Abstract
Epidemiological surveillance of porcine rotavirus (PoRV) strains was carried out in Chiang Mai Province, Thailand, from 2002 to 2003, and eight rotavirus isolates could not be completely typed by PCR. Of these, six were G3 and one was G4 and displayed a P-nontypeable genotype, while another isolate was both G and P nontypeable. Analysis of a partial VP4 gene of all eight P-nontypeable strains revealed a high degree of amino acid sequence identities (94.7% to 100%), suggesting that they belonged to the same P genotype. Comparison of the amino acid sequences of two representative strains (namely, strains CMP178 and CMP213) with those of 27 other known P genotypes revealed a high degree of amino acid sequence identity with those of P[13] porcine rotavirus reference strains HP113 and HP140, which were recently isolated in India. However, amino acid sequence comparison with non-P[13] rotavirus strains revealed relatively low identities, ranging from 58.2% to 84.8% for full-length VP4 sequences and 35.1% to 80.6% for VP8* sequences. Phylogenetic analysis revealed that CMP178 and CMP213 clustered together in a monophyletic branch with P[13]-like genotypes HP113 and HP140 which was clearly separated from the other lineages of P[13] or P[22] strains. Altogether, these findings indicate that PoRV strains CMP178 and CMP213 should be considered the P[13]-like VP4 genotype, a rare genotype that has been identified only in pigs. This study provides additional evidence of increasing genetic diversity among group A rotaviruses in nature.
Group A rotaviruses are the most important etiologic agents of severe diarrhea in young children and young animals worldwide. In developing countries, these severe diarrhea cases lead to an estimated 454,000 to 705,000 deaths annually among children under 5 years of age (32). Group A rotaviruses are members of the Reoviridae family with nonenveloped icosahedral particles. The mature virion is formed by three concentric layers of proteins that enclosed a genome of 11 segments of double-stranded RNA. Rotaviruses are classified according to the genetic and antigenic diversity of the two outer capsid proteins, VP4 and VP7. These proteins independently induce type-specific neutralizing antibodies and form the basis of the dual classification of group A rotaviruses into the P (protease sensitive) and the G (glycoprotein) serotypes, respectively (5, 17).
Rotaviruses express extensive antigenic and genomic diversities. To date, at least 15 G genotypes and 26 P genotypes have been identified from humans and a variety of animal species (5, 22, 26, 28, 30, 35, 36). Most recently, several groups of investigators have proposed a novel genotype, P[27], which was isolated from diarrheic piglets (18, 25, 38). Generally, rotaviruses of the same G genotypes share at least 90 to 91% VP7 amino acid sequence identity (11, 12, 13, 16). Rotavirus strains sharing ≥89% VP4 amino acid sequence identities are considered to belong to the same P genotype, while those sharing VP4 amino acid sequence identities of <89% belong to different genotypes (2, 5, 7). Moreover, the VP8* trypsin-cleavage product of VP4 coding for amino acids (aa) 13 to 250, including the greatest sequence divergent region (aa 71 to 204), correlates well with VP4 genotype specificity (20, 21).
Several epidemiological studies have demonstrated that among porcine rotaviruses (PoRVs), G3, G4, and G5, are the most common G genotypes and usually associate with the P[6] or P[7] genotype (5, 23, 39, 40). In addition, other G and P types, such as G1, G2, G6, G8, G9, and G10 and P[13], P[19], P[23], P[26], and P[27], have also been identified in various geographical settings (1, 3, 5, 9, 10, 16, 18, 27, 29, 33, 34, 37, 39, 40, 41). Accordingly, a comprehensive genotypic characterization of the rotavirus strains circulating in domestic animal populations, especially pigs, is important to define the extent of rotavirus diversity.
Rotavirus strains bearing P[13] genotype specificity are host restricted. The P[13] rotavirus genotype is commonly detected among pigs and has not been identified from other animal sources or humans (5, 7, 39, 16). Nevertheless, only a few studies have reported on the distribution of the P[13] genotypes circulating worldwide. Most recently, genome characterization of two PoRV strains, HP113 and HP140, isolated from eastern India revealed that their VP4 sequences were similar to those of P[13] genotypes, while their VP7 sequences were closely related to those of the G6 genotype reference strains (7). In our study, eight strains of P[13] in combination with either G3, G4, or G5 were isolated from diarrheic piglets raised in several farms in Chiang Mai, Thailand. The VP4 genes of these strains were most closely related to those of P[13]-like PoRV strains HP113 and HP140. These findings affirm the evidence that rotavirus strains bearing the P[13] genotype are frequently detected in pigs.
MATERIALS AND METHODS
Fecal specimens and rotavirus detection.
A total of 250 fecal specimens were collected from diarrheic piglets on six different farms in Chiang Mai, Thailand, in 2002 and 2003. The specimens were evaluated for the presence of group A rotavirus by enzyme-linked immunosorbent assay (ELISA) with a polyclonal antibody against group A rotavirus as the capture antibody (15). The samples that were positive for group A rotavirus by ELISA were investigated further for their G and P genotypes by multiplex reverse transcription-PCR (RT-PCR), as described elsewhere (4, 6, 9, 10, 11, 24, 31, 40).
RNA extraction and RT-PCR genotyping.
Viral double-stranded RNA was extracted from a 20% fecal sample suspension with a QIAamp viral RNA mini kit (Qiagen, Hilden, Germany), according to the manufacturer's protocol. For G typing, the VP7 gene was reverse transcribed and amplified by using the consensus primer pair Beg9-End9 (11). Then, the G type was determined by using different pools of primers currently reported to be specific for human and animal G types (G1 to G6 and G8 to G11) (4, 10, 11, 40). For P typing, the consensus primer pair Con2-Con3 was used to generate a 876-bp fragment (VP8*) of VP4, and the P type was determined by using different pools of primers currently reported to be specific for human and animal P types (P[1], P[3] to P[11], P[14], and P[19]) (6, 9, 24, 31, 40).
Nucleotide sequencing.
Rotavirus strains that failed to be genotyped by multiple sets of primers were further investigated by nucleotide sequencing. The full-length VP7 (1,062-bp) and partial VP4 (876-bp) genes of these strains were reverse transcribed and were amplified by using the primer pairs Beg9-End9 (10) for VP7 and Con2-Con3 (8) for partial VP4 genes. In addition, the full-length VP4 genes of two representative strains, CMP178 and CMP213, were also amplified by using reverse primer 170 (nucleotides [nt] 2344 to 2368; 5′-GGTCACAWCCTCTAGMMRYTRCTTA-3′) (26) in combination with forward primer VP4-5F (nt 1 to 23; 5′-GGCTATAAAATGGCTTCDCTCAT-3′). The PCR amplicons were purified with a QIAquick PCR purification kit (Qiagen) and sequenced in both directions by using a BigDye Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) on an automated sequencer (ABI 3100; Applied Biosystems). For sequencing of the full-length VP4 genes, four additional primers were used as internal sequencing primers. The nucleotide positions and sequences of these primers (5′ to 3′) were as follows: forward primer P28F650, nt 685 to 704, CTTCCACCAATGCAGAATAC; forward primer P28In1, nt 1262 to 1283, CGGATTATGTATCTCTTAACTC; forward primer P28In2, nt 2007 to 2027, GGAGAAATTCATACCAAATAG; and reverse primer P28In3, nt 375 to 396, CACTAGGTTAACTTGTTGACCG.
Sequences and phylogenetic analysis.
The nucleotide sequences were assembled and analyzed by using Bioedit software packages (14). The sequences were compared with those available in the GenBank database by use of the BLAST program in order to determine their genotypes. Phylogenetic and molecular evolutionary analyses were conducted by using the MEGA program (version 3.1) (19). A phylogenetic tree was elaborated by both the parsimony and the distance methods and by supplying statistical support with bootstrapping over 100 replicates.
Nucleotide sequence accession numbers.
The full lengths of the VP7 and VP4 gene sequences of strains CMP178 and CMP213 described in this study have been deposited in the GenBank sequence database under accession numbers DQ515961 (VP7) and DQ536362 (VP4) for CMP178 and DQ786576 (VP7) and DQ786578 (VP4) for CMP213.
RESULTS
G and P genotypes.
Of a total of 250 fecal specimens tested, 43 (17.2%) were positive for group A rotavirus by ELISA. Characterization of their G and P genotypes by multiplex RT-PCR revealed that 29 isolates were G3, 1 was G4, 4 were G8, 2 were G9, and 7 were G nontypeable. These seven G-nontypeable strains were further characterized by nucleotide sequence analysis, and six were identified to be G3 and 1 was G5. The P genotype was also characterized; and 20 were P[6], 10 were P[7], 5 were P[19], and 8 were P nontypeable. Among these P-nontypeable strains, six were found in combination with G3 (strains CMP213, CMP214, CMP215, CMP225, CMP234, and CMP239), one was found to carry G4 genotype (strain CMP077), while another one was found to be both G and P nontypeable (strain CMP178). Therefore, representative strains CMP178 and CMP213 were selected and their full-length VP4 and VP7 gene sequences were characterized further.
VP7 sequence analysis.
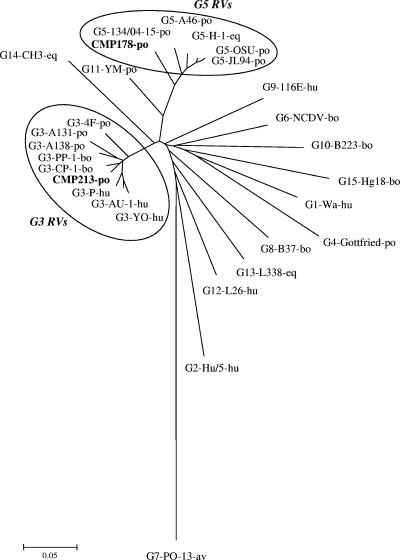
The deduced amino acid sequences of strains CMP178 and CMP213 were compared with those of representative strains of 15 other known G genotypes. The VP7 amino acid sequence of CMP178 was the most closely related to the amino acid sequences of G5 rotavirus strains, with the amino acid sequence identities ranging from 92.3% to 93.2%, while the VP7 amino acid sequence of CMP213 was the most closely related to the amino acid sequences of the G3 genotypes (94.4% to 96.8%) (Table 1). The data indicated that CMP178 belongs to the G5 genotype and also confirmed that CMP213 has the G3 genotype. Phylogenetic analysis of the deduced amino acid sequences corroborated the results of the analysis of the VP7 sequences by demonstrating that CMP178 and CMP213 clustered with the reference strains of the G5 and G3 rotaviruses, respectively (Fig. 1).
TABLE 1.
Comparison of the VP7 deduced amino acid sequence identities of strains CMP178 and CMP213 with the amino acid sequences of 15 known G genotypes
| Strain (origin)a | G genotype | % VP7 amino acid identityb
|
|
|---|---|---|---|
| CMP178 | CMP213 | ||
| Wa (human) | 1 | 78.2 | 80.7 |
| Hu/5 (human) | 2 | 72.3 | 74.1 |
| YO (human) | 3 | 84.6 | 94.4 |
| AU-1 (human) | 3 | 84.6 | 95.4 |
| P (human) | 3 | 85.5 | 95.1 |
| CP-1 (bovine) | 3 | 85.5 | 96.8 |
| PP-1 (bovine) | 3 | 85.5 | 96.8 |
| A131 (porcine) | 3 | 85.8 | 94.7 |
| A138 (porcine) | 3 | 84.9 | 96.0 |
| 4F (porcine) | 3 | 86.1 | 95.0 |
| Gottfried (porcine) | 4 | 74.5 | 77.2 |
| OSU (porcine) | 5 | 92.3 | 86.0 |
| JL49 (porcine) | 5 | 92.6 | 86.3 |
| H-1 (equine) | 5 | 92.9 | 86.3 |
| A46 (porcine) | 5 | 92.6 | 86.7 |
| 134/04-15 (porcine) | 5 | 93.2 | 86.3 |
| NCDV (bovine) | 6 | 81.5 | 83.5 |
| PO-13 (avian) | 7 | 58.9 | 56.7 |
| B37 (bovine) | 8 | 78.2 | 82.5 |
| 116E (human) | 9 | 80.9 | 83.5 |
| B223 (bovine) | 10 | 78.8 | 81.1 |
| YM (porcine) | 11 | 88.3 | 86.7 |
| L26 (human) | 12 | 79.4 | 80.7 |
| L338 (equine) | 13 | 77.6 | 80.7 |
| CH3 (equine) | 14 | 78.5 | 84.2 |
| Hg18 (bovine) | 15 | 77.6 | 77.2 |
The GenBank accession numbers of the VP7 genes are as indicated for the following strains: Wa, K02033; Hu/5, A01028; YO, D86284; AU-1, D86271; P, AB118024; CP-1, AF448852; PP-1, AF427124; Gottfried, X06386; OSU, X04613; JL49, AY538665; H-1, AF242393; A46, L35054; 134/04-15, DQ062572; NCDV, M12394; PO-13, D82979; B37, J04334; 116E, L14072; B223, X57852; YM, M23194; L26, M58290; L338, D13549; CH3, D25229; and Hg18, AF237666.
Boldface data indicate percentages of the highest sequence identity that CMP178 and CMP213 shared with the corresponding reference genotypes.
FIG. 1.
Phylogenetic tree of the full-length VP7 amino acid sequences displaying the relationships between PoRV strains CMP178, CMP213, and representative strains of all the VP7 genotypes recognized to date. The following abbreviations are used to identify the species of strain origin: hu, human; eq, equine; po, porcine; bo, bovine; av, avian.
VP4 sequence analysis.
In order to determine the P genotypes of all eight P-nontypeable PoRV strains, their partial VP4 amplicons (876 bp), which had been generated by the consensus primer pair Con2-Con3, were first subjected to nucleotide sequencing. Comparison of the partial VP4 amino acid sequences of these strains revealed high degrees of amino acid sequence identity (94.7% to 100%) among themselves (data not shown), suggesting that they all belong to the same P genotype.
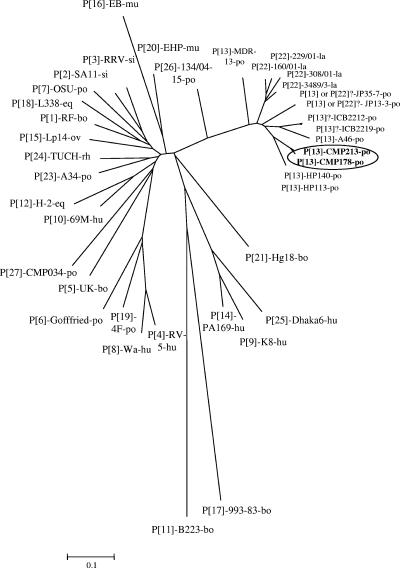
Analysis of the full-length VP4 amino acid sequences revealed that both representative strains, CMP178 and CMP213, were almost identical (98.4% amino acid identity; data not shown). In addition, when the amino acid sequences of CMP178 and CMP213 were compared with those of 27 known P genotypes (Table 2), the highest level of amino acid sequence identity was found to be with P[13]-like PoRV strains HP113 and HP140 (88.1% and 87.2% for the VP8* portion and 92.2 and 92.4% for the complete VP4 sequences for CMP178 and CMP213, respectively). In contrast, they showed somewhat lower levels of VP8* amino acid sequence identity with other P[13] reference strains, strains MDR-13, JP35-7, JP13-3, ICB2212, ICB2219, and A46, ranging from 79.2% to 83.7% amino acid identity for CMP178 and 79.2% to 83.2% amino acid identity for CMP213. Additionally, they also showed lower levels of amino acid sequence identity with P[22] lapine rotavirus reference strains 160/01, 229/01, 308/01, and 3489/3, ranging from 77.9% to 81.0% for CMP178 and 78.4% to 81.0% for CMP213. Therefore, all eight isolates were considered to be P[13]-like viruses since they had the highest level of sequence identity with P[13] PoRV strains and revealed remarkably lower levels of amino acid sequence identity with other reference strains of non-P[13] rotavirus (35.1% to 81.0% for both CMP178 and CMP213). Phylogenetic analysis supported the amino acid sequence alignments by showing that strains CMP178 and CMP213 clustered in the same branch with strains HP113 and HP140. Of note, although strains CMP178 and CMP213 were closely related to PoRV strains HP113 and HP140, they were rather distantly related to other P[13] PoRV strains and formed a distinct lineage (Fig. 2).
TABLE 2.
Comparison of the VP4 and VP8* deduced amino acid sequence identities of strains CMP178 and CMP213 with the amino acid sequences of 27 known P genotypesa
| Strain (origin) | P genotype | % Amino acid identityb
|
|||
|---|---|---|---|---|---|
| VP4
|
VP8*
|
||||
| CMP178 | CMP213 | CMP178 | CMP213 | ||
| RF (bovine) | P[1] | 76.9 | 77.1 | 59.8 | 59.3 |
| SA11 (simian) | P[2] | 78.1 | 78.2 | 60.5 | 60.9 |
| RRV (simian) | P[3] | 77.3 | 77.3 | 59.7 | 59.7 |
| RV-5 (human) | P[4] | 68.8 | 68.8 | 51.4 | 51.4 |
| UK (bovine) | P[5] | 70.5 | 71.0 | 53.9 | 55.1 |
| Gottfried (porcine) | P[6] | 71.5 | 71.8 | 54.3 | 54.7 |
| OSU (porcine) | P[7] | 77.7 | 78.0 | 62.6 | 63.0 |
| Wa (human) | P[8] | 69.1 | 69.1 | 51.8 | 51.8 |
| K8 (human) | P[9] | 65.9 | 66.0 | 49.8 | 49.3 |
| 69M (human) | P[10] | 76.3 | 76.7 | 57.2 | 57.6 |
| B223 (bovine) | P[11] | 58.2 | 58.2 | 35.1 | 35.1 |
| H-2 (equine) | P[12] | 74.8 | 74.8 | 58.0 | 57.2 |
| MDR-13 (porcine) | P[13] | 87.5 | 87.9 | 79.2 | 79.2 |
| JP35-7 (porcine) | P[13] or P[22]? | —c | — | 81.9 | 83.2 |
| JP13-3 (porcine) | P[13] or P[22]? | — | — | 81.9 | 83.2 |
| ICB2212 (porcine) | P[13]? | — | — | 83.2 | 82.3 |
| ICB2219 (porcine) | P[13]? | — | — | 82.3 | 81.4 |
| A46 (porcine) | P[13] | 89.3 | 89.7 | 83.7 | 83.2 |
| HP113 (porcine) | P[13] | 92.2 | 92.2 | 88.1 | 87.2 |
| HP140 (porcine) | P[13] | 92.4 | 92.4 | 88.1 | 87.2 |
| PA169 (human) | P[14] | 68.1 | 68.3 | 51.8 | 52.2 |
| Lp14 (ovine) | P[15] | 76.7 | 76.9 | 59.3 | 59.7 |
| EB (murine) | P[16] | 72.4 | 72.7 | 52.6 | 52.2 |
| 993-83 (bovine) | P[17] | 59.9 | 59.9 | 60.0 | 60.0 |
| L338 (equine) | P[18] | 74.6 | 75.0 | 56.4 | 56.8 |
| 4F (porcine) | P[19] | 71.8 | 71.8 | 52.6 | 52.6 |
| EHP (murine) | P[20] | 74.1 | 74.5 | 57.2 | 57.6 |
| Hg18 (bovine) | P[21] | 75.1 | 75.8 | 55.4 | 58.0 |
| 160/01 (lapine) | P[22] | — | — | 80.6 | 80.6 |
| 229/01 (lapine) | P[22] | — | — | 79.7 | 79.7 |
| 308/01 (lapine) | P[22] | — | — | 77.9 | 78.4 |
| 3489/3 (lapine) | P[22] | — | — | 81.0 | 81.0 |
| A34 (porcine) | P[23] | — | — | 61.8 | 62.2 |
| TUCH (rhesus macaque) | P[24] | 77.6 | 77.7 | 61.8 | 61.8 |
| Dhaka6 (human) | P[25] | 66.1 | 66.0 | 51.4 | 51.0 |
| 134/04-15 (porcine) | P[26] | 84.7 | 84.8 | 69.2 | 69.2 |
| CMP034 (porcine) | P[27] | 71.0 | 71.0 | 57.1 | 56.0 |
The GenBank accession numbers of VP4 genes are as indicated for the following strains: RF, U65924; SA11, M23188; RRV, M18736; RV-5, M32559; UK, M22306; Gottfried, M33516; OSU, X13190; Wa, M96825; K8, D90260; 69M, M60600; B223, D13394; H-2, L04638; MDR-13, L07886; PA169, D14724; Lp14, L11599; EB, U08419; 993-83, D16352; L338, D13399; 4F, L10359; EHP, U08424; Hg18, AF237665; 160/01, AF526374; A34, AY174094; TUCH, AY596189; Dhaka6, AY773004; 134/04-15, DQ061053; and CMP034, DQ534016.
Boldface data indicate percentages of the highest sequence identity that CMP178 and CMP213 shared with the corresponding reference genotypes.
—, no full-length VP4 sequence is available in the GenBank database.
FIG. 2.
Phylogenetic tree of the partial VP4 amino acid sequences displaying the relationships between PoRV strains CMP178, CMP213, and selected strains of the 27 P genotypes recognized to date. The following abbreviations are used to identify the species of strain origin: hu, human; eq, equine; po, porcine; bo, bovine; av, avian; pi, pigeon; mu, murine; si, simian; ov, ovine; la, lapine.
Taken together, the eight strains of P[13] PoRV described in this study comprised six strains of G3P[13] genotype and one strain each of the G4P[13] genotype and the G5P[13] genotype.
DISCUSSION
The eight strains of PoRV described in the present study were isolated from diarrheic piglets on several farms in Chiang Mai, Thailand, during epidemiological surveillance in 2002 and 2003. The initial characterization of the G and P genotypes of these strains by PCR by the use of multiple specific primer sets reported previously (6, 9, 24, 31, 40) revealed that six were G3, one was G4, and the other one was G nontypeable, while all were P nontypeable, as determined by PCR-based genotyping. We therefore further characterized those nontypeable strains by sequencing their VP7 and partial VP4 (VP8*) genes. The amino acid sequence of the VP8* fragment of these strains showed high degrees of identity, ranging from 94.7% to 100%, suggesting that they all belong to the same P genotype. In contrast, when the VP8* amino acid sequences of these strains were compared with those of the existing 27 P genotypes, only a low level of identity (35.1% to 88.1%) was observed (Table 2). In order to verify whether these strains belong to a novel P genotype or a variant strain of an existing known P genotype, the entire VP4 amino acid sequences were analyzed. Comparison of the full-length VP4 amino acid sequences of two representative strains, CMP178 and CMP213, with those of previously reported 27 P genotypes revealed sequence identities that ranged from 58.2% to 92.4% (Table 2). The high levels of amino acid identity were shared with two strains of PoRV recently isolated in India (7), strains HP113 and HP140, at 92.2% and 92.4%, respectively. On the basis of full-length VP4 amino acid sequence analysis, our strains most likely belong to the P[13] genotype. Phylogenetic analysis of the full-length amino acid sequences of the VP4 genes of strains CMP178 and CMP213 supported the sequence alignment data. CMP178 and CMP213 clustered tightly together with strains HP113 and HP140 in a single branch separated from other P[13] PoRV strains (Fig. 2).
Although strain CMP178 was initially identified as a G- and P-nontypeable strain by PCR-based genotyping, it was identified as a P[13] genotype strain (Table 2) that associated with the G5 genotype (Table 1) as a G5P[13] strain by amino acid sequence analyses of the VP4 and VP7 genes. Alignment of the G5-specific primer (SG 5) sequence (40) with the VP7 nucleotide sequence of CMP178 revealed four point mutations in the primer binding region (nt 180 to 200) at positions 183 (A to T), 186 (G to A), 187 (C to A), and 196 (G to A) (data not shown). These point mutations may lead to the failure of the binding of primer SG 5 at the target sequence in the VP4 gene of strain CMP178. This may explain why CMP178 was initially identified as G nontypeable by PCR-based genotyping. Additionally, CMP178 was also initially identified by PCR as P nontypeable, even though it was later identified as being of the P[13] genotype by VP4 sequence analysis, because no primer specific for P[13] has previously been reported in the literature.
Strain CMP213 was initially identified by PCR-based genotyping, and it was confirmed by VP7 sequence analysis (Table 1) to be genotype G3 with a P-nontypeable genotype and was later identified by VP4 sequence analysis to have the P[13] genotype (Table 2). On the basis of these data, CMP213 has a G3 genotype that exists in association with the P[13] genotype. In addition, among the P[13] strains of PoRV isolated in this study, one was found to have P[13] in combination with a G4 genotype (strain CMP077).
Altogether, the strains of PoRV isolated in the present study were P[13] in combination with several G types, including G3, G4, and G5. This finding implies multiple reassortment events and shows the diversity of the P[13] PoRV strains circulating in the pig population in Chiang Mai, Thailand.
Acknowledgments
This study was supported by the Endowment Fund for Medical Research of the Faculty of Medicine and Graduate School of Chiang Mai University, Chiang Mai, Thailand. Additionally, this study was supported in part by the Core University System Exchange Program under the Japan Society for the Promotion of Science, coordinated by the Graduate School of Medicine, University of Tokyo, Tokyo, Japan, and Mahidol University, Thailand.
Footnotes
Published ahead of print on 30 January 2008.
REFERENCES
- 1.Burke, B., M. A. McCrae, and U. Desselberger. 1994. Sequence analysis of two porcine rotaviruses differing in growth in vitro and in pathogenicity: distinct VP4 sequences and conservation of NS53, VP6 and VP7 genes. J. Gen. Virol. 752205-2212. [DOI] [PubMed] [Google Scholar]
- 2.Ciarlet, M., M. K. Estes, and M. E. Corner. 1997. Comparative amino acid sequence analysis of outer capsid protein VP4 from four lapine rotavirus strains reveal identity with genotype P[14] human rotavirus. Arch. Virol. 1421059-1069. [DOI] [PubMed] [Google Scholar]
- 3.Ciarlet, M., and F. Liprandi. 1994. Serological and genomic characterization of two porcine rotaviruses with serotype G1 specificity. J. Clin. Microbiol. 32269-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das, B. K., J. R. Gentsch, H. G. Cicirello, P. A. Woods, A. Gupta, M. Ramachandran, R. Kumar, M. K. Bhan, and R. I. Glass. 1994. Characterization of rotavirus strains from newborns in New Delhi, India. J. Clin. Microbiol. 321820-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estes, M. K. 2001. Rotaviruses and their replication, p. 1747-1786. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 6.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 301365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghosh, S., V. Varghese, S. Samajdar, S. K. Bhattacharya, N. Kobayashi, and T. N. Naik. 2007. Evidence for independent segregation of the VP6- and NSP4-encoding genes in porcine group A rotavirus G6P[13] strains. Arch. Virol. 152423-429. [DOI] [PubMed] [Google Scholar]
- 8.Gorziglia, M., G. Larralde, A. Z. Kapikian, and R. M. Chanock. 1990. Antigenic relationships among human rotaviruses as determined by outer capsid protein VP4. Proc. Natl. Acad. Sci. USA 877155-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gouvea, V., N. Santos, and M. C. Timenetsky. 1994. VP4 typing of bovine and porcine group A rotaviruses by PCR. J. Clin. Microbiol. 321333-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gouvea, V., N. Santos, and M. C. Timenetsky. 1994. Identification of bovine and porcine rotavirus G types by PCR. J. Clin. Microbiol. 321338-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green, K. Y., Y. Hoshino, and N. Ikegami. 1989. Sequence analysis of the gene encoding the serotype-specific glycoprotein (VP7) of two new human rotavirus serotypes. Virology 168429-433. [DOI] [PubMed] [Google Scholar]
- 13.Green, K. Y., J. F. Sears, K. Taniguchi, K. Midthun, Y. Hoshino, M. Gorziglia, K. Nishikawa, S. Urasawa, A. Z. Kapikian, and R. M. Chanock. 1988. Prediction of human rotavirus serotype by nucleotide sequence analysis of the VP7 protein gene. J. Virol. 621819-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment and analysis program for Windows 95/98/NT. Nucleic. Acids Symp. Ser. 4195-98. [Google Scholar]
- 15.Hasegawa, A., A. Mukoyama, H. Suzuki, S. Inouye, S. Chearskul, and P. Thongkrajai. 1987. Rotavirus infection of Thai infants: antigen detection, RNA electrophoresis and virus cultivation. J. Diarrhoeal Dis. Res. 5165-170. [PubMed] [Google Scholar]
- 16.Huang, J. A., H. S. Nagesha, and I. H. Holmes. 1993. Comparative sequence analysis of VP4s from five Australian porcine rotaviruses: implication of an apparent new P-type. Virology 196319-327. [DOI] [PubMed] [Google Scholar]
- 17.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 18.Khamrin, P., N. Maneekarn, S. Peerakome, W. Chan-it, F. Yagyu, S. Okitsu, and H. Ushijima. 2007. Novel porcine rotavirus of the genotype P[27] shares new phylogenetic lineage with G2 porcine rotavirus strain. Virology 361243-252. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 20.Larralde, G., and M. Gorziglia. 1992. Distribution of conserved and serotype specific epitopes on the VP8* subunit of rotavirus VP4. J. Virol. 72117-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larralde, G., L. B. Kapikian, and M. Gorziglia. 1991. Serotype-specific epitope(s) present on the VP8* subunit of rotavirus VP4. J. Virol. 653213-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liprandi, F., M. Gerder, Z. Bastidas, J. A. López, F. H. Pujol, J. E. Ludert, D. B. Joelsson, and M. Ciarlet. 2003. A novel type of VP4 carried by a porcine rotavirus strain. Virology 315373-380. [DOI] [PubMed] [Google Scholar]
- 23.Liprandi, F., I. Rodriguez, C. I. Pina, G. Larralde, and M. Gorziglia. 1991. VP4 monotype specificities among porcine rotaviruses strains of the same VP4 serotype. J. Virol. 651658-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maneekarn, N., P. Khamrin, W. Chan-it, S. Peerakome, S. Suckchai, K. Pringprao, and H. Ushijima. 2006. Detection of rare G3P[19] porcine rotavirus strains in Chiang Mai, Thailand, provides evidence for origin of the VP4 genes of Mc323 and Mc345 human rotaviruses. J. Clin. Microbiol. 444113-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martella, V., M. Ciarlet, K. Banyai, E. Lorusso, S. Arista, A. Lavazza, G. Pezzotti, N. Decaro, A. Cavalli, M. S. Lucente, M. Corrente, G. Elia, M. Camero, M. Tempesta, and C. Buonavoglia. 2007. Identification of group A porcine rotavirus strains bearing a novel VP4 (P) genotype in Italian swine herds. J. Clin. Microbiol. 45577-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martella, V., M. Ciarlet, K. Banyai, E. Lorusso, A. Cavalli, M. Corrente, G. Elia, S. Arista, M. Camero, C. Desario, N. Decaro, A. Lavazza, and C. Buonavoglia. 2006. Identification of a novel VP4 genotype carried by a serotype G5 porcine rotavirus strain. Virology 346301-311. [DOI] [PubMed] [Google Scholar]
- 27.Martella, V., M. Ciarlet, R. Baselga, S. Arista, G. Elia, E. Lorusso, K. Banyai, V. Terio, A. Madio, F. M. Ruggeri, E. Falcone, M. Camero, N. Decaro, and C. Buonavoglia. 2005. Sequence analysis of the VP7 and VP4 genes identifies a novel VP7 gene allele of porcine rotaviruses, sharing a common evolutionary origin with human G2 rotaviruses. Virology 337111-123. [DOI] [PubMed] [Google Scholar]
- 28.Martella, V., M. Ciarlet, A. Camarda, A. Pratelli, M. Tempesta, G. Greco, A. Cavalli, G. Elia, N. Decaro, V. Terio, G. Bozzo, M. Camero, and C. Buonavoglia. 2003. Molecular characterization of the VP4, VP6, VP7, and NSP4 genes of lapine rotaviruses identified in Italy: emergence of a novel VP4 genotype. Virology 314358-370. [DOI] [PubMed] [Google Scholar]
- 29.Martella, V., A. Pratelli, G. Greco, M. Tempesta, M. Ferrari, M. N. Losio, and C. Buonavoglia. 2001. Genomic characterization of porcine rotaviruses in Italy. Clin. Diagn. Lab. Immunol. 8129-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNeal, M. M., K. Sestak, A. Choi, M. Basu, M. J. Cole, P. P. Aye, R. P. Bohm, and R. L. Ward. 2005. Development of a rotavirus-shedding model in rhesus macaques, using a homologous wild-type rotavirus of a new P genotype. J. Virol. 79944-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mphahlele, M. J., I. Peenze, and A. D. Steele. 1999. Rotavirus strains bearing the VP4P[14] genotype recorded from South African children with diarrhea. Arch. Virol. 1441027-1034. [DOI] [PubMed] [Google Scholar]
- 32.Parashar, U. D., C. J. Gibson, J. S. Bresee, and R. I. Glass. 2006. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 12304-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pongsuwanna, Y., K. Taniguchi, M. Chiwakul, T. Urasawa, F. Wakasugi, C. Jayavasu, and S. Urasawa. 1996. Serological and genomic characterization of porcine rotaviruses in Thailand: detection of a G10 porcine rotavirus. J. Clin. Microbiol. 341050-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Racz, M. L., S. S. Kroeff, V. Munford, T. A. R. Caruzo, E. L. Durigon, Y. Hayashi, V. Gouvea, and E. A. Palombo. 2000. Molecular characterization of porcine rotaviruses from the southern region of Brazil: characterization of an atypical genotype G[9] strain. J. Clin. Microbiol. 382443-2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahman, M., J. Matthijnssens, S. Nahar, G. Podder, D. A. Sack, T. Azim, and M. Van Ranst. 2005. Characterization of a novel P[25],G11 human group A rotavirus. J. Clin. Microbiol. 433208-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rao, C. D., K. Gowda, and B. S. Y. Reddy. 2000. Sequence analysis of VP4 and VP7 genes of nontypeable strains identifies a new pair of outer capsid proteins representing novel P and G genotypes in bovine rotaviruses. Virology 267104-113. [DOI] [PubMed] [Google Scholar]
- 37.Santos, N., R. C. C. Lima, C. M. Nozawa, R. E. Linhares, and V. Gouvea. 1999. Detection of porcine rotavirus type G9 and of a mixture of types G1 and G5 associated with Wa-like VP4 specificity: evidence for natural human-porcine genetic reassortment. J. Clin. Microbiol. 372734-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steyer, A., M. Poljsak-Prijatelj, D. Barlic-Maganja, U. Jamnikar, J. Z. Mijovski, and J. Marin. 2007. Molecular characterization of a new porcine rotavirus P genotype found in an asymptomatic pig in Slovenia. Virology 359275-282. [DOI] [PubMed] [Google Scholar]
- 39.Teodoroff, T. A., H. Tsunemitsu, K. Okamoto, K. Katsuda, M. Kohmoto, K. Kawashima, T. Nakagomi, and O. Nakagomi. 2005. Predominance of porcine rotavirus G9 in Japanese piglets with diarrhea: close relationship of their VP7 genes with those of recent human G9 strains. J. Clin. Microbiol. 431377-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winiarczyk, S., P. S. Paul, S. Mummidi, R. Panek, and Z. Gradzki. 2002. Survey of porcine rotavirus G and P genotype in Poland and in the United States using RT-PCR. J. Vet. Med. 49373-378. [DOI] [PubMed] [Google Scholar]
- 41.Zaberezhny, A. D., Y. S. Lyoo, and P. S. Paul. 1994. Prevalence of P types among porcine rotaviruses using subgenomic VP4 gene probe. Vet. Microbiol. 3997-110. [DOI] [PubMed] [Google Scholar]