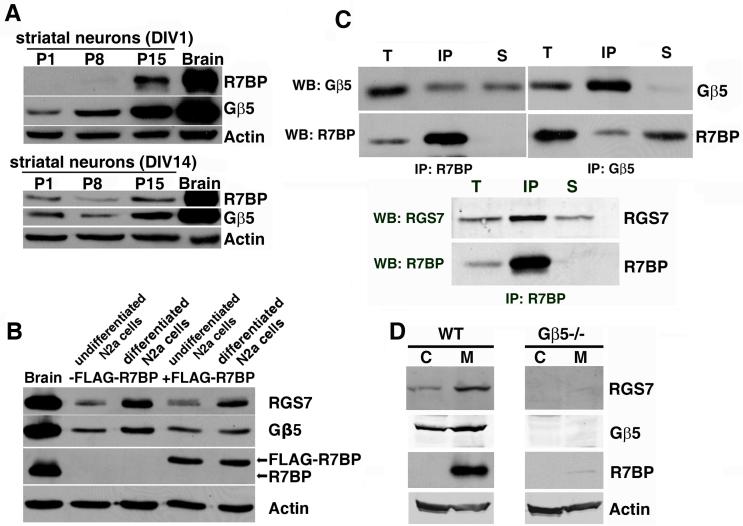
To determine whether R7-Gβ5 complexes are obligate binding partners of R7BP, we used several approaches. First, we investigated whether changes in R7BP expression affect the levels of Gβ5 and RGS7. With the goal of exploring the consequences of R7BP knockdown on R7 and Gβ5 expression, we initially isolated primary neurons from several regions of neonatal rat brain and determined whether they express R7BP at levels similar to that in adult rat brain. However, this approach proved unsuitable because, as shown in Figure 2A, primary striatal neurons expressed R7BP and Gβ5 at levels much lower than in adult brain, even when neurons were cultured for 1, 14 or 21 days in vitro (DIV; DIV21 results were identical to DIV14 data and are not shown) and were morphologically well differentiated. Equivalent results were obtained using primary neurons from hippocampus or thalamus, and when primary neurons were co-cultured with astrocytes (data not shown).
Figure 2.
Expression, subcellular fractionation and complex formation by R7BP, RGS7 and Gβ5 in differentiated N2a cells and brain. A) Immunoblot of R7BP, Gβ5 and β-actin expression in adult rat brain as compared to primary rat striatal neurons isolated at postnatal day P1, P8 or P15 and cultured 1 or 14 days in vitro (DIV). B) Immunoblots detecting expression of R7BP, RGS7, Gβ5 and β-actin in undifferentiated or differentiated N2a cells stably transfected or not with wild type FLAG-tagged R7BP. Results shown are representative of those obtained with three independently derived N2a cell lines stably expressing FLAG-R7BP. C) Reciprocal co-immunoprecipitation of R7BP, Gβ5 and RGS7 from detergent-extracted mouse brain membranes. The indicated R7BP (chicken), Gβ5 or RGS7 antibodies were used for immnoprecipitation and subsequent immunoblotting of the immunoprecipitates. The proportion of the total extract (T), immunoprecipitated material (IP), and non-immunoprecipitated material (S) analyzed in each lane was 6%, 25% and 6%, respectively. D) Immunoblots detecting expression of R7BP, Gβ5 and RGS7 in brain cytosol (C) and membrane (M) fractions from wild type and Gβ5-/- mice. Results shown in each panel are representative of 2-5 independent experiments.
As an alternative approach, we examined several commonly used cell lines (PC12, N2a, NG108-15) that can be differentiated into neuronal cells. When differentiated or undifferentiated, none of these cell lines expressed R7BP at levels detectable by immunoblotting, although undifferentiated and differentiated N2a cells did express detectable levels of endogenous RGS7 and Gβ5 (Figure 2B), thereby providing an R7BP-deficient cell type that we could stably transfect with a functional wild type FLAG-R7BP construct (Drenan et al., 2005). We generated three stably transfected N2a lines that, when undifferentiated or differentiated, expressed FLAG-R7BP at levels similar to endogenous R7BP in brain extracts as indicated by immunoblotting. As shown in Figure 2B, we found that expression of FLAG-R7BP modestly increased the levels of endogenous RGS7 in undifferentiated N2a cells. In contrast, FLAG-R7BP expression in undifferentiated or differentiated N2a cells did not appear to increase the levels of endogenous Gβ5. Therefore, in N2a cells R7BP appears to be an accessory rather than an essential binding partner of RGS7-Gβ5 complexes.
Next, we determined whether endogenous R7BP, RGS7 and Gβ5 form obligatory complexes in vivo. We used co-immunoprecipitation of R7BP with RGS7 or Gβ5 as indicators of heterotrimeric complex formation in brain detergent extracts. As shown in Figure 2C, Gβ5 and RGS7 were detected in R7BP immunoprecipitates, and R7BP was detected in Gβ5 immunoprecipitates, consistent with heterotrimer formation. We also determined the extent to which endogenous R7BP, RGS7 and Gβ5 co-fractionate in mouse brain extracts. As shown in Figure 2D, Gβ5 and RGS7 were detected in both the membrane and cytosol fractions, as reported previously (Witherow et al., 2000, Zhang and Simonds, 2000). In contrast, R7BP was detected only in the membrane fraction of brain extracts. Therefore, the membrane-bound but not the cytosolic pool of R7-Gβ5 complexes co-fractionates with R7BP. Lastly, we analyzed R7BP protein expression in brain extracts from wild type and Gβ5-/- mice (Chen et al., 2003). As shown in Figure 2D, R7BP and RGS7 protein levels were dramatically reduced in brain membrane fractions isolated from Gβ5-/- mice. Taken together, these lines of evidence indicated that R7-Gβ5 complexes are obligate binding partners of R7BP in vivo.
R7BP is sufficient to recruit endogenous RGS7-Gβ5 complexes to the plasma membrane
We hypothesized previously that palmitoylated R7BP functions as a membrane-anchoring subunit for R7-Gβ5 complexes (Drenan et al., 2005). However, this hypothesis was based on overexpression studies in non-neuronal cells. Furthermore, other processes, such as Goα overexpression or RGS7 palmitoylation (Rose et al., 2000, Takida et al., 2005), have also been proposed as plasma membrane recruitment mechanisms.
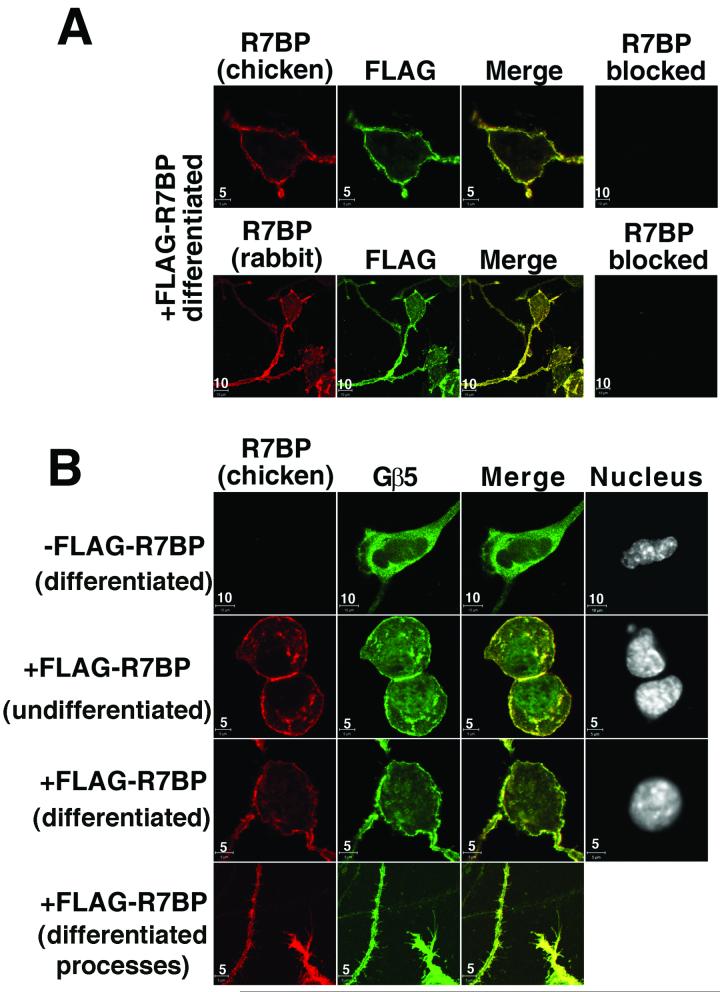
We therefore used confocal immunofluorescence microscopy of N2a cells described above that do or do not stably express wild type FLAG-R7BP to assess the ability of R7BP to function as a membrane anchoring protein for endogenous RGS7-Gβ5 complexes. First, we determined whether R7BP antibodies from chicken and/or rabbit could detect the subcellular localization of FLAG-R7BP expressed in N2a cells at levels approximating those of endogenous R7BP in brain. As shown in Figure 3A, extensive co-staining with R7BP and FLAG antibodies indicated that FLAG-R7BP is present mainly on the somatic plasma membrane and processes of differentiated N2a cells. Weak cytoplasmic staining was also observed but intranuclear staining was not, indicating that FLAG-R7BP is efficiently palmitoylated and membrane targeted in N2a cells. Staining obtained with R7BP antibodies from chicken or rabbit was specific because it was not detected when the antibodies were blocked or when cells lacking FLAG-R7BP were probed (Figure 3).
Figure 3.
R7BP recruits endogenous RGS7-Gβ5 complexes to the plasma membrane in differentiated N2a cells. A) R7BP antibodies from chicken or rabbit specifically detect FLAG-R7BP expressed in differentiated N2a cells, as indicated by co-staining with FLAG antibodies. B) R7BP recruits endogenous RGS7-Gβ5 heterodimers to the plasma membrane and dendrites in N2a cells. Undifferentiated or differentiated N2a cells stably transfected or not with FLAG-R7BP were stained with R7BP chicken antibodies, Gβ5 antibodies, and DAPI to detect nuclei. Results shown are representative of at least three independent experiments. Scale bars (5 or 10μm) are indicated.
Next, because Gβ5 and R7BP interact indirectly by binding independently to different domains of the R7 subunit (Drenan et al., 2005, Martemyanov et al., 2005, Drenan et al., 2006, Song et al., 2006), and because we showed above that RGS7-Gβ5 complexes co-immunoprecipitate with R7BP, we could use Gβ5 localization as a specific assay for recruitment of endogenous RGS7-Gβ5 complexes to the plasma membrane by FLAG-R7BP. By using Gβ5 antibodies in confocal immunofluorescence microscopy experiments, we found that Gβ5 was cytoplasmic in differentiated N2a cells lacking FLAG-R7BP (Figure 3B). However, in differentiated N2a cells expressing FLAG-R7BP, Gβ5 co-localized extensively with FLAG-R7BP on the somatic plasma membrane and cell processes, and also apparently on intracellular membranes (Figure 3B). Therefore, expression of FLAG-R7BP at levels approximating those of endogenous R7BP in brain tissue was sufficient to recruit endogenous RGS7-Gβ5 complexes in neuronal N2a cells mainly to the plasma membrane, and also possibly to intracellular organelles.
Analysis of R7BP association with detergent-resistant lipid “rafts”
Many palmitoylated signaling and scaffold proteins in the nervous system associate with specialized domains of the plasma membrane enriched in cholesterol and sphingolipids that can be isolated as detergent-resistant membrane (DRM) or lipid “raft” fractions on sucrose density gradients. Therefore, we determined whether R7-Gβ5-R7BP complexes co-fractionate as DRM-associated proteins in mouse brain extracts. We analyzed the fractionation of R7BP, Gβ5, RGS7, several palmitoylated DRM marker proteins (PSD-95, flotillin, caveolin, Giα subunits), and a non-DRM marker (transferrin receptor) on sucrose gradients of total brain membranes extracted at 4°C with 1% Triton X-100. Low or undetectable amounts of the total R7BP, RGS7 and Gβ5 pool co-fractionated with palmitoylated DRM marker proteins (Figure 4). In contrast, nearly all of the total R7BP, RGS7 and Gβ5 pool co-fractionated with a non-raft marker (transferrin receptor) under the conditions employed. Similar results were obtained when the detergent:membrane protein ratio was varied (data not shown).
Figure 4.
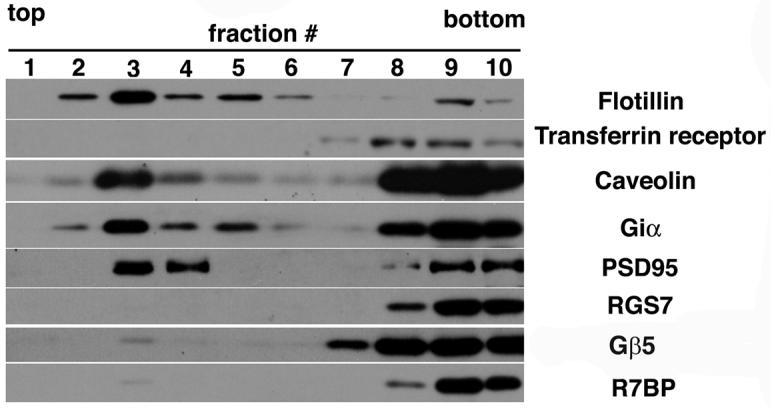
R7BP, RGS7 and Gβ5 in brain associate inefficiently with detergent-resistant lipid raft fractions. Immunoblot analysis of sucrose gradient fractions derived from total mouse brain membranes following extraction with Triton X-100. Blots were probed with antibodies for palmitoylated marker proteins (PSD95, Giα. flotillin and caveolin) known to associate with detergent-resistant lipid rafts, a non-lipid raft marker protein (transferrin receptor), as well as R7BP, Gβ5 and RGS7. The results shown are representative of three independent experiments.
We also investigated the effects of G protein activation on the association of RGS7-Gβ5-R7BP complexes with lipid raft-enriched fractions of brain membranes. We did so because in retinal rods an analogous heterotrimeric complex containing RGS9-1, Gβ5 and the R7BP-like protein R9AP exhibits augmented association with detergent-resistant lipid raft fractions following light stimulation or G protein (transducin) activation in vitro with Mg-GDP and AlF4- (Nair et al., 2002). We therefore determined whether the distribution of R7-Gβ5-R7BP complexes between raft and non-raft fractions in brain membrane extracts was affected when detergent extraction and fractionation experiments were performed in the presence versus the absence of Mg-GDP and AlF4-. No difference in the distributions of RGS7, Gβ5 or R7BP between raft and non-raft fractions was apparent under these two conditions (data not shown), even though endogenous Gi/o was activated by Mg-GDP and AlF4- treatment as indicated by its ability to co-immunoprecipitate with R7BP-containing complexes (data not shown). Therefore, under the conditions employed, R7-Gβ5-R7BP complexes associated inefficiently with lipid raft fractions whether their substrate G proteins were inactive or active.
R7BP and Gβ5 protein levels are upregulated during postnatal brain development
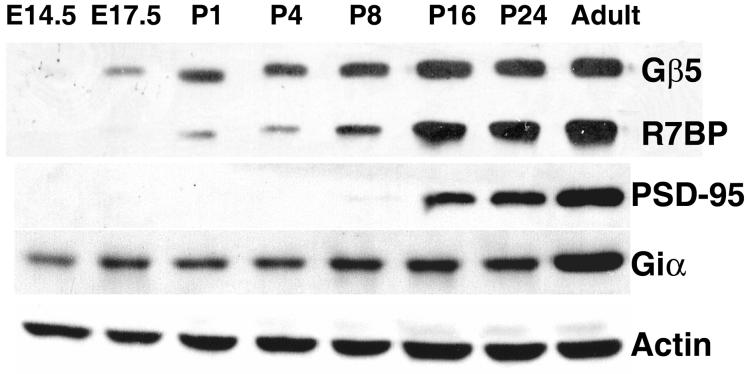
One potential reason why we noted above that R7BP is expressed at low levels in cultured primary neurons relative to adult brain is that R7BP expression may be developmentally regulated, which may occur inefficiently in vitro. Therefore, we analyzed R7BP and Gβ5 protein expression by performing Western blots of brain extracts obtained from embryonic, postnatal and adult mice. We also analyzed expression of PSD-95 (a marker of postnatal nervous system development (Sans et al., 2000)), Giα subunits (substrates of R7-Gβ5 complexes (Hooks et al., 2003)), and actin as controls. As shown in Figure 5, R7BP and Gβ5 were not detected at E14.5. Low-level expression of Gβ5 and R7BP was first detected at E17.5 and P1, respectively. Later during postnatal brain development (P16 and P24) Gβ5 and R7BP increased to near-adult levels. Upregulation of R7BP and Gβ5 was observed earlier in postnatal brain development than PSD-95, and was in marked contrast to the relatively constant expression of Giα subunits. These results therefore indicated that induction of R7-Gβ5-R7BP complexes is a previously unrecognized feature of postnatal brain development.
Figure 5.
R7BP and Gβ5 protein expression in developing mouse brain. Immunoblots of total protein (25μg/lane) extracts from whole mouse brains prepared at the indicated stages of embryonic or postnatal development were probed with antibodies directed against R7BP (chicken antibody), Giα, PSD95, Gβ5, and β-actin. Results shown are representative of three experiments using five mouse pups for each developmental stage.
R7BP is expressed in neurons throughout much of the CNS
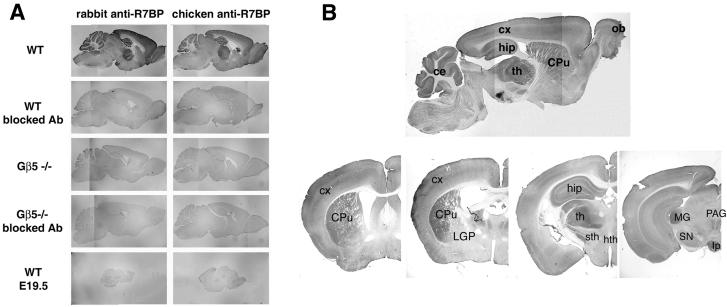
Although previous studies have shown that R7BP mRNA is widely expressed in brain (Drenan et al., 2005), the detailed regional, cellular and subcellular localization of R7BP protein in intact brain has yet to be reported. Accordingly, we used R7BP antibodies in immunohistochemistry and immunoelectron microscopy experiments to assess the distribution of R7BP immunoreactivity. We employed several specificity controls, including the use of affinity-purified R7BP antibodies from both chicken and rabbit, blocked antibodies, as well as developmental (embryonic E19.5 brain) and genetic (Gβ5-/- mice) controls where R7BP is absent or poorly expressed, as established by results presented above.
To analyze the regional expression of R7BP in brain, we probed free-floating frozen sections of adult mouse brains from three animals with affinity purified chicken or rabbit R7BP antibodies followed by DAB staining. With both antibodies, R7BP immunoreactivity was detected in many brain regions (Figure 6A and B; Table 1), including cerebral cortex, hippocampus, striatum, thalamus and cerebellum. In contrast, low background staining with either antibody was observed in all brain regions under all negative control conditions, including adult brain probed with blocked antibodies, adult brain from Gβ5-/- mice probed with blocked or unblocked antibodies, and E19.5 brains stained with unblocked antibodies.
Figure 6.
Regional distribution of R7BP protein expression in adult mouse brain. Free-floating frozen sections of brain were probed as indicated with unblocked or blocked anti-R7BP antibodies (chicken), followed by anti-chicken-HRP conjugates and visualization by DAB staining. A) Specificity of R7BP antibody staining. Sagittal sections of brains from adult wild type mice that express R7BP were compared with sections from brains of adult Gβ5-/- mice or E19.5 wild type mice, which do not express R7BP. B) Anti-R7BP antibody staining of sagittal sections and a series of coronal sections (0.5 mm to 3.28 mm from the bregma suture). Abbreviations: ec, cerebellum; CPu, caudate-putamen; cx, cortex; hip, hippocampus; th, thalamus; LGP, lateral globus pallidus; sth, subthalamic nucleus; hth, hypothalamus; PAG, periaqueductal grey; MG, medial geniculate nucleus; SN, substantia nigra; Ip, interpeduncular nucleus. Results shown are representative of more than five independent experiments in which sections of brains from 3 animals were used.
Table 1.
Expression pattern of R7BP in adult mouse brain detected by DAB staining
| Neocortex | |
| Layer I/ II/III | +++ |
| Layer IV | ++ |
| Layer V | ++ |
| Layer VI | +++ |
| Basal ganglia | |
| Caudate-putamen | +++ |
| Globus pallidus | + |
| Subthalamic nucleus | +++ |
| Ventral striatum | + |
| Brainstem | |
| Periaqueductal grey | + |
| substantia nigra pars reticulate | ++ |
| substantia nigra pars compacta | - |
| Ventral tegmental area | - |
| Hippocampus | |
| CA1/ CA2 | +++ |
| CA3 | +++ |
| DG | +++ |
| Thalamus | |
| Ventrobasal nuclei | +++ |
| Geniculate bodies | +++ |
| Anterior nuclei | + |
| Midline nuclei | + |
| Hypothalamus | + |
| Cerebellum | |
| Purkinje cells | ++++ |
| Granular cell layer | +++ |
| Molecular cell layer | ++ |
| Olfactory bulb | |
| Olfactory nerve layer | +++ |
| Glomerular layer | ++ |
| Mitral cell layer | ++ |
| Granular cell layer | ++ |
Semi-quantitive evaluation of R7BP staining was done by visual inspecting serial coronal and sagittal sections probed with anti-R7BP antibodies, followed by anti-chicken-HRP conjugates and visualized by DAB staining. Two investigators independently scored the same immunohistochemical sections. Staining intensities relative to the highest level observed (Purkinje cell layer) are indicated as follows: very dense (equivalent to that in the Purkinje cell layer), ++++; dense, +++; moderately dense, ++; light (visible above background), +; not detectable (indistinguishable from nonspecific background), -.
The intensity of R7BP immunoreactivity differed from region to region (Figure 6A and B; Table 1), in a pattern similar to that of Gβ5 (Brunk et al., 1999, Liang et al., 2000). R7BP staining appeared to be most intense in the Purkinje cell layer of the cerebellum, which was used as a reference for other brain regions. Dense R7BP immunoreactivity was observed in the subthalamic nucleus, thalamus, hippocampus, cerebellum, cerebral cortex, striatum and olfactory bulb. In hippocampus, strong immunoreactivity was found in all CA fields (CA1-CA3) and in the dentate gyrus. Analysis of the thalamus revealed moderate to high R7BP expression level in most regions examined, except for midline and anterior nuclei that had significantly less immunoreactivity than other nuclear groups. In contrast, the ventrobasal nuclei and geniculate bodies were the most strongly labeled thalamic structures (Figure 6B). In the cerebellum, the Purkinje and granule cell layers exhibited intense R7BP staining, whereas the molecular layer stained less strongly. Within the basal ganglia and associated regions, intense R7BP antibody staining was observed in the caudate putamen, but significantly less labeling was found in the ventral striatum. The subthalamic nucleus was also intensely labeled, while weaker immunoreactivity was found in the substantia nigra pars reticulata and the globus pallidus (Figure 6B). In the olfactory bulb, R7BP expression was detected predominantly in the olfactory nerve layer; other areas of the olfactory bulb showed weaker expression of R7BP. R7BP immunoreactivity was intense in all layers of cerebral cortex, except for moderate labeling in layers IV and V. The white matter was devoid of R7BP immunoreactivity.
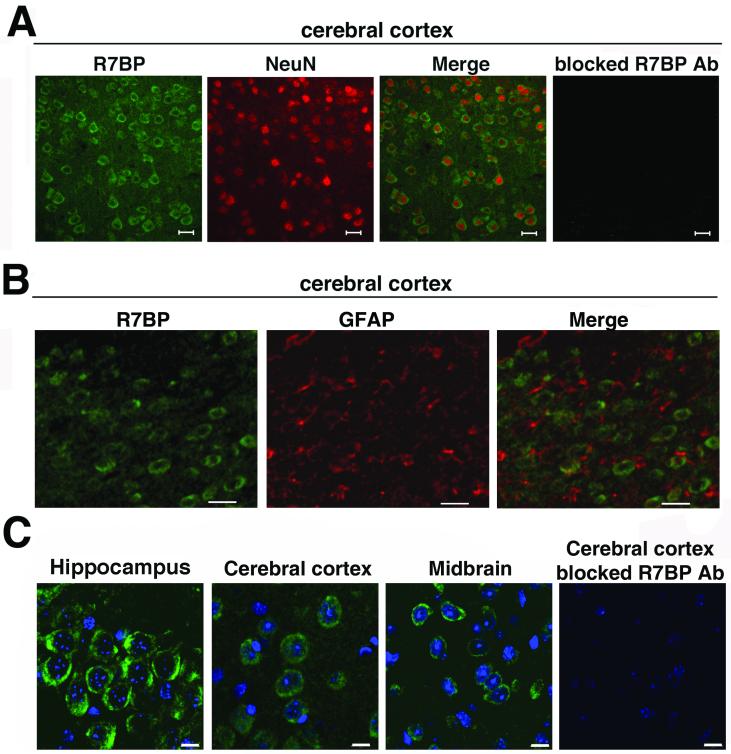
To determine whether R7BP is expressed in neurons, we co-stained paraffin-embedded brain sections from three mice with R7BP chicken antibodies and an antibody for the neuron-specific marker NeuN, followed by incubation with fluorescent secondary antibodies and visualization by confocal fluorescence microscopy. This approach proved to be somewhat difficult because conditions that resulted in strong NeuN staining reduced the intensity of R7BP staining, and vice versa. We therefore used conditions that preserved specific R7BP staining, which reduced the intensity of NeuN staining. Under these conditions, the clearest results were obtained with cerebral cortical sections including lamina I-III, possibly due to a combination of the lamellar organization and relatively high levels of R7BP expression in this region. As shown in Figure 7A, fluorescence confocal microscopy indicated that R7BP immunoreactivity was detected in soma. Analysis of R7BP and NeuN co-staining indicated that 98% of all cells that were NeuN-positive (n=100) were also R7BP-positive. Because under the conditions employed NeuN staining was relatively weak, we could not accurately quantify the fraction of all R7BP-positive cells that were also NeuN-positive. However, it was clear that not all cells in this region of the cerebral cortex expressed R7BP, because only 60% of all cells (n=115; marked by the DNA dye; Figure 7C) displayed R7BP-positive staining. itive cells failed to stain with an antibody specific for GFAP (Figure 7B), an astrocyte-specific markeresults indicated that R7BP in cortex is expressed mainly and perhaps exclusively in neurons, and not in astrocytes.
Figure 7.
R7BP is expressed in neurons but not astrocytes. R7BP protein expression in neurons or astrocytes was analyzed by performing fluorescence confocal microscopy of sections co-stained with chicken anti-R7BP antibody (green) and NeuN antibody (red; panel A), which specifically marks neuronal cell bodies, or GFAP antibody (red; panel B), which s astro Adjacent cortical sections stained with blocked R7BP antibodies are shown. C) High magnification images of R7BP immunoreactivity detected in hippocampal, midbrain, and cerebellar sections stained with chicken anti-R7BP antibodies (green), and the DNA dye DAPI (blue). Scale bars =20 μm (A and B), 10 μm (C). Results shown are representative of 2-5 independent experiments in which sections of brains from 3 animals were used.
Analysis of sections from cerebral cortex, hippocampus and midbrain (Figure 7) indicated strong somatic R7BP staining. In no case did we detect R7BP staining within the nucleus, even at high magnification (Figure 7C and data not shown). Because R7BP staining was diffuse, the results could not address whether R7BP localizes to the somatic plasma membrane, neuronal processes or intracellular organelles.
Analysis of R7BP localization by immunoelectron microscopy
To determine the subcellular pattern of R7BP labeling at higher resolution, we used immunoelectron microscopy to analyze the ultrastructural localization of R7BP immunoreactivity in adult rat brain sections. Rats were used because we have extensive experience with this system, because our affinity purified polyclonal antibodies can specifically detect rat R7BP in Western blots (Figure 2A), and because the regional pattern of R7BP immunoreactivity in rat brain (data not shown) proved to be the same as described above in mice (Figure 6).
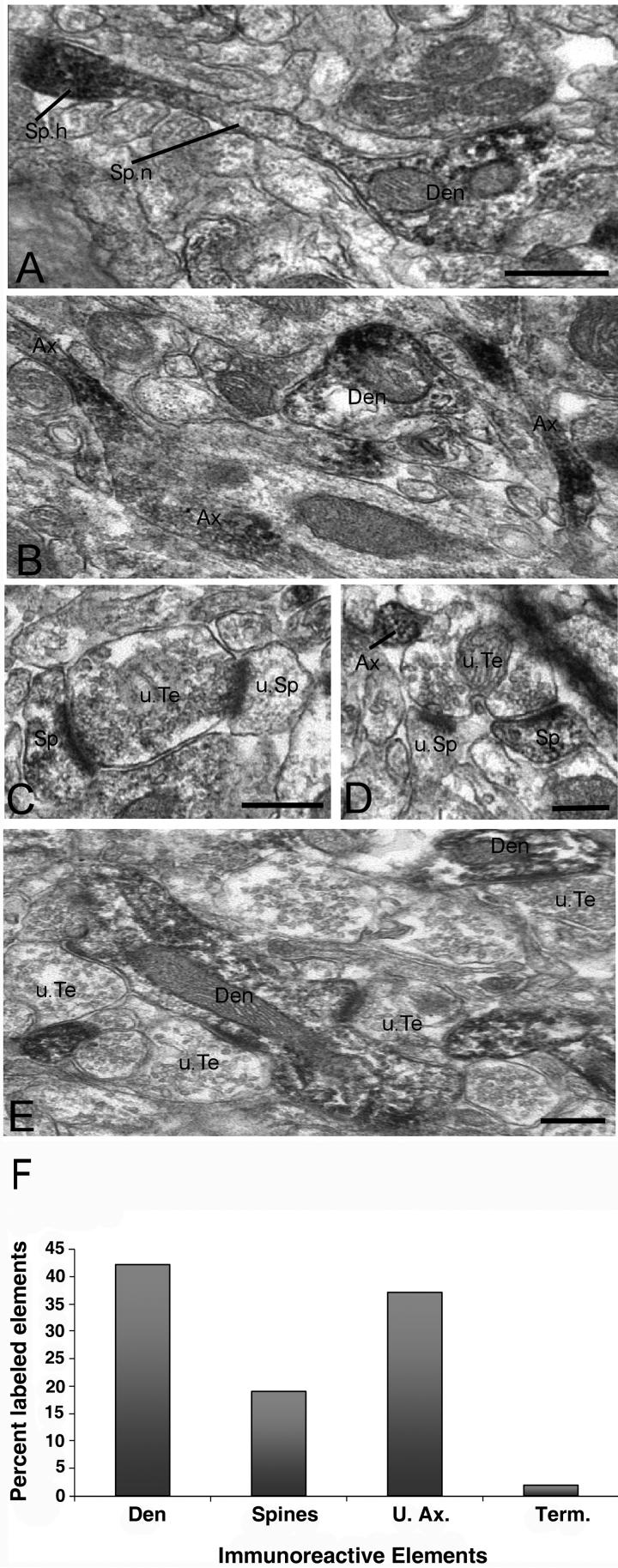
At the electron microscopic level, we focused on R7BP-immunoreactive elements in the striatum and ventrolateral thalamus. R7BP-positive elements were easily differentiated from unlabeled structures by the presence of amorphous peroxidase deposit (Figure 8). In general, the DAB deposit homogeneously filled labeled structures without obvious association with the plasma membrane, synapses or specific intracellular organelles (Figure 8). Labeling was present within somatic and non-somatic neuronal structures. In perikarya, labeling was diffuse but undetectable within the nucleus (data not shown), similar to the results of immunofluorescence experiments described above. Excluding somatic elements, quantitative data collected from three rats showed that dendrites (42%) and unmyelinated axons (37%) accounted for the largest pool of R7BP-immunoreactive elements in the striatum (Figure 8). R7BP immunoreactivity was also detected significantly in spines (19%), whereas axon terminals and myelinated axons were largely devoid of labeling (Figure 8).
Figure 8.
Ultrastrutctural analysis of R7BP-immunoreactive neuronal elements in the rat striatum (A-D) and ventrolateral thalamus (E). A) Immunoreactive dendritic shaft (Den) from which grows a dendritic spine (Sp). Note the increased R7BP expression in the spine head (Sp.h.) compared to the relatively low level of labeling along the neck (Sp.n.). B) Examples of immunoreactive unmyelinated axons (Ax). A labeled dendrite is seen in the same field. C-D) Examples of immunoreactive spines (Sp) contacted by putative unlabeled glutamatergic terminals (u.Te). Note that the same terminals also contact non-immunoreactive spines (u.Sp). An immunoreactive unmyelinated axon is depicted in D. E) Typical example of R7BP labeling in the ventrolateral thalamus. Dendritic shafts (Den) contacted by a multitude of non-immunoreactive boutons that form asymmetric synapses (u.Te) account for most of R7BP-immunoreactive elements in the thalamus. Scale bars: 0.5 μm in panels A (also valid for panel B); 0.5 μm in panels C-E. F) Histogram showing the relative percentage of R7BP-immunoreactive neuronal elements in the rat striatum. A total of 1262 immunoreactive elements were photographed in the dorsolateral striatum of three rats. Abbreviations: Den, dendrites; U.Ax, unmyelinated axons; Term, terminals.
Knowing that striatal spines display a significant level of neurochemical specificity and receive glutamatergic inputs from different sources (Lei et al., 2004, Raju et al., 2006), we assessed the degree of heterogeneity of R7BP expression associated with striatal spines through quantitative analysis of the relative percentages of immunoreactive and non-immunoreactive spine heads in the rat caudate-putamen complex. The analysis of 521 spines in striatal tissue from three rats revealed that 46% of striatal spines displayed R7BP immunoreactivity, while 54% were non-immunoreactive. Each of the R7BP-positive spines received asymmetric synapses from non-immunoreactive axon terminals (Figure 8C and D). In some instances where single terminals formed synapses with two spines, one spine displayed strong R7BP labeling while the other was devoid of immunoreactivity (Figure 8C and D), clearly indicating a high degree of specificity in striatal R7BP distribution. In contrast, thalamic R7BP labeling was found exclusively in dendrites of projection neurons (Figure 8E), some of which were heavily contacted by putative glutamatergic inputs.
Due to the limited spatial resolution of the immunoperoxidase deposit, these findings did not provide detailed information about the subsynaptic localization of R7BP. However, in both striatal and thalamic tissue, glial processes were devoid of R7BP labeling, reinforcing the conclusion that R7BP is expressed only in neurons.
DISCUSSION
Members of the RGS7 (R7) family and Gβ5 form heterodimeric complexes that are required for normal regulation of GPCR function in retina and the central nervous system (Chen et al., 2000, Chen et al., 2003, Krispel et al., 2003, Rahman et al., 2003, Zachariou et al., 2003, Keresztes et al., 2004, Nishiguchi et al., 2004, Krispel et al., 2006). To advance understanding of R7-Gβ5 complexes in brain, we have focused on R7BP, a recently identified palmitoylated protein that, in heterologous expression systems, binds to and regulates the localization and function of R7-Gβ5 complexes (Drenan et al., 2005, Drenan et al., 2006, Song et al., 2006). Here we have provided evidence indicating that R7 proteins, Gβ5 and R7BP form obligate heterotrimers in vivo, that R7BP is sufficient for targeting endogenous RGS7-Gβ5 complexes to membranes of neuronal cells, that R7BP and Gβ5 are strikingly upregulated during postnatal brain development, and that R7BP is present principally in neuronal soma, dendrites and axons, but is present at low levels or absent in nerve terminals, glia and detergent-resistant lipid raft fractions. These findings and their implications for understanding the roles of R7-Gβ5-R7BP complexes in brain are discussed below.
R7-Gβ5 complexes appear to be obligatory binding partners of R7BP in brain, because we have found that: 1) R7BP and RGS7 protein expression is dramatically reduced in Gβ5-/- mice, probably due to proteolytic degradation (Chen et al., 2003); 2) R7BP in brain membrane extracts co-immunoprecipitates with RGS7 and Gβ5; and 3) the regional distributions of R7BP, Gβ5 and the combined R7 family are highly coincident (Gold et al., 1997, Brunk et al., 1999, Liang et al., 2000). However, it also appears that R7-Gβ5 dimers free of R7BP also exist in brain, because we found that R7BP associates with brain membrane fractions, whereas RGS7 and Gβ5 are readily detected in both membrane and cytoplasmic fractions. Indeed, we found in neuronal N2a cells, which do not express endogenous R7BP, that endogenously expressed Gβ5 is cytoplasmic, indicating that R7BP is not an essential binding partner of R7-Gβ5 complexes. Consistent with this hypothesis, R7-Gβ5 complexes can function in heterologous expression systems as G protein regulators with or without R7BP (Drenan et al., 2005, Drenan et al., 2006, Narayanan et al., 2007). Therefore, it is possible that R7-Gβ5 complexes in brain may function with R7BP on the membrane and free of R7BP in the cytoplasm.
A second main conclusion is that R7BP is sufficient for plasma membrane recruitment of endogenously expressed RGS7-Gβ5 complexes in neuronal cells. This conclusion is supported by our finding that expression of FLAG-R7BP in neuronal N2a cells at levels approximating that of endogenous R7BP in adult brain was sufficient to recruit endogenous RGS7-Gβ5 complexes to the plasma membrane and also possibly to intracellular organelles.
Although R7BP is a membrane-bound protein that can be palmitoylated, we found that RGS7-Gβ5-R7BP complexes in brain extracts associate inefficiently with detergent-resistant lipid raft fractions. Furthermore, G protein activation did not promote raft association of RGS7-Gβ5-R7BP heterotrimers. These findings are in contrast to the properties of complexes in retina containing RGS9-1, Gβ5 and the R7BP-like protein R9AP, which exhibit lipid raft association that is augmented following activation of the G protein transducin (Nair et al., 2002). The properties of complexes containing R7BP also appear to be in contrast to many palmitoylated proteins in brain, including PSD-95 and Gi/o-class α subunits, which are enriched in lipid rafts (reviewed in (Pike, 2004, Smotrys and Linder, 2004)). The mechanism responsible for the apparently inefficient association of RGS7-Gβ5-R7BP complexes with raft fractions is unclear. One possibility is that R7BP in brain is not palmitoylated, but instead is modified by unsaturated fatty acids, which are known to reduce protein association with lipid rafts (Liang et al., 2001, Zacharias et al., 2002). Regardless of the mechanism, the association of R7-Gβ5-R7BP complexes with non-raft domains would not preclude their interaction with raft-associated G protein substrates, because lipid rafts are thought to be dynamic rather than static structures and proteins can shuttle between raft and non-raft domains (Kenworthy et al., 2004, Nicolau et al., 2006).
Although many GPCRs and Gi/o-class Gα subunits that can be regulated by the GAP activity of R7-Gβ5-R7BP complexes are well expressed in brain both before and after birth (Turgeon and Albin, 1994, Wang et al., 2003a, Wang et al., 2003b, Lopez-Bendito et al., 2004, Bianchi et al., 2005, Lujan and Shigemoto, 2006), we found that R7BP and Gβ5 proteins are not expressed significantly in embryonic brain. Instead, R7BP and Gβ5 protein levels in brain are strikingly upregulated to near-adult levels during the first two to three weeks of postnatal life. R7 proteins would also be expected to be upregulated postnatally because their stable accumulation requires Gβ5 (He et al., 2000, Witherow et al., 2000, Chen et al., 2003). Postnatal induction of R7-Gβ5-R7BP complexes may occur at the level of mRNA accumulation, as suggested by studies of Gβ5 (Zhang et al., 2000). We speculate that R7-Gβ5-R7BP complex induction is functionally important because it occurs during the same time period in which Gβ5-/- mice fail to gain weight (postnatal day 15-20 (Chen et al., 2003).
Our immunofluorescence and immunoelectron microscopic analyses in brain have revealed diverse patterns of R7BP immunoreactivity. Both approaches indicated that R7BP is expressed in neurons but not glia. Immunoelectron microscopy indicated that R7BP staining is readily detected in neuronal soma, dendrites, spines and unmyelinated axons, but not in myelinated axons and axon terminals. This pattern is consistent with that described for many Gi/o-coupled GPCRs in the striatum (Smith et al., 2000, Smith et al., 2001, Galvan et al., 2006). Interestingly, less than half of all spines in the striatum appeared to be R7BP-positive, possibly resembling the overall distribution of D1 and D2 dopamine receptors in striatal spines which mark, respectively, direct and indirect pathway neurons (Gerfen et al., 1990). In this case, we speculate that R7BP might associate preferentially with one striatofugal pathway versus another.
Another notable finding of our ultrastructural studies is that R7BP immunoreactivity was detected in striatal but not thalamic axons. This pattern suggests that R7-Gβ5-R7BP complexes could have different roles in striatum vs. thalamus, which express various Gi/o-coupled receptors pre- or post-synaptically (Charara et al., 2000, Charara et al., 2004, Galvan et al., 2004, Galvan et al., 2006, Villalba et al., 2006). The mechanisms that determine the differential targeting of R7BP to certain axons, dendrites or spines remain to be elucidated.
In conclusion, our findings suggest that R7-Gβ5-R7BP complexes in brain have the potential to regulate signaling by modulatory GPCRs. Further work promises to establish the precise physiologic and mechanistic roles of R7 family members, Gβ5 and R7BP in brain development and/or function.
ACKNOWLEDGEMENTS
D.G. and M.J. are co-first authors who made equal contributions to this study. D.G. was supported in part by a postdoctoral fellowship from the American Heart Association (0520080Z). Grant support from the NIH (GM44592 and HL075632 to K.J.B.; NS037423 and NS042937 to Y.S.; GM060019 to V.S.; and RR00165 to the Yerkes National Primate Center) is gratefully acknowledged. We thank Benedict Kolber, and Drs. Stephen J. Gold, Lou Muglia, Joshua B. Rubin and for providing advice, Dr. William Simonds for the generous gift of affinity purified Gβ5 antibodies, and Dr. Jason Chen for providing Gβ5-/+ mice to V.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Berman DM, Wilkie TM, Gilman AG. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- Bianchi MS, Lux-Lantos VA, Bettler B, Libertun C. Expression of gamma-aminobutyric acid B receptor subunits in hypothalamus of male and female developing rats. Developmental Brain Research. 2005;160:124. doi: 10.1016/j.devbrainres.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Brunk I, Pahner I, Maier U, Jenner B, Veh RW, Nurnberg B, Ahnert-Hilger G. Differential distribution of G-protein beta-subunits in brain: an immunocytochemical analysis. Eur J Cell Biol. 1999;78:311–322. doi: 10.1016/s0171-9335(99)80065-x. [DOI] [PubMed] [Google Scholar]
- Cabrera JL, de Freitas F, Satpaev DK, Slepak VZ. Identification of the Gbeta5-RGS7 protein complex in the retina. Biochem Biophys Res Commun. 1998;249:898–902. doi: 10.1006/bbrc.1998.9218. [DOI] [PubMed] [Google Scholar]
- Charara A, Galvan A, Kuwajima M, Hall RA, Smith Y. An electron microscope immunocytochemical study of GABA(B) R2 receptors in the monkey basal ganglia: a comparative analysis with GABA(B) R1 receptor distribution. J Comp Neurol. 2004;476:65–79. doi: 10.1002/cne.20210. [DOI] [PubMed] [Google Scholar]
- Charara A, Heilman TC, Levey AI, Smith Y. Pre- and postsynaptic localization of GABA(B) receptors in the basal ganglia in monkeys. Neuroscience. 2000;95:127–140. doi: 10.1016/s0306-4522(99)00409-1. [DOI] [PubMed] [Google Scholar]
- Chen CK, Burns ME, He W, Wensel TG, Baylor DA, Simon MI. Slowed recovery of rod photoresponse in mice lacking the GTPase accelerating protein RGS9-1. Nature. 2000;403:557–560. doi: 10.1038/35000601. [DOI] [PubMed] [Google Scholar]
- Chen CK, Eversole-Cire P, Zhang H, Mancino V, Chen YJ, He W, Wensel TG, Simon MI. Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc Natl Acad Sci U S A. 2003;100:6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhami GK, Ferguson SS. Regulation of metabotropic glutamate receptor signaling, desensitization and endocytosis. Pharmacol Ther. 2006;111:260–271. doi: 10.1016/j.pharmthera.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Drenan RM, Doupnik CA, Boyle MP, Muglia LJ, Huettner JE, Linder ME, Blumer KJ. Palmitoylation regulates plasma membrane-nuclear shuttling of R7BP, a novel membrane anchor for the RGS7 family. J Cell Biol. 2005;169:623–633. doi: 10.1083/jcb.200502007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenan RM, Doupnik CA, Jayaraman M, Buchwalter AL, Kaltenbronn KM, Huettner JE, Linder ME, Blumer KJ. R7BP augments the function of RGS7*Gbeta5 complexes by a plasma membrane-targeting mechanism. J Biol Chem. 2006;281:28222–28231. doi: 10.1074/jbc.M604428200. [DOI] [PubMed] [Google Scholar]
- Gainetdinov RR, Premont RT, Bohn LM, Lefkowitz RJ, Caron MG. Desensitization of G protein-coupled receptors and neuronal functions. Annu Rev Neurosci. 2004;27:107–144. doi: 10.1146/annurev.neuro.27.070203.144206. [DOI] [PubMed] [Google Scholar]
- Galvan A, Charara A, Pare JF, Levey AI, Smith Y. Differential subcellular and subsynaptic distribution of GABA(A) and GABA(B) receptors in the monkey subthalamic nucleus. Neuroscience. 2004;127:709–721. doi: 10.1016/j.neuroscience.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Galvan A, Kuwajima M, Smith Y. Glutamate and GABA receptors and transporters in the basal ganglia: what does their subsynaptic localization reveal about their function? Neuroscience. 2006;143:351–375. doi: 10.1016/j.neuroscience.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, Engber TM, Mahan LC, Susel Z, Chase TN, Monsma FJ, Jr., Sibley DR. D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science. 1990;250:1429–1432. doi: 10.1126/science.2147780. [DOI] [PubMed] [Google Scholar]
- Gold SJ, Ni YG, Dohlman HG, Nestler EJ. Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J Neurosci. 1997;17:8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorodinsky A, Harris DA. Glycolipid-anchored proteins in neuroblastoma cells form detergent-resistant complexes without caveolin. J Cell Biol. 1995;129:619–627. doi: 10.1083/jcb.129.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Lu L, Zhang X, El-Hodiri HM, Chen CK, Slep KC, Simon MI, Jamrich M, Wensel TG. Modules in the photoreceptor RGS9-1.Gbeta 5L GTPase-accelerating protein complex control effector coupling, GTPase acceleration, protein folding, and stability. J Biol Chem. 2000;275:37093–37100. doi: 10.1074/jbc.M006982200. [DOI] [PubMed] [Google Scholar]
- Hooks SB, Waldo GL, Corbitt J, Bodor ET, Krumins AM, Harden TK. RGS6, RGS7, RGS9, and RGS11 Stimulate GTPase Activity of Gi Family G-proteins with Differential Selectivity and Maximal Activity. J Biol Chem %R 101074/jbcM211382200. 2003;278:10087–10093. doi: 10.1074/jbc.M211382200. [DOI] [PubMed] [Google Scholar]
- Hunt TW, Fields TA, Casey PJ, Peralta EG. RGS10 is a selective activator of G alpha i GTPase activity. Nature. 1996;383:175–177. doi: 10.1038/383175a0. [DOI] [PubMed] [Google Scholar]
- Jones MB, Siderovski DP, Hooks SB. The G{beta}{gamma} DIMER as a NOVEL SOURCE of SELECTIVITY in G-Protein Signaling: GGL-ing AT CONVENTION. Mol Interv. 2004;4:200–214. doi: 10.1124/mi.4.4.4. [DOI] [PubMed] [Google Scholar]
- Kenworthy AK, Nichols BJ, Remmert CL, Hendrix GM, Kumar M, Zimmerberg J, Lippincott-Schwartz J. Dynamics of putative raft-associated proteins at the cell surface. J Cell Biol. 2004;165:735–746. doi: 10.1083/jcb.200312170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keresztes G, Martemyanov KA, Krispel CM, Mutai H, Yoo PJ, Maison SF, Burns ME, Arshavsky VY, Heller S. Absence of the RGS9.Gbeta5 GTPase-activating complex in photoreceptors of the R9AP knockout mouse. J Biol Chem. 2004;279:1581–1584. doi: 10.1074/jbc.C300456200. [DOI] [PubMed] [Google Scholar]
- Kovoor A, Chen CK, He W, Wensel TG, Simon MI, Lester HA. Co-expression of Gbeta5 enhances the function of two Ggamma subunit-like domain-containing regulators of G protein signaling proteins. J Biol Chem. 2000;275:3397–3402. doi: 10.1074/jbc.275.5.3397. [DOI] [PubMed] [Google Scholar]
- Krispel CM, Chen CK, Simon MI, Burns ME. Prolonged photoresponses and defective adaptation in rods of Gbeta5-/- mice. J Neurosci. 2003;23:6965–6971. doi: 10.1523/JNEUROSCI.23-18-06965.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krispel CM, Chen D, Melling N, Chen YJ, Martemyanov KA, Quillinan N, Arshavsky VY, Wensel TG, Chen CK, Burns ME. RGS expression rate-limits recovery of rod photoresponses. Neuron. 2006;51:409–416. doi: 10.1016/j.neuron.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- Levay K, Cabrera JL, Satpaev DK, Slepak VZ. Gbeta5 prevents the RGS7-Galphao interaction through binding to a distinct Ggamma-like domain found in RGS7 and other RGS proteins. Proc Natl Acad Sci U S A. 1999;96:2503–2507. doi: 10.1073/pnas.96.5.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JJ, Chen HH, Jones PG, Khawaja XZ. RGS7 complex formation and colocalization with the Gbeta5 subunit in the adult rat brain and influence on Gbeta5gamma2-mediated PLCbeta signaling. J Neurosci Res. 2000;60:58–64. doi: 10.1002/(SICI)1097-4547(20000401)60:1<58::AID-JNR6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Liang X, Nazarian A, Erdjument-Bromage H, Bornmann W, Tempst P, Resh MD. Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J Biol Chem. 2001;276:30987–30994. doi: 10.1074/jbc.M104018200. [DOI] [PubMed] [Google Scholar]
- Linder ME, Middleton P, Hepler JR, Taussig R, Gilman AG, Mumby SM. Lipid modifications of G proteins: alpha subunits are palmitoylated. Proc Natl Acad Sci U S A. 1993;90:3675–3679. doi: 10.1073/pnas.90.8.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bendito G, Shigemoto R, Kulik A, Vida I, Fairen A, Lujan R. Distribution of metabotropic GABA receptor subunits GABAB1a/b and GABAB2 in the rat hippocampus during prenatal and postnatal development. Hippocampus. 2004;14:836–848. doi: 10.1002/hipo.10221. [DOI] [PubMed] [Google Scholar]
- Lujan R, Shigemoto R. Localization of metabotropic GABA receptor subunits GABAB1 and GABAB2 relative to synaptic sites in the rat developing cerebellum. Eur J Neurosci. 2006;23:1479–1490. doi: 10.1111/j.1460-9568.2006.04669.x. [DOI] [PubMed] [Google Scholar]
- Makino ER, Handy JW, Li T, Arshavsky VY. The GTPase activating factor for transducin in rod photoreceptors is the complex between RGS9 and type 5 G protein beta subunit. Proc Natl Acad Sci U S A. 1999;96:1947–1952. doi: 10.1073/pnas.96.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Wang JQ. Upregulation of preprodynorphin and preproenkephalin mRNA expression by selective activation of group I metabotropic glutamate receptors in characterized primary cultures of rat striatal neurons. Brain Res Mol Brain Res. 2001;86:125–137. doi: 10.1016/s0169-328x(00)00276-x. [DOI] [PubMed] [Google Scholar]
- Martemyanov KA, Yoo PJ, Skiba NP, Arshavsky VY. R7BP, a novel neuronal protein interacting with RGS proteins of the R7 family. J Biol Chem. 2005;280:5133–5136. doi: 10.1074/jbc.C400596200. [DOI] [PubMed] [Google Scholar]
- Miller MW, Vogt BA. Direct connections of rat visual cortex with sensory, motor, and association cortices. J Comp Neurol. 1984;226:184–202. doi: 10.1002/cne.902260204. [DOI] [PubMed] [Google Scholar]
- Misgeld U, Dietzel I. Synaptic potentials in the rat neostriatum in dissociated embryonic cell culture. Brain Res. 1989;492:149–157. doi: 10.1016/0006-8993(89)90897-4. [DOI] [PubMed] [Google Scholar]
- Nair KS, Balasubramanian N, Slepak VZ. Signal-dependent translocation of transducin, RGS9-1-Gbeta5L complex, and arrestin to detergent-resistant membrane rafts in photoreceptors. Curr Biol. 2002;12:421–425. doi: 10.1016/s0960-9822(02)00691-7. [DOI] [PubMed] [Google Scholar]
- Narayanan V, Sandiford SL, Wang Q, Keren-Raifman T, Levay K, Slepak VZ. Intramolecular interaction between the DEP domain of RGS7 and the Gbeta5 subunit. Biochemistry. 2007;46:6859–6870. doi: 10.1021/bi700524w. [DOI] [PubMed] [Google Scholar]
- Nicolau DV, Jr., Burrage K, Parton RG, Hancock JF. Identifying optimal lipid raft characteristics required to promote nanoscale protein-protein interactions on the plasma membrane. Mol Cell Biol. 2006;26:313–323. doi: 10.1128/MCB.26.1.313-323.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi KM, Sandberg MA, Kooijman AC, Martemyanov KA, Pott JW, Hagstrom SA, Arshavsky VY, Berson EL, Dryja TP. Defects in RGS9 or its anchor protein R9AP in patients with slow photoreceptor deactivation. Nature. 2004;427:75–78. doi: 10.1038/nature02170. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotaxic coordinates. Academic Press; New York: 1982. [DOI] [PubMed] [Google Scholar]
- Peters A, Palay SL, Webster HD. The Fine Structure of the Nervous System: Neurons and their Supporting Cells. Oxford University Press; New York: 1991. [Google Scholar]
- Pike LJ. Lipid rafts: heterogeneity on the high seas. Biochem J. 2004;378:281–292. doi: 10.1042/BJ20031672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman Z, Schwarz J, Gold SJ, Zachariou V, Wein MN, Choi KH, Kovoor A, Chen CK, DiLeone RJ, Schwarz SC, Selley DE, Sim-Selley LJ, Barrot M, Luedtke RR, Self D, Neve RL, Lester HA, Simon MI, Nestler EJ. RGS9 modulates dopamine signaling in the basal ganglia. Neuron. 2003;38:941–952. doi: 10.1016/s0896-6273(03)00321-0. [DOI] [PubMed] [Google Scholar]
- Rose JJ, Taylor JB, Shi J, Cockett MI, Jones PG, Hepler JR. RGS7 is palmitoylated and exists as biochemically distinct forms. J Neurochem. 2000;75:2103–2112. doi: 10.1046/j.1471-4159.2000.0752103.x. [DOI] [PubMed] [Google Scholar]
- Sans N, Petralia RS, Wang YX, Blahos J, 2nd, Hell JW, Wenthold RJ. A developmental change in NMDA receptor-associated proteins at hippocampal synapses. J Neurosci. 2000;20:1260–1271. doi: 10.1523/JNEUROSCI.20-03-01260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert V, Da Silva JS, Dotti CG. Localized recruitment and activation of RhoA underlies dendritic spine morphology in a glutamate receptor-dependent manner. J Cell Biol. 2006;172:453–467. doi: 10.1083/jcb.200506136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogomori H, Hammond AT, Ostermeyer-Fay AG, Barr DJ, Feigenson GW, London E, Brown DA. Palmitoylation and intracellular domain interactions both contribute to raft targeting of linker for activation of T cells. J Biol Chem. 2005;280:18931–18942. doi: 10.1074/jbc.M500247200. [DOI] [PubMed] [Google Scholar]
- Smith Y, Charara A, Hanson JE, Paquet M, Levey AI. GABA(B) and group I metabotropic glutamate receptors in the striatopallidal complex in primates. J Anat. 2000;196(Pt 4):555–576. doi: 10.1046/j.1469-7580.2000.19640555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Y, Charara A, Paquet M, Kieval JZ, Pare JF, Hanson JE, Hubert GW, Kuwajima M, Levey AI. Ionotropic and metabotropic GABA and glutamate receptors in primate basal ganglia. J Chem Neuroanat. 2001;22:13–42. doi: 10.1016/s0891-0618(01)00098-9. [DOI] [PubMed] [Google Scholar]
- Smotrys JE, Linder ME. PALMITOYLATION OF INTRACELLULAR SIGNALING PROTEINS: Regulation and Function. Annual Review of Biochemistry. 2004;73:559–587. doi: 10.1146/annurev.biochem.73.011303.073954. [DOI] [PubMed] [Google Scholar]
- Snow BE, Krumins AM, Brothers GM, Lee SF, Wall MA, Chung S, Mangion J, Arya S, Gilman AG, Siderovski DP. A G protein gamma subunit-like domain shared between RGS11 and other RGS proteins specifies binding to Gbeta5 subunits. Proc Natl Acad Sci U S A. 1998;95:13307–13312. doi: 10.1073/pnas.95.22.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JH, Song H, Wensel TG, Sokolov M, Martemyanov KA. Localization and differential interaction of R7 RGS proteins with their membrane anchors R7BP and R9AP in neurons of vertebrate retina. Mol Cell Neurosci. 2007;35:311–319. doi: 10.1016/j.mcn.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Song JH, Waataja JJ, Martemyanov KA. Subcellular targeting of RGS9-2 is controlled by multiple molecular determinants on its membrane anchor, R7BP. J Biol Chem. 2006;281:15361–15369. doi: 10.1074/jbc.M600749200. [DOI] [PubMed] [Google Scholar]
- Takida S, Fischer CC, Wedegaertner PB. Palmitoylation and plasma membrane targeting of RGS7 are promoted by alpha o. Mol Pharmacol. 2005;67:132–139. doi: 10.1124/mol.104.003418. [DOI] [PubMed] [Google Scholar]
- Turgeon SM, Albin RL. Postnatal ontogeny of GABAB binding in rat brain. Neuroscience. 1994;62:601–613. doi: 10.1016/0306-4522(94)90392-1. [DOI] [PubMed] [Google Scholar]
- Villalba RM, Raju DV, Hall RA, Smith Y. GABA(B) receptors in the centromedian/parafascicular thalamic nuclear complex: an ultrastructural analysis of GABA(B)R1 and GABA(B)R2 in the monkey thalamus. J Comp Neurol. 2006;496:269–287. doi: 10.1002/cne.20950. [DOI] [PubMed] [Google Scholar]
- Wang H, Cuzon VC, Pickel VM. Postnatal development of mu-opioid receptors in the rat caudate-putamen nucleus parallels asymmetric synapse formation. Neuroscience. 2003a;118:695–708. doi: 10.1016/s0306-4522(02)00926-0. [DOI] [PubMed] [Google Scholar]
- Wang H, Cuzon VC, Pickel VM. Ultrastructural localization of delta-opioid receptors in the rat caudate-putamen nucleus during postnatal development: relation to synaptogenesis. J Comp Neurol. 2003b;467:343–353. doi: 10.1002/cne.10920. [DOI] [PubMed] [Google Scholar]
- Watson N, Linder ME, Druey KM, Kehrl JH, Blumer KJ. RGS family members: GTPase-activating proteins for heterotrimeric G-protein alpha-subunits. Nature. 1996;383:172–175. doi: 10.1038/383172a0. [DOI] [PubMed] [Google Scholar]
- Witherow DS, Wang Q, Levay K, Cabrera JL, Chen J, Willars GB, Slepak VZ. Complexes of the G protein subunit gbeta 5 with the regulators of G protein signaling RGS7 and RGS9. Characterization in native tissues and in transfected cells. J Biol Chem. 2000;275:24872–24880. doi: 10.1074/jbc.M001535200. [DOI] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- Zachariou V, Georgescu D, Sanchez N, Rahman Z, DiLeone R, Berton O, Neve RL, Sim-Selley LJ, Selley DE, Gold SJ, Nestler EJ. Essential role for RGS9 in opiate action. Proc Natl Acad Sci U S A. 2003;100:13656–13661. doi: 10.1073/pnas.2232594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Lai Z, Simonds WF. Differential expression of the G protein beta(5) gene: analysis of mouse brain, peripheral tissues, and cultured cell lines. J Neurochem. 2000;75:393–403. doi: 10.1046/j.1471-4159.2000.0750393.x. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Simonds WF. Copurification of brain G-protein beta5 with RGS6 and RGS7. J Neurosci. 2000;20:RC59. doi: 10.1523/JNEUROSCI.20-03-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]