Abstract
RecAX53 is a chimeric variant of the Escherichia coli RecA protein (RecAEc) that contains a part of the central domain of Pseudomonas aeruginosa RecA (RecAPa), encompassing a region that differs from RecAEc at 12 amino acid positions. Like RecAPa, this chimera exhibits hyperrecombination activity in E. coli cells, increasing the frequency of recombination exchanges per DNA unit length (FRE). RecAX53 confers the largest increase in FRE observed to date. The contrasting properties of RecAX53 and RecAPa are manifested by in vivo differences in the dependence of the FRE value on the integrity of the mutS gene and thus in the ratio of conversion and crossover events observed among their hyperrecombination products. In strains expressing the RecAPa or RecAEc protein, crossovers are the main mode of hyperrecombination. In contrast, conversions are the primary result of reactions promoted by RecAX53. The biochemical activities of RecAX53 and its ancestors, RecAEc and RecAPa, have been compared. Whereas RecAPa generates a RecA presynaptic complex (PC) that is more stable than that of RecAEc, RecAX53 produces a more dynamic PC (relative to both RecAEc and RecAPa). The properties of RecAX53 result in a more rapid initiation of the three-strand exchange reaction but an inability to complete the four-strand transfer. This indicates that RecAX53 can form heteroduplexes rapidly but is unable to convert them into crossover configurations. A more dynamic RecA activity thus translates into an increase in conversion events relative to crossovers.
The bacterial RecA protein is multifunctional (11, 17, 25). In Escherichia coli, the RecA protein is first a recombinase, promoting homologous recombination and recombinational DNA repair. Second, RecA filaments formed on DNA facilitate the autocatalytic cleavage of the LexA protein to allow induction of the SOS response. This activity is referred to as the RecA coprotease function. Finally, RecA is directly involved in the activation of DNA polymerase V, the polymerase responsible for SOS mutagenesis (11, 17, 25). The present study focused on RecA function in recombination and recombinational DNA repair.
The key steps in homologous recombination promoted by a RecA nucleoprotein filament are (i) formation of the presynaptic filament (a ternary complex including RecA, ATP, and single-stranded DNA [ssDNA]) in the presence of magnesium ion, (ii) pairing of the presynaptic filament with homologous double-stranded DNA (dsDNA), and (iii) DNA strand exchange between ssDNA and dsDNA that results in a new heteroduplex dsDNA and a displaced ssDNA (11, 15, 17, 25, 37). To promote these reactions, RecA has two main activities, the DNA-dependent ATPase, activated with both ssDNA and dsDNA, and DNA strand transferase. The latter activity can be broken down into several characteristics commonly used for the comparison of different RecA proteins. These include RecA protein binding to ssDNA to form a presynaptic complex (PC), RecA protein-mediated displacement of SSB protein from ssDNA, the stability of PCs (for example, in solutions with increasing salt concentrations), and the rate and efficiency of three- or four-strand transfer reactions. Obviously, if any combination of these properties is enhanced or impaired, it will presumably result in higher or lower RecA protein activity in the overall DNA strand exchange reaction.
In E. coli, homologous recombination promoted by the RecA protein comes about by means of two genetic pathways, RecBCD and RecFOR (8, 9). According to a current view, the RecBCD pathway initiates recombination mainly on dsDNA breaks or dsDNA ends by the mechanism known as double-strand break repair (DSBR). What is important is that the RecBCD helicase-endonuclease loads RecA protein onto ssDNA and thus forms the presynaptic filament (3, 7). The RecFOR pathway uses mainly ssDNA gaps or nicks as a trigger for the mechanism of single-strand gap repair (SSGR) (12, 36). It is noteworthy that, in this case, binding of RecA to ssDNA proceeds in competition with the SSB protein. As a rule, DSBR results in the formation of the crossover-type recombinants (that means recombination exchanges proceeding at the dsDNA level) while SSGR tends to produce recombinants of the conversion type (implying recombination exchanges at the ssDNA level, i.e., via heteroduplex formation) (12). However, neither crossovers nor conversions can be completely excluded from recombination events managed by SSGR or DSBR, respectively, as well as by other possible models of homologous recombination (36).
E. coli cells have long- and short-patch mismatch repair systems (28) in which the MutS protein recognizes and binds to a mismatched base pair and thus creates a MutS-DNA complex that allows the initiation of repair. A more general long-patch repair MutSLH system is methyl directed. This means that it corrects mismatches to the sequence of the methylated strand in heteroduplex DNA (26). In conjugal recombination, proceeding via DSBR or especially via the SSGR mechanism, hemimethylated heteroduplexes are possible recombinant products which can be corrected by MutSLH to the phenotype of the methylated strand. In fact, during conjugation, only one methylated donor strand is transferred while the other is synthesized in the recipient cell (18) and thus is transiently undermethylated. On the other side, the recipient chromosome can also be undergoing replication and thus contain newly synthesized regions of DNA in the unmethylated state. Therefore, if recombinational heteroduplexes are formed, they can be corrected to either the donor or the recipient sequence. Correction to the former fixes the donor genotype in the recipient strain, while correction to the latter eliminates the donor genotype and thus has a so-called antirecombination effect (38).
In vivo genetic analysis of recombinants obtained after bacterial conjugation allows us to quantitatively estimate RecA recombinogenic activity by measuring the frequency of recombination exchanges (FRE) per DNA unit length (19, 20) Such measurements have revealed that RecA from E. coli (RecAEc) has a relatively weak recombinase activity (16, 17), although this is presumably optimal for this bacterium. This optimal FRE level provides a baseline against which to measure the increase in FRE values that reflects hyperrecombination. Abnormal recombination levels have been described for some RecAEc mutants, including RecA730 (22), RecA803 (26), and RecAX21 (6); for Pseudomonas aeruginosa RecA (RecAPa), expressed in E. coli (5, 29); and for a set of E. coli-P. aeruginosa chimeric RecA (RecAX) proteins (4). Interestingly, there is no direct correlation between the amount of different RecA proteins within the cell and the intracellular recombination activity measured through FRE values. As shown previously (29), the ratio of the intracellular amounts of RecAEc produced by the recAEc gene in the E. coli chromosome, RecAEc produced by the recAEc gene located in plasmid pUC19, RecAPa produced in E. coli under the control of the E. coli lac promoter, and RecAPa produced in E. coli under the control of the recAPa promoter can be presented as 1:3.4:0.9:0.05, whereas the corresponding FRE ratio is 1:1:6.5:8. This means that the cell recombination activity is an intrinsic property of a given RecA protein and is usually not affected greatly by its intracellular concentration.
The in vitro biochemical activities of all of the hyperrecombination RecA proteins enumerated above appear to be substantially enhanced, though differently, relative to the same activities promoted by RecAEc (4, 6, 29). The increases in activities which seem to be important for recombination imply that there is considerable plasticity in the robustness of a bacterial RecA protein. In the light of these observations, it seems reasonable to suggest the existence of certain critical characteristics of RecA protein complexes that might be responsible for their recombinogenic activities.
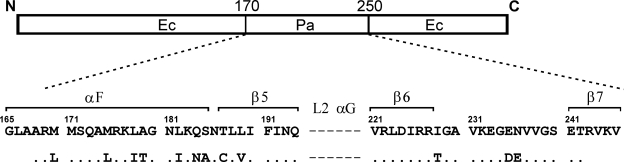
Here, we characterize the genetic and biochemical recombination activities of the chimeric RecAX53 protein (30). This protein, expressed in E. coli, produces the highest measured FRE values of 30 chimeras (E. coli-P. aeruginosa) analyzed (5). Relative to wild-type RecAEc, RecAX53 contains 12 amino acid residue substitutions in the middle of the RecAEc central domain, all present in one α helix (αF), two β strands (β5 and β6), and one unstructured loop (Fig. 1). All of the substitutions reflect the amino acid residues present at the same positions in RecAPa. The distinguishing genetic characteristics of RecAX53 presented here allow us to look at the problem of hyperrecombination from a new perspective in an attempt to understand the type of recombination events (crossovers or conversions) promoted by different RecA proteins and correlate the events with the properties of the responsible RecA variants.
FIG. 1.
Structure of the chimeric RecAX53 protein (30). The alignment shown at the bottom shows the 12 amino acid residues from RecAPa which substitute for residues of RecAEc at the positions indicated.
As shown earlier (20), in normal RecAEc-promoted recombination, the inactivation of MutS protein by the mutS215 mutation reveals additional recombination exchanges concealed by the correction of recombinant heteroduplexes (the antirecombination effect). Formally, for each mutS215-independent recombination event, there are approximately five mutS215-dependent events. In a normal cell, the MutS protein suppresses hyperrecombination and thus the inactivation of MutS formally increases the FRE value (FRE-up effect). In principle, one can imagine the opposite situation in which a mutS215 mutation results in a FRE decrease (FRE-down effect). Here, we show that these FRE-up and FRE-down effects can reflect two different modes (crossover and conversion, respectively) of hyperrecombination.
Comparative genetic analysis of RecAX53 and its ancestors, RecAEc and RecAPa, suggests that the conversion is probably the primary path of hyperrecombination managed by the RecAX53 protein while the crossovers are a main part of that promoted by RecAPa. These properties correlate with several critical biochemical characteristics of RecAX53.
MATERIALS AND METHODS
Enzymes and biochemicals.
The RecAEc, RecAPa, and RecAX53 proteins were purified as previously described (16). RecAEc and RecAX53 protein concentrations were determined by absorbance at 280 nm by using an extinction coefficient of 2.23 × 104 M−1 cm−1. The RecAPa protein concentration was determined with the help of a protein Coomassie Plus Protein Assay Reagent kit (Pierce) with the RecAEc protein as the standard. The SSB protein was either purchased (Sigma, USB) or purified as previously described (23). Its concentration was determined by absorbance at 280 nm by using an extinction coefficient (ɛ280) of 2.83 × 104 M−1 cm−1 (24). NADH, lactate dehydrogenase, pyruvate kinase, phosphoenolpyruvate, and ATP were all purchased from Sigma. All of the other reagents used in this study were research grade and were commercially available.
Strains and plasmids.
Donor strain KL227 (HfrP4x metB) and recipient strains AB1157 (thr-1 leuB6 proA2 hisG4 argE3 thi-1 supE44 rpsL31) and JC10289 (recombination-deficient; same as AB1157 but Δ[recA-srlR306]::Tn10 = ΔrecA306) were from A. J. Clark's collection. Recombination-proficient strains JC10289-Ec-S, JC10289-Pa-S, and JC10289-53-S were constructed by P1 transduction to transfer mutS215::Tn10, respectively, into JC10289-Ec (same as JC10289 but with plasmid pRecAEc expressing a wild-type recAEc gene under the control of its own promoter), JC10289-Pa (same as JC10289 but with plasmid pRecAPa expressing a wild-type recAPa gene under the control of its own promoter), and JC10289-53 (same as JC10289 but with plasmid pRecAX53 expressing a chimeric recAX53 gene under the control of the lac promoter) (5). Plasmid P200 F′-lac was used to standardize the conjugation abilities of recipient strains.
DNA.
Circular M13mp8 ssDNA was from New England BioLabs. Its nucleotide molar concentration was determined by absorbance at 260 nm with an extinction coefficient of 6.5 × 103 M−1 cm−1. The etheno-modified calf thymus ssDNA used was as previously described (2). Its nucleotide molar concentration was determined by absorbance at 260 nm with an extinction coefficient of 8.325 × 103 M−1 cm−1. The blunt-ended 34-mer double-stranded oligonucleotides containing the emitter fluorophore FAM (fluorescein phosphoramidite) positioned at the 5′ ends had the following compositions: FAM-TCA CCA ATG AAA CCA TCG ATA GCA GCA CCG TAA T-3′-PO4 and 3′-AGT GGT TAC TTT GGT AGC TAT CGT CGT GGC ATT A.
The oligonucleotides were synthesized by Integrated DNA Technologies, Inc., and used in the fluorescence assay for pairing with M13mp8 circular ssDNA. The poly(dT) used was purchased from Sigma. An extinction coefficient at 260 nm of 8.54 × 103 M−1 cm−1 was used for determination of the concentration of poly(dT). Concentrations of ssDNA, dsDNA, and single- and double-stranded oligonucleotides are expressed in moles of nucleotide residues.
Conjugation was carried out essentially as previously described (20). Both Hfr and F− were grown, crossed, and selected for recombinants at 37°C in mineral salts 56/2 medium supplied with all necessary growth factors at pH 7.5. The ratio of donors to recipients in mating mixture was 1:10, 2 × 107 to 4 × 107 donors and 2 × 108 to 4 × 108 recipients per ml. The yield of Ara+ Strr recombinants in all independent crosses was normalized by the mating ability of each recipient used. The latter was determined by the yield of F′-lac+ transconjugants in crosses between the recipients and the donor P200 F′-lac.
FRE value calculations were carried out as previously described (5, 20). Quantitative estimations of FRE alterations (ΔFRE) promoted by the RecAX53 and RecAPa proteins relative to the FRE value promoted by RecAEc were done by use of the following formula: ΔFRE = ln(2μ1 − 1)/(2μ2 − 1), where μ2 is the linkage of selected ara+ and unselected leu+ markers in a cross with AB1157 and μ1 is a similar linkage in the cross analyzed. Donor strain KL227 transfers leu+ and ara+ as proximal and distal markers, respectively. Calculations of uncertainty in determinations of relative values of FRE were done as deviations from the average values by making use of the program Excel-97 with the formula [=2*STDEV] and by inputting the values from independent repeats of three experiments.
DNA-dependent hydrolysis of ATP.
ssDNA-dependent ATP hydrolysis was measured by using a spectrophotometric assay that couples the production of ADP to the oxidation of NADH at 380 nm by using an extinction coefficient for NADH of 1.21 mM−1 cm−1. Unless otherwise indicated, reactions were carried out at 37°C in TMD buffer (25 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 1 mM dithiothreitol) containing 1 mM ATP, an ATP-regenerating system (5 mM phosphoenolpyruvate and 30 U ml−1 pyruvate kinase), and a coupling system (3 mM NADH and 30 U ml−1 lactate dehydrogenase).
ɛDNA fluorescence measurement.
The binding of RecA proteins to ɛDNA was measured by fluorescence quenching as previously described (27). The fluorescence measurements were monitored with a Hitachi F-4000 spectrofluorometer (Hitachi, Tokyo, Japan) at excitation and emission wavelengths of 305 and 410 nm, respectively. Emission spectra were recorded in correction mode by subtracting the spectrum of the TMD buffer used.
Electrophoresis-fluorescence assay for pairing and strand displacement of M13mp8 circular ssDNA and double-stranded oligonucleotides.
All reactions were carried out at 37°C in TMD buffer (25 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 1 mM dithiothreitol) containing 2 mM ATP, an ATP-regenerating system (5 mM phosphoenolpyruvate and 30 U ml−1 pyruvate kinase), and RecA protein as indicated. After addition of 21 μM M13mp8 ssDNA in the TMD-ATP-RecA mixture (100 μl), the mixture was incubated for 3 min to preform a PC, and then 2 μM SSB was added and after 5 min the pairing reaction was started (t = 0 min) by adding 0.3 μM double-stranded oligonucleotide probes. At the indicated times, aliquots (16 μl) were removed, the reaction was stopped by addition of 1.6 μl sodium dodecyl sulfate (0.9%) and placing the mixture on ice. The samples were treated with 1 μl proteinase K (2 mg/ml), loaded onto a 1% agarose gel with TAE buffer, and developed and scanned with the Typhoon 9410 Variable Mode Imager. The software package TotalLab v1.10 (Phoretix) was used for quantitation of the data.
Agarose gel assay for the four-stranded DNA exchange reaction.
The reaction conditions and DNA substrates used were as previously described (13). All reactions were carried out at 37°C in TMD buffer containing 5% (wt/vol) glycerol and an ATP regeneration system (8 mM phosphocreatine and 8 U/ml phosphocreatine kinase). A combination of 5 μM RecA, 2 mM ATP, and 10 μM circular M13mp7 dsDNA with a 793-nucleotide gap was preincubated in TMD buffer for 10 min to preform a PC in the gap. The reaction was started (t = 0 min) by addition of 0.3 μM SSB and 10.6 μM M13mp7 dsDNA linearized with BsmBI. After 100 min of incubation, aliquots (10 μl) were removed and the reaction was stopped by addition of 5 μl of a solution containing 60 mM EDTA, 6% sodium dodecyl sulfate, 25% (wt/vol) glycerol, and 0.2% bromphenol blue. Samples were subjected to electrophoresis in 1.0% agarose gels with TAE buffer, stained with ethidium bromide, and exposed to UV light.
Uncertainty of the data.
All of the data presented in Fig. 2, 3, 4, 5, and 6 were reproduced at least three times. The curves presented in these figures are from one typical experiment with uncertainty in measurements that did not exceed 5 to 8%.
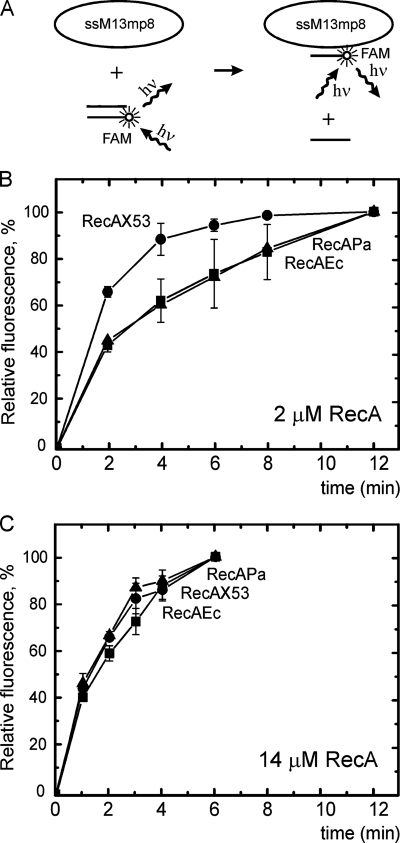
FIG. 2.
RecA-dependent pairing and strand displacement enhancement by RecAX53. Reactions between M13mp8 ssDNA (targets) and 34-mer double-stranded oligonucleotides (probes) were carried out and monitored by electrophoresis-fluorescence assays. (A) Scheme of the electrophoresis-fluorescence assay. FAM is the fluorophore. (B and C) Time course of strand displacement promoted by the RecAX53, RecAEc, or RecAPa protein. All reactions were performed at 37°C in TMD buffer containing 2 mM ATP, an ATP-regenerating system, 2.0 μM RecA (B) or 14 μM RecA (C), and 21 μM M13mp8 ssDNA. A ternary PC, RecA/ATP/M13mp8 ssDNA, was formed in the presence of 2 μM SSB. At time zero, 0.3 μM 34-mer double-stranded oligonucleotides (described in Materials and Methods) were added and the kinetics of strand transfer reactions was followed by quantitating the increase in the fluorescence of the M13mp8 ssDNA-34mer single-stranded oligonucleotide electrophoretic band as indicated in Materials and Methods. The fluorescence of the completed reaction (a plateau of the curves) was taken as 100 arbitrary units. The fluorescence in all samples was normalized to this level. hν, quantum energy of UV excitation (inward arrow) and that of fluorescent irradiation (outward arrow).
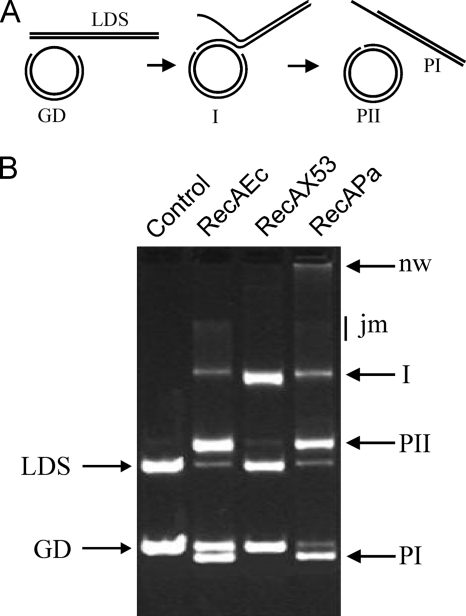
FIG. 3.
Four-strand exchange reactions managed by bacterial RecA proteins monitored by the agarose gel assay. (A) Scheme of the reaction showing its intermediate and final products. (B) Products of the reaction revealed by agarose gel assay. All reactions were performed at 37°C in TMD buffer containing an ATP-regenerating system; 5 μM RecAEc, RecAX53, or RecAPa protein (as indicated); 0.3 μM E. coli SSB; 10 μM gapped DNA with a 793-nucleotide gap; and 10.6 μM linear DNA. Following preincubation (see Materials and Methods), reactions were carried out for 100 min. The products of strand exchange were revealed by electrophoresis as described in Materials and Methods. The control lane shows the composition of the reaction mixture without RecA protein after 100 min of incubation. The other lanes show the reaction products produced in the presence of RecAEc, RecAX53, and RecAPa, respectively. I, intermediate stage; PI, linear dsDNA with an ssDNA tail; PII, nicked circular duplex DNA; nw, networks; jm, joint molecules; LDS, linear dsDNA; GD, gapped dsRNA.
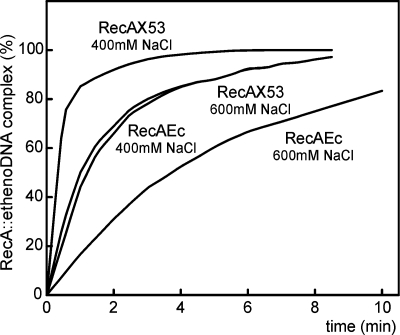
FIG. 4.
Time course of RecAEc and RecAX53 PC formation on calf thymus ɛDNA in the presence of ATPγS. Two high concentrations of NaCl were used. Reactions were monitored by measuring the change in ɛDNA fluorescence as described in Materials and Methods. Experiments were performed at 37°C in TMD buffer containing 0.1 mM ATPγS, 1.9 μM ɛDNA, and 0.8 μM RecA protein (for details, see Materials and Methods). At time zero, RecA protein was added.
FIG. 5.
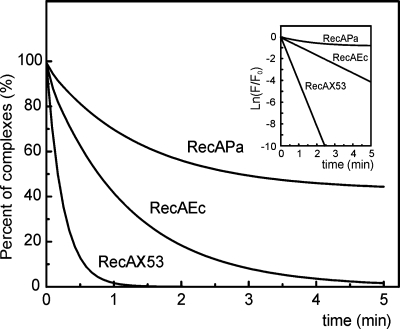
Time course of the disassembly of the RecA/ATP/ɛDNA complex formed by the RecAX53, RecAEc, and RecAPa proteins. Reaction mixtures were challenged with an excess of poly(dT) and monitored by measuring the change in ɛDNA fluorescence. Experiments were performed at 37°C in TMD buffer containing 1 mM ATP and its regenerating system, 3 μM ɛDNA, 2.5 μM RecA, and 50 μM poly(dT). At time zero, poly(dT) was added. The inset shows ln(F/F0) versus time. F is fluorescence at a given time; F0 is the original fluorescence.
FIG. 6.
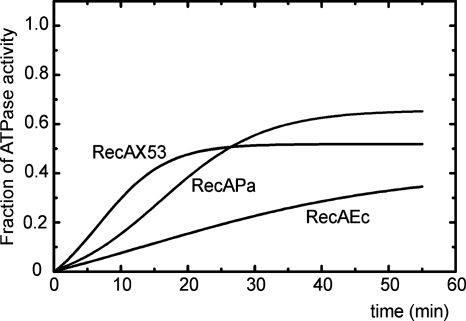
Kinetics of SSB protein displacement from ssDNA by RecAX53, RecAEc, and RecAPa. Reactions were monitored in real time by following RecA-mediated (and DNA-dependent) ATP hydrolysis. Reactions were performed at 37°C in TMD buffer containing 2 μM RecA, 3 μM circular M13 ssDNA, 0.6 μM SSB, 1 mM ATP and an ATP-regenerating system, and a coupling system (for details, see Materials and Methods). The ssDNA was preincubated with SSB for 2 min, and the reactions were initiated by the addition of RecA. The fraction of recovered ATPase activity was determined by dividing the rate of ATP hydrolysis at each time point by the rate of ATP hydrolysis observed in reaction mixtures where the SSB protein is added to ssDNA after the RecA protein. These control conditions bring about maximum RecA coating of the ssDNA under these conditions.
RESULTS
Inactivation of MutS protein reveals two modes of hyperrecombination.
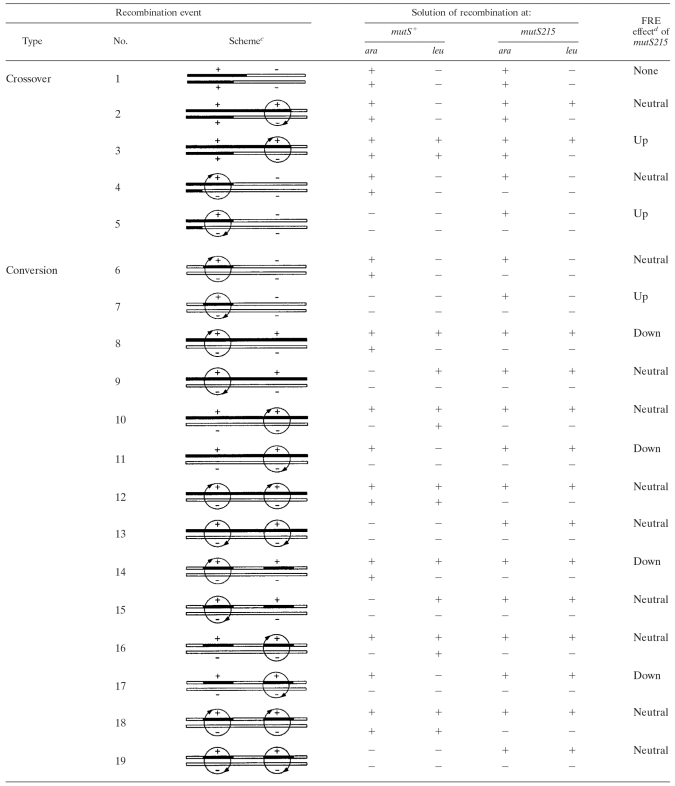
Table 1 illustrates 19 possible conjugal recombination events (between selected ara+ and unselected leu markers) formed under conditions of either a wild-type MutS protein or MutS inactivated by the mutS215 mutation. Five events result from a crossover configuration of donor and recipient markers, and the other 14 are presented in a pure heteroduplex configuration and are ready for the conversion of donor markers (selected, unselected, or both) to either the donor or the recipient sequence. The five crossover events are naturally complicated by conversions at the edge of the crossovers in the proximity of the selected donor marker. A relatively long distance between the selected ara+ and unselected leu markers (approximately 13 kb) was chosen to practically exclude the possibility of finding ara+ and leu included in one heteroduplex at the edge of crossovers. Therefore, this possibility is considered only in the case of conversion events.
TABLE 1.
Effect of the mutS215 mutation on measured FRE values in conjugal crosses where the ara+ marker is selecteda and leu is unselectedb
Left in schemes.
Right in schemes.
The donor and recipient DNAs in possible recombinants are represented by black and white bars, respectively. The circles denote mismatch repair events, with the arrowhead on each circle indicating the methylated strand that guides the repair (the arrowhead pointing up or down on the circle means correction to the donor or recipient sequence, respectively). The effects listed are predicted. All possible events are shown, including the antirecombination effect of the ara+ marker corrected to ara (no. 5, 7, 13, and 19; predicted but not detected).
Up or down means the possible appearance or disappearance of the FRE effect because of the mutS215 mutation. Neutral means no change in FRE status after correction. None means no possible corrections.
As shown, of these 19 events, only 7 (no. 3, 5, 7, 8, 11, 14, and 17) are mutS215 dependent and only 3 (no. 3, 5, and 7) require MutS inactivation to be revealed. Therefore, mutS215 formally increases the overall FRE value (FRE-up) for those three events. Four other events require MutS integrity, and mutS215 causes a FRE decrease (FRE-down) for these. As is also evident, six events (no. 5 among crossovers and no. 7, 9, 13, 15, and 19 among conversions) can be fixed only because of the mutS215 mutation. (Note that four of them, no. 5, 7, 13, and 19, demonstrate the mutS+-dependent antirecombination effect when correction of the selected marker ara+ to ara mimics the absence of the recombination event.) However, this yield-up effect of mutS215 cannot be observed in a real experiment because the increments are too small to see above the background. The analysis we present allows us to make one prediction: if recombination events promoted by RecA are preferentially crossovers, the mutS215-dependent recombinants will exhibit an increase in the FRE value, whereas in the same genetic background the conversion events will mostly result in a FRE decrease. Thus, in principle, the mutS dependence of FRE allows us to discriminate between the two main modes of recombination.
Contrasting mutS dependence of FRE promoted by RecAX53 protein and its ancestors.
Table 2 presents an examination of the consequences of mutS deficiency for RecAX53 and its ancestors, RecAEc and RecAPa. The ΔFRE values were calculated relative to the FRE found in the cross HfrP4x × JC10289-Ec as described in Materials and Methods. The actual baseline FRE was about five exchanges per E. coli genome (or one exchange per 20-min region of the genome), as determined by measurement of the linkage μ (see Materials and Methods).
TABLE 2.
Comparison of FRE values promoted by different RecA proteins and a specific role for MutS protein inactivation
| Recipient | Relevant genotype | Yield of Ara+ recombinants (%)a | Linkage μb (between ara+ and leu+) | ΔFREc | FRE effect |
|---|---|---|---|---|---|
| JC10289-Ec | recAEc+ | 3.7 ± 0.8 | 0.986 ± 0.007 (600) | 1.0 | |
| JC10289-Ec-S | recAEc+mutS215 | 3.1 ± 0.8 | 0.923 ± 0.017 (900) | 5.6 ± 0.6 | 5.6 up |
| JC10289-Pa | recAPa+ | 4.5 ± 1.0 | 0.896 ± 0.032 (900) | 7.8 ± 0.5 | |
| JC10289-Pa-S | recAPa+mutS215 | 2.9 ± 0.6 | 0.801 ± 0.015 (1,200) | 16.9 ± 3.4 | 2.2 up |
| JC10289-53 | recAX53 | 1.4 ± 0.3 | 0.883 ± 0.025 (600) | 8.9 ± 0.6 | |
| JC10289-53-S | recAX53 mutS215 | 0.8 ± 0.2 | 0.951 ± 0.006 (900) | 3.4 ± 0.1 | 2.6 down |
The yield of recombinants is presented as a percentage of the number of transconjugants counted in crosses between the donor P200 F′-lac and these recipients under identical conditions.
The data shown are based on a genetic analysis of random recombinant Ara+ Strr clones taken from three independent crosses between Ara+ Strs strain HfrP4x and the set of Ara− Strr JC10289 recipients shown. The linkage between selective ara and unselective leu markers was analyzed. The values in parentheses are the number of Ara+ clones analyzed.
ΔFRE was determined relative to the FRE value found for the cross HfrP4x × JC10289-Ec.
As expected, the mutation mutS215 produced a 5.6-fold increase in FRE for the RecAEc protein. This mutation also resulted in a more-than-twofold increase in the case of RecAPa. A different picture was found in the same FRE analysis for recombination managed by RecAX53 protein. In this case, mutS215 decreased the FRE value by a factor of 2.6. These data are in good agreement with the FRE-up and FRE-down analyses presented in the previous section and consign RecAPa and RecAX53 to two different predicted modes of hyperrecombination. The data also suggest that RecAEc protein preferentially uses the same crossover mode of recombination as RecAPa.
RecAX53 protein is very active in the initiation of strand exchange.
Pairing and strand displacement are two core phases of the strand exchange reaction between the presynaptic ssDNA filament and homologous dsDNA. In principle, both phases can be observed in fluorescence assays using ssDNA and double-stranded oligonucleotides as DNA substrates and a fluorescent strand of a double-stranded oligonucleotide (that is complementary to the ssDNA) as a reporter.
A 34-mer blunt-ended duplex oligonucleotide probe labeled with FAM at the appropriate 5′ end interacted with a PC, RecA/ATP/ssDNA M13mp8, bearing a 34-mer target sequence for complementary pairing. This pairing, followed by the strand displacement, resulted in the fluorescence induction of oligonucleotide molecules transferred to the M13mp8 phage DNA. These molecules were separated by electrophoresis in agarose gel, the gel was developed and scanned, and the molecules were quantitated as described in Materials and Methods. This procedure allowed us to monitor the target hybridization process. Figure 2A illustrates the procedure described, and Fig. 2B and C compare the 34-mer duplex pairing and strand displacement with the target in M13 ssDNA promoted by RecAX53 and its ancestors, RecAEc and RecAPa. As shown in Fig. 2B, when the M13 ssDNA (21 μM) is present in excess relative to the RecA concentration (2 μM), RecAX53 promoted the initial phases of the short oligonucleotide exchange reaction more efficiently (approximately 1.5 times faster) than both of its ancestors, RecAEc and RecAPa. Under conditions of DNA excess, a significant fraction of the ssDNA would not have RecA protein coating the target DNA sequence. Pairing could not occur for these DNAs until the RecA protein disassembled and reassembled to coat the target. The results indicate that the chimeric mutant protein RecAX53 carries out the required filament redistribution faster than its ancestor proteins. In contrast, under conditions in which the RecA protein (14 μM) was in excess over M13 ssDNA (21 μM), the efficiency of the oligonucleotide exchange initiation became practically the same for all three proteins (Fig. 2C).
The data so far are consistent with the idea that RecAX53 is more efficient in the early steps of DNA strand exchange under conditions in which the PC can be formed only on a small part of the target DNA at a given moment.
RecAX53 is efficient in the three-strand exchange but inefficient in the four-strand reaction.
The scheme presented in Fig. 3A shows the strand exchange reaction between preformed PC RecA/ATP/gapped dsDNA and homologous linear dsDNA. As shown, the reaction passes through an intermediate stage (I) that is effectively a three-strand reaction between ssDNA (793-nucleotide gap) and a long, linear dsDNA molecule (7,238 bp). Finally, the reaction can be completed as a four-strand exchange by the generation of linear dsDNA with an ssDNA tail (product PI) and nicked circular duplex DNA (product PII). As often occurs in RecA reactions, the reaction can be complicated by the formation of complex DNA networks. The latter becomes possible when joint molecules are involved in new rounds of the reaction when free RecA molecules displace SSB on ssDNA to form new active PCs (21).
Under the conditions used (see Materials and Methods), RecAEc completed the four-strand reaction successfully. After 100 min, it converted essentially all of the linear dsDNA into the final products PI and PII (Fig. 3B), additionally producing a small amount of joint molecules. As expected, under the same conditions, RecAPa produced a certain amount of the final products but converted a part of the linear dsDNA into joint molecules and networks (29). This known property of RecAPa is based on its capacity to displace SSB and form stable PCs. As shown in Fig. 3, RecAX53 was very effective in the conversion of a part of the linear dsDNA into intermediate products without any complication with joint molecules or networks, but it appeared to be absolutely ineffective in the further conversion of these molecules into final products.
These data are in good agreement with the capacity of RecAX53 PCs to initiate three-strand exchange and even to process it to some extent. However, the RecAX53 PCs do not promote productive four-strand exchange reactions. The further biochemical analysis of RecAX53 presented below reveals that the reason for these unusual strand exchange characteristics is that the RecAX53 protein filament is more dynamic and less persistent than its wild-type progenitors.
RecAX53 protein is more dynamic in the assembly and disassembly of its PC.
According to one prominent model (11, 25), the formation of a ternary PC, RecA/ATP/ssDNA, proceeds very rapidly after a rate-limiting nucleation event. In order to compare the kinetics of RecAX53 and RecAEc nucleoprotein filament formation, one suggestion and three findings were taken into consideration. First, we suggested that the kinetics of RecA-DNA binding can be retarded in the presence of high salt concentrations (400 to 600 mM NaCl). Second, such salt concentrations are known to inhibit ssDNA binding to a secondary DNA-binding site of the RecA filament (4). Third, the ternary complex with ATPγS (the nearly nonhydrolyzable ATP analog) is extremely stable, even at 2.5 M NaCl (27), and this concerns only the primary DNA-binding site (4). Fourth, because the salt titration midpoint (STMP) for RecAX53 and that for its main ancestor, RecAEc (but not that for RecAPa), are similar (Table 3), RecAX53 and RecAEc can be used for comparative experiments in which a high salt concentration is an important parameter.
TABLE 3.
Comparison of kinetic and structural parameters of poly(dT)- dependent ATP hydrolysis catalyzed by the RecAX53 protein and its ancestors, RecAEc and RecAPa
| Protein | S05 (μM) | kcat (min−1) with poly(dT) | kcat (min−1) in 2 M sodium acetate | Site size (nucleotides) | STMP |
|---|---|---|---|---|---|
| RecAX53 | 79.0 ± 5.8a | 45.2 ± 2.2b | 43.0 ± 2.1b | 3.3 ± 0.4 | 300 ± 30 |
| RecAEc | 76.0 ± 9.5 | 29.2 ± 0.2 | 30.0 ± 1.2 | 3.0 ± 0.2 | 375 ± 30 |
| RecAPa | 71.5 ± 6.9 | 31.5 ± 2.9 | 31.1 ± 2.0 | 3.1 ± 0.3 | 600 ± 40 |
The values shown are averages; the error limit was determined as a standard deviation.
These results were repeated with four different independent purifications of the RecAX53 protein.
As shown previously, RecA binding to etheno-modified ssDNA (ɛDNA) results in a proportional increase in fluorescence upon the formation of a RecA-ɛDNA complex (27). Figure 4 illustrates the kinetics of RecAX53/ATPγS/ɛDNA and RecAEc/ATPγS/ɛDNA complex formation in the presence of 400 and 600 mM NaCl. Comparison of the rates of complex formation, especially in the initial stage of the reactions, shows a threefold increase in the rate of RecAX53 relative to that of RecAEc at both salt concentrations.
The RecA PC formed on linear ssDNA is subject to a 5′-end-dependent disassembly of RecA protomers from DNA requiring a concomitant ATP hydrolysis (33). To measure this process quantitatively, we analyzed the change in fluorescence of RecA/ATP/ɛDNA complexes, expecting the fluorescence to decrease when the complex disassembles. To make the process irreversible, the complexes formed were challenged with an excess of poly(dT), which binds both free RecA and disassembled RecA molecules. Figure 5 shows the time-dependent drop in fluorescence proceeding exponentially for the three proteins compared (RecAX53, RecAEc, and RecAPa), but at different rates. Being plotted as ln(F/F0) versus time, where F is fluorescence at a given time and F0 is the original fluorescence, all three dependences are converted into linear functions. The half-time for the drop in fluorescence changed with a RecAEc/RecAPa/RecAX53 ratio of 1:3:0.2. As expected, the RecAPa complex was much more stable than RecAEc, whereas the chimeric RecAX53 protein differs from both of its ancestors by a substantially increased rate of filament disassembly.
The data presented in this section indicate that RecAX53 filaments are highly dynamic, assembling and disassembling at enhanced rates as they promote recombination events.
RecAX53 is more active in SSB displacement from ssDNA.
When a PC is forming without preliminary operation of the RecBCD helicase-nuclease enzyme (15), the RecA protein has to displace SSB before loading onto ssDNA. This process can be monitored by measuring the rate of DNA-dependent ATP hydrolysis promoted by the RecA protein. When ssDNA was completely covered by the SSB protein, RecA and ATP with its regenerating system were added to the reaction mixture at concentrations sufficient to replace SSB entirely. Figure 6 shows the time course of such reactions for RecAX53 and its ancestors. As we will show in the next section, the ATPase activities of all three comparable RecA proteins are their intrinsic characteristics because the same activities are kept even in the absence of DNA (at the high salt concentration). Since RecAX53 has an increased ATPase activity, the three experimental curves presented in Fig. 6 were normalized relative to the maximum ATPase values found in reaction mixtures where we added the same amount of RecA to ssDNA but added RecA first and SSB second (a situation used to measure the intrinsic kcat).
As expected from earlier observations (6, 29), a profound difference between the RecAPa and RecAEc proteins was revealed: RecAPa displaced SSB much more actively. However, RecAX53 was even more robust in this activity; its initial lag period (before establishing a constant acceleration of ATP hydrolysis) was fourfold less than that seen with RecAPa, and a steady state was achieved 25 min after the start of the reaction—twofold faster than that seen with RecAPa. On the other hand, the maximum extent of RecA protein binding, as reflected by the fraction of the maximum rate of ATP hydrolysis achieved, appears higher for RecAPa than for RecAX53. This lower steady state of RecAX53 binding again reflects the more dynamic nature of this protein, which disassembles, as well as assembles, faster than the other RecA proteins.
In sum, comparative to RecAPa, the displacement of SSB from ssDNA by RecAX53 proceeds much more actively, at least at the initial step of the reaction of PC formation. These results are in concordance with the high initial reactivity and dynamics of the RecAX53 protein PC described above.
Basal characteristics of ssDNA-dependent ATP hydrolysis promoted by RecAX53 protein: comparison with its ancestors.
Two steady-state kinetic and two structural characteristics of ATP hydrolysis promoted by the PC of RecAX53 and its ancestors are summarized in Table 3. The first parameter, S05 (the substrate concentration for the half-maximal observed rate of hydrolysis), was similar for all three proteins at 10 mM MgCl2, which is the optimal magnesium concentration for the RecA protein in E. coli in vitro (1). The second parameter, kcat (the rate constant for the rate-determining step of ATP hydrolysis), appeared to be significantly increased for the chimeric protein relative to its ancestors. This was true both for the PC bound to ssDNA and for a DNA-free multimer at a high salt concentration (2 M sodium acetate), which is known to mimic the PC state (31).
The structural parameters presented in Table 3 include the number of nucleotide residues of ssDNA per RecA subunit (the apparent site size based on titrations) and the STMP or the NaCl concentration required to dissociate half of the RecA protein present in PCs and thus halve its ATPase activity. As shown, all three proteins interact with poly(dT) with identical and standard stoichiometry, three nucleotides per subunit in the RecA polymer structure. As for the STMP, the salt stability of the ternary complex RecA/ATP/poly(dT) formed by RecAX53 was quite similar to that of RecAEc (within the limits of error) and twofold less than that of RecAPa.
The data indicate that among the four general parameters analyzed, there is one, the rate of ATP hydrolysis, which distinguishes RecAX53 from its ancestors, RecAEc and RecAPa. The 12 amino acid substitutions, or perhaps a subset of them, enhance the ATPase potential in the structure of RecAEc. However, the same residues do not change the ATPase activity in the structure of RecAPa, probably because their effects are compensated for by other amino acid residues unique to RecAPa.
DISCUSSION
The chimeric RecAX53 protein is a hybrid protein which contains 12 amino acid residues from a central part of RecAPa in RecAEc (Fig. 1). These residues, or probably only a subset of them, change three main RecAX53 functional characteristics relative to those in its ancestors. First, they enhance the ATPase potential in the structure of RecAEc, but not in RecAPa (Table 3), because in the latter their effects are probably compensated by other unique residues. Second, they change the mode of hyperrecombination from crossovers to conversions (Tables 1 and 2). Finally, they create a RecA variant that is more dynamic in terms of binding to ssDNA (and displacing SSB), as well as dissociating from ssDNA. The overall effect of the increased dynamics is to improve the initiation of DNA strand exchange but to compromise the efficiency of extended DNA strand exchange.
In conjugal recombination managed by the RecAEc protein, internal recombination exchanges are very infrequent (one per 20 min or per about 930 kb) because a significant number of the extra exchanges (five versus one) are suppressed, or rather autosuppressed, by the MutS protein-dependent correction in heteroduplexes (14, 38). This is perhaps one of the pathways for the regulation of excessive recombination activity in E. coli. A similar situation is observed for RecAPa expressed in E. coli. In fact, relative to RecAEc, RecAPa promotes hyperrecombination, giving an eightfold increase in the FRE value in normal mutS+ cells (Table 2). However, an additional number of exchanges (approximately nine versus eight) was revealed in the mutS215 genetic background (Table 2). As shown for RecAEc, the intermediate exchanges promoted by RecAPa are corrected to zero by the same mechanism.
Quite the opposite situation was described for RecAX53. In this case, MutS protein inactivation resulted in a 2.6-fold decrease in the FRE value. This means that of 9 hyperrecombination events, 5.5 appeared to be also mutS dependent, but this time they were absolutely dependent on MutS integrity (Table 2). In other words, there are two different types of RecA proteins: the RecAEc/RecAPa type with a FRE-up effect in the mutS215 background and the RecAX53 type with a contrasting FRE-down effect under the same conditions. These two very different effects almost certainly reflect different mechanisms of hyperrecombination.
Table 1 gives us predictions from the two modes of recombination (crossovers and conversions) widely discussed in the literature. The genetic analysis presented in Table 2 unambiguously shows that RecAX53 differs from its ancestors by the preferential use of the SSGR pathway of recombination, especially for hyperrecombination events.
This hypothesis finds support in the biochemical analysis of the complex and elementary properties of RecAX53 and its ancestors. In effect, the RecAX53 protein is more active in the initiation of the strand exchange reaction (Fig. 2); it exhibits at least normal efficiency in the three-strand exchange but is completely defective in the four-strand reaction (Fig. 3). In other words, RecAX53 is effective at getting things started but ineffective in the extended reactions that lead to transfer of heteroduplexes into dsDNA exchanges (crossovers). The genetic observations are in accord with the dynamic biochemical properties of the RecAX53 protein, its ability to be more active in SSB displacement and PC formation (Fig. 6) but to form very unstable PCs (Fig. 4 and 5). In other words, RecAX53 is particularly robust in the initiation of recombination that leads to conversion but ineffective in promoting the more extensive recombination required for crossovers.
Although ATP binding rather than ATP hydrolysis is obligatory for initiation of the strand exchange reaction (16), the latter has been found to confer several important properties to the DNA strand exchange. ATP hydrolysis is required in particular for a lengthy exchange, unidirectional branch migration, bypassing structural barriers in DNA, and four-strand exchanges (10, 11, 34), as well as for overcoming some other limitations expected from a low level of presynaptic filament flexibility (32). However, much depends on how ATP hydrolysis is coupled to strand exchange.
In contrast to RecAEc and RecAPa, RecAX53 demonstrated a significant increase in its ATPase activity though some other structural and functional characteristics appeared to be similar (Table 3). In principle, the increased ATPase activity suggests an enhanced dynamic of RecA association with and dissociation from DNA (35) and enhanced dynamics in presynaptic filaments registered during RecAEc-mediated ATP hydrolysis (32) that is likely to be important for efficient strand exchange. Thus, though indirectly, the increased ATPase activity can improve the frequency of initiation of strand exchanges observed in experiments described here. However, in RecAX53, it also leads to a defect in the extended strand exchange reactions.
Further investigations are needed to check these conclusions and suggestions. However, one statement is clear now: hyperrecombination can occur via different pathways that directly depend on the combination of critical RecA functional and structural characteristics.
Acknowledgments
We are grateful to Leonid M. Firsov (PNPI) for fruitful discussion. Y.V.K. acknowledges with gratitude M. M. Cox's research group for providing assistance with the part of the work carried out in the United States.
This work was supported by Russian Foundation for Basic Research grant 08-04-00235 to V.A.L., Fogarty International Research Collaboration Award 2R03 TW00 1319-04 to M.M.C., and Russian Ministry of Education and Science grant RNP 2.2.1.1.4663. Work carried out in the Cox laboratory was further supported by National Institutes of Health grant GM32335.
Footnotes
Published ahead of print on 22 February 2008.
REFERENCES
- 1.Alatossava, T., H. Jutte, A. Kuhn, and E. Kellenberger. 1985. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J. Bacteriol. 162413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexseyev, A. A., D. M. Baitin, S. Kuramitsu, T. Ogawa, H. Ogawa, and V. A. Lanzov. 1997. A recombinational defect in the C-terminal domain of Escherichia coli RecA2278-5 protein is compensated by protein binding to ATP. Mol. Microbiol. 23255-265. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, D. A., and S. C. Kowalczykowski. 2000. Facilitated loading of RecA protein is essential to recombination by RecBCD enzyme. J. Biol. Chem. 27512261-12265. [DOI] [PubMed] [Google Scholar]
- 4.Baitin, D. M., E. N. Zaitsev, and V. A. Lanzov. 2003. Hyper-recombinogenic RecA protein from Pseudomonas aeruginosa with enhanced activity of its primary DNA binding site. J. Mol. Biol. 3281-7. [DOI] [PubMed] [Google Scholar]
- 5.Bakhlanova, I. V., T. Ogawa, and V. A. Lanzov. 2001. Recombinogenic activity of chimeric recA genes (Pseudomonas aeruginosa/Escherichia coli): a search for RecA protein regions responsible for this activity. Genetics 1597-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chervyakova, D., A. Kagansky, M. Petukhov, and V. Lanzov. 2001. [L29M] substitution in the interface of subunit-subunit interactions enhances Escherichia coli RecA protein properties important for its recombinogenic activity. J. Mol. Biol. 314923-935. [DOI] [PubMed] [Google Scholar]
- 7.Churchill, J. J., and S. C. Kowalczykowski. 2000. Identification of the RecA protein-loading domain of RecBCD enzyme. J. Mol. Biol. 297537-542. [DOI] [PubMed] [Google Scholar]
- 8.Clark, A. J. 1971. Toward a metabolic interpretation of genetic recombination of E. coli and its phages. Annu. Rev. Microbiol. 25437-464. [DOI] [PubMed] [Google Scholar]
- 9.Clark, A. J., and S. J. Sandler. 1994. Homologous genetic recombination: the pieces begin to fall into place. Crit. Rev. Microbiol. 20125-142. [DOI] [PubMed] [Google Scholar]
- 10.Cox, M. M. 1994. Why does RecA protein hydrolyse ATP? Trends Biochem. Sci. 19217-222. [DOI] [PubMed] [Google Scholar]
- 11.Cox, M. M. 2003. The bacterial RecA protein as a motor protein. Annu. Rev. Microbiol. 57551-577. [DOI] [PubMed] [Google Scholar]
- 12.Cromie, G. A., and D. R. Leach. 2000. Control of crossing over. Mol. Cell 6815-826. [DOI] [PubMed] [Google Scholar]
- 13.Haruta, N., X. Yu, S. Yang, E. H. Egelman, and M. M. Cox. 2003. A DNA pairing-enhanced conformation of bacterial RecA proteins. J. Biol. Chem. 27852710-52723. [DOI] [PubMed] [Google Scholar]
- 14.Jones, M., R. Wagner, and M. Radman. 1987. Mismatch repair and recombination in E. coli. Cell 50621-626. [DOI] [PubMed] [Google Scholar]
- 15.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25156-165. [DOI] [PubMed] [Google Scholar]
- 16.Kowalczykowski, S. C., and R. A. Krupp. 1995. DNA-strand exchange promoted by RecA protein in the absence of ATP: implications for the mechanism of energy transduction in protein-promoted nucleic acid transactions. Proc. Natl. Acad. Sci. USA 923478-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanka, E., and B. M. Wilkins. 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64141-169. [DOI] [PubMed] [Google Scholar]
- 19.Lanzov, V. A. 2002. Hyper-recombination in Escherichia coli with and without SOS response, p. 21-38. In M. Ruiz-Rubio, E. Alejandre-Duran, and T. Roldan-Arjona (ed.), Recent research development in DNA repair and mutagenesis. Research Signpost, Kerala, India.
- 20.Lanzov, V. A., I. V. Bakhlanova, and A. J. Clark. 2003. Conjugational hyperrecombination achieved by derepressing the LexA regulon, altering the properties of RecA protein and inactivating mismatch repair in Escherichia coli K-12. Genetics 1631243-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavery, P. E., and S. C. Kowalczykowski. 1990. Properties of RecA441 protein-catalyzed DNA strand exchange can be attributed to an enhanced ability to compete with SSB protein. J. Biol. Chem. 2654004-4010. [PubMed] [Google Scholar]
- 22.Lavery, P. E., and S. C. Kowalczykowski. 1992. Biochemical basis of the constitutive repressor cleavage activity of RecA730 protein: a comparison to recA441 and recA803 proteins. J. Biol. Chem. 26720648-20658. [PubMed] [Google Scholar]
- 23.Lohman, T. M., J. M. Green, and R. S. Beyer. 1986. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product: expression of the ssb gene under lambda PL control. Biochemistry 2521-25. [DOI] [PubMed] [Google Scholar]
- 24.Lohman, T. M., and L. B. Overman. 1985. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes: modulation by NaCl concentration. J. Biol. Chem. 2603594-3603. [PubMed] [Google Scholar]
- 25.Lusetti, S. L., and M. M. Cox. 2002. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 7171-100. [DOI] [PubMed] [Google Scholar]
- 26.Madiraju, M. V., P. E. Lavery, S. C. Kowalczykowski, and A. J. Clark. 1992. Enzymatic properties of the RecA803 protein, a partial suppressor of recF mutations. Biochemistry 3110529-10535. [DOI] [PubMed] [Google Scholar]
- 27.Menetski, J. P., and S. C. Kowalczykowski. 1985. Interaction of recA protein with single-stranded DNA: quantitative aspects of binding affinity modulation by nucleotide cofactors. J. Mol. Biol. 181281-295. [DOI] [PubMed] [Google Scholar]
- 28.Modrich, P. 1991. Mechanisms and biological effects of mismatch repair. Annu. Rev. Genet. 25229-253. [DOI] [PubMed] [Google Scholar]
- 29.Namsaraev, E. A., D. Baitin, I. V. Bakhlanova, A. A. Alexseyev, H. Ogawa, and V. A. Lanzov. 1998. Biochemical basis of hyper-recombinogenic activity of Pseudomonas aeruginosa RecA protein in Escherichia coli cells. Mol. Microbiol. 27727-738. [DOI] [PubMed] [Google Scholar]
- 30.Ogawa, T., A. Shinohara, H. Ogawa, and J. Tomizawa. 1992. Functional structures of the RecA protein found by chimera analysis. J. Mol. Biol. 226651-660. [DOI] [PubMed] [Google Scholar]
- 31.Pugh, B. F., and M. M. Cox. 1988. High salt activation of RecA protein ATPase in the absence of DNA. J. Biol. Chem. 26376-83. [PubMed] [Google Scholar]
- 32.Ramreddy, T., S. Sen, B. J. Rao, and G. Krishnamoorthy. 2003. DNA dynamics in RecA-DNA filaments: ATP hydrolysis-related flexibility in DNA. Biochemistry 4212085-12094. [DOI] [PubMed] [Google Scholar]
- 33.Shan, Q., J. M. Bork, B. L. Webb, R. B. Inman, and M. M. Cox. 1997. RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J. Mol. Biol. 265519-540. [DOI] [PubMed] [Google Scholar]
- 34.Shan, Q., and M. M. Cox. 1996. RecA protein dynamics in the interior of RecA nucleoprotein filaments. J. Mol. Biol. 257756-774. [DOI] [PubMed] [Google Scholar]
- 35.Shivashankar, G. V., M. Feingold, O. Krichevsky, and A. Libchaber. 1999. RecA polymerization on double-stranded DNA by using single-molecule manipulation: the role of ATP hydrolysis. Proc. Natl. Acad. Sci. USA 967916-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, G. R. 2001. Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu. Rev. Genet. 35243-274. [DOI] [PubMed] [Google Scholar]
- 37.West, S. C. 2003. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4435-445. [DOI] [PubMed] [Google Scholar]
- 38.Worth, L., Jr., S. Clark, M. Radman, and P. Modrich. 1994. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc. Natl. Acad. Sci. USA 913238-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]