Abstract
Legionella pneumophila is a facultative intracellular pathogen responsible for severe lung disease in humans, known as legionellosis or Legionnaires' disease. Previously, we reported on the ∼60-kDa glucosyltransferase (Lgt1) from Legionella pneumophila, which modified eukaryotic elongation factor 1A. In the present study, using L. pneumophila Philadelphia-1, Lens, Paris, and Corby genome databases, we identified several genes coding for proteins with considerable sequence homology to Lgt1. These new enzymes form three subfamilies, termed Lgt1 to -3, glucosylate mammalian elongation factor eEF1A at serine-53, inhibit its activity, and subsequently kill target eukaryotic cells. Expression studies on L. pneumophila grown in broth medium or in Acanthamoeba castellanii revealed that production of Lgt1 was maximal at stationary phase of broth culture or during the late phase of Legionella-host cell interaction, respectively. In contrast, synthesis of Lgt3 peaked during the lag phase of liquid culture and at early steps of bacterium-amoeba interaction. Thus, the data indicate that members of the L. pneumophila glucosyltransferase family are differentially regulated, affect protein synthesis of host cells, and represent potential virulence factors of Legionella.
The protein synthesis machinery of eukaryotic cells is a well-known target for pathogenic microorganisms during host-pathogen interaction. Examples of bacterial protein toxins targeting host protein synthesis include Shiga- and Shiga-like toxins, which act as rRNA N-glycosidases, and diphtheria toxin (DT) and Pseudomonas aeruginosa exotoxin A, which ADP-ribosylate the modified histidine residue diphthamide in eukaryotic elongation factor 2 (eEF2). Both types of enzymatic activities result in inhibition of protein synthesis and death of target cells (26, 34).
Recently, an ∼60-kDa glucosyltransferase, referred to here as Lgt1 (Legionella pneumophila glucosyltransferase 1), was identified in L. pneumophila cultures. Well-studied examples of bacterial glucosylating enzymes targeting eukaryotic proteins are the large clostridial cytotoxins. They glucosylate 20- to 25-kDa small GTPases of the Rho family, thereby inhibiting the regulatory functions of these switch proteins (1, 16). In contrast, the Legionella enzyme modified an ∼50-kDa component in mammalian cell extracts, which was identified subsequently as eEF1A. This elongation factor also represents a GTP-binding protein, possessing GTPase activity. Lgt1 modifies serine-53 of eEF1A, located in the GTPase domain near the switch 1 region of the GTPase. This modification results in inhibition of protein synthesis both in vitro and in vivo and causes death of intoxicated eukaryotic cells (3, 4).
Many Legionella proteins occur in a set of redundant molecules, executing apparently closely related functions (6, 12, 18, 27, 28). Therefore, we screened Legionella genome databases for Lgt1 analogs. Here we report that L. pneumophila strains Philadelphia-1, Lens, Paris, and Corby (GenBank accession numbers NC_002942, NC_006369, NC_006368, and NC_009494, respectively) contain open reading frames (ORFs) encoding proteins related to Lgt1 and representing a novel family of Legionella glucosyltransferases.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The following L. pneumophila strains were used throughout this study: the sequenced Philadelphia-1, Lens, and Paris strains (8), as well as strains 130b (serogroup 1), ATCC 33823 (serogroup 7), ATCC 35096 (serogroup 8), ATCC 43136 (serogroup 13), and 1169-MN-H (serogroup 14). The non-L. pneumophila representatives Legionella longbeachae ATCC 33462, Legionella gormanii ATCC 33297, and Legionella steigerwaltii ATCC 35302 were also used. Bacteria were grown on N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered charcoal yeast extract (BCYE) agar for 2 days at 37°C (13) or in ACES-buffered yeast extract broth (BYEB) or ACES-buffered proteose-peptone no. 3 broth (BPPB) (24, 33) at 37°C on a shaker for up to 3 days. Escherichia coli strain BL21(DE3) and the expression vectors pGEX-4T1, pGEX-4T2, pGEX-4T3 (GE Healthcare, Moscow, Russia), pET28a, and pET28c (Novagen, Madison, WI) were used for cloning and the production of recombinant proteins. E. coli was maintained on Luria-Bertani (LB) broth or agar supplemented with 200 μg/ml of ampicillin or 50 μg/ml of kanamycin when necessary.
Cloning of genes and purification of proteins.
Based upon the data from the L. pneumophila Philadelphia-1 genome (GenBank accession number AE017354) and the sequence of DT (GenBank accession number K01723), DNA primers were synthesized for the amplification of the corresponding ORFs, i.e., lgt2, lgt3, and the gene for the A subunit of DT (dt-A) (Table 1) (Litekh, Moscow, Russia). PCR (94°C-55°C-68°C; 25 cycles) was performed in a Sprint thermocycler (Hybaid, Moscow, Russia) with FideliTaq polymerase (GE Healthcare). As a template, L. pneumophila genomic DNA or plasmid p28-DT (a generous gift of Y. Vertiev, Gamaleya Institute), carrying the gene for whole DT, was used. For cloning of the amplified genes, the PCR products were digested with EcoRI and SalI (for lgt2), BamHI and SalI (for lgt3), or SmaI and XhoI (for dt-A) and then ligated into similarly digested pGEX-4T1 (for dt-A), pGEX-4T2 and pET28c (for lgt2), or pGEX-4T3 and pET28a (for lgt3) vectors. E. coli BL21(DE3) was transformed with the resulting plasmids by electroporation using a model 2510 electroporator (Eppendorf, Hamburg, Germany). Construction of site-mutated eEF1A molecules with S53T (i.e., exchange of serine-53 with threonine) and S53A (i.e., exchange of serine-53 with alanine) substitutions as well as Lgt1 mutated in the DXD region (i.e., double D246N/D248N mutant) was described earlier (4). DNA sequencing was carried out with ABI Prism BigDye Terminator v. 3.1 reagents on an ABI Prism 3100 Avant sequencer.
TABLE 1.
Primers used for amplification of nucleotide sequences
| Primer name | Primer sequencea | Target for amplification |
|---|---|---|
| 163 sense | TTTGGAGGAATTCTGAGCGAACAATATTGGCGT | lgt2 |
| 164 antisense | ATTAGTCGACTATCTAATCCAAGGATTATGAG | lgt2 |
| 152 sense | GGTGGGATGGATCCATGAAAGAGCAACAAAAGGCAA | lgt3 |
| 153 antisense | CTATGTCGACCCTAATTCCCTAACAGTTTTTTT | lgt3 |
| 320 sense | GCATCCAGAATTCGAAAAAGCCCAAAG | ralF |
| 321 antisense | CCCAGATTTGTCGACTCCAAACACTATG | ralF |
| 220 sense | GCTAGCCCGGGCGCTGATGATGTTGTTGAT | dt-A |
| 221 antisense | GCCACCTCGAGATCGCCTGACACGATTTCC | dt-A |
| 56 sense | CTCCATTATCAGTCGACCCATAGCTCCACA | 542-bp internal fragment of lgt1 |
| 150 antisense | AAATGGCACCTGGCCCTGAAATCT | 542-bp internal fragment of lgt1 |
| 275 sense | TCTGTAACATGGAGGTAGTTGCC | 803-bp internal fragment of lgt2 |
| 274 antisense | GTCCAAGAGACTTATTCAGGTGAG | 803-bp internal fragment of lgt2 |
| 276 sense | GTCGTGAAGATAAATACGCATTT | 1,176-bp internal fragment of lgt3 |
| 277 antisense | CACACCATTTAGGAATGTTATCAG | 1,176-bp internal fragment of lgt3 |
| 333 sense | AGCGATCAAGAATGTTATTCATCCG | lpg1368 (RT-PCR) |
| 334 antisense | TATTTCGGGTATGCTTTGGCAAC | lpg1368 (RT-PCR) |
| 367 sense | TCATAGAAGGCAAGAAGCTGTTTGT | lpg1368 (RT-PCR) |
| 368 antisense | GCGACCGTATCCATCATTTTTTG | lpg1368 (RT-PCR) |
| 335 sense | AGAGGTTGCTTTAGATTGGGTTCG | lpg1488 (RT-PCR) |
| 336 antisense | CATCTTGAGCAGGCTGATTTTCG | lpg1488 (RT-PCR) |
| 337 sense | TTAACACATGCAAGTCGAACGGC | 16S rRNA (RT-PCR) |
| 338 antisense | CGTTACTCACCCGTTCGCCA | 16S rRNA (RT-PCR) |
Engineered restriction endonuclease site sequences are underlined.
For purification of recombinant proteins, the E. coli clones were grown in LB broth supplemented with ampicillin or kanamycin on a shaker at 37°C until the optical density at 600 nm was 0.5. Expression of the cloned proteins was then induced by supplementation of the culture with 0.1 mM of isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma, Moscow, Russia) overnight at 22°C. The bacterial cells from 2 liters of culture were harvested by centrifugation at 6,000 × g for 15 min, resuspended in 15 ml of 20 mM Tris-HCl-buffered saline, pH 7.4 (TBS), and then lysed by French press. Following clarification by centrifugation, the bacterial extracts were subjected to chromatography on a glutathione-Sepharose Fast Flow (pGEX vectors) or nickel-equilibrated chelating Sepharose Fast Flow (pET vectors) column according to the manufacturer's instructions (GE Healthcare). Because the pGEX plasmid-encoded Legionella enzymes were unstable following thrombin cleavage, glutathione S-transferase (GST)-tagged (i.e., eluted from glutathione-Sepharose by 20 mM reduced glutathione) Lgt1, Lgt2, and Lgt3 were used throughout the study.
Enzymatic assays.
Eukaryotic cell extracts, used as substrates in the reaction mixtures, were prepared from EBL (embryonic bovine lung), Caco2 (human colon carcinoma), or HeLa (human cervical cancer) cells by brief sonication, with the total protein concentration being in the range of 7 to 10 mg/ml (4). Protein concentrations were determined by Coomassie brilliant blue G250 assay (Serva, Heidelberg, Germany), using bovine serum albumin as a standard (5). The glucosylation reaction was carried out in a 20-μl mixture consisting of 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM MnCl2, 2 to 5 μg of recombinant Legionella proteins, 50 to 70 μg of crude ultrasonic cell extract or 2 to 4 μg of purified recombinant eEF1A1 (the elongation factor-encoding plasmid was a generous gift from Charlotte R. Knudsen, Aarhus University, Denmark), and 10 μM of UDP-[14C]glucose (ARC, St. Louis, MO). ADP-ribosylation was carried out in a 20-μl mixture consisting of 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 2 μg of recombinant purified DT-A, 50 to 70 μg of crude ultrasonic cell extract, and 0.5 mCi [32P]NAD (GE Healthcare). The mixtures were incubated at 37°C for 1 h, after which the reactions were stopped by the addition of Laemmli sample buffer and heating at 100°C for 5 min. The samples were then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and scanned on a Storm 820 phosphorimager (Molecular Dynamics, Vienna, Austria). In experiments on preglucosylation of eEF1A, 0.5 ml of EBL or HeLa cell extract was treated with 200 μg of purified wild-type or DXD-mutated GST-tagged Lgt1 in reaction buffer with 2 mM of unlabeled UDP-glucose for 40 min at 37°C. Thereafter, the reaction mix was dialyzed against TBS with 10% glycerol overnight at 4°C and used for reglucosylation by GST-Lgt1, GST-Lgt2, or GST-Lgt3 in the presence of 10 μM of UDP-[14C]glucose, as described above.
Intoxication of eukaryotic cells by electroporation.
EBL cells were grown in an atmosphere of 5% CO2 at 37°C until confluence on a 9-cm petri dish in modified Eagle medium (MEM) supplemented with antibiotic solution and 15% fetal calf serum (11). Cells from one petri dish were trypsinized and resuspended in 2 to 3 ml of fresh supplemented MEM (the resulting density was 2 × 106 to 3 × 106 cells/ml), to which recombinant GST-tagged L. pneumophila proteins or DT-A was added at different concentrations (denoted in Fig. 4 and 5). Electroporation settings were 200 V and 950 mF for a 4-mm standard electroporation cuvette (GenePulser; Bio-Rad, Austria). Immediately following a 15- to 25-ms pulse, cells were seeded into 24-well cell culture plates and incubated for 2 h in a CO2 incubator. Afterwards, cells were washed with TBS and incubation was continued for up to 3 days in fresh supplemented MEM. At daily time points, cells were subjected to phase-contrast microscopy. For enumeration of live cells, cells were trypsinized, stained by 0.25% trypan blue dye solution, and counted in a hemocytometer.
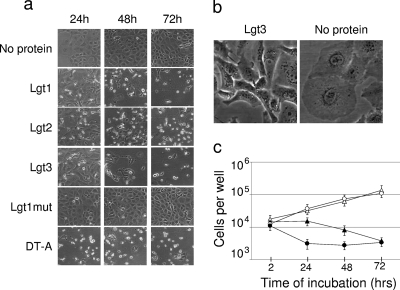
FIG. 4.
Cytotoxic activity of L. pneumophila glucosyltransferases and DT-A. EBL cells were electroporated without added protein or in the presence of fully active GST-Lgt1, GST-Lgt2, GST-Lgt3, or GST-DT-A or inactive GST-Lgt1mut at 40 μg/ml. (a) Microscopic pictures of electroporated cells taken after 24 h, 48 h, and 72 h of incubation. (b) Microscopic pictures of cells taken after 24 h (enlarged view). (c) Counts of live cells (means [n = 3] and standard deviations). Electroporation without added protein is represented by empty circles, that with Lgt1mut is represented by empty triangles, that with DT-A is represented by filled circles, and that with Lgt3 is represented by filled triangles. Patterns of intoxication with Lgt1 and Lgt2 were very similar to that with Lgt3 and therefore are not shown in panel c.
FIG. 5.
Inhibition of protein synthesis by L. pneumophila glucosyltransferases and DT-A. (a) Inhibition of in vitro transcription/translation. Transcription/translation reactions were performed in the presence of recombinant GST-Lgt1, GST-Lgt2, GST-Lgt3, or GST-DT-A. After SDS gel electrophoresis, gels were dried and scanned on a phosphorimager. Concentrations of used proteins, given in μg per ml, are indicated on the top. The matrix DNA represents a luciferase gene-containing plasmid coding for an ∼60-kDa luciferase. Very similar results were obtained with a β-actin-encoding plasmid (not shown). NT, concentration was not tested. (b) Inhibition of in vivo [35S]methionine incorporation. EBL cells were electroporated with GST-Lgt1, GST-Lgt2, GST-Lgt3, or GST-DT-A (black columns) in methionine-free MEM. Concentrations in each series were 30 ng/ml and 0.3, 3.0, and 30 μg/ml, shown below the x axis. Two hours following intoxication, cells were pulsed with 0.5 μCi of [35S]methionine for 3 h, lysed, and assayed for incorporation of radioactivity into proteins (means [n = 3] and standard deviations). In control experiments (columns C), cells were electroporated either without Legionella proteins (white column) or with the double D246N/D248N Lgt1 mutant at 30 μg/ml (gray column).
In vitro transcription/translation assay.
In vitro transcription/translation assays were performed with a rabbit reticulocyte lysate system as suggested by the manufacturer (L4610; Promega, Mannheim, Germany). As a target DNA for transcription/translation experiments, a luciferase gene-containing plasmid (included in the kit) or a β-actin gene-containing plasmid (a generous gift of John C. Sparrow, University of York, United Kingdom) was used. To investigate the influence of the recombinant L. pneumophila proteins and DT-A on protein synthesis, different amounts of purified proteins were added to the reaction mixture. Reaction with DT-A was performed in the presence of 20 μM NAD. The mixtures were incubated for 90 min at 30°C, subjected to SDS-PAGE, and scanned on a phosphorimager.
Methionine incorporation assay.
For methionine incorporation assay, EBL cells were harvested in supplemented MEM without l-methionine (MEM-M) and were intoxicated by electroporation (as described above) with recombinant L. pneumophila proteins or DT-A at 33 to 0.03 μg/ml. After 2 h of incubation at 37°C, cells were washed and pulsed with [35S]methionine (0.5 μCi per well of a 24-well plate) for 3 h in MEM-M (at this time, >95% of cells in control and intoxication experiments were alive, as confirmed by microscopy and trypan blue assay; differences in cell densities in each well were insignificant, as determined by cell counting). Following this time, cells were washed with TBS and lysed by 0.1% SDS supplemented with 0.2 mg/ml of bovine serum albumin. Proteins were precipitated by 10% trichloroacetic acid, and the amounts of incorporated [35S]methionine were measured using a filter assay in a liquid scintillation counter.
Acanthamoeba castellanii infection studies.
A. castellanii C3 was cultivated routinely in PYG medium in 250-ml flasks at room temperature (21). For intracellular infection studies, amoebae were washed once and transferred into 6-cm petri plates with 5 ml of amoeba buffer to achieve a concentration of 105 cells/ml. A. castellanii amoebae were allowed to adhere for 30 to 40 min at room temperature, and a suspension of L. pneumophila Philadelphia-1 was added at a multiplicity of infection of 0.1 (i.e., 1 bacterium per 10 amoebae). Afterwards, the coinfection proceeded for up to 72 h at 37°C. At certain time points, the corresponding plate was washed with the buffer and treated with 0.2% Triton X-100 solution to release intracellular bacteria, which were titrated subsequently on BCYE agar for CFU counting. At the 72-h time point, a washing step was omitted.
RT-PCR.
Total RNA from L. pneumophila Philadelphia-1 cells, grown in proteose-peptone broth or in A. castellanii for certain periods, was isolated using an RNeasy mini kit with RNAprotect bacterial reagent (Qiagen, Hilden, Germany). For reverse transcription (RT), a QuantiTect Rev transcription kit (Qiagen) was utilized as suggested by the manufacturer. Real-time PCR was performed with a QuantiTect SYBR green PCR kit (Qiagen) on an Mx3000P cycler controlled by MxPro software (Stratagene, Amsterdam, Netherlands). Data analysis was done using MxPro software (Stratagene), and data were normalized to 16S rRNA levels. The following equation was used to calculate induction in the RT-PCR experiments: ratio = 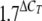 , where ΔCT = CT(sample 1) − CT(sample 2) (12) and CT means the cycle threshold at which the fluorescence level reaches the cutoff value, determined by the computer program.
, where ΔCT = CT(sample 1) − CT(sample 2) (12) and CT means the cycle threshold at which the fluorescence level reaches the cutoff value, determined by the computer program.
Immunoblot analysis.
Mouse monospecific sera against purified polyhistidine-tagged recombinant Lgt1, Lgt2, and Lgt3 were used for Western blotting. Following SDS-PAGE, immunoblot analysis was performed according to a standard procedure (17, 31), using a 1/50,000 dilution of each anti-Lgt serum, a 1/10,000 dilution of anti-mouse-horseradish peroxidase conjugate, and an ECL detection system (GE Healthcare) on an LAS-3000 mini device (Fujifilm, Dusseldorf, Germany).
RESULTS
Identification of Lgt1-like sequences in L. pneumophila.
Our previous data suggested that by inhibiting protein synthesis in target cells, the glucosyltransferase Lgt1 plays an important role in the virulence of L. pneumophila (3, 4). Therefore, we studied whether the L. pneumophila genome database contains more than one copy of the corresponding gene or any other homologous sequences. Using a BLAST search, we identified several ORFs exhibiting considerable similarities with the Lgt1 gene in four different L. pneumophila strains. In particular, strain Philadelphia-1 contains the homologous coding sequences lpg1488 and lpg2862 in addition to lpg1368 (the gene for Lgt1). Moreover, strain Lens harbors the genes lpl1319 and lpl1540, strain Paris contains lpp1322 and lpp1444, and strain Corby possesses the genes lpc_0784 and lpc_0903, which are all related to lpg1368 (Fig. 1). Based on the amino acid sequence similarities, the corresponding proteins could be grouped into three subfamilies. We called them Lgt1 (translated lpg1368, lpp1322, lpl1319, and lpc_0784), Lgt2 (translated lpg2862), and Lgt3 (translated lpg1488, lpp1444, lpl1540, and lpc_0903) (Fig. 2). Representatives within the family were >80% identical, while the identities between different groups were in the range of 20 to 40% (data not shown).
FIG. 1.
Amino acid alignment of Lgt-like proteins as deduced from sequenced L. pneumophila genomes. Lpg1368, Lpl1319, Lpp1322, and Lpc_0784 represent the Lgt1 group, Lpg2862 represents the Lgt2 group, and Lpg1488, Lpl1540, Lpp1444, and Lpc_0903 represent the Lgt3 group proteins. Amino acid residues that are similar among at least two different Lgt groups are darkened. A putative DXD motif is indicated above the corresponding line. The region of repeats in Lgt3 group proteins is marked by italics. A repeat unit (octapeptide KXEEEQRI) is underlined in lpg1488. The fragment of toxin B of C. difficile (GenBank accession number X53138) encompassing the DXD motif (the region of highest similarity to the corresponding L. pneumophila proteins) is shown in bold.
FIG. 2.
Guide tree demonstrating similarities among Lgt-like proteins. Coding numbers for the corresponding sequences (in italics) are shown according to L. pneumophila genome database nomenclature.
Alignment of Lgt1-, Lgt2-, and Lgt3-like sequences demonstrated the highest homology in a DXD motif-containing region, which is typical for type A glucosyltransferases (10). In this region, the putative Legionella enzymes exhibit significant similarity with the DXD motif of the active site of clostridial glucosylating cytotoxins (this region of toxin B of Clostridium difficile is shown in bold in Fig. 1).
ORFs coding for Lgt1- and Lgt3-like proteins could be found in all four sequenced L. pneumophila genomes. The corresponding products have molecular masses in the ranges of 60 to 65 and 87 to 100 kDa, respectively. Interestingly, Lgt3-like proteins possessed a C-terminal domain having high similarity with another group of proteins (lpg1491 and lpp1447 in strains Philadelphia-1 and Paris, respectively) (not shown) and a unique region of repeats with the repeat sequence KXEEEQRI (Fig. 1, italics). In contrast, Lgt2-encoding genes were missing in three of the four L. pneumophila strains for which genome data are available (i.e., in strains Paris, Corby, and Lens). In strain Philadelphia-1, the molecular mass of translated lpg2862 (single representative of the Lgt2 group) was ∼70 kDa.
The apparent irregular distribution of lgt genes prompted us to investigate their existence in a broader number of Legionella strains by PCR with specific internal primers. In these experiments, lgt1- and lgt3-containing sequences were detected in all tested L. pneumophila representatives, i.e., Philadelphia-1, 130b (serogroup 1), ATCC 33823 (serogroup 7), ATCC 35096 (serogroup 8), ATCC 43136 (serogroup 13), and 1169-MN-H (serogroup 14). In contrast, lgt2-specific sequences could be detected in only three of the six strains, i.e., Philadelphia-1, ATCC 35096, and ATCC 43136 (data not shown). This suggests that Lgt1 and Lgt3 represent an indispensable pair, while the role of Lgt2 is not essential, at least for some strains. In no case were we able to detect by PCR lgt-like sequences in a limited number of tested non-L. pneumophila legionellae (L. longbeachae, L. gormanii, and L. steigerwaltii) (data not shown).
Using sequence information from the L. pneumophila Philadelphia-1 genome, two sets of primers were synthesized in order to amplify and clone lgt genes from various strains of L. pneumophila (Table 1). To this end, lgt2 sequences were cloned using the chromosomal DNAs obtained from L. pneumophila Philadelphia-1, ATCC 35096, and ATCC 43136, while lgt3 was cloned from strains Philadelphia-1, Lens, and 130b. Lgt2 of strain ATCC 35096 and Lgt3 of strain 130b were 97% and 95% identical to the corresponding translated genes of L. pneumophila Philadelphia-1 (lpg2862 and lpg1488, respectively) (not shown). The enzymes matched the proposed scheme of Lgt proteins (Fig. 2) and have been used in subsequent experiments as representatives of Lgt2 and Lgt3 family proteins.
Glucosyltransferase activity of Lgt2 and Lgt3.
Lgt2 and Lgt3 were expressed as GST fusion proteins in E. coli (Fig. 3a, lanes 1 to 3). As shown in Fig. 3b, all three proteins (Lgt1, Lgt2, and Lgt3) glucosylated eukaryotic proteins with similar molecular masses of ∼50 kDa in the presence of UDP-[14C]glucose (Fig. 3b, lanes 5 to 7). The intensities of glucosylation were approximately the same with all three enzymes. Recently, we have shown that the glucosyltransferase Lgt1 modifies eEF1A at serine-53 (4). To confirm that Lgt2 and Lgt3 also modify eEF1A, we tested purified recombinant wild-type eEF1A (Fig. 3c). Moreover, we studied whether Lgt2 and Lgt3 share the same target site in eEF1A as that for Lgt1. To this end, we employed two mutants, S53T eEF1A and S53A eEF1A (i.e., threonine and alanine substitutions for serine-53), as substrates for glucosylation by Lgt2 and Lgt3. As shown in Fig. 3c, all three L. pneumophila glucosyltransferases modified purified wild-type eEF1A (Fig. 3c, lanes 1, 4, and 7) and the S53T eEF1A mutant did so to a lesser extent, but the mutant with the S53A substitution did not (Fig. 3c, lanes 3, 6, and 9 and lanes 2, 5, and 8, respectively), suggesting that the enzymes share the same acceptor amino acid residue.
FIG. 3.
(a) SDS-PAGE analysis of purified preparations of GST-tagged Lgt1, Lgt2, Lgt3, and DT-A (lanes 1, 2, 3, and 4, respectively). Approximately 2 μg of each purified protein was loaded onto the gel, separated by electrophoresis, and stained with Coomassie R-250. Molecular masses of proteins, in kilodaltons, are shown on the left (K). (b to d) Enzymatic activities of Lgt1, Lgt2, and Lgt3. Purified GST-tagged L. pneumophila proteins were tested in a [14C]glucosyltransferase assay. After SDS gel electrophoresis, gels were dried and scanned on a phosphorimager. (b) Lane 1, Lgt1 without eukaryotic substrate; lane 2, Lgt2 without eukaryotic substrate; lane 3, Lgt3 without eukaryotic substrate; lane 4, EBL cell extract without L. pneumophila proteins; lane 5, Lgt1 plus EBL cell extract; lane 6, Lgt2 plus EBL cell extract; lane 7, Lgt3 plus EBL cell extract. Similar results were obtained with Caco2 and HeLa cell extracts (not shown). (c) Lane 1, Lgt1 plus eEF1A1; lane 2, Lgt1 plus eEF1A1 with S53A mutation; lane 3, Lgt1 plus eEF1A1 with S53T mutation; lane 4, Lgt2 plus eEF1A1; lane 5, Lgt2 plus eEF1A1 with S53A mutation; lane 6, Lgt2 plus eEF1A1 with S53T mutation; lane 7, Lgt3 plus eEF1A1; lane 8, Lgt3 plus eEF1A1 with S53A mutation; lane 9, Lgt3 plus eEF1A1 with S53T mutation. (d) EBL cell extracts, nontreated (lane 1; NT) or treated previously with “cold” UDP-glucose and wild-type (WT; lanes 2 to 4) or doubly mutated (Mut; lanes 5 to 7) Lgt1, were reglucosylated with UDP-[14C]glucose and Lgt1 (lanes 1, 2, and 5), Lgt2 (lanes 3 and 6), or Lgt3 (lanes 4 and 7). Molecular masses of modified eEF1A molecules (∼50 kDa for native protein and ∼80 kDa for GST-tagged protein) are shown on the right, in kilodaltons (K). Note the strong automodification of Lgt2 (panel b, lanes 2 and 6, and panel c, lanes 4 to 6).
To additionally confirm the target site identity for all three enzymes, EBL cell extracts were treated initially with either wild-type Lgt1 or inactive DXD-mutated Lgt1 in the presence of 2 mM unlabeled UDP-glucose in a standard reaction mix. The reaction mixture was then dialyzed (see Materials and Methods) and used for reglucosylation by Lgt1, Lgt2, or Lgt3 in the presence of UDP-[14C]glucose as a cofactor. As shown in Fig. 3d, labeling of proteins pretreated by wild-type Lgt1 was marginal in the second round of glucosylation by Lgt1, Lgt2, or Lgt3 with radioactive UDP-glucose (Fig. 3d, lanes 2 to 4). In contrast, in cell extracts pretreated with the inactive DXD mutant of Lgt1, the three glucosyltransferases catalyzed significant labeling of the 50-kDa protein in the second glucosylation reaction (Fig. 3d, lanes 5 to 7). Thus, all of these data are in accordance with the view that the three related but not identical transferases modify a single serine residue in eEF1A with similar efficiencies.
Cytotoxic activity of L. pneumophila glucosyltransferases.
eEF1A, which is a substrate of L. pneumophila glucosyltransferases, plays a pivotal role in protein synthesis (7). To study whether Lgt2 and Lgt3 have cytotoxic activity, we compared their effects with that of another elongation factor-targeting molecule, DT. For this purpose, DT-A was used as a GST fusion protein (Fig. 3a, lane 4). EBL cells electroporated with recombinant enzymatically active proteins failed to develop a monolayer and died 48 to 72 h (earlier with DT-A treatment) after electroporation (Fig. 4a). However prominent changes in morphology could be seen already after 24 h of intoxication (Fig. 4b). It should be noted that the observed major morphological changes and ultimate cell death were caused by minute amounts of protein, which were undetectable by Western blotting with monospecific anti-Lgt sera (not shown). Addition of the enzymes (Lgt1 to -3 and DT-A) to cell culture medium did not cause any cytotoxicity (not shown). Also, electroporation of cells with the inactive DXD (D246N/D248N) Lgt1 mutant or electroporation in the absence of any bacterial enzymes was not toxic (Fig. 4a).
The results of the above morphological studies were in good agreement with direct counting of residual live EBL cells 24, 48, and 72 h following intoxication. As shown in Fig. 4, intoxication by Lgt1, Lgt2, and Lgt3 was considerably slower than that with DT-A. With the Lgts, a significant decrease in cell quantity was observed after 48 h, whereas DT-A reduced the number of live cells already at the 24-h time point. However, differences in numbers of live cells after 3 days were insignificant for all tested enzymes (Fig. 4c).
Inhibition of protein synthesis by Lgt2 and Lgt3.
To study whether the observed cytotoxic activity of L. pneumophila glucosyltransferases was due to effects on transcription/translation, we employed two protein synthesis assays, i.e., in vitro transcription/translation and in vivo methionine incorporation. As shown in Fig. 5a, whereas Lgt1 and Lgt2 completely suppressed in vitro luciferase translation at 2 μg/ml, Lgt3 and DT-A were even more potent and blocked luciferase production at 0.2 μg/ml. Considering the differences in molecular masses of the enzymes, Lgt3 was the most efficient inhibitory protein in this assay. To prove that inhibition of protein synthesis takes place under in vivo conditions, [35S]methionine incorporation into newly synthesized proteins was studied. As shown in Fig. 5b, intoxication of EBL cells by electroporation with increasing concentrations of glucosyltransferases Lgt1, Lgt2, and Lgt3 and the ADP-ribosyltransferase DT-A resulted in significant reductions of radioactivity incorporated into trichloroacetic acid-precipitated material. The inhibitory activities of all the glucosyltransferases were roughly the same and were approximately 10 times lower than that of DT-A.
Production of glucosyltransferases by L. pneumophila.
In contrast to Lgt1, which was initially identified in and isolated from L. pneumophila cells (3), Lgt2 and Lgt3 were studied here initially as recombinant proteins expressed in E. coli. Therefore, it was of interest to investigate the production of these molecules during growth of L. pneumophila. For this purpose, we utilized Western blotting with monospecific sera raised in mice by immunization with purified His-tagged Lgt1, Lgt2, and Lgt3. As shown in Fig. 6a, each anti-Lgt antibody reacted specifically with the corresponding transferase (e.g., Lgt1, Lgt2, and Lgt3). However, when a crude ultrasonic extract of agar-grown L. pneumophila strain Philadelphia-1 was used in the assay, only anti-Lgt1 and anti-Lgt2 sera produced positive signals. The antibodies recognized bands with the expected molecular masses (i.e., ∼60 kDa for Lgt1 and ∼70 kDa for Lgt2). Anti-Lgt3 serum failed to react with the crude preparation of L. pneumophila. These results suggested that only Lgt1 and Lgt2, not Lgt3, were produced by L. pneumophila Philadelphia-1 at detectable quantities under the growth conditions used.
FIG. 6.
Analysis of glucosyltransferase production by L. pneumophila Philadelphia-1. (a) An ultrasonic extract of L. pneumophila strain Philadelphia-1, grown on BCYE agar for 48 h, or corresponding purified glucosyltransferases were subjected to SDS gel electrophoresis, transferred to a nitrocellulose membrane, and probed with anti-Lgt1 (lanes 1 and 2), anti-Lgt2 (lanes 3 and 4), and anti-Lgt3 (lanes 5 and 6) sera. Lanes 1, 3, and 5 contained purified GST-Lgt1, GST-Lgt2, and GST-Lgt3, respectively (100 ng per lane). Lanes 2, 4, and 6 contained a crude extract of L. pneumophila (extract; 40 μg per lane). (b) Ultrasonic extracts of L. pneumophila strain Philadelphia-1, grown in BPPB for 3, 6, 12, 24, 36, and 48 h, were processed with sera against Lgt1, Lgt2, Lgt3, and RalF (a type IV secretion system effector protein) or subjected to glucosylation assay with UDP-[14C]glucose for 10 min at 37°C, using EBL cell extract as a substrate (panel 14C). A time point characterized by the appearance of a dark-brown pigment (characteristic of late stationary phase) is shown on the plot. Molecular masses of the corresponding bands are indicated on the right. OD660, optical density at 660 nm. (c) RNAs obtained from L. pneumophila Philadelphia-1 cells at 6, 12, 24, 36, and 48 h were subjected to RT-PCR in order to estimate levels of lgt1 (white columns) and lgt3 (black columns) transcription. Induction ratios, calculated as proportions of mRNA levels at certain time points to that at the time point with the minimal value (12 h for lgt1 and 36 h for lgt3), are shown.
To study whether the production of glucosyltransferases is dependent upon the growth phase of Legionella, we cultivated microorganisms in BYEB (24) or in BPPB (14, 25, 29, 33). In order to allow bacteria to adapt to the proteose-peptone nourishing base, legionellae from yeast extract agar plates were transferred into BPPB and cultivated subsequently for 96 h on a shaker. These late-stationary-phase cultures were used to inoculate fresh broth (time point “0 h”), and incubation proceeded thereafter for up to 48 h. Later, we found that the production of proteins was identical in the first and second rounds of cultivation, and thus, such “adaptation” seemed to be unnecessary. However, we still used this approach throughout the investigations.
Strong production of Lgt1, Lgt2, and the type IV secretion system effector RalF (this protein has been shown previously [22] to be repressed at exponential phase and induced in post-exponential-phase bacteria and thus served as a control in our experiments) could be detected at lag phase, at the beginning of logarithmic phase, and maximally at the stationary period of growth in both types of medium. Actively replicating L. pneumophila cells obtained at exponential stage produced small amounts of the enzymes. Neither BYEB nor BPPB stimulated significant production of Lgt3 at all time points. The enzymatic activity of cell extracts correlated well with expression of the two glucosyltransferases and was maximal at lag and exponential phases (Fig. 6b; also, Fig. S1A in the supplemental material [only experiments with proteose-peptone medium are shown]).
Since a lack of detection of Lgt3 could be caused by the low sensitivity of the assay (Western blotting), we utilized real-time PCR to study a time course of expression of L. pneumophila glucosyltransferase genes. These experiments were done with lgt1 and lgt3 genes as targets, obtained from bacteria grown in BPPB.
As shown, transcription of lgt1 was elevated at lag phase and especially elevated at postexponential phase, while being repressed during logarithmic growth of legionellae. Thus, these results correlated well with the data obtained earlier in Western blots with anti-Lgt1 sera. However, in contrast to previous negative Western blot experiments with anti-Lgt3 serum, RT-PCR with lgt3-targeting primers was positive. Yet the expression pattern in the latter case was quite different from that for lgt1. Synthesis of lgt3 mRNA was strongly induced at lag phase and declined with the onset of bacterial multiplication (Fig. 6c). Estimation of the melting temperatures of the resultant PCR products confirmed that these were amplified fragments but not products of side reactions (e.g., primer dimers, nonspecific amplification, etc. [not shown]).
To study further the expression dynamics of glucosyltransferases, we investigated the production of Lgt1 and Lgt3 in two other L. pneumophila strains, Paris and Lens, cultivated in BPPB (the lgt2 gene is lacking in these strains), using Western blotting. Growth curves for both strains were similar to growth of the Philadelphia-1 strain. As with the Philadelphia-1 strain, induction of Lgt1 synthesis could be observed at lag and stationary phases, while synthesis of Lgt3 peaked at preexponential phase and was repressed in bacteria entering logarithmic growth (Fig. 7a; also, Fig. S1B in the supplemental material).
FIG. 7.
Analysis of glucosyltransferase production by L. pneumophila Paris grown in proteose-peptone-based medium. (a) Ultrasonic extracts of L. pneumophila strain Paris, grown in BPPB for 3, 6, 10, 12, 24, 36, and 48 h, were processed with sera against Lgt1 and Lgt3 as described above or subjected to glucosylation assay with UDP-[14C]glucose for 10 min at 37°C, using EBL cell extract as a substrate (panel 14C). A representative bacterial growth curve is shown at the top. A time point characterized by the appearance of a dark-brown pigment (characteristic of late stationary phase) is shown on the plot. Molecular masses are indicated on the right. OD660, optical density at 660 nm. (b) Ultrasonic extracts of L. pneumophila strain Paris, cultivated in ACES-K buffer alone (A) or in BPPB (B) for 1, 2, 3, and 4 h, were processed with sera against Lgt1 and Lgt3. Molecular masses of the corresponding bands are indicated on the right.
To investigate Lgt1 and Lgt3 production at earlier time points, we incubated L. pneumophila Paris in ACES-K buffer or BPPB for up to 4 h and investigated the resulting samples by Western blotting with the corresponding sera. Whereas the amount of Lgt1 was not changed during the cultivation (Fig. 7b; also, Fig. S1C in the supplemental material), the level of Lgt3 was increased significantly in BPPB already after 1 to 2 h of incubation, remaining stable in ACES-K solution for up to 4 h (observation period). No growth of bacteria occurred in ACES-K or BPPB during this time interval (not shown).
Such characteristic patterns of Lpg1 and Lpg3 synthesis during cultivation of L. pneumophila in liquid medium in vitro prompted us to study the expression of the corresponding genes during infection of A. castellanii, one of the natural hosts of this bacterium. In these investigations, we infected amoebae with agar-grown cultures of L. pneumophila and studied specific mRNA levels at different time points by RT-PCR. As shown in Fig. 8, maximal expression of lgt1 was observed early in the infection (3 h) and at a very late time point (72 h), when the majority of microorganisms remain in the extracellular space. In contrast, maximal expression of lgt3 mRNA could be observed only at the 3-h time point. Thereafter, with the onset of bacterial multiplication, synthesis of the product was suppressed.
FIG. 8.
Expression of lgt1 and lgt3 during L. pneumophila infection of A. castellanii. Amoeba cells, infected at a multiplicity of infection of 0.1 with agar-grown L. pneumophila Philadelphia-1 at 0 h, were cultivated for different time periods, lysed, and sampled for RT-PCR with primers specific for lgt1 (white columns) and lgt3 (black columns) (a) or for plate counting (b). The corresponding induction ratios, calculated as proportions of mRNA levels at certain time points to that at the time point with the minimal value (24 h for both lgt1 and lgt3), are shown in panel a. Growth of L. pneumophila (b) is shown as changes in log CFU/ml over time. Cultures were extensively washed after 3, 24, and 48 h of incubation before being sampled in order to remove extracellular bacteria. Data obtained demonstrate induction of the genes (a) and changes in numbers (b), predominantly of intracellular bacteria. In contrast, the culture at 72 h was sampled without being washed. This was done due to decreasing numbers of attached amoebae. Therefore, data obtained at 0 h and 72 h demonstrate induction of the genes mainly in extracellular bacteria, which are starting to infect eukaryotes (0 h) or exiting amoebae (72 h). CFU numbers at these time points are shown by single filled circles in panel b. The figure represents data from three independent cell culture experiments.
Taken together, our data demonstrate that synthesis of all Lgt enzymes is repressed in actively proliferating microorganisms. In contrast, a strong increase in production of glucosyltransferases takes place in slowly replicating or nondividing bacteria under both in vitro and in vivo conditions.
DISCUSSION
For interference with the metabolism of eukaryotic host cells, protein synthesis is an attractive target for bacterial pathogens. At least two mechanisms have been shown to be used by microorganisms to inhibit translation processes in eukaryotic systems. These are RNA cleavage by N-glycosidases, known as Shiga- and Shiga-like toxins, and eEF2 inactivation, accomplished by ADP-ribosylating toxins of Clostridium diphtheriae and P. aeruginosa (26, 34). Recently, we showed that the 60-kDa glucosyltransferase Lgt1 from L. pneumophila inhibits protein synthesis by modification of eEF1A. Here we demonstrate that Lgt1 belongs to a family of Legionella glucosyltransferases. This family can be divided into three subfamilies, encompassing ∼60-kDa Lgt1 glucosyltransferases, ∼70-kDa Lgt2 glucosyltransferases, and ∼100-kDa Lgt3 glucosyltransferases. Our results confirm that all of these related proteins glucosylate eEF1A of mammalian host cells. Moreover, we show that the same serine residue (serine-53) was modified. The modification resulted in protein synthesis termination and produced profound changes in cellular morphology and, ultimately, cell death. Replacement of serine-53 by an alanine residue abolished glucosylation, while replacement by threonine caused a strong decrease in labeling. The residual modification in the latter case was not unexpected, since a threonine residue, as shown for clostridial cytotoxins, could also represent a target site for glucosylation (16).
In in vitro transcription/translation experiments, Lgt3 was the most active enzyme. However, its in vivo activities in eukaryotic cells (tested in cytotoxic and methionine incorporation assays) did not differ considerably from those of Lgt1 and Lgt2. The reason for this discrepancy is not clear, but electroporation experiments might be influenced by many additional factors (e.g., the size of the delivered molecule, its charge, hydrophobicity, etc.) which are different for the three glucosyltransferases and thus probably define the different accessibilities of the enzymes in target cells.
Lgt1 and Lgt2 were detected in agar and liquid cultures of L. pneumophila Philadelphia-1 by positive reaction with monospecific sera in Western blots. In agreement, we were able to demonstrate induction of Lgt1 and Lgt3 mRNA synthesis by RT-PCR. All approaches used showed that transcription/translation of glucosyltransferase genes was markedly growth phase dependent, being higher at periods when proliferation of bacteria was low, i.e., at the lag (for Lgt1, Lgt2, and Lgt3) and stationary (for Lgt1 and Lgt2) phases. The results obtained with strain Philadelphia-1 were confirmed for Lgt1 and Lgt3 by using the Paris and Lens strains of L. pneumophila. It is not clear, however, if elevated production of Lgt1 (and Lgt2) during lag phase results from actual induction at early time points or simply represents the lack of an active shutdown. According to the experimental protocol, bacterial cells from the late stationary phase were used to inoculate fresh medium. Therefore, theoretically, production of virulence traits that are up-regulated at stationary phase (e.g., Lgt1 and Lgt2) could continue for some time during the lag phase. Thereafter, their production was decreased. Apparently, this was not the case with Lgt3, whose synthesis was induced only during a short period of the lag phase and was repressed thereafter.
This type of regulation prompted us to investigate lgt1 and lgt3 expression in the amoeba model of infection by RT-PCR. The results of these experiments are in general agreement with the in vitro studies and reveal that at an early infection time (3 h following the start of the infection), L. pneumophila exhibits a high level of lgt1 expression and, additionally, activates lgt3 for a limited period of time. Thereafter, bacteria which are able to multiply actively inside phagocytes (at 24 h and 48 h) repress synthesis of both Lgt1 and Lgt3. After successful intracellular multiplication, bacteria go out into the surrounding medium. These microorganisms (taken at 72 h) induce the expression of lgt1. Thus, the data obtained suggest the importance of L. pneumophila glucosyltransferases for transmission of microorganisms from an old host to a new one and for establishing a new reservoir for proliferation.
Functional redundancy of virulence factors is typical for Legionella (6, 12, 18, 28) and appears to be true for Lgt proteins. It has been speculated that the bacterium may optimize the adaptation for colonization of different hosts by producing several redundant isoforms of proteins important in infectious processes (19, 23). Our data suggest that this redundancy is functional not only during infection of different hosts but also at different stages of single-host colonization. Different isoforms of enzymes with distinct enzymatic and biochemical properties could adapt more efficiently to environmental changes occurring during host-pathogen interactions.
In a previous report, we demonstrated that in eukaryotic cells infected with L. pneumophila, modification of eEF1A took place (4). On the other hand, in the present study we failed to detect Lgts in extracellular liquid of Legionella cultures grown for 3 h or 24 h (not shown). These data suggested that the bacteria are able to deliver glucosylating enzymes directly into the cytoplasm of target cells. The best-studied transporting device in L. pneumophila is the dot/icm type IV secretion system (20, 32). Using bioinformatic screening, de Felipe and coworkers recently searched for eukaryotic protein motifs in L. pneumophila (12). They were especially interested in identifying gene products of potential eukaryotic origin, which are substrates of Legionella type IV secretion and may be acquired by horizontal gene transfer. Interestingly, among several proteins which met their criteria, they detected Lgt3 and Lgt2 (LegC5 [ORF lpg1488] and LegC8 [ORF lpg2862]) as proteins possessing coiled-coil motifs (9). Both coding sequences were induced at the stationary phase of growth, as shown by RT-PCR. Moreover, LegC5 (i.e., Lgt3) was translocated into Dictyostelium discoideum, in a DotA-dependent manner, as a fusion protein with adenylate cyclase of Bordetella pertussis (12). Thus, Lgt3 could represent a novel substrate of the type IV dot/icm secretion apparatus.
Whether the coiled-coil structure of these enzymes is essential for targeting the eukaryotic substrate or important for transport remains an open question. It is not clear, however, why maximal induction of Lgt3 in three different L. pneumophila strains was detected by us by two assays during lag phase, in contrast to postexponential phase, as observed for a single strain by de Felipe and coworkers. It appears that they did not study the expression of the investigated genes in lag phase, where we observed an almost 20-fold increase in the lgt3 mRNA level. Additionally, this discrepancy may also be explained by usage of different L. pneumophila strains.
The coding sequences of the glucosyltransferases investigated in our studies are located in diverse areas of the L. pneumophila chromosome. Since the identification numbers of the corresponding sequences in the Philadelphia-1 genome are lpg1368, lpg1488, and lpg2862 for Lgt1, Lgt3, and Lgt2, respectively, the distance between lpg1368 and lpg1488 is 120 genes, while the gap between lpg1488 and lpg2862 is even larger, at more than 1,300 genes. This raises a question concerning the orchestration of their expression. As shown in recent publications, the regulation of expression of virulence-associated genes is governed by a network of numerous interactions (2, 6, 15, 30, 36). In a recent report, a major response regulator of the icm/dot type IV secretion system, PmrA, was identified (35). As shown in motility shift experiments, it bound to a consensus motif present within the promoter areas of a number of sequences coding for several already studied effectors of the type IV secretion system as well as for some new proteins, including LegC5/Lgt3, and stimulated their expression. These data indicated that lgt3 could be regulated in concert with a certain group of type IV secretion system-related genes. Concerning other glucosyltransferases of L. pneumophila, we failed to find such a consensus motif in the promoter region of lpg1368 (the gene for Lgt1) or lpg2862 (the gene for Lgt2). Such an observation suggested that the corresponding genes (lgt3 versus lgt1 and lgt2) should be regulated differently. This speculation seems to be in line with our results of expression experiments (Fig. 6 to 8). As shown by Zusman and coworkers (35), the translocated substrate RalF of the type IV secretion system also does not contain a PmrA-specific sequence in its promoter region, yet this protein has been shown by us to be regulated similarly to Lgt1 and Lgt2 in Philadelphia-1 (Fig. 6a) and similarly to Lgt1 in strain Lens (not shown). It would be interesting to study whether other type IV effectors (e.g., sidE, sdhA, sidG, sidB, and others) which are known to contain PmrA-specific sequences are strongly induced during preexponential phase of liquid culture, similar to Lgt3.
Taken together, our data demonstrate that Lgt glucosyltransferases represent a family of redundant molecules which are capable of modifying and inactivating eEF1A. All investigated L. pneumophila strains possess a pair of Lgt1 and Lgt3 proteins, while some other strains additionally carry Lgt2. Synthesis of all these enzymes is strictly and differentially regulated, suggesting a specialized role of each protein in the Legionella life cycle.
Supplementary Material
Acknowledgments
This work was financially supported by the BMBF, INTAS (project no. 05-1000004-7756 to Y.B. and K.A.), and the DFG (K.A.). DNA sequencing was carried out in the Genome Centre, Institute of Molecular Biology, RAS (RFBR grant 00-04-55000).
We thank Ralf Gilsbach and Nadine Beetz (Albert-Ludwigs-University, Freiburg, Germany) for help with RT-PCR experiments, Carmen Buchrieser (Institut Pasteur, Paris, France) for L. pneumophila strains Lens and Paris, and Can Uenal (Unversity of Wuerzburg, Germany) for the A. castellanii strain.
Footnotes
Published ahead of print on 15 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aktories, K., and J. T. Barbieri. 2005. Bacterial cytotoxins: targeting eukaryotic switches. Nat. Rev. Microbiol. 3397-410. [DOI] [PubMed] [Google Scholar]
- 2.Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 401201-1214. [DOI] [PubMed] [Google Scholar]
- 3.Belyi, I., M. R. Popoff, and N. P. Cianciotto. 2003. Purification and characterization of a UDP-glucosyltransferase produced by Legionella pneumophila. Infect. Immun. 71181-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belyi, Y., R. Niggeweg, B. Opitz, M. Vogelsgesang, S. Hippenstiel, M. Wilm, and K. Aktories. 2006. Legionella pneumophila glucosyltransferase inhibits host elongation factor 1A. Proc. Natl. Acad. Sci. USA 10316953-16958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 6.Broich, M., K. Rydzewski, T. L. McNealy, R. Marre, and A. Flieger. 2006. The global regulatory proteins LetA and RpoS control phospholipase A, lysophospholipase A, acyltransferase, and other hydrolytic activities of Legionella pneumophila JR32. J. Bacteriol. 1881218-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browne, G. J., and C. G. Proud. 2002. Regulation of peptide-chain elongation in mammalian cells. Eur. J. Biochem. 2695360-5368. [DOI] [PubMed] [Google Scholar]
- 8.Bruggemann, H., C. Cazalet, and C. Buchrieser. 2006. Adaptation of Legionella pneumophila to the host environment: role of protein secretion, effectors and eukaryotic-like proteins. Curr. Opin. Microbiol. 986-94. [DOI] [PubMed] [Google Scholar]
- 9.Burkhard, P., J. Stetefeld, and S. V. Strelkov. 2001. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 1182-88. [DOI] [PubMed] [Google Scholar]
- 10.Busch, C., F. Hofmann, J. Selzer, S. Munro, D. Jeckel, and K. Aktories. 1998. A common motif of eukaryotic glycosyltransferases is essential for the enzyme activity of large clostridial cytotoxins. J. Biol. Chem. 27319566-19572. [DOI] [PubMed] [Google Scholar]
- 11.Busch, C., J. Orth, N. Djouder, and K. Aktories. 2001. Biological activity of a C-terminal fragment of Pasteurella multocida toxin. Infect. Immun. 693628-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Felipe, K. S., S. Pampou, O. S. Jovanovic, C. D. Pericone, S. F. Ye, S. Kalachikov, and H. A. Shuman. 2005. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol. 1877716-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edelstein, P. H. 1981. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J. Clin. Microbiol. 14298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman, R. L., B. H. Iglewski, and R. D. Miller. 1980. Identification of a cytotoxin produced by Legionella pneumophila. Infect. Immun. 29271-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gal-Mor, O., T. Zusman, and G. Segal. 2002. Analysis of DNA regulatory elements required for expression of the Legionella pneumophila icm and dot virulence genes. J. Bacteriol. 1843823-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Just, I., J. Selzer, M. Wilm, C. Von Eichel-Streiber, M. Mann, and K. Aktories. 1995. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 375500-503. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 18.Laguna, R. K., E. A. Creasey, Z. Li, N. Valtz, and R. R. Isberg. 2006. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc. Natl. Acad. Sci. USA 10318745-18750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo, Z. Q., and R. R. Isberg. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 101841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marra, A., S. J. Blander, M. A. Horwitz, and H. A. Shuman. 1992. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc. Natl. Acad. Sci. USA 899607-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moffat, J. F., and L. S. Tompkins. 1992. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 60296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295679-682. [DOI] [PubMed] [Google Scholar]
- 23.Ninio, S., and C. R. Roy. 2007. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 15372-380. [DOI] [PubMed] [Google Scholar]
- 24.Ristroph, J. D., K. W. Hedlund, and R. G. Allen. 1980. Liquid medium for growth of Legionella pneumophila. J. Clin. Microbiol. 1119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saito, A., R. D. Rolfe, P. H. Edelstein, and S. M. Finegold. 1981. Comparison of liquid growth media for Legionella pneumophila. J. Clin. Microbiol. 14623-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandvig, K., and B. Van Deurs. 1996. Endocytosis, intracellular transport, and cytotoxic action of Shiga toxin and ricin. Physiol. Rev. 76949-966. [DOI] [PubMed] [Google Scholar]
- 27.Segal, G., M. Feldman, and T. Zusman. 2005. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol. Rev. 2965-81. [DOI] [PubMed] [Google Scholar]
- 28.Shohdy, N., J. A. Efe, S. D. Emr, and H. A. Shuman. 2005. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc. Natl. Acad. Sci. USA 1024866-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, M. R., R. D. Miller, and B. H. Iglewski. 1981. In vitro production of an extracellular protease by Legionella pneumophila. Infect. Immun. 34299-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiaden, A., T. Spirig, S. S. Weber, H. Bruggemann, R. Bosshard, C. Buchrieser, and H. Hilbi. 2007. The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell. Microbiol. 92903-2920. [DOI] [PubMed] [Google Scholar]
- 31.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets. Proc. Natl. Acad. Sci. USA 764350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279873-876. [DOI] [PubMed] [Google Scholar]
- 33.Warren, W. J., and R. D. Miller. 1979. Growth of Legionnaires' disease bacterium (Legionella pneumophila) in chemically defined medium. J. Clin. Microbiol. 1050-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson, B. A., and R. J. Collier. 1992. Diphtheria toxin and Pseudomonas aeruginosa exotoxin A: active-site structure and enzymic mechanism. Curr. Top. Microbiol. Immunol. 17527-42. [DOI] [PubMed] [Google Scholar]
- 35.Zusman, T., G. Aloni, E. Halperin, H. Kotzer, E. Degtyar, M. Feldman, and G. Segal. 2007. The response regulator PmrA is a major regulator of the Icm/Dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol. Microbiol. 631508-1523. [DOI] [PubMed] [Google Scholar]
- 36.Zusman, T., O. Gal-Mor, and G. Segal. 2002. Characterization of a Legionella pneumophila relA insertion mutant and roles of RelA and RpoS in virulence gene expression. J. Bacteriol. 18467-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.