Abstract
Campylobacter jejuni, a spiral-shaped gram-negative bacterium, is a leading bacterial cause of human food-borne illness. Acute disease is associated with C. jejuni invasion of the intestinal epithelium. Further, maximal host cell invasion requires the secretion of proteins termed Campylobacter invasion antigens (Cia). As bile acids are known to alter the pathogenic behavior of other gastrointestinal pathogens, we hypothesized that the virulence potential of Campylobacter may be triggered by the bile acid deoxycholate (DOC). In support of this hypothesis, culturing C. jejuni with a physiologically relevant concentration of DOC significantly altered the kinetics of cell invasion, as shown by gentamicin protection assays. In contrast to C. jejuni harvested from Mueller-Hinton (MH) agar plates, C. jejuni harvested from MH agar plates supplemented with DOC secreted the Cia proteins, as judged by metabolic labeling experiments. DOC was also found to induce the expression of the ciaB gene, as determined by β-galactosidase reporter, real-time reverse transcription-PCR, and microarray analyses. Microarray analysis further revealed that DOC induced the expression of virulence genes (ciaB, cmeABC, dccR, and tlyA). In summary, we demonstrated that it is possible to enhance the pathogenic behavior of C. jejuni by modifying the culture conditions. These results provide a foundation for identifying genes expressed by C. jejuni in response to in vivo-like culture conditions.
Campylobacter jejuni is recognized as one of the leading bacterial causes of gastrointestinal disease in humans (1, 2). An estimated 2.4 million persons are infected by C. jejuni each year in the United States. Infection with C. jejuni results in symptoms that range from mild watery diarrhea to more severe diarrhea with blood and leukocytes. The most notable complication of campylobacteriosis is the development of Guillain-Barré syndrome, an acute demyelinating polyneuropathy. Approximately 1 in 1,000 diagnosed C. jejuni infections result in Guillain-Barré syndrome (6).
Bile is a digestive secretion that plays a major role in fat dispersion and absorption. Approximately 50% of organic bile consists of bile acids, which are synthesized in the liver from cholesterol by a multienzyme process. Bile acids, including cholates and deoxycholates (DOCs), are amphipathic molecules that act as detergents and possess potent antimicrobial activity. The average concentration of bile acids in the human intestine ranges from 0.2 to 2%, and DOC accounts for about 15% of the bile acids (7).
Bile has been shown to regulate virulence gene expression in several gastrointestinal pathogens (5, 12, 31-34, 38-40, 46). For example, Shigella spp. grown in the presence of bile show increased secretion of invasion plasmid antigens (Ipa) and enhanced invasion potential (36). Specifically, DOC stimulates the localization of IpaB to the tip of the type III secretion apparatus needle, where IpaB, in association with IpaD, is hypothesized to act as a sensor of host cell contact (31). In Vibrio parahaemolyticus, bile acids enhance the production of thermostable direct hemolysin (32, 33) and capsule and adherence to epithelial cells (34).
The ability of Campylobacter to cause illness is related to its ability to invade epithelial cells lining the intestinal tract (3, 9, 13, 48, 51). The proteins known to promote entry of the bacteria into eukaryotic cells are different from those that facilitate binding (15). In contrast to cellular adhesion, C. jejuni must be metabolically active to invade human epithelial cells. When it is cultured with epithelial cells, C. jejuni synthesizes and secretes a set of proteins that are required for maximal invasion of host epithelial cells (15, 16, 18, 44, 45). These proteins are termed Campylobacter invasion antigens (Cia). The secretion of the Cia proteins is dependent on a functional flagellum, indicating that this organelle has a dual function in motility and as a type III secretion system (17). To date, only one Cia, termed CiaB, has been identified. In contrast to the C. jejuni F38011 wild-type isolate, the host cell invasion of a ciaB null mutant is impaired and the mutant is secretion deficient (16). Further, the severity and time of onset of disease in piglets inoculated with a C. jejuni ciaB null mutant are retarded compared with the severity and time of onset of disease in piglets inoculated a C. jejuni wild-type isolate. Piglets inoculated with a C. jejuni ciaB null mutant developed diarrhea 3 days postinoculation, whereas piglets inoculated with a C. jejuni wild-type isolate developed diarrhea within 24 h (19).
Although a number of studies have highlighted the mechanism of resistance of Campylobacter to bile (20, 21, 24, 41), little is known about the effect of bile on Campylobacter virulence determinants. Bile acids, including DOC, cholate, and chenodeoxycholate, have previously been shown to induce synthesis of the Cia proteins (44). This study was undertaken to determine the role that bile plays in the temporal expression of ciaB and its effect on Campylobacter pathogenesis. More specifically, we studied the effect of a physiologically relevant concentration of DOC on Campylobacter invasion potential, which is an important virulence determinant and contributes to the development of severe disease. We demonstrated that compared to bacteria grown on Mueller-Hinton (MH) agar, C. jejuni grown in the presence of DOC show (i) an increase in the kinetics of host cell invasion, (ii) an increase in ciaB gene expression, and (iii) an alteration in the expression of genes that play a role in Campylobacter pathogenesis. In summary, we demonstrated that it is possible to enhance the pathogenic behavior of C. jejuni in the laboratory by culturing this organism under conditions that resemble the in vivo environment.
(A portion of this work was presented at the Conference of Research Workers in Animal Diseases, Chicago, IL, 3 to 5 December 2006.)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. jejuni F38011 was recovered from an individual with clinical signs of campylobacteriosis. The C. jejuni F38011 strain was cultured on MH agar plates supplemented with 5% bovine citrated blood (MH-blood) under microaerobic conditions (5% O2, 10% CO2, 85% N2) at 37°C and was subcultured to a fresh plate every 48 h. MH-blood agar plates were supplemented with 200 μg/ml of kanamycin (Kan) when appropriate. Where indicated, C. jejuni was cultured on plates containing MH agar and MH agar supplemented with 0.1% sodium DOC (MHD agar) (1 mg/ml; Sigma, St. Louis, MO). Escherichia coli INVαF′ (Invitrogen, Carlsbad, CA) was cultured in Luria-Bertani (LB) broth and on LB agar plates at 37°C. LB agar plates were supplemented with 50 μg/ml Kan when appropriate.
Tissue culture.
Stock cultures of INT 407 epithelial cells (human embryonic intestinal; ATCC CCL 6) were grown in minimal essential medium (MEM) supplemented with 10% (vol/vol) fetal bovine serum (FBS) (HyClone Laboratories Inc., Logan, UT) and were maintained at 37°C in a humidified, 5% CO2 incubator.
Binding assay.
For experimental assays, a 24-well tissue culture tray was seeded with 2 × 105 INT 407 cells per well and incubated for 18 h at 37°C in a humidified, 5% CO2 incubator. C. jejuni F38011 was cultured on MH and MHD agar plates for 18 h. The bacteria were harvested in MEM supplemented with 1% FBS and washed twice. The optical density at 540 nm (OD540) of the bacterial suspension was adjusted to 0.2. Tenfold serial dilutions of the initial suspension were used to inoculate INT 407 cells as described previously (29). Briefly, the INT 407 cells were washed with MEM, and 1 ml of a bacterial suspension was added to each well. The bacteria were centrifuged at 600 × g to facilitate bacterium-host cell interaction. After 1 h of incubation, the nonadherent cells were removed by rinsing with phosphate-buffered saline (PBS). The INT 407 cells were lysed with a 0.1% Triton X-100 solution in PBS. The suspensions were 10-fold serially diluted, and the number of viable, adherent bacteria was determined by counting the resultant colonies on MH-blood agar plates. The initial inocula were also plated to determine the multiplicity of infection. The values reported below are the mean counts ± standard deviations derived from quadruplicate wells. The assay was repeated three times to ensure reproducibility.
Secretion assay.
C. jejuni F38011 was cultured for 18 h on MH agar and MHD agar plates. The bacteria were harvested in MEM without methionine (labeling medium) (ICN Biomedicals, Inc., Aurora, OH), pelleted by centrifugation at 6,000 × g, and washed twice in MEM. For metabolic labeling, approximately 5 × 108 CFU was suspended in MEM without methionine. Six milliliters of the bacterial suspension was then used for labeling with [35S]methionine (Perkin Elmer Life Sciences, Inc., Boston, MA) at a concentration of 50 μCi/ml (15). Both cultures were incubated for 30 min at 37°C under microaerobic conditions to allow incorporation of [35S]methionine. To each bacterial suspension (harvested from MH and MHD agar plates), chloramphenicol (Cm) was added at a final concentration of 128 μg/ml (44). This concentration of Cm immediately halts protein synthesis, as judged by the absence of [35S]methionine incorporation. The flasks were incubated for 30 min at 37°C under microaerobic conditions. The suspensions were divided into two flasks and incubated with and without 1% FBS (HyClone Laboratories Inc., Logan, UT) for 30 min at 37°C under microaerobic conditions to stimulate C. jejuni protein secretion. Prior to each secretion assay, the albumin was removed from the FBS using a SwellGel Blue albumin removal kit (Pierce, Rockford, IL). Following an additional 30 min of incubation, the supernatant fluids were harvested and samples were processed as described previously (15). The bacterial pellets were resuspended in water and mixed with an equal volume of double-strength sample solubilization buffer. The secreted proteins and whole-cell lysates (WCLs) (OD540, 0.1) were electrophoretically transferred to polyvinylidene fluoride membranes (Immobilon P; Millipore Corp., Bedford, MA). The membranes were exposed to phosphorimaging screens to detect secreted proteins and WCL proteins.
The C. jejuni secreted proteins and WCL were also probed with goat anti-CadF serum. Membranes were washed three times in PBS and incubated for 18 h at 4°C with a 1:500 dilution of the goat anti-C. jejuni CadF antibody in PBS (pH 7.4)-0.01% Tween 20 containing 5% dried milk. Bound antibodies were detected using peroxidase-conjugated rabbit anti-goat immunoglobulin G (Sigma, St. Louis, MO) at a 1:1,000 dilution and 4-chloro-1-naphthol (Sigma) as the chromogenic substrate.
Internalization kinetics.
The wells of 24-well tissue culture trays were seeded with INT 407 cells as described above for the binding assay. For the internalization assay, each well of a tray was inoculated with 1 ml of a bacterial suspension. The tissue culture tray was centrifuged at 600 × g for 5 min and incubated at 37°C in a humidified, 5% CO2 incubator for various periods of time. After 15, 30, 60, 90, and 180 min of incubation, the C. jejuni-infected INT 407 cells were washed with MEM and incubated with MEM supplemented with 1% FBS containing gentamicin at a concentration of 250 μg/ml. After 3 h of incubation, the cells were lysed, and the number of internalized bacteria was determined as described above.
Construction of PciaB-pMW10 and PporA-pMW10 reporter vectors.
The ciaB and porA promoter regions were PCR amplified from the C. jejuni NCTC 11168 strain using primers designed with BglII and BamHI sites, respectively, at the 5′ end and were cloned into pCR2.1 using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA). The ligated vectors were electroporated into E. coli INVαF′, and the transformants were selected on LB agar supplemented with Kan (50 μg/ml). The PciaB-pCR2.1 and PporA-pCR2.1 vectors were digested with BglII and BamHI to obtain 568- and 377-bp fragments, respectively. The fragments were gel purified using a QIAquick gel extraction kit (Qiagen, Valencia, CA). The promoter shuttle vector pMW10 (52) was digested with BamHI, gel purified, and ligated with the BglII PciaB and PporA fragments. The ligation mixtures were electroporated into E. coli INVαF′. Transformants were selected on LB agar supplemented with Kan using blue-white screening. The sequences of positive clones were verified using gene-specific primers as described elsewhere (52). The PciaB-pMW10 and PporA-pMW10 reporter vectors were electroporated into C. jejuni F38011, and the resultant colonies were screened on MH-blood agar plates with Kan (200 μg/ml). The presence of each vector was confirmed by PCR using primers designed to amplify a portion of the aphA-3 kanamycin gene.
β-Galactosidase assay.
To determine the levels of ciaB and porA promoter activities in the presence and absence of DOC, overnight cultures of the C. jejuni F38011 strain harboring the PciaB-pMW10 and PporA-pMW10 constructs were subcultured in MH broth at an initial OD540 of 0.05. The cultures were then incubated and allowed to reach an OD540 of 0.2. Five-milliliter portions of the log-phase cultures were then used to inoculate MH and MHD agar plates. The plates were incubated for 9, 12, 15, and 20 h. At each time point the bacteria were harvested in cold PBS and washed twice. To determine the effect of DOC concentration on the promoter activities of ciaB and porA, 5-ml log-phase cultures were used to inoculate MH agar plates supplemented with 0.05, 0.1, 0.2, and 0.4% DOC. The bacteria were harvested after 15 h with cold PBS and washed twice. The β-galactosidase assays were performed as described previously (28).
RNA isolation.
Total cellular RNA was isolated from C. jejuni F38011 cultured on MH and MHD agar plates after incubation for 3, 6, 9, 12, and 15 h using a RiboPure-bacterial kit (Ambion, Austin, TX) according to the manufacturer's instructions. The extracted RNA was treated twice with DNase at 37°C for 30 min to remove genomic DNA. The absence of genomic DNA was confirmed by PCR using C. jejuni ciaB gene sequence-specific primers CiaB-F (CTATGCTAGCCATACTTAGGC) and CiaB-R (GCCCGCCTTAGAACTTAC).
Real-time RT-PCR.
To determine the temporal expression of ciaB, real-time quantitative reverse transcription-PCR (RT-PCR) was performed using RNA isolated from the C. jejuni F38011 strain cultured on MH agar and MHD agar plates for 3, 6, 9, 12, and 15 h. cDNA was synthesized using the ThermoScript RT-PCR system (Invitrogen, Carlsbad, CA) according to the manufacturer's directions and 500 ng of RNA. Real-time RT-PCR amplification of 2.5 μl of cDNA (1:100 dilution) was performed using a reaction mixture containing Power SYBR green PCR master mixture (Applied Biosystems, Foster City, CA), 300 nM forward primer, 300 nM reverse primer, and diethyl pyrocarbonate-treated water. The real-time RT-PCR analysis was performed using a Gene Amp 7000 thermocycler (Applied Biosystems) with the following PCR parameters: 2 min at 50°C and then 40 cycles of denaturation at 95°C for 15 s and annealing at 55°C for 1 min. Threshold cycle values were determined using Prism SDS software, version 1.0 (Applied Biosystems). The comparative threshold cycle method was used to calculate changes, and samples were normalized to glyA, as this gene is a housekeeping gene that is not differentially expressed in the presence of DOC. We also performed real-time RT-PCR analysis to determine the change in porA in the presence of DOC as a control. The primers used for the analyses are shown in Table 1. Duplicate reactions were performed, and three biological replicates were used for each sample.
TABLE 1.
Primers used for real-time RT-PCR analysis
| Primer | Gene | Sequence (5′-3′) |
|---|---|---|
| Cj0181 RT-F | tonB | CTCAAGAAAAATCAAGTGGTGTTG |
| Cj0181 RT-R | tonB | CGATAGGAAACTCTGATACCATC |
| Cj0323 RT-F | Cj0323 | TATACTCAAATAACTTCAAATCATAGTG |
| Cj0323 RT-R | Cj0323 | CTCTTCTTGATTCTGTTCTAAAATTG |
| Cj0367c RT-F | cmeA | GTGTTGATTCGGCTTACGGAC |
| Cj0367cRT-R | cmeA | TCTAGCACTTGCTAGACTAGC |
| Cj0402 RT-F | glyA | CGATGGAACGGATAATCACC |
| Cj0402RT-R | glyA | AATACCTGCATTTCCAAGAGC |
| Cj0561 RT-F | Cj0561 | TTGGCAAACAGTGATTATCTAAGC |
| Cj0561 RT-R | Cj0561 | TAAAGTTCTGCACCGATAAAAGG |
| Cj0588 RT-F | tlyA | AATTTATGTTTCAAGAGCAGCTTT |
| Cj0588 RT-R | tlyA | CGTACTAGATCCTATATCAAGAC |
| Cj0706 RT-F | Cj0706 | AATTAGACGCTGCAAATGATGAG |
| Cj0706 RT-R | Cj0706 | CTTACTCTAATCTCGTTAATATTTTGC |
| Cj0786 RT-F | Cj0786 | GGTGTTATTTTTGGAATTGATTATGTG |
| Cj0786 RT-R | Cj0786 | CTATATATTCTTTTTTTTCTTTCTCTAAGC |
| Cj0793 RT-F | flgS | TGTTGCCTAGTGCGCTTTGG |
| Cj0793 RT-R | flgS | ATAAAACCTACCTTCAAATTCAAGC |
| Cj0862c RT-F | pabB | AAATGATACAAAAAATCTGAGTGAAAATG |
| Cj0862c RT-R | pabB | TTTGGTTTTTAAGCTTTTTTTTGTGATG |
| Cj0863c RT-F | xerD | AAGCAAAATGAAGAAGATGAAAAAGC |
| Cj0863c RT-R | xerD | TAATTTTACCCCTTTAGAACCTGC |
| Cj0914c RT-F | ciaB | AGACAAAGAAGATGTGGGTGA |
| Cj0914c RT-R | ciaB | AATCAATCAAACGCCTAAGTATGG |
| Cj0989 RT-F | Cj0989 | TCTTTATCATCGTTACTCGCTATG |
| Cj0989 RT-R | Cj0989 | TATCTTCTTTCATATTTTGTATGTTTTGG |
| Cj1212c RT-F | rbn | AGCAGCGCTTAGTTTTTATACTG |
| Cj1212c RT-R | rbn | GGAAATTTGCGTAAAAACAGAAAAAC |
| Cj1223 RT-F | dccR | GATATTTTGATCTTTGGATTTTAGATGT |
| Cj1223 RT-R | dccR | GGAGTTTGCTTTCCGCTTTTTC |
| Cj1224 RT-F | Cj1224 | CAAAATGGGATAACAGCTATAGTG |
| Cj1224 RT-R | Cj1224 | CTTCATTTTTACTTACAGATCTATCTG |
| Cj1314c RT-R | hisF | AATGCACGCAATGTTGATGAGC |
| Cj1314c RT-F | hisF | GCAGCCCTTGAGCCATCG |
| Cj1458c RT-F | thiL | GAACAAAGAAGATTTTATTATCAAAGC |
| Cj1458c RT-R | thiL | AAATCCTTACTAAAACACCAATCATC |
| Cj1530 RT-F | coaA | AACCGCTTCAATTGCTTGTGG |
| Cj1530 RT-R | coaA | CGATTTTGTCTGCGCTAATGC |
| Cj1531 RT-F | dapF | AGGTGCGGATGGCTTTATCG |
| Cj1531 RT-R | dapF | GCAGCCCTTGAGCCATCG |
Construction of the C. jejuni DNA microarray.
DNA fragments of individual open reading frames (ORFs) were amplified with the Sigma-Genosys (The Woodlands, TX) C. jejuni ORFmer primer set specific for strain NCTC 11168 coding sequences and with primers from Operon Technologies (Alameda, CA) specific for strain RM1221 unique sequences, as described previously (35). The PCR products were purified with a Qiagen 8000 robot by using a QIAquick 96-well Biorobot kit (Qiagen, Valencia, CA). A total of 1,530 ORFs from strain NCTC 11168 and 227 ORFs from strain RM1221 were PCR amplified, purified, and spotted in duplicate onto Ultra-GAPS glass slides (Corning Inc., Corning, NY) using an OmniGrid Accent (GeneMachines, Ann Arbor, MI), as described previously (35). Immediately after printing, the microarrays were UV cross-linked at 300 mJ using a Stratalinker UV Cross-linker 1800 (Stratagene, La Jolla, CA) and stored in a desiccator. Before use, the microarrays were blocked with Pronto! prehybridization solution (Corning Inc.) used according to the manufacturer's specifications.
Microarray hybridization and analysis.
For the expression profiling arrays, an indirect comparison of gene expression was performed, in which the expression profiles of C. jejuni F38011 cultured in the presence and absence of DOC were determined separately on different slides as described previously (26). Briefly, Cy5-labeled reference DNA from the C. jejuni F38011 strain was mixed with Cy3-labeled test cDNA (C. jejuni F38011 cultured in the presence and absence of DOC) and hybridized to the Campylobacter cDNA array (26) on separate slides. DNA microarrays were scanned using an Axon GenePix 4000B microarray laser scanner (Axon Instruments, Union City, CA), and the data for spot and background intensities were processed using the GenePix 4.0 software. To compensate for differences in the amount of template and uneven Cy3 or Cy5 dye incorporation, data normalization was performed as previously described (26).
Normalized data that passed the quality controls were analyzed using GENESPRING 7.3 software (Silicon Genetics, Palo Alto, CA). To compare genes differentially expressed in the presence and absence of DOC, at least four hybridization measurements were generated for each biological experiment (two technical replicate arrays and two replicate features per array), and the experiment was repeated two times (biological replicate). The significance of the centered data at a P value of <0.05 was determined using a parametric statistical t test, adjusting the individual P value with the Benjamini-Hochberg false discovery rate multiple test correction in the GeneSpring analysis package.
The microarray results were confirmed by real-time RT-PCR analysis. The genes upregulated in the presence of DOC were categorized into functional classes as described by the Sanger Center website (http://www.sanger.ac.uk/Projects/C_jejuni/). Genes in each functional class (a total of 19 genes) were selected, and the change in gene expression of the C. jejuni F38011 strain cultured in the presence and absence of DOC was confirmed using real-time RT-PCR analysis as described previously. The primers used for the analyses are shown in Table 1.
Microarray accession numbers.
Details of the microarray have been deposited in the NCBI GEO repository (http://www.ncbi.nlm.nih.gov/geo/) under platform accession number GPL6265. The microarray data set has been deposited in the NCBI Gene Expression Omnibus (GEO) repository (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE10110 (samples GSM255693, GSM255694, GSM255695, GSM255696, GSM255697, and GSM255698).
RESULTS
Culture with sodium DOC does not alter adherence or motility of C. jejuni.
To determine if DOC affects the ability of C. jejuni to adhere to INT 407 cells, a binding assay was performed with bacteria harvested from an MH agar plate or an MH agar plate supplemented with DOC. DOC did not alter the ability of C. jejuni to bind to the INT 407 cells (Fig. 1A). In addition, DOC did not alter the motility of the C. jejuni F38011 strain, as judged by motility assays (Fig. 1B). Collectively, these results indicate that culturing C. jejuni in the presence of DOC does not alter the adherence potential or motility of this bacterium.
FIG. 1.
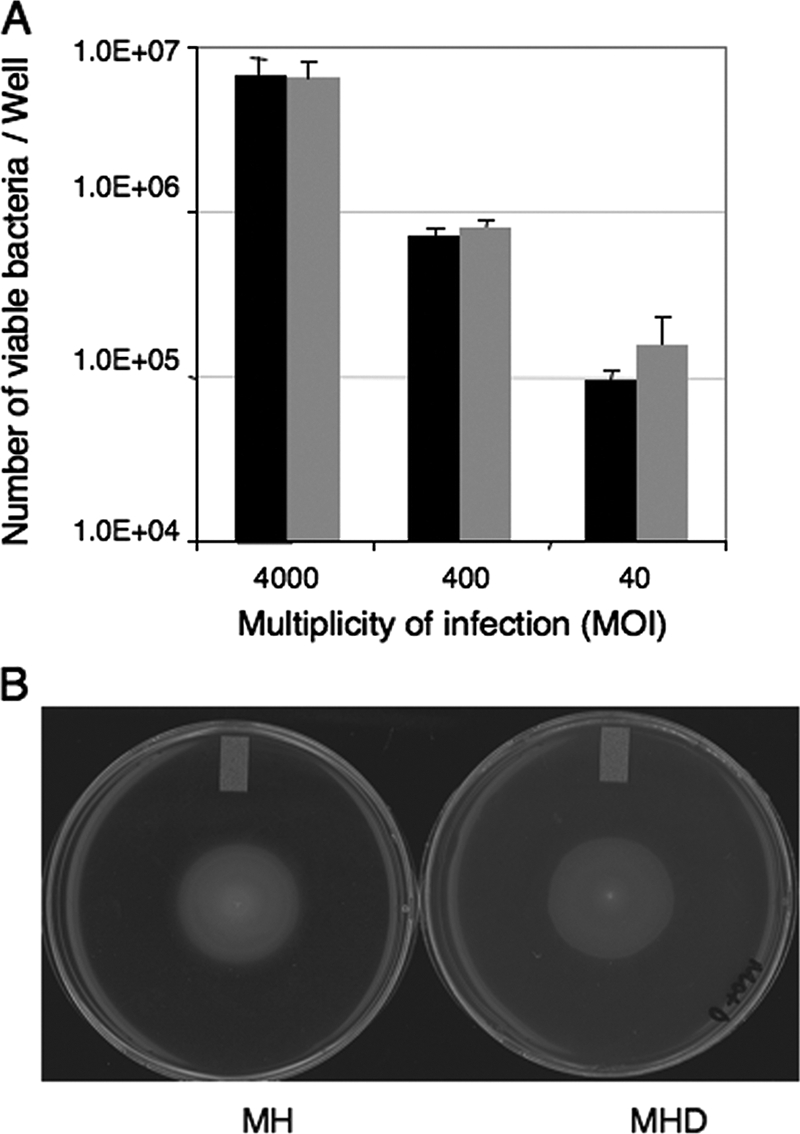
Sodium DOC does not alter the adherence (A) or motility (B) of the C. jejuni F38011 strain. (A) Tenfold serial dilutions of the C. jejuni F38011 strain, cultured on MH and MHD agar plates for 18 h, were used to inoculate INT 407 cells. The data indicate the numbers of bacteria from MH agar plates (black bars) and MHD agar plates (gray bars) that bound to INT 407 cells 30 min postinoculation. The bars indicate the mean numbers of viable bacteria recovered per well of a 24-well tissue culture tray, and the error bars indicate the standard deviations. (B) C. jejuni cultures on MH and MHD agar plates with 0.4% agar displayed equivalent zones of migration.
DOC stimulates the synthesis but not secretion of Campylobacter invasion antigens.
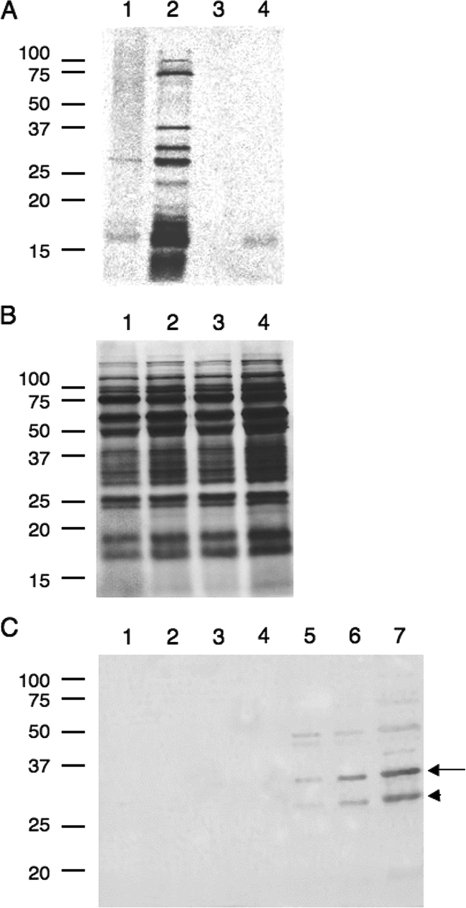
Invasion of the host cell is required for maximal disease. To determine if DOC alters the phenotypic behavior of C. jejuni with respect to invasion, we initially examined the effect of DOC on the secretion of the Cia proteins (Fig. 2). The bacteria were harvested from MH and MHD agar plates and labeled with [35S]methionine for 30 min. Following the metabolic labeling of proteins, Cm was added to inhibit protein synthesis. After an additional 30 min of incubation, FBS was added to the labeling medium. Previous studies have demonstrated that it is necessary to add FBS to the medium to induce secretion of the Cia proteins from C. jejuni (44). In contrast to the supernatants of the bacteria harvested from MH agar plates, the Cia proteins were clearly visible in the supernatants of bacteria harvested from MHD agar plates (Fig. 2A, lanes 1 and 2). To ensure that this finding was due to active protein secretion and reflected a bona fide difference in the phenotypic behaviors of C. jejuni harvested from MH and MHD agar plates, several controls were performed in parallel. First, samples in which FBS was not added to the labeling medium were included in the assay. Consistent with previous work, the Cia proteins were not present in the supernatants of bacteria harvested from both MH and MHD agar plates in the absence of FBS (Fig. 2A, lanes 3 and 4). Second, we harvested the bacteria after the labeling assay and assessed their metabolic state by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis coupled with autoradiography of the WCLs (Fig. 2B). As expected, the intensities of the protein bands were similar for the bacterial samples, indicating that the bacteria were metabolically active (Fig. 2B, lanes 1 to 4). Third, we probed the secreted proteins with an antibody prepared against the C. jejuni CadF outer membrane protein to ensure that the proteins detected in the supernatants were not due to bacterial cell lysis (Fig. 2C). In contrast to the C. jejuni WCLs, the CadF protein was not detected in the supernatants of any of the samples. Based on these results, we concluded that DOC is capable of stimulating the synthesis of the Cia proteins.
FIG. 2.
Sodium DOC stimulates the synthesis of Campylobacter invasion antigens. (A) Proteins secreted by C. jejuni as determined by SDS-PAGE coupled with autoradiography of the supernatants. (B) Proteins synthesized by C. jejuni as judged by SDS-PAGE coupled with autoradiography of the WCLs (OD540, 0.1). (C) Control, in which the proteins secreted by C. jejuni were probed with an antibody prepared against the C. jejuni CadF outer membrane protein. The metabolic labeling assays, preparation of supernatants and WCLs, and autoradiography were performed as described in Materials and Methods. Lane 1, C. jejuni F38011 harvested from an MH agar plate and radioactively labeled in medium supplemented with 1% FBS; lane 2, C. jejuni F38011 harvested from an MHD agar plate and radioactively labeled in medium supplemented with 1% FBS; lane 3, C. jejuni F38011 harvested from an MH agar plate and radioactively labeled in medium without 1% FBS; lane 4, C. jejuni F38011 harvested from an MHD agar plate and radioactively labeled in medium without 1% FBS; lane 5, 1:4 dilution of C. jejuni WCL; lane 6, 1:2 dilution of C. jejuni WCL; lane 7, 1:1 dilution of C. jejuni WCL. The migration of CadF as two protein species, indicated by the arrow (37-kDa species) and the arrowhead (32-kDa species), was due to the protein's heat-modifiable property.
DOC alters the invasion kinetics of the C. jejuni F38011 strain.
We tested the effect of DOC on the ability of C. jejuni F38011 to invade INT 407 cells as judged by a gentamicin protection assay. Here, the bacteria were incubated with INT 407 cells for 3 h, after which medium containing gentamicin was added for a 3-h period to kill the extracellular bacteria. Compared to C. jejuni harvested from MH agar plates, C. jejuni harvested from MHD agar plates was more invasive with the INT 407 cells (not shown). To further assess the specificity of DOC for stimulating the synthesis of the C. jejuni Cia proteins, an additional gentamicin assay was performed, in which the bacteria harvested from MH and MHD agar plates were suspended in medium containing Cm prior to inoculation of the INT 407 cells. The concentration of Cm used in these assays immediately halted C. jejuni protein synthesis, thereby preventing additional protein synthesis by C. jejuni in the presence of epithelial cells (not shown). In the presence of Cm, an 11-fold increase in the number of intracellular C. jejuni cells harvested from MHD agar plates was observed compared with bacteria harvested from MH agar plates. Collectively, these data suggest that culture of C. jejuni with DOC induces the synthesis of proteins that facilitate the organism's invasion of epithelial cells.
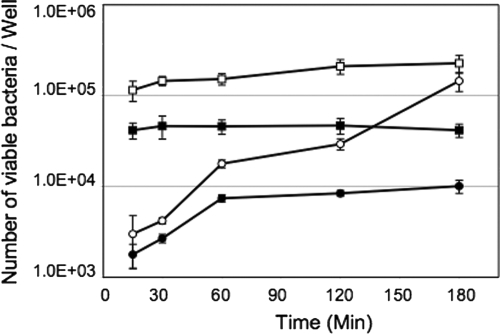
Since culture with DOC stimulates Cia protein synthesis and enhances the ability of C. jejuni to invade INT 407 cells, an invasion assay was performed to determine the effect of DOC on the kinetics of invasion. Based on the observation that DOC stimulates the bacterium to synthesize the Cia proteins, we hypothesized that culturing C. jejuni on MHD agar plates would increase the invasive potential compared to that of bacteria harvested from MH agar plates. As predicted, C. jejuni F38011 harvested from MHD agar plates was able to maximally invade INT 407 cells within 15 min after infection (Fig. 3). An increase in INT 407 cell invasion was also observed at 15 min after infection with C. jejuni F38011 harvested from MHD agar plates and suspended in medium with Cm compared to the invasion efficiency of C. jejuni F38011 harvested from plates containing MH agar alone. For C. jejuni cultured on MH agar plates, the number of intracellular bacteria increased over time, and the level reached was similar to that observed for the bacteria harvested from MHD plates after a 3-h period. As expected, the invasion of INT 407 cells was significantly reduced when bacteria that were harvested from MH agar plates and resuspended in medium containing Cm were used. Based on these results, we concluded that culturing C. jejuni in the presence of DOC alters the invasion kinetics of C. jejuni such that maximal invasion is achieved as early as 15 min postinfection.
FIG. 3.
Culturing the C. jejuni F38011 strain with sodium DOC alters the kinetics of invasion of INT 407 cells. Gentamicin protection assays were performed as described in Materials and Methods with C. jejuni F38011 cultured on MH and MHD agar plates for 18 h. The bacteria were incubated with INT 407 cells in the absence and presence of 128 μg/ml of Cm to inhibit protein synthesis. The numbers of internalized bacteria are indicated for C. jejuni F38011 cultured on an MH agar plate (○), C. jejuni F38011 cultured on an MHD agar plate (□), C. jejuni F38011 cultured on an MH agar plate and then incubated for 30 min with Cm (•), and C. jejuni F38011 cultured on an MHD agar plate and then incubated for 30 min with Cm (▪). The symbols indicate the mean numbers of viable bacteria recovered per well of a 24-well tissue culture tray, and the error bars indicate standard deviations.
DOC induces the ciaB promoter.
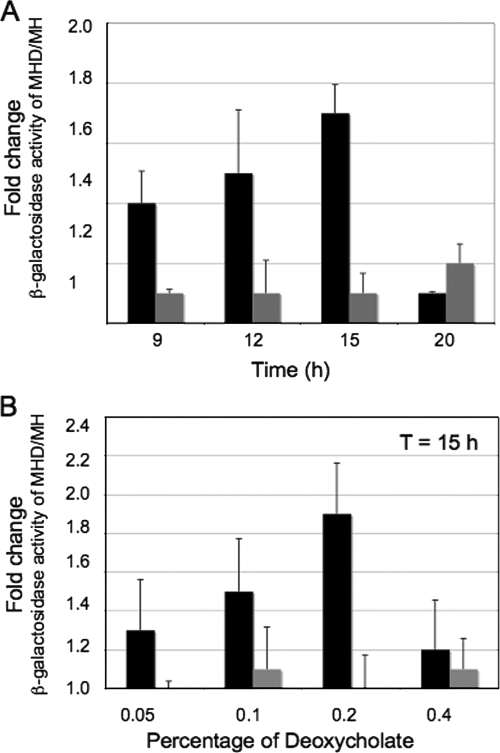
To determine if DOC induces ciaB gene expression, the ciaB promoter was cloned upstream of the β-galactosidase gene (ciaB promoter-β-galactosidase) in the pMW10 vector. A log-phase culture of C. jejuni F38011 harboring the PciaB-pMW10 construct was then used to inoculate MH and MHD agar plates, and β-galactosidase assays were performed. The ciaB promoter activity gradually increased in the presence of 0.1% DOC and reached a maximum at 15 h postinoculation, after which it decreased (Fig. 4A). The ability of DOC to induce the promoter activity of porA was also measured. In contrast to the effect on the activity of the ciaB promoter, DOC did not significantly alter porA promoter activity over time.
FIG. 4.
Stimulation of ciaB promoter activity by sodium DOC is time (A) and dose (B) dependent. The data shown represent the ratios of the β-galactosidase activity of ciaB (black bars) and porA (gray bars) for the C. jejuni F38011 strain cultured on an MHD agar plate to the β-galactosidase activity of ciaB and porA for the C. jejuni F38011 strain cultured on an MH agar plate. The bars indicate the means of two separate experiments, and the error bars indicate the standard deviations; each experiment was comprised of triplicate samples.
We also measured the activities of the ciaB and porA promoters in response to different concentrations of DOC. Here, MH and MH agar plates supplemented with various concentrations of DOC were inoculated with a log-phase culture of C. jejuni F38011 harboring PciaB-pMW10 and PporA-pMW10. The β-galactosidase activity was measured at 15 h postinoculation. In contrast to the activity of the porA promoter, the activity of the ciaB promoter was influenced directly by the concentration of DOC. Specifically, the maximum increase in ciaB promoter activity observed when the C. jejuni F38011 strain was cultured on an MH agar plate supplemented with 0.2% DOC was twofold (Fig. 4B). Similar to the results of previous experiments, the ciaB promoter activity was also found to be maximal at 15 h postinoculation with 0.2% DOC (not shown). These results indicate that DOC stimulates ciaB promoter activity in a dose- and time-dependent manner.
Exposure to DOC stimulates the expression of ciaB.
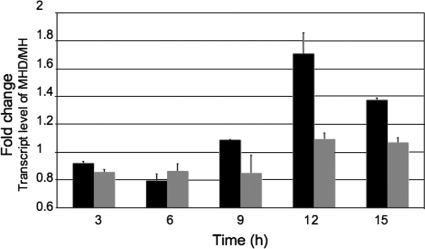
To identify genes that are differentially expressed in response to DOC, we performed microarray experiments with RNA extracted from C. jejuni cultured in the presence and absence of DOC. Real-time RT-PCR was performed using RNA isolated at 3, 6, 9, 12, and 15 h to determine the difference in the ciaB transcript levels in C. jejuni when it was cultured with DOC. The maximum increase in the ciaB transcript level was observed when C. jejuni was cultured with 0.1% DOC for 12 h (Fig. 5). Based on this result, we performed microarray experiments with RNA extracted from C. jejuni cultured with 0.1% DOC for 12 h to determine if genes were coexpressed with ciaB. A total of 202 genes were differentially expressed in response to culture of C. jejuni F38011 with 0.1% DOC for 12 h. A total of 156 genes, including ciaB, were upregulated ≥1.5-fold (Table 2), while 46 genes were downregulated ≥1.5-fold (Table 3) in the presence of DOC. To confirm the microarray data, genes representing each functional class (Fig. 6) were selected, and real-time RT-PCR was performed (Table 4). The genes found to be upregulated in the presence of DOC by microarray analyses were also found to be upregulated by real-time RT-PCR analyses.
FIG. 5.
Temporal expression of ciaB and porA in C. jejuni F38011 cultured in the presence of DOC. Real-time RT-PCR analysis was performed with total RNA extracted from the C. jejuni F38011 strain grown on MH and MHD agar plates for 3, 6, 9, 12, and 15 h. The changes in the ciaB (black bars) and porA (gray bars) transcript levels were measured using the comparative threshold cycle method, and glyA was used as the internal control.
TABLE 2.
Transcripts upregulated in the C. jejuni F38011 strain in the presence of 0.1% sodium DOC
| Gene | Common name | Change (fold) | Functional classification |
|---|---|---|---|
| Cj0002 | dnaN | 1.6 | DNA replication |
| Cj0008 | Cj0008 | 1.6 | Conserved hypothetical proteins |
| Cj0037c | Cj0037c | 1.9 | Electron transport |
| Cj0040 | Cj0040 | 2.9 | Unknown |
| Cj0073c | Cj0073c | 1.7 | Conserved hypothetical proteins |
| Cj0080 | Cj0080 | 5.1 | Cell envelope (membranes, lipoproteins, and porins) |
| Cj0100 | Cj0100 | 2.2 | Cell division |
| Cj0114 | Cj0114 | 1.6 | Cell envelope (miscellaneous periplasmic proteins) |
| Cj0155c | rpmE | 1.5 | Ribosomal protein synthesis and modification |
| Cj0179 | exbB1 | 1.6 | Transport/binding proteins (others) |
| Cj0181 | tonB1 | 5.1 | Transport/binding proteins (others) |
| Cj0188c | Cj0188c | 1.5 | Conserved hypothetical proteins |
| Cj0189c | Cj0189c | 1.6 | Conserved hypothetical proteins |
| Cj0190c | Cj0190c | 2.2 | Miscellaneous |
| Cj0193c | tig | 1.9 | Chaperones, chaperonins, heat shock |
| Cj0199c | Cj0199c | 7.5 | Cell envelope (miscellaneous periplasmic proteins) |
| Cj0207 | infC | 1.9 | Protein translation and modification |
| Cj0237 | cynT | 1.5 | Central intermediary metabolism |
| Cj0246c | Cj0246c | 2.0 | Signal transduction |
| Cj0247c | Cj0247c | 3.0 | Unknown |
| Cj0295 | Cj0295 | 2.0 | Miscellaneous |
| Cj0301c | modB | 1.8 | Transport/binding proteins (anions) |
| Cj0309c | Cj0309c | 3.3 | Drug sensitivity |
| Cj0323 | Cj0323 | 1.6 | Unknown |
| Cj0324 | ubiE | 1.8 | Biosynthesis of cofactors, prosthetic groups, and carriers |
| Cj0346 | trpD | 1.7 | Amino acid biosynthesis (aromatic amino acid family) |
| Cj0352 | Cj0352 | 1.6 | Cell envelope (membranes, lipoproteins, and porins) |
| Cj0356c | Cj0356c | 2.0 | Conserved hypothetical proteins |
| Cj0365c | cmeA | 1.9 | Antibiotic resistance |
| Cj0366c | cmeB | 1.6 | Antibiotic resistance |
| Cj0367c | cmeC | 2.1 | Antibiotic resistance |
| Cj0381c | pyrF | 2.1 | Pyrimidine biosynthesis |
| Cj0382c | nusB | 1.5 | RNA synthesis, RNA modification, and DNA transcription |
| Cj0395c | Cj0395c | 2.2 | Unknown |
| Cj0397c | Cj0397c | 1.9 | Conserved hypothetical proteins |
| Cj0413 | Cj0413 | 2.8 | Cell envelope (miscellaneous periplasmic proteins) |
| Cj0484 | Cj0484 | 1.9 | Transport/binding proteins (others) |
| Cj0512 | purC | 1.5 | Purine biosynthesis |
| Cj0526c | fliE | 2.0 | Cell envelope (surface structures) |
| Cj0539 | Cj0539 | 1.7 | Unknown |
| Cj0561c | Cj0561c | 3.5 | Cell envelope (miscellaneous periplasmic proteins) |
| Cj0573 | Cj0573 | 3.9 | Conserved hypothetical proteins |
| Cj0579c | Cj0579c | 1.6 | Conserved hypothetical proteins |
| Cj0580c | Cj0580c | 1.5 | Miscellaneous |
| Cj0588 | tlyA | 1.7 | Pathogenicity |
| Cj0609c | Cj0609c | 1.6 | Cell envelope (miscellaneous periplasmic proteins) |
| Cj0618 | Cj0618 | 2.2 | Unknown |
| Cj0659c | Cj0659c | 2.4 | Cell envelope (miscellaneous periplasmic proteins) |
| Cj0667 | Cj0667 | 1.6 | Conserved hypothetical proteins |
| Cj0679 | kdpD′ | 2.2 | Transport/binding proteins (cations) |
| Cj0683 | Cj0683 | 2.7 | Cell envelope (miscellaneous periplasmic proteins) |
| Cj0690c | Cj0690c | 2.1 | DNA replication |
| Cj0692c | Cj0692c | 1.5 | Cell envelope (membranes, lipoproteins, and porins) |
| Cj0705 | Cj0705 | 1.8 | Conserved hypothetical proteins |
| Cj0706 | Cj0706 | 1.9 | Conserved hypothetical proteins |
| Cj0707 | kdtA | 1.5 | Cell envelope (surface polysaccharides, lipopolysaccharides, and antigens) |
| Cj0713 | trmD | 1.7 | Aminoacyl tRNA synthetases and their modification |
| Cj0724 | Cj0724 | 1.5 | Unknown |
| Cj0729 | Cj0729 | 2.7 | Unknown |
| Cj0733 | Cj0733 | 1.7 | Conserved hypothetical proteins |
| Cj0734c | hisJ | 2.5 | Transport/binding proteins (amino acids and amines) |
| Cj0742 | Cj0742 | 1.9 | Cell envelope (membranes, lipoproteins, and porins) |
| Cj0777 | Cj0777 | 1.6 | DNA replication |
| Cj0778 | peb2 | 1.9 | Cell envelope (miscellaneous periplasmic proteins) |
| Cj0784 | Cj0784 | 1.5 | Cell envelope (miscellaneous periplasmic proteins) |
| Cj0785 | napD | 2.3 | Electron transport |
| Cj0786 | Cj0786 | 14.0 | Unknown |
| Cj0787 | Cj0787 | 1.6 | Conserved hypothetical proteins |
| Cj0788 | Cj0788 | 1.8 | Conserved hypothetical proteins |
| Cj0789 | Cj0789 | 1.5 | RNA synthesis, RNA modification, and DNA transcription |
| Cj0793 | flgS | 1.5 | Signal transduction |
| Cj0798c | ddlA | 1.7 | Cell envelope (murein sacculus and peptidoglycan) |
| Cj0837c | Cj0837c | 2.0 | Unknown |
| Cj0838c | metS | 1.5 | Aminoacyl tRNA synthetases and their modification |
| Cj0842 | Cj0842 | 2.6 | Cell envelope (membranes, lipoproteins, and porins) |
| Cj0843c | Cj0843c | 1.6 | Protein degradation |
| Cj0848c | Cj0848c | 3.3 | Conserved hypothetical proteins |
| Cj0852c | Cj0852c | 1.7 | Cell envelope (membranes, lipoproteins, and porins) |
| Cj0862c | pabB | 1.5 | Biosynthesis of cofactors, prosthetic groups and carriers |
| Cj0863c | xerD | 2.0 | DNA replication |
| Cj0881c | Cj0881c | 1.5 | Conserved hypothetical proteins |
| Cj0884 | rpsO | 1.6 | Ribosomal protein synthesis and modification |
| Cj0914c | ciaB | 1.5 | Pathogenicity |
| Cj0926 | Cj0926 | 1.5 | Cell envelope (membranes, lipoproteins, and porins) |
| Cj0960c | rnpA | 1.6 | RNA synthesis, RNA modification, and DNA transcription |
| Cj0962 | Cj0962 | 1.5 | Miscellaneous |
| Cj0963 | Cj0963 | 1.7 | Conserved hypothetical proteins |
| Cj0967 | Cj0967 | 1.8 | Cell envelope (miscellaneous periplasmic proteins) |
| Cj0983 | Cj0983 | 1.7 | Cell envelope (membranes, lipoproteins, and porins) |
| Cj0987c | Cj0987c | 1.6 | Cell envelope (membranes, lipoproteins, and porins) |
| Cj0989 | Cj0989 | 4.4 | Cell envelope (membranes, lipoproteins, and porins) |
| Cj1006c | Cj1006c | 3.0 | Conserved hypothetical proteins |
| Cj1028c | Cj1028c | 1.8 | Miscellaneous nucleoside/nucleotide reactions |
| Cj1038 | ftsW | 2.4 | Cell division |
| Cj1053c | Cj1053c | 6.3 | Cell envelope (membranes, lipoproteins, and porins) |
| Cj1056c | Cj1056c | 1.5 | Conserved hypothetical proteins |
| Cj1070 | rpsF | 1.7 | Ribosomal protein synthesis and modification |
| Cj1071 | ssb | 1.5 | DNA replication |
| Cj1079 | Cj1079 | 2.1 | Cell envelope (miscellaneous periplasmic proteins) |
| Cj1103 | csrA | 2.0 | Broad regulatory function |
| Cj1180c | Cj1180c | 2.0 | Transport/binding proteins (others) |
| Cj1181c | tsf | 1.7 | Protein translation and modification |
| Cj1191c | Cj1191c | 1.5 | Signal transduction |
| Cj1201 | metE | 2.3 | Amino acid biosynthesis (aspartate family) |
| Cj1204c | atpB | 1.7 | ATP-proton motive force |
| Cj1212c | rbn | 1.8 | Aminoacyl tRNA synthetases and their modification |
| Cj1217c | Cj1217c | 2.5 | Conserved hypothetical proteins |
| Cj1223c | Cj1223c | 1.5 | Signal transduction |
| Cj1224 | Cj1224 | 5.3 | Transport/binding proteins (cations) |
| Cj1242 | Cj1242 | 5.2 | Unknown |
| Cj1289 | Cj1289 | 1.8 | Cell envelope (miscellaneous periplasmic proteins) |
| Cj1349c | Cj1349c | 2.2 | Pathogenicity |
| Cj1355 | ceuE | 1.5 | Transport/binding proteins (cations) |
| Cj1384c | Cj1384c | 1.6 | Unknown |
| Cj1385 | katA | 1.6 | Detoxification |
| Cj1388 | Cj1388 | 1.5 | Conserved hypothetical proteins |
| Cj1416c | Cj1416c | 1.7 | Cell envelope (surface polysaccharides, lipopolysaccharides, and antigens) |
| Cj1417c | Cj1417c | 1.5 | Miscellaneous |
| Cj1418c | Cj1418c | 1.8 | Miscellaneous |
| Cj1442c | Cj1442c | 1.6 | Unknown |
| Cj1450 | Cj1450 | 1.7 | Unknown |
| Cj1457c | Cj1457c | 1.6 | Conserved hypothetical proteins |
| Cj1458c | thiL | 1.5 | Biosynthesis of cofactors, prosthetic groups, and carriers |
| Cj1463 | Cj1463 | 2.4 | Conserved hypothetical proteins |
| Cj1472c | Cj1472c | 2.3 | Cell envelope (membranes, lipoproteins, and porins) |
| Cj1473c | Cj1473c | 1.7 | Unknown |
| Cj1475c | Cj1475c | 2.2 | Unknown |
| Cj1484c | Cj1484c | 1.5 | Cell envelope (membranes, lipoproteins, and porins) |
| Cj1492c | Cj1492c | 1.7 | Signal transduction |
| Cj1495c | Cj1495c | 1.5 | Conserved hypothetical proteins |
| Cj1530 | Cj1530 | 1.6 | Conserved hypothetical proteins |
| Cj1531 | dapF | 1.8 | Amino acid biosynthesis (aspartate family) |
| Cj1540 | Cj1540 | 1.5 | Cell envelope (miscellaneous periplasmic proteins) |
| Cj1547 | Cj1547 | 2.3 | Conserved hypothetical proteins |
| Cj1556 | Cj1556 | 2.8 | Conserved hypothetical proteins |
| Cj1581c | dppD | 1.8 | Transport/binding proteins (others) |
| Cj1589 | Cj1589 | 2.8 | Conserved hypothetical proteins |
| Cj1603 | hisF | 6.9 | Amino acid biosynthesis (histidine) |
| Cj1611 | rpsT | 1.6 | Ribosomal protein synthesis and modification |
| Cj1614 | chuA | 3.0 | Transport/binding proteins (cations) |
| Cj1621 | Cj1621 | 5.9 | Cell envelope (miscellaneous periplasmic proteins) |
| Cj1640 | Cj1640 | 2.0 | Conserved hypothetical proteins |
| Cj1646 | iamB | 1.9 | Transport/binding proteins (others) |
| Cj1675 | fliQ | 2.4 | Cell envelope (surface structures) |
| Cj1679 | Cj1679 | 1.8 | Unknown |
| Cj1695c | rplE | 1.5 | Ribosomal protein synthesis and modification |
| Cj1724c | Cj1724c | 1.7 | Conserved hypothetical proteins |
| CJE1111 | CJE1111 | 2.7 | Miscellaneous |
| CJE1278 | CJE1278 | 1.5 | Miscellaneous |
| CJE1470 | CJE1470 | 2.5 | Miscellaneous |
| CJE1472 | CJE1472 | 1.7 | Miscellaneous |
| ORF00215 | CJE0220 | 2.0 | Miscellaneous |
| ORF00225 | CJE0230 | 1.5 | Miscellaneous |
| ORF00226 | CJE0231 | 1.9 | Miscellaneous |
| ORF00236 | CJE0241 | 3.4 | Miscellaneous |
| ORF00237 | CJE0243 | 3.8 | Miscellaneous |
TABLE 3.
Transcripts downregulated in the C. jejuni F38011 strain in the presence of 0.1% sodium DOC
| Gene | Common name | Change (fold) | Functional classification |
|---|---|---|---|
| Cj0056c | Cj0056c | 1.6 | Unknown |
| Cj0069 | Cj0069 | 1.6 | Unknown |
| Cj0076c | lctP | 1.8 | Transport/binding proteins |
| Cj0105 | atpA | 1.6 | Respiration |
| Cj0107 | atpD | 1.5 | Respiration |
| Cj0136 | infB | 1.5 | Synthesis and modification of macromolecules |
| Cj0146c | trxB | 2.1 | Biosynthesis of cofactors, prosthetic groups, and carriers |
| Cj0153c | Cj0153c | 1.7 | Synthesis and modification of macromolecules |
| Cj0453 | thiC | 3.5 | Biosynthesis of cofactors, prosthetic groups, and carriers |
| Cj0471 | rpmG | 2.3 | Synthesis and modification of macromolecules |
| Cj0472 | secE | 2.4 | Protein and peptide secretion |
| Cj0473 | nusG | 2.9 | Synthesis and modification of macromolecules |
| Cj0475 | rplA | 1.7 | Synthesis and modification of macromolecules |
| Cj0499 | Cj0499 | 1.6 | Conserved hypothetical proteins |
| Cj0537 | oorB | 1.7 | Respiration |
| Cj0698 | flgG | 1.7 | Cell envelope |
| Cj0720c | flaC | 2.0 | Cell envelope |
| Cj0855 | folD | 1.5 | Biosynthesis of cofactors, prosthetic groups, and carriers |
| Cj0864 | dsbA | 1.5 | Cell envelope |
| Cj0913c | hup | 1.8 | Synthesis and modification of macromolecules |
| Cj0982c | Cj0982 | 1.5 | Transport/binding proteins |
| Cj0998c | Cj0998c | 2.2 | Cell envelope |
| Cj1014c | livF | 1.6 | Transport/binding proteins |
| Cj1015c | livG | 1.6 | Transport/binding proteins |
| Cj1060c | Cj1060c | 1.5 | Unknown |
| Cj1068 | Cj1068 | 1.5 | Cell envelope |
| Cj1163c | Cj1163c | 1.7 | Transport/binding proteins |
| Cj1168c | Cj1168c | 1.7 | Cell envelope |
| Cj1198 | luxS | 1.7 | Conserved hypothetical proteins |
| Cj1229 | cbpA | 1.5 | Chaperones, chaperonins, heat shock |
| Cj1274c | pyrH | 1.6 | Purines, pyrimidines, nucleosides, and nucleotides |
| Cj1290c | accC | 1.5 | Fatty acid biosynthesis |
| Cj1291c | accB | 1.8 | Fatty acid biosynthesis |
| Cj1293 | flmA | 1.8 | Cell envelope |
| Cj1309c | Cj1309c | 1.5 | Unknown |
| Cj1364c | fumC | 1.5 | Energy metabolism |
| Cj1400c | fabI | 1.5 | Fatty acid biosynthesis |
| Cj1403c | gapA | 1.6 | Energy metabolism |
| Cj1502c | putP | 1.8 | Transport/binding proteins |
| Cj1548c | Cj1548c | 1.8 | Miscellaneous |
| Cj1567c | nuoM | 1.6 | Respiration |
| Cj1628 | exbB2 | 1.8 | Transport/binding proteins |
| Cj1658 | Cj1658 | 1.6 | Cell envelope |
| Cj1659 | Cj1659 | 2.0 | Cell envelope |
| Cj1682c | gltA | 1.7 | Energy metabolism |
| Cj1719c | leuA | 1.8 | Amino acid biosynthesis |
FIG. 6.
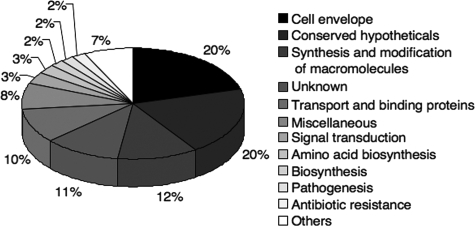
Functional classification of the C. jejuni F38011 genes upregulated ≥1.5-fold in the presence of DOC. The chart shows the percentages of genes belonging to functional classes based on the total number of genes upregulated in the presence of DOC. The percentages are the values for functional categories containing more than 3% of the total number of genes upregulated. Functional categories with two or fewer genes were grouped together as “Others”.
TABLE 4.
Real-time RT-PCR analysis of transcripts upregulated in the C. jejuni F38011 strain in the presence of 0.1% sodium DOC
| Gene | Increase (fold) |
|---|---|
| Pathogenesis | |
| ciaB | 3.4 |
| tlyA | 2.2 |
| Antibiotic resistance | |
| cmeA | 4.0 |
| Amino acid biosynthesis | |
| dapF | 1.7 |
| hisF | 2.0 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | |
| pabB | 2.0 |
| thiL | 1.7 |
| Cell envelope | |
| Cj0989 | 1.9 |
| Cj0561 | 4.5 |
| Conserved hypothetical proteins | |
| coaA | 3.2 |
| Cj0706 | 2.9 |
| Signal transduction | |
| Cj0246 | 2.5 |
| dccR | 1.7 |
| Synthesis and modification of macromolecules | |
| xerD | 1.7 |
| rbn | 2.1 |
| Transport/binding proteins | |
| Cj1224 | 2.4 |
| tonB1 | 1.8 |
| Unknown | |
| Cj0786 | 3.3 |
| Cj0323 | 3.8 |
Not surprisingly, the cmeABC operon, which encodes an efflux pump that participates in the resistance of C. jejuni to the deleterious effects of bile salts (21-24), was upregulated in response to culture with DOC. Moreover, approximately 20% of the genes upregulated in the presence of DOC were genes whose products were associated with the cell envelope of C. jejuni. Twelve percent of the genes upregulated in the presence of DOC were involved in synthesis and modification of RNA and DNA molecules. Ten percent of the genes upregulated in the presence of DOC were genes that encode components of transport systems (ceuE, chuA, exbB1, modB, and tonB). Perhaps most relevant to this study, genes known to play an important role in C. jejuni pathogenesis were upregulated (ciaB, dccR, flgS, and tlyA). Finally, a large number of genes upregulated in response to DOC were conserved hypothetical protein genes (∼20%) and genes with unknown functions (11%). Overall, the results of the microarray experiments indicated that expression of virulence genes in C. jejuni is increased in the presence of DOC.
DISCUSSION
Invasion is an important virulence determinant in C. jejuni pathogenesis. Newell et al. (30) reported that clinical isolates are more invasive than environmental isolates. Further, Everest et al. (8) observed that C. jejuni isolates recovered from individuals with colitis were significantly more invasive than isolates recovered from individuals with noninflammatory diarrhea. Evidence from a number of animal studies also supports the hypothesis that disease development is related to the invasive potential of the C. jejuni strain (3, 9, 13, 48, 51). Here we report that culturing C. jejuni with the bile acid DOC triggers its invasive potential by stimulating the expression and synthesis of the Cia proteins. Specifically, we monitored the effect of DOC on the expression of ciaB and found that the expression was upregulated by three independent assays, the β-galactosidase reporter assay, the real-time RT-PCR assay, and microarrays.
The ability of C. jejuni to invade epithelial cells is dependent on several bacterial properties, including motility, adherence, and protein secretion. Thus, the effect of DOC on all three of these virulence factors was investigated. We found that culturing C. jejuni in the presence of DOC did not alter the organism's motility or adherence potential. The zones of migration measured for the C. jejuni F38011 strain on semisolid MH and MHD agar were comparable. In addition, the abilities of bacteria harvested from MH and MHD agar plates to bind to epithelial cells did not differ; this finding was reproducible regardless of the multiplicity of infection used. However, there was a marked difference in the amount of the Cia proteins secreted by C. jejuni when it was cultured in the presence of DOC.
To determine if DOC plays a role in Cia synthesis, the bacteria were labeled with [35S]methionine and incubated in the presence of Cm prior to induction of Cia secretion. Addition of Cm helped distinguish the proteins synthesized in the presence of DOC from the proteins synthesized subsequently in presence of FBS, which was added to stimulate Cia protein secretion. While the Cia proteins were detected in the supernatants of C. jejuni harvested from MHD agar plates, we did not detect any secreted proteins in the supernatants of C. jejuni harvested from MH agar plates. This finding indicated that presynthesized Cia proteins were present in the bacteria cultured in the presence of DOC.
The Cia proteins are required for maximal invasion of host cells. Thus, bacteria cultured in the presence of DOC are “primed” to invade epithelial cells as they harbor presynthesized Cia proteins. This conclusion is supported by the difference in the invasion kinetics of bacteria harvested from MH and MHD agar plates. While bacteria harvested from MH agar plates require 3 h to achieve maximal invasion, bacteria harvested from MHD agar plates are able to invade INT 407 cells within 15 min. Taken together, the data indicate that culturing C. jejuni in the presence of a physiological concentration of DOC results in global changes in gene expression and alteration of the bacterium's phenotype which significantly enhances its invasive potential.
A number of in vitro studies have been performed to better understand how bacteria modulate gene expression when they encounter hostlike conditions and to determine the growth conditions that alter an organism's invasive behavior (4, 11, 14, 37, 47, 49, 50). We hypothesized that we could use the expression of the ciaB gene as a marker to better define the kinetics of cia induction by DOC and use the data to identify when the other cia genes are maximally expressed. Using a ciaB promoter-lacZ fusion construct, we found that DOC was able to induce ciaB promoter activity. The ciaB promoter activity increased moderately over time; however, higher levels of induction were observed with increasing concentrations of bile. We observed maximal induction of the ciaB promoter in the presence of 0.2% DOC at 15 h. It is noteworthy that bile has been reported to have a similar effect on the promoter that drives the expression of the genes encoding the multidrug efflux pump CmeABC (21), which is inducible by bile acids in a dose- and time-dependent manner.
Based on the data from the β-galactosidase assays, we performed a real-time RT-PCR analysis with the C. jejuni F38011 strain cultured with DOC for 3, 6, 9, 12, and 15 h to determine when the ciaB gene was maximally expressed. The RT-PCR analysis revealed that there was maximal expression of ciaB following 12 h of incubation with DOC, results which are complementary to the results of the β-galactosidase assay. Although the temporal expression of the ciaB gene with DOC was slower than one might predict, the induction kinetics are similar to the induction kinetics of the cmeABC operon in response to bile (21). The kinetics of ciaB induction in vivo are not known, nor are the other factors that contribute to ciaB expression.
Because the genes coregulated with ciaB might code for Cia proteins and other unidentified virulence factors, RNA was extracted from C. jejuni cultured with DOC, and microarray experiments were performed to determine the entire transcriptome. DOC differentially regulated a total of 202 genes in the C. jejuni F38011 strain, 150 of which were upregulated and 46 of which were downregulated. A number of C. jejuni genes predicted to play a role in signal transduction were upregulated by DOC, including flgS and dccR. The FlgS sensor kinase is a part of the FlgSR two-component system known to play an important role in C. jejuni motility (53). The dccRS two-component system has been shown to play a role in the in vivo colonization of immunocompetent limited flora (I-LF) mice, severe combined immunodeficient limited flora (SCID-LF) mice, and 1-day-old chicks (25). Taken together, the evidence indicated that DOC acts as a stimulus to trigger a global regulatory response. Whether DOC acts directly or indirectly to induce the signal transduction pathways requires additional study.
Since bile salts are surface-active, amphipathic molecules that act as detergents, exerting their effect primarily on cell membranes (5), it is not surprising that 31 genes encoding cell envelope-associated proteins were significantly upregulated when C. jejuni was cultured with DOC. In addition, genes encoding members of the ABC transporter family, including exbB1 and modB, along with genes involved in iron transport, including chuA and ceuE (42, 43), were upregulated. The gene encoding hemolysin A (TlyA), a member of a family of contact-dependent hemolysins found in Helicobacter, Serpulina, and Mycobacterium, was also found to be upregulated. In Helicobacter pylori, TlyA is homologous to a pore-forming cytolysin, and a tlyA defined mutant showed reduced in vitro hemolytic activity and reduced adherence to human gastric adenocarcinoma cells and did not colonize the gastric mucosa of mice (27). Also of interest was the finding that culturing C. jejuni with DOC resulted in upregulation of a number of conserved hypothetical protein genes and genes with unknown functions. Some of these genes may code for virulence factors, including the other Cia proteins.
It is noteworthy that Fox et al. (10) studied the effect of culturing C. jejuni with a concentration of bile that exceeds the concentration normally found in human and chicken intestinal tracts. Using a proteomic approach, these workers found that culturing C. jejuni with 2.5% ox bile for 18 h increased the synthesis of GroEL, GalU, and bacterioferritin proteins (10). These proteins are indicative of a bacterial stress response. We did not observe that these genes were upregulated in C. jejuni in response to DOC, as judged by the microarray analysis, indicating that the conditions used in this study did not induce a bacterial stress response.
We show here that the bile acid DOC acts as a signal for C. jejuni, triggering its pathogenic behavior, as indicated by its ability to invade epithelial cells. More specifically, we show that culture with DOC “primes” C. jejuni to invade epithelial cells by stimulating the synthesis of the Cia proteins. This is a significant finding as it highlights the effect of in vivo-like culture conditions on virulence factors of C. jejuni. Harvesting C. jejuni from MHD agar plates should help shorten the time required for invasion assays currently performed from 3 h to 15 min. It should also allow researchers to synchronize the infection and to dissect the early events in Campylobacter invasion of host cells. Studies are currently being performed to characterize the proteins encoded by the genes whose expression mirrors that of ciaB, which encodes a known virulence factor. These genes may code for unidentified virulence factors that may play a role in bile resistance and/or invasion of the intestinal tract.
Acknowledgments
We thank Brian Raphael for construction of the PciaB-pMW10 and PporA-pMW10 reporter vectors. We thank Jeffrey Christensen for technical assistance with the real-time RT-PCR. We also thank Jeffrey Christensen, Charlie Larson, Jason Neal-McKinney, Sophia Pacheco, and Kari Sweeney (School of Molecular Biosciences, Washington State University) for critical reviews of the manuscript.
This work was supported by funds awarded to M.E.K. from grant 2005-35212-15287 from the Food Safety Coordinated Agricultural Project.
Footnotes
Published ahead of print on 25 January 2008.
REFERENCES
- 1.Allos, B. M. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 321201-1206. [DOI] [PubMed] [Google Scholar]
- 2.Altekruse, S. F., N. J. Stern, P. I. Fields, and D. L. Swerdlow. 1999. Campylobacter jejuni—an emerging foodborne pathogen. Emerg. Infect. Dis. 528-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babakhani, F. K., G. A. Bradley, and L. A. Joens. 1993. Newborn piglet model for campylobacteriosis. Infect. Immun. 613466-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22703-714. [DOI] [PubMed] [Google Scholar]
- 5.Begley, M., C. G. Gahan, and C. Hill. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29625-651. [DOI] [PubMed] [Google Scholar]
- 6.Crushell, E., S. Harty, F. Sharif, and B. Bourke. 2004. Enteric campylobacter: purging its secrets? Pediatr. Res. 553-12. [DOI] [PubMed] [Google Scholar]
- 7.Elliot, W. H. 1985. Metabolism of bile acids in liver and extrahepatic tissues, p. 3003-3329. In H. Danielsson and J. Sjovall (ed.), Sterols and bile acids. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 8.Everest, P. H., H. Goossens, J. P. Butzler, D. Lloyd, S. Knutton, J. M. Ketley, and P. H. Williams. 1992. Differentiated Caco-2 cells as a model for enteric invasion by Campylobacter jejuni and C. coli. J. Med. Microbiol. 37319-325. [DOI] [PubMed] [Google Scholar]
- 9.Field, L. H., V. L. Headley, J. L. Underwood, S. M. Payne, and L. J. Berry. 1986. The chicken embryo as a model for campylobacter invasion: comparative virulence of human isolates of Campylobacter jejuni and Campylobacter coli. Infect. Immun. 54118-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox, E. M., M. Raftery, A. Goodchild, and G. L. Mendz. 2007. Campylobacter jejuni response to ox-bile stress. FEMS Immunol. Med. Microbiol. 49165-172. [DOI] [PubMed] [Google Scholar]
- 11.Galan, J. E., and R. Curtiss III. 1990. Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect. Immun. 581879-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta, S., and R. Chowdhury. 1997. Bile affects production of virulence factors and motility of Vibrio cholerae. Infect. Immun. 651131-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humphrey, C. D., D. M. Montag, and F. E. Pittman. 1985. Experimental infection of hamsters with Campylobacter jejuni. J. Infect. Dis. 151485-493. [DOI] [PubMed] [Google Scholar]
- 14.Jones, B. D., and S. Falkow. 1994. Identification and characterization of a Salmonella typhimurium oxygen-regulated gene required for bacterial internalization. Infect. Immun. 623745-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konkel, M. E., and W. Cieplak, Jr. 1992. Altered synthetic response of Campylobacter jejuni to cocultivation with human epithelial cells is associated with enhanced internalization. Infect. Immun. 604945-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konkel, M. E., B. J. Kim, V. Rivera-Amill, and S. G. Garvis. 1999. Bacterial secreted proteins are required for the internalization of Campylobacter jejuni into cultured mammalian cells. Mol. Microbiol. 32691-701. [DOI] [PubMed] [Google Scholar]
- 17.Konkel, M. E., J. D. Klena, V. Rivera-Amill, M. R. Monteville, D. Biswas, B. Raphael, and J. Mickelson. 2004. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J. Bacteriol. 1863296-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konkel, M. E., D. J. Mead, and W. Cieplak, Jr. 1993. Kinetic and antigenic characterization of altered protein synthesis by Campylobacter jejuni during cultivation with human epithelial cells. J. Infect. Dis. 168948-954. [DOI] [PubMed] [Google Scholar]
- 19.Konkel, M. E., M. R. Monteville, V. Rivera-Amill, and L. A. Joens. 2001. The pathogenesis of Campylobacter jejuni-mediated enteritis. Curr. Issues Intest. Microbiol. 255-71. [PubMed] [Google Scholar]
- 20.Lin, J., M. Akiba, O. Sahin, and Q. Zhang. 2005. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob. Agents Chemother. 491067-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, J., C. Cagliero, B. Guo, Y. W. Barton, M. C. Maurel, S. Payot, and Q. Zhang. 2005. Bile salts modulate expression of the CmeABC multidrug efflux pump in Campylobacter jejuni. J. Bacteriol. 1877417-7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, J., and A. Martinez. 2006. Effect of efflux pump inhibitors on bile resistance and in vivo colonization of Campylobacter jejuni. J. Antimicrob. Chemother. 58966-972. [DOI] [PubMed] [Google Scholar]
- 23.Lin, J., L. O. Michel, and Q. Zhang. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 462124-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, J., O. Sahin, L. O. Michel, and Q. Zhang. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect. Immun. 714250-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacKichan, J. K., E. C. Gaynor, C. Chang, S. Cawthraw, D. G. Newell, J. F. Miller, and S. Falkow. 2004. The Campylobacter jejuni dccRS two-component system is required for optimal in vivo colonization but is dispensable for in vitro growth. Mol. Microbiol. 541269-1286. [DOI] [PubMed] [Google Scholar]
- 26.Malik-Kale, P., B. H. Raphael, C. T. Parker, L. A. Joens, J. D. Klena, B. Quinones, A. M. Keech, and M. E. Konkel. 2007. Characterization of genetically matched isolates of Campylobacter jejuni reveals that mutations in genes involved in flagellar biosynthesis alter the organism's virulence potential. Appl. Environ. Microbiol. 733123-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martino, M. C., R. A. Stabler, Z. W. Zhang, M. J. Farthing, B. W. Wren, and N. Dorrell. 2001. Helicobacter pylori pore-forming cytolysin orthologue TlyA possesses in vitro hemolytic activity and has a role in colonization of the gastric mucosa. Infect. Immun. 691697-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Monteville, M. R., and M. E. Konkel. 2002. Fibronectin-facilitated invasion of T84 eukaryotic cells by Campylobacter jejuni occurs preferentially at the basolateral cell surface. Infect. Immun. 706665-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newell, D. G., H. McBride, F. Saunders, Y. Dehele, and A. D. Pearson. 1985. The virulence of clinical and environmental isolates of Campylobacter jejuni. J. Hyg. 9445-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olive, A. J., R. Kenjale, M. Espina, D. S. Moore, W. L. Picking, and W. D. Picking. 2007. Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect. Immun. 752626-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osawa, R., E. Arakawa, T. Okitsu, S. Yamai, and H. Watanabe. 2002. Levels of thermostable direct hemolysin produced by Vibrio parahaemolyticus O3:K6 and other serovars grown anaerobically with the presence of a bile acid. Curr. Microbiol. 44302-305. [DOI] [PubMed] [Google Scholar]
- 33.Osawa, R., and S. Yamai. 1996. Production of thermostable direct hemolysin by Vibrio parahaemolyticus enhanced by conjugated bile acids. Appl. Environ. Microbiol. 623023-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pace, J. L., T. J. Chai, H. A. Rossi, and X. Jiang. 1997. Effect of bile on Vibrio parahaemolyticus. Appl. Environ. Microbiol. 632372-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker, C. T., B. Quinones, W. G. Miller, S. T. Horn, and R. E. Mandrell. 2006. Comparative genomic analysis of Campylobacter jejuni strains reveals diversity due to genomic elements similar to those present in C. jejuni strain RM1221. J. Clin. Microbiol. 444125-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pope, L. M., K. E. Reed, and S. M. Payne. 1995. Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect. Immun. 633642-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porter, M. E., and C. J. Dorman. 1994. A role for H-NS in the thermo-osmotic regulation of virulence gene expression in Shigella flexneri. J. Bacteriol. 1764187-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prouty, A. M., I. E. Brodsky, S. Falkow, and J. S. Gunn. 2004. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology 150775-783. [DOI] [PubMed] [Google Scholar]
- 39.Prouty, A. M., I. E. Brodsky, J. Manos, R. Belas, S. Falkow, and J. S. Gunn. 2004. Transcriptional regulation of Salmonella enterica serovar Typhimurium genes by bile. FEMS Immunol. Med. Microbiol. 41177-185. [DOI] [PubMed] [Google Scholar]
- 40.Prouty, A. M., and J. S. Gunn. 2000. Salmonella enterica serovar Typhimurium invasion is repressed in the presence of bile. Infect. Immun. 686763-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raphael, B. H., S. Pereira, G. A. Flom, Q. Zhang, J. M. Ketley, and M. E. Konkel. 2005. The Campylobacter jejuni response regulator, CbrR, modulates sodium deoxycholate resistance and chicken colonization. J. Bacteriol. 1873662-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richardson, P. T., and S. F. Park. 1995. Enterochelin acquisition in Campylobacter coli: characterization of components of a binding-protein-dependent transport system. Microbiology 1413181-3191. [DOI] [PubMed] [Google Scholar]
- 43.Ridley, K. A., J. D. Rock, Y. Li, and J. M. Ketley. 2006. Heme utilization in Campylobacter jejuni. J. Bacteriol. 1887862-7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivera-Amill, V., B. J. Kim, J. Seshu, and M. E. Konkel. 2001. Secretion of the virulence-associated Campylobacter invasion antigens from Campylobacter jejuni requires a stimulatory signal. J. Infect. Dis. 1831607-1616. [DOI] [PubMed] [Google Scholar]
- 45.Rivera-Amill, V., and M. E. Konkel. 1999. Secretion of Campylobacter jejuni Cia proteins is contact dependent. Adv. Exp. Med. Biol. 473225-229. [DOI] [PubMed] [Google Scholar]
- 46.Schuhmacher, D. A., and K. E. Klose. 1999. Environmental signals modulate ToxT-dependent virulence factor expression in Vibrio cholerae. J. Bacteriol. 1811508-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sen, A., M. A. Leon, and S. Palchaudhuri. 1991. Environmental signals induce major changes in virulence of Shigella spp. FEMS Microbiol. Lett. 68231-236. [DOI] [PubMed] [Google Scholar]
- 48.Sosula, L., E. M. Nicholls, and M. Skeen. 1988. Ultrastructure of Campylobacter jejuni in gamma-irradiated mouse jejunum. Am. J. Pathol. 131125-131. [PMC free article] [PubMed] [Google Scholar]
- 49.Tobe, T., S. Nagai, N. Okada, B. Adler, M. Yoshikawa, and C. Sasakawa. 1991. Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol. Microbiol. 5887-893. [DOI] [PubMed] [Google Scholar]
- 50.Tobe, T., M. Yoshikawa, and C. Sasakawa. 1994. Deregulation of temperature-dependent transcription of the invasion regulatory gene, virB, in Shigella by rho mutation. Mol. Microbiol. 12267-276. [DOI] [PubMed] [Google Scholar]
- 51.Welkos, S. L. 1984. Experimental gastroenteritis in newly-hatched chicks infected with Campylobacter jejuni. J. Med. Microbiol. 18233-248. [DOI] [PubMed] [Google Scholar]
- 52.Wosten, M. M., M. Boeve, M. G. Koot, A. C. van Nuene, and B. A. van der Zeijst. 1998. Identification of Campylobacter jejuni promoter sequences. J. Bacteriol. 180594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wosten, M. M., J. A. Wagenaar, and J. P. van Putten. 2004. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J. Biol. Chem. 27916214-16222. [DOI] [PubMed] [Google Scholar]