Abstract
The surface of polyhydroxybutyrate (PHB) storage granules in bacteria is covered mainly by proteins referred to as phasins. The layer of phasins stabilizes the granules and prevents coalescence of separated granules in the cytoplasm and nonspecific binding of other proteins to the hydrophobic surfaces of the granules. Phasin PhaP1Reu is the major surface protein of PHB granules in Ralstonia eutropha H16 and occurs along with three homologues (PhaP2, PhaP3, and PhaP4) that have the capacity to bind to PHB granules but are present at minor levels. All four phasins lack a highly conserved domain but share homologous hydrophobic regions. To identify the region of PhaP1Reu which is responsible for the binding of the protein to the granules, N-terminal and C-terminal fusions of enhanced green fluorescent protein with PhaP1Reu or various regions of PhaP1Reu were generated by recombinant techniques. The fusions were localized in the cells of various recombinant strains by fluorescence microscopy, and their presence in different subcellular protein fractions was determined by immunodetection of blotted proteins. The fusions were also analyzed to determine their capacities to bind to isolated PHB granules in vitro. The results of these studies indicated that unlike the phasin of Rhodococcus ruber, there is no discrete binding motif; instead, several regions of PhaP1Reu contribute to the binding of this protein to the surface of the granules. The conclusions are supported by the results of a small-angle X-ray scattering analysis of purified PhaP1Reu, which revealed that PhaP1Reu is a planar, triangular protein that occurs as trimer. This study provides new insights into the structure of the PHB granule surface, and the results should also have an impact on potential biotechnological applications of phasin fusion proteins and PHB granules in nanobiotechnology.
Many biopolymers are important reserve compounds that are stored in the cytoplasm as insoluble inclusions (14). The best-studied storage compounds in bacteria are polyhydroxyalkanoates (PHAs), and poly(3-hydroxybutyrate) (PHB) is the most prominent example. In most bacteria accumulation of PHAs occurs if an excess amount of a carbon source is present and if growth is concomitantly limited by the lack of another essential nutrient, like nitrogen, phosphorus, oxygen, or something else (45, 49). Therefore, PHAs serve as storage compounds for energy and carbon under starvation conditions. PHA synthesis results in the formation of multiple cytoplasmic inclusions that are generally 200 to 500 nm in diameter in the final accumulation phase (1). The first investigations of PHB granules in bacteria were performed by Williamson and Wilkinson (57) and by Griebel et al. (8). Isolated PHB granules contain approximately 97.5% PHA, 2% protein, and 0.5% phospholipids (8), although some estimates of the lipid contents have been considerably higher (50).
Phasins are small amphiphilic proteins which are localized at the surface of PHB granules and are synthesized by PHB-accumulating bacterium under conditions permissive for PHB synthesis (41, 42). The gram-negative bacterium Ralstonia eutropha is used as a model organism to study all aspects of PHB metabolism and has four different phasin proteins (39, 36). PhaP1Reu is the major phasin, and the cells produce large amounts of this protein. Expression of PhaP1Reu is strictly regulated by PhaRReu at the transcription level (38). The phasins sensu stricto, which includes PhaP1Reu, stabilize the dispersion of the hydrophobic PHB granules in the cytoplasm on the one hand and prevent the nonspecific binding of other proteins to the surface of the PHB granules on the other hand. The cells of PhaP1Reu-negative mutants of R. eutropha have only a single large PHB granule, whereas the wild type has several medium-size granules (56). It was previously shown that lysozyme of Gallus gallus binds to PHB granules during isolation of the granules from cells of Allochromatium vinosum (23). It was also shown that the β-lactamase (24) and heat shock protein HspA (51) bind to PHB granules in recombinant strains of Escherichia coli accumulating PHB. Binding of the latter protein is suppressed in E. coli if the PHB biosynthesis enzymes are coexpressed together with PhaP1Reu. Besides PhaP1Reu, minor amounts of three additional phasin proteins (PhaP2Reu, PhaP3Reu, and PhaP4Reu) occur in R. eutropha. Due to the small amounts, it is unlikely that these minor phasins have the same functions as PhaP1Reu; however, their physiological or structural functions are not known yet.
Therefore, phasins sensu stricto are interesting structural proteins with unique properties and possibly structures. Since they may also be used for various biotechnological applications, it is necessary to understand the binding of the phasins to PHB granules. In the 14-kDa phasin of the gram-positive organism Rhodococcus ruber (34) two short stretches comprising about 8 hydrophobic amino acids were identified close to the C-terminal region. If one or both stretches were removed from this protein, the truncated phasin was no longer capable of binding to the granules and occurred in the cytoplasm (35). If, on the other hand, the short region of this phasin comprising the two hydrophobic stretches was fused with an acetaldehyde dehydrogenase of R. eutropha, the fusion protein bound in vivo as well as in vitro to PHB granules (35). This was the first demonstration that phasins can be used as anchors for binding of other proteins to the surface of PHB granules. This was later confirmed and also demonstrated for other fusion proteins with PhaP1Reu (3, 2) and also with the PHA granule-associated protein PhaFPpu of Pseudomonas putida (26) in other laboratories. Furthermore, it was recently shown that other proteins involved in PHA metabolism, like a fusion of the substrate-binding domain of a PHA depolymerase (22, 30) or of PHA synthases (32, 33), can be used to immobilize other proteins with PHA. Similarly, PhaP1Reu fused to other proteins also binds intracellularly to triacylglycerol inclusions in oleogenous actinomycetes (12) like eukaryotic lipid body proteins bind to these hydrophobic inclusions (13).
The aims of this study were to identify the region in PhaP1Reu of R. eutropha which mediates binding of this phasin to PHB granules and to obtain some information about the structure of PhaP1, thereby also contributing to our understanding of phasins.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are shown in Table 1.
TABLE 1.
Bacteria and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80 lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| E. coli S17-1 | thi proA hsdR17 hsdM+recA RP4-tra function | 47 |
| R. eutropha H16 | Wild type | DSM 428a |
| R. eutropha ΔphaP1 | phaP1 precise deletion gene replacement strain (Re1052), derived from R. eutropha H16 | 58 |
| Plasmids | ||
| pEGFP-N3 | Kmr Neoregfp, pMB1 ori | BD Biosciences |
| pBBR1MCS-2 | Broad-host-range, KmrlacPOZ mobRP4 | 19 |
| pJAM2::phaP1-egfp | pJAM2 harboring phaP1-egfp fusion | 12 |
| pBBR1MCS-2::phaP1Reu | pBBR1MCS-2 harboring PCR product comprising phaP1 | This study |
| pBBR1MCS-2::egfp | pBBR1MCS-2 harboring PCR product comprising egfp | This study |
| pBBR1MCS-2::phaP1Reu-egfp | pBBR1MCS-2 harboring PCR product comprising phaP1Reu-egfp | This study |
| pBBR1MCS-2::egfp-phaP1Reu | pBBR1MCS-2 harboring PCR product comprising egfp-phaP1Reu | This study |
| pBBR1MCS-2::phaP1Reu[52>C]-egfp | pBBR1MCS-2 harboring PCR product comprising phaP1Reu[52>C]-egfp | This study |
| pBBR1MCS-2::phaP1Reu[113>C]-egfp | pBBR1MCS-2 harboring PCR product comprising phaP1Reu[113>C]-egfp | This study |
| pBBR1MCS-2::phaP1Reu[ΔM]-egfp | pBBR1MCS-2 harboring PCR product comprising phaP1Reu[ΔM]-egfp | This study |
| pBBR1MCS-2::phaP1Reu[52-112]-egfp | pBBR1MCS-2 harboring PCR product comprising phaP1Reu[52-112]-egfp | This study |
| pBBR1MCS-2::phaP1Reu[1-85]-egfp | pBBR1MCS-2 harboring PCR product comprising phaP1Reu[1-85]-egfp | This study |
| pBBR1MCS-2::egfp-phaP1Reu[N>85] | pBBR1MCS-2 harboring PCR product comprising egfp-phaP1Reu[N>85] | This study |
| pBBR1MCS-2::egfp-phaP1Reu[N>112] | pBBR1MCS-2 harboring PCR product comprising egfp-phaP1Reu[N>112] | This study |
| pBBR1MCS-2::egfp-phaP1Reu[ΔM] | pBBR1MCS-2 harboring PCR product comprising egfp-phaP1Reu[ΔM] | This study |
| pBBR1MCS-2::egfp-phaP1Reu[52-112] | pBBR1MCS-2 harboring PCR product comprising egfp-phaP1Reu[52-112] | This study |
DSM, Deutsche Sammlung für Mikroorganismen und Zellkulturen.
Media and cultivation of cells.
Cells of R. eutropha were grown at 30°C in mineral salts medium supplemented with 1.0% (wt/vol) sodium gluconate (46). Solid media contained 1.5% (wt/vol) purified agar. Cells of E. coli were cultivated at 37°C in Luria-Bertani medium (44).
Isolation of PHB granules.
For isolation of PHB granules a modification of the method of Wieczorek et al. (56) was used. Cells of an R. eutropha ΔphaP1 mutant were grown in mineral salts medium under storage conditions. The cells were cultivated for 72 h, harvested by centrifugation (20 min, 6,000 × g, 4°C), washed and resuspended in Tris-HCl buffer (10 mM, pH 7.0), and disrupted by a three passages through a French press (100 × 106 Pa). The disrupted cells were layered on top of a discontinuous glycerol gradient consisting of 5 ml of (vol/vol) glycerol 90% and 5 ml of 60% (vol/vol) glycerol. After ultracentrifugation (60 min, 100,000 × g, 4°C) the PHB granules formed a layer between the two phases and could be separated. The isolated granules were washed three times with Tris-HCl buffer (10 mM, pH 7.0) and stored at −20°C.
Purification of PhaP1Reu.
PhaP1Reu was purified from PHB granules isolated from R. eutropha cells by detergent treatment as described previously (56).
Polyacrylamide gel electrophoresis and Western immunoblotting.
Protein samples were resuspended in gel loading buffer (0.6% [wt/vol] sodium dodecyl sulfate [SDS], 1.25% [wt/vol] β-mercaptoethanol, 0.25 mM EDTA, 10% [vol/vol] glycerol, 0.001% [wt/vol] bromophenol blue, 12.5 mM Tris-HCl; pH 6.8) and separated in 12.5% (wt/vol) SDS-polyacrylamide gels, as described by Laemmli (21). Proteins were stained with Coomassie brilliant blue R-250 (54). Immunological detection of enhanced green fluorescent protein (EGFP) fusion proteins blotted from the SDS-polyacrylamide gel onto polyvinylidene difluoride-nitrocellulose membranes was performed exactly as described by Towbin et al. (53), using polyclonal EGFP antibodies (BD Biosciences).
Molecular weight determination by gel filtration.
A Superdex 200 HP column (XK 26/60; GE Healthcare) was equilibrated with 50 ml of 100 mM potassium phosphate buffer (pH 7.0). Purified protein and calibration proteins (1.0 mg each) were applied to the column and eluted at a flow rate of 0.5 ml/min. Relative molecular masses were calculated from semilogarithmic plots of the molecular masses of calibration proteins versus elution volume.
Isolation, amplification, and manipulation of DNA.
Chromosomal DNA of R. eutropha H16 was isolated by the method of Marmur (25). Plasmid DNA was isolated by the method of Birnboim and Doly (5). DNA restriction fragments were purified with a Perfectprep gel cleanup kit, as described by the manufacturer (Eppendorf). Restriction enzymes, ligases, and other enzymes used for DNA manipulation were used according to the manufacturers' instructions.
All PCR amplifications were carried out as described by Sambrook et al. (44), employing Pfx DNA polymerase (Invitrogen) and an Omnigene HBTR3CM DNA thermal cycler (Hybaid). All oligonucleotides which were used as primers for PCR amplification or other purposes are shown in Table 2.
TABLE 2.
Oligonucleotides used for PCR amplification and other purposes in this study
| Oligonucleotide | Sequence | Location and orientation |
|---|---|---|
| PhaP1_N_XhoI | 5′-AAA CTC GAG AAG GAG GGA TCC ATG ATC CTC ACC CCG G-3′ | 5′ region of phaP1Reu |
| EGFP_C_HindIII | 5′-AAA AAG CTT TTA CTT GTA CAG CTC GTC CAT GCC-3′ | 3′ region of egfp |
| EGFP_C_EcoRI | 5′-G GCC GAA TTC CTT GTA CAG CTC GTC CAT GCC G-3′ | 5′ region of egfp |
| EGFP_N_MCS(SalI) | 5′-T TCT GCA GTC GAC GGT ACC GC-3′ | 3′ region of egfp |
| PhaP(Re)_C_HindIII | 5′-GC AGA AGC TTA TCA GGC AGC CGT CGT CTT C-3′ | 3′ region of phaP1Reu |
| PhaP(Re)_N_EcoRI | 5′-G AGA GAA TTC ATG ATC CTC ACC CCG GAA CAA G-3′ | 5′ region of phaP1Reu |
| PhaP-52C | 5′-A GAA CTC GAG GAG AAG GCC ATG AAG GCG CTG TCG GCC AAG-3′ | 5′ region of truncated phaP1Reu |
| PhaP-N85-rev | 5′-GA AAA GCT TTC TTA CAG GTG GCG GGT GTA GGC-3′ | 3′ region of truncated phaP1Reu |
| PhaP-113C | 5′-C GAA CTC GAG AAG AAC GTG CAA ATG CTG GTC GAG AAC CTC GCC-3′ | 5′ region of truncated phaP1Reu |
| PhaP-N112-rev | 5′-AC CAA AGC TTA CAC GTT CTT CGA GCC TTC GG-3′ | 3′ region of truncated phaP1Reu |
| P-EGFP-EcoRI-N | 5′-G AGG GAA TTC ATG GTG AGC AAG GGC GAG GAG C-3′ | 5′ region of egfp |
| PhaP(112)_EcoRI_C | 5′-A AAA GAA TTC CAC GTT CTT CGA GCC TTC GG-3′ | 3′ region of egfp |
| PhaP(52C)_EcoRI_N | 5′-A AAA GAA TTC AAG GCG CTG TCG GCC AAG-3′ | 3′ region of truncated phaP1Reu |
| PhaP1-85_C_EcoRI | 5′-C CGA GAA TTC GCC GCC GCC GTG GCG GGT GTA GGC CAG G-3′ | 3′ region of truncated phaP1Reu |
Construction of hybrid plasmids encoding fusions of the egfp gene with the phaP1 gene or parts of the phaP1 gene.
The phaP1Reu-egfp fusion fragment was amplified by PCR using plasmid pJAM2::phaP1-egfp (12) as the template and oligonucleotides PhaP1_N_XhoI and EGFP_C_HindIII (Table 2) as the primers. The resulting PCR product (1.3 kbp) was restricted with XhoI and HindIII and cloned into vector pBBR1MCS-2, which was treated with the same restriction endonucleases, yielding hybrid plasmid pBBR1MCS-2::phaP1Reu-egfp. The egfp-phaP1Reu fusion was obtained by amplification of both fragments. The egfp-fragment (0.8 kbp) was amplified from plasmid pEGFP-N3 by performing PCR using oligonucleotides EGFP_N_MCS(SalI) and EGFP_C_EcoRI. The phaP1Reu fragment (0.6 kbp) was amplified with oligonucleotides PhaP(Re)_N_EcoRI and PhaP(Re)_C_HindIII from genomic DNA of R. eutropha H16. The PCR products obtained were then digested with EcoRI and ligated. The ligation product was then amplified by PCR using oligonucleotides EGFP_N_MCS(SalI) and PhaP(Re)_C_HindIII. The resulting fusion product (1.4 kbp) was restricted with SalI and HindIII and cloned into the vector pBBR1MCS-2, yielding hybrid plasmid pBBR1MCS-2::egfp-phaP1Reu.
For control experiments the genes encoding EGFP and PhaP1Reu were cloned using the same vector, which yielded plasmids pBBR1MCS-2::egfp and pBBR1MCS-2::phaP1Reu. The phaP1Reu gene was amplified from genomic DNA of R. eutropha H16 using oligonucleotides PhaP1_N_XhoI and PhaP1(Re)_C_HindIII (Table 2). The resulting fragment (0.6 kbp) was digested with XhoI und HindIII and cloned into plasmid pBBR1MCS-2, which was restricted with the same enzymes. The egfp fragment was obtained by performing PCR with pEGFP-N3 as the template and oligonucleotides EGFP_N_MCS(SalI) and EGFP_C_HindIII. The PCR product (0.8 kbp) was restricted with SalI and HindIII and ligated to SalI- and HindIII-digested pBBR1MCS-2 DNA, yielding pBBR1MCS-2::egfp.
Truncated versions of PhaP1Reu fused to EGFP (PhaP1Reu[52>C]-EGFP, PhaP1Reu[113>C]-EGFP, EGFP-PhaP1Reu[N>112], EGFP-PhaP1Reu[N>85]) were prepared by performing PCR using different primer combinations (Table 2) and plasmids pBBR1MCS-2::egfp-phaP1Reu and pBBR1MCS-2::phaP1Reu-egfp PCR as the templates. Hybrid plasmids pBBR1MCS-2::phaP1Reu[ΔM]-egfp and pBBR1MCS-2::egfp-phaP1Reu[ΔM] were constructed by digestion of the PCR products (phaP1Reu-egfp and egfp-phaP1Reu) with MscI, which deleted the nucleotides coding for amino acids 65 to 131 in frame. Hybrid plasmids pBBR1MCS-2::phaP1Reu[1-85]-egfp, pBBR1MCS-2::phaP1Reu[52-112]-egfp, and pBBR1MCS-2::egfp-phaP1Reu[52-112] were constructed by PCR amplification employing the oligonucleotides shown in Table 2 for both fragments and the fusion product as described above for the egfp-phaP1Reu fusion.
DNA sequencing.
DNA sequencing was performed by using a SequiTherm EXCEL TM II long read cycle sequencing kit (Epicenter Technologies), IRD800-labeled oligonucleotides (MWG-Biotech), and a Li-Cor 4000L (Li-Cor Biosciences) automated sequencer (MWG-Biotech).
Transfer of DNA.
Competent cells of E. coli were prepared and transformed by using the CaCl2 procedure described by Hanahan (11). Transfer of DNA to R. eutropha strains by conjugation was performed by spot agar mating as described by Friedrich et al. (7) using E. coli S17-1 harboring the desired plasmid as the donor.
Theory of SAXS analyses.
The small-angle X-ray scattering technique (SAXS) is a powerful tool able to detect the shape and conformational and aggregational states of proteins in solution for a broad range of conditions and sizes. In a SAXS experiment, a protein solution is exposed to a focused X-ray beam, and the scattered intensity is collected as a function of the scattering angle. The theoretical background, basic equations, and methods used for SAXS data analysis have been fully described previously (10, 15, 16, 18, 28, 31, 37, 48, 54).
SAXS analyses.
SAXS experiments were performed using a conventional laboratory X-ray Kratky camera. The wavelength of the X-rays was 1.54 Å, and the sample-to-detector distance was 1.5 m. The scattering vector range was 0.022 to 0.2 Å−1. A PhaP1Reu sample at a concentration of 6 mg/ml in 10 mM Tris-HCl buffer (pH 7.0) was analyzed using 1.5-mm glass capillaries. The experimental intensity was corrected for background, buffer contribution, detector inhomogeneities, and sample transmission.
Secondary structure predictions for proteins.
For PhaP1Reu, phasin homologues, and the substrate-binding domain of the Pseudomonas stutzeri PHB depolymerase, predictions were made as described by Deleage et al. (6; http://npsa-pbil.ibcp.fr). For predictions of the secondary structure of PhaP1Reu the following algorithms were used: MLRC (9), DSC (17), and PHD (43).
Microscopic analysis of cells containing EGFP fusion proteins.
For microscopic localization of EGFP and of EGFP fusion proteins, an Olympus BX51 microscope (Olympus Europe) was used. The fluorescence of the EGFP used could be excited at a wavelength of 488 nm and detected at 507 nm (data for plasmid pEGFP-N3 according to the manufacturer [BD Biosciences]) using a U-MNIBA3 filter combination (excitation wavelength, 470 to 495 nm; emission wavelength, 510 to 550 nm).
RESULTS
Comparison of primary structures of phasins and secondary structure predictions.
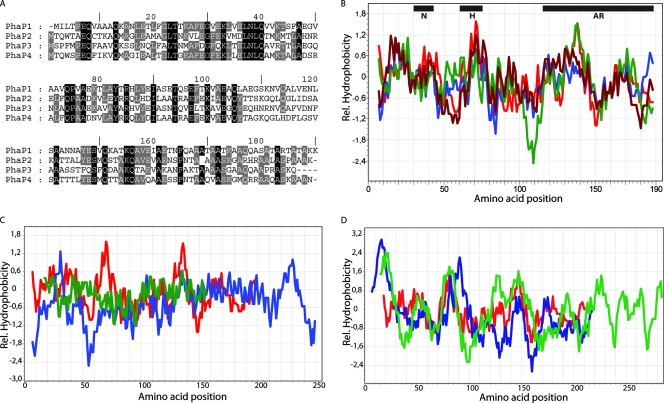
Sequence and hydrophobicity plot analysis of the four PhaP1Reu phasin homologues in R. eutropha H16 revealed a high degree of similarity and several highly conserved amino acids in PhaP1Reu, PhaP2Reu, PhaP3Reu, and PhaP4Reu (Fig. 1A). In contrast, comparison of the primary structures of R. eutropha PhaP1Reu, P. putida PhaFPpu, and R. ruber GA14Rru revealed only a few highly conserved amino acids in these proteins (data not shown). Therefore, in general, there is not a conserved sequence motif representing a “PHB granule-binding box” in phasins. Binding could be caused by short stretches of hydrophobic proteins, as such stretches were identified in GA14Rru of R. ruber (35). Hydrophobicity plots obtained for the four R. eutropha phasins (Fig. 1B), for PhaP1Reu, PhaFPpu from P. putida, and GA14Rru from R. ruber (Fig. 1C), and for PhaP1Reu, lysozyme from G. gallus, and β-lactamase from E. coli (Fig. 1D) did not reveal conserved hydrophobic regions like those that occur, for example, in membrane proteins; in addition, motifs representing minima and maxima occur in different regions of these proteins. PhaFPpu exhibits a hydrophilic minimum at amino acid positions 50 to 60, and the entire sequence is more hydrophilic than the sequences of PhaP1Reu and GA14Rru. GA14Rru is less hydrophobic than PhaP1Reu.
FIG. 1.
Primary structure and hydrophobicity of phasins and other proteins that bind to PHB granules. (A) Primary structures of the R. eutropha H16 phasins PhaP1Reu, PhaP2Reu, PhaP3Reu, and PhaP4Reu. The gray and black backgrounds indicate identities of the amino acids (black background, conserved in all four phasins; gray background, conserved in at least three of the four phasins). (B) Hydrophobicity plot (20) of the four R. eutropha H16 phasins. Red line, Phap1Reu; blue line, PhaP2Reu; green line, PhaP3Reu; brown line, PhaP4Reu. The regions of interest are an N-terminal conserved part (N), a hydrophobic patch (H), and the alanine-rich C terminus (AR). (C) Hydrophobicity plot (20) of the phasin homologues PhaFPpu of P. putida and GA14Rru of R. ruber compared to PhaP1Reu. Red line, PhaP1Reu; blue line, PhaFPpu; green line, GA14Rru. (D) Hydrophobicity plot (20) of the PHB-binding proteins lysozyme (G. gallus) and β-lactamase (E. coli) compared to PhaP1Reu. Red line, PhaP1Reu; blue line, lysozyme; green line, β-lactamase. Rel., relative.
The binding of phasins to PHB granules could be caused not only by a discrete motif in the primary structure but also by the secondary, tertiary, or quaternary structure of the protein. Therefore, we performed a secondary structure prediction analysis for various phasins as described by Deleage et al. (6; http://npsa-pbil.ibcp.fr) and in Materials and Methods. We obtained very similar predictions for the phasins sensu stricto, such as the four phasins occurring in R. eutropha H16, PhaP1 of Ralstonia solanacearum, and Ralstonia metallidurans. These phasins share a very high fraction (about 90%) of α-helical structure that could be a characteristic feature of the proteins. Interestingly, the putative PHB-binding domain in the C-terminal region of the regulator PhaRReu is also α-helical, in contrast to the N-terminal putative DNA-binding domain of this protein (the Pfam version 18.0 domains are PHB_acc_N [amino acids 10 to 73] and PHB_acc [amino acids 75 to 115]) (4; http://www.sanger.ac.uk/Software/Pfam). On the other hand, the substrate-binding domain of the extracellular PHB depolymerase from P. stutzeri does not have an analogous α-helical structure. The PHA-binding motif of this protein and that of the P. putida phasin PhaFPpu might be different, as shown by Ohura et al. (29) and Moldes et al. (26).
In vivo binding capacity of EGFP-PhaP1Reu fusion proteins in E. coli and R. eutropha.
Various fusions of phaP1Reu and egfp were constructed by PCR amplification of different DNA fragments, digestion, ligation, and amplification of the fusion products exactly as described in Materials and Methods.
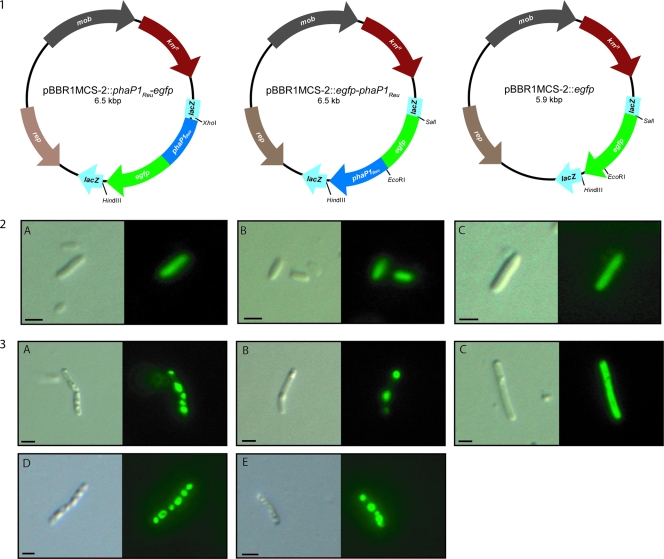
The capacities of the resulting PhaP1Reu-EGFP fusion proteins to bind to PHB granules in cells were then investigated by fluorescence microscopy. The expression of all proteins could be analyzed in the absence of PHB using cells of E. coli harboring the different plasmids (Fig. 2, panel 2). In such cells the fusion proteins showed a diffuse fluorescence that was caused by the homogeneous distribution of the fluorescent proteins inside the entire cytoplasm (Fig. 2, panels 2A and 2B), like cells harboring the EGFP control (Fig. 2, panel 2C). The binding capacities of the fusion proteins were visible in R. eutropha ΔphaP1 and H16 recombinant cells expressing the fusion protein PhaP1Reu-EGFP or EGFP-PhaP1Reu (Fig. 2, panels 3A to 3E). In these cells the fluorescence was restricted to the PHB granules.
FIG. 2.
Distribution of EGFP-PhaP1Reu and PhaP1Reu-EFGP fusion proteins in cells of recombinant strains of E. coli Top10 and an R. eutropha Δphap1 mutant. (Panel 1) Plasmids constructed for fluorescence microscopic studies encoding the EGFP-PhaP1Reu and PhaP1Reu-EGFP fusion proteins and the EGFP control. kmR, kanamycin resistance; lacZ, α-fragment of the β-galactosidase gene; mob, mobilization site; rep, origin of replication. (Panel 2) Fluorescence microscopy of E. coli Top10 expressing the following fusion proteins: (A) PhaP1Reu-EGFP, (B) EGFP-PhaP1Reu, and (C) the EGFP control. (Panel 3) Fluorescence microscopy of R. eutropha Δphap1 mutant (A to C) and R. eutropha H16 (D to E) expressing the following fusion proteins: (A) PhaP1Reu-EGFP, (B) EGFP-PhaP1Reu, (C) EGFP control, (D) PhaP1Reu-EGFP, and (E) EGFP-PhaP1Reu. Each pair of images consists of a conventional microscopic image (left) and the corresponding fluorescent image (right). The corresponding plasmids are indicated above the images in panel 1. Bars, 1 μm.
In vivo binding capacities of truncated PhaP1Reu-EGFP fusion proteins in E. coli and R. eutropha.
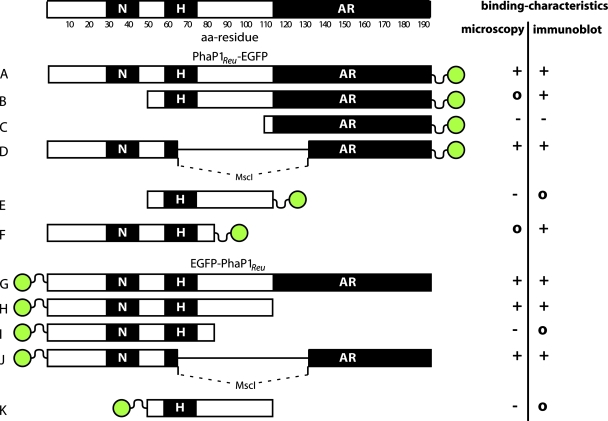
Various fusions of EGFP to truncated PhaP1Reu proteins lacking either parts of the N-terminal region (PhaP1Reu[52>C]-EGFP and PhaP1Reu[113>C]-EGFP), parts of the C-terminal region (PhaP1Reu[1-85]-EGFP, EGFP-PhaP1Reu[N>112], and EGFP-PhaP1Reu[N>85]), parts of the N-terminal region plus parts of the C-terminal region (PhaP1Reu[52-112]-EGFP and EGFP-PhaP1Reu[52-112]), or parts of the central region (PhaP1Reu[ΔM]-EGFP and EGFP-PhaP1Reu[ΔM]) of the phasin were constructed by PCR as described in Materials and Methods. A schematic overview of all fusion proteins generated in this study is shown in Fig. 3. The binding of these fusion proteins to PHB granules was then studied by fluorescence microscopy and compared to the binding of PhaP1Reu-EGFP and EGFP-PhaP1Reu to PHB granules. We were especially interested to see whether the binding was mediated by the N-terminal conserved part, the hydrophobic patch, or the alanine-rich C terminus of PhaP1Reu (Fig. 1) and whether one of these regions is responsible for the binding of the whole protein to the PHB granules.
FIG. 3.
Summary of EGFP fusions with various regions of PhaP1Reu. The position of the EGFP is indicated by a green circle, and the corresponding region of PhaP1Reu used to create the fusion is shown in a schematic diagram. Regions of interest are described in the legend to Fig. 1. Binding characteristics were determined by the microscopic studies and immunoblotting (+, binding; −, no binding; o, unclear). (A) PhaP1Reu-EGFP; (B) PhaP1Reu[52>C]-EGFP; (C) PhaP1Reu[113>C]-EGFP; (D) PhaP1Reu[ΔM]-EGFP; (E) PhaP1Reu[52-112]-EGFP; (F) PhaP1Reu[1-85]-EGFP; (G) EGFP-PhaP1Reu; (H) EGFP-PhaP1Reu[N>112]; (I) EGFP-PhaP1Reu[N>85]; (J) EGFP-PhaP1Reu[ΔM]; (K) EGFP-PhaP1Reu[52-112]. aa, amino acid.
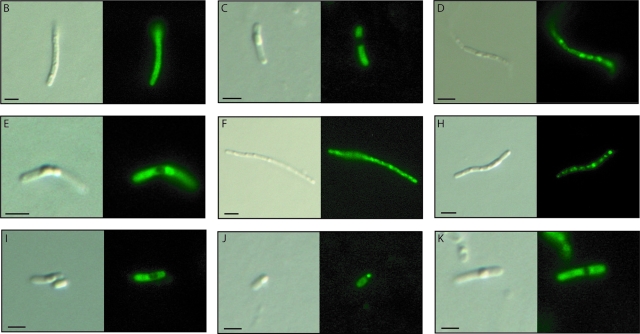
The results of the microscopic analyses of the capacities of truncated fusion proteins to bind to PHB granules are shown in Fig. 4. For the PhaP1Reu[ΔM]-EGFP, EGFP-PhaP1Reu[N>112], and EGFP-PhaP1Reu[ΔM] fusions the fluorescence was restricted to the region containing PHB granules in the cells, thus indicating that the PHB affinities of the PhaP1Reu regions of these fusions were high. In contrast, the PhaP1Reu[113>C]-EGFP, PhaP1Reu-[52-112]-EGFP, EGFP-PhaP1Reu[N>85], and EGFP-PhaP1Reu[52-112] fusions showed only diffuse fluorescence, indicating that the regions of PhaP1Reu could obviously not mediate binding of the fusion protein to PHB granules. The other two fusion proteins, PhaP1Reu[52>C]-EGFP and PhaP1Reu[1-85]-EGFP, gave no clear images.
FIG. 4.
Distribution of EGFP fusion proteins with various regions of PhaP1Reu in cells of recombinant strains of the R. eutropha H16 ΔphaP1 mutant. Each pair of images consists of a conventional microscopic image (left) and the corresponding fluorescent image (right). The letters correspond to the letters shown on the left in Fig. 3. Bars, 1 μm.
In summary, the binding of the whole PhaP1Reu protein to PHB granules appears to be greater than the binding of any truncated PhaP1Reu, because the fluorescence of the fusions with complete PhaP1 was completely restricted to the granules, whereas truncated fusions always showed higher levels of cytoplasmic fluorescence.
To check the capacity of fusion proteins to bind to PHB granules and to verify the microscopic images for the other proteins, polyacrylamide gel electrophoresis and Western immunoblotting of crude cell extracts and isolated granules were performed (not shown). In the immunoblots of proteins of the crude cell extracts all expressed fusion proteins should have been detectable. In contrast, only PHB-bound fusion proteins should have been present in the immunoblots prepared for isolated granules. The EGFP-PhaP1Reu[N>85], EGFP-PhaP1Reu[52-112], and PhaP1Reu[52-112]-EGFP fusion proteins could not be detected either in crude extracts or in granule fractions. The PhaP1Reu[113>C]-EGFP fusion was detected only in the crude extracts and not in the isolated granules. The PhaP1Reu[52>C]-EGFP, PhaP1Reu[ΔM]-EGFP, PhaP1Reu[1-85]-EGFP, EGFP-PhaP1Reu[N>112], and EGFP-PhaP1Reu[ΔM] fusion proteins were detected in the crude extracts, as well as in the isolated PHB granules. The results of these experiments are summarized in Fig. 3, which provides an overview of the results obtained with all fusion proteins in this study.
SAXS analysis of purified PhaP1Reu.
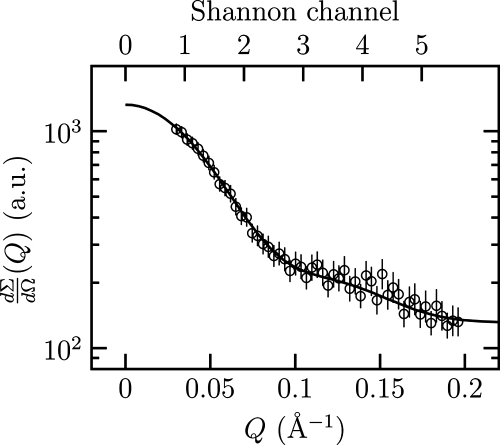
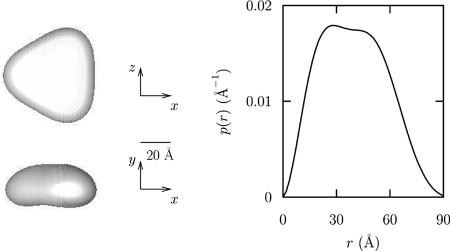
To obtain the first information concerning the quaternary and three-dimensional structure of PhaP1Reu, the protein was purified from a recombinant strain of E. coli and subjected to SAXS analysis. Experimental SAXS data for PhaP1 are shown in Fig. 5 as a semilogarithmic plot. To assess the particle shape, the SAXS curve was analyzed using the multipole expansion method described in Materials and Methods. The maximum rank was fixed at 4, and the width of the hydration layer at the particle border was fixed at 2 Å. Considering the quality of the data, the final fitting curve, also shown in Fig. 5, seems particularly good. The recovered fitting parameters of the multipole expansion analysis for PhaP1 for an M value of 5 (al,m) are as follows: Re a0,0, 10.7 ± 0.1; Re a1,0, −0.3 ± 0.1; Re a2,0, −1.05 ± 0.03; Re a3,0, 0.3 ± 0.1; and Re a3,3, 0.10 ± 0.05. It should be noted that the point group symmetry obtained is C3, with a threefold symmetry axis. The corresponding blunt-end triangular shape function is shown in Fig. 6, which suggests a trimeric aggregation number. The side length of the triangles and the height were calculated to be 80 ± 3 and 26 ± 1 Å, respectively. Further evidence for the hypothesis that PhaP1Reu occurs as a homotrimer was obtained by gel filtration of the purified PhaP1, which revealed a molecular mass of 60 ± 5 kDa for purified PhaP1Reu (data not shown). Further confirmation was obtained from the protein volume, 1.07 × 105 ± 0.05 × 105 Å3; by assuming that the standard specific volume of proteins is as great as 0.77 cm3/g, calculations of the molecular mass gave a value of 74 ± 1 kDa, which is about three times the known molecular mass (24 kDa) of monomeric PhaP1Reu. The distance distribution function is also shown in Fig. 6. It was observed that the maximum distance corresponds to a D value of 86 ± 3 Å, a value that, for the effect of the width of the hydration layer, is slightly greater than the side length of the triangle. Finally, we noticed that the number of Shannon channels, which are shown in Fig. 5, was greater than an M value of 5, confirming that the present analysis did not introduce an arbitrary high number of parameters.
FIG. 5.
Semilogarithmic SAXS profile of PhaP1. The line indicates the best fit obtained with the multipole expansion method. Q, scattering vector; a.u., arbitrary units.
FIG. 6.
Structure proposed for PhaP1. (Left panel) Shape reconstruction of PhaP1Reu. (Right panel) Distance distribution function [p(r)] of PhaP1Reu calculated from the reconstructed shape.
DISCUSSION
The secondary structure predictions for the phasins sensu stricto showed comparable high proportions (about 90%) of α-helical structure. This structure could be a general characteristic of phasins. In this case the whole protein is a binding protein and probably has no additional function. The binding capacity is obviously not determined by a short motif conserved at the level of the primary structure but is probably due to the secondary structure and may also be due to the tertiary or quaternary structure of the protein.
The microscopic analyses and Western immunoblotting did not produce clear results. Both analyses indicated that there was binding of the PhaP1Reu[ΔM]-EGFP, EGFP-PhaP1Reu[N>112], and EGFP-PhaP1Reu[ΔM] fusions. The PhaP1Reu[113>C]-EGFP fusion did not bind to PHB granules in either experiment. The other fusions, such as PhaP1Reu-[52-112]-EGFP, PhaP1Reu[52>C]-EGFP, PhaP1Reu[1-85]-EGFP, EGFP-PhaP1Reu[N>85], and EGFP-PhaP1Reu[52-112], gave no clear images or immunoblot results. Because of these binding characteristics, no region of PhaP1 that is obviously responsible for the binding of the fusions could be identified. None of the interesting regions, such as the N-terminal conserved part, the hydrophobic patch, or the alanine-rich C terminus, seem to represent PHB-binding domains, because all of the parts could be found in PHB-binding fusion proteins as well as in nonbinding fusion proteins (Fig. 3).
If the function of phasins is solely to stabilize the granules (i.e., dispersion of the hydrophobic PHB in the cytoplasm) and to prevent the coalescence of individual granules, like the coalescence that occurs in a phaP1Reu mutant (56, 40), it is likely that a certain structure of the whole protein is necessary and that no part can be eliminated without a loss of binding capacity. A high α-helix content throughout the whole protein could be a hint for similar structure and thus function of the complete protein. We found that the binding of the whole PhaP1Reu protein to PHB granules was better than the binding of any truncated PhaP1Reu protein. The SAXS analyses showed that PhaP1Reu does not bind to PHB as a monomer but binds as a homotrimer with a triangular and planar structure. This structure seems to be optimal to tightly cover the entire granule. This correlates very well with the results of Banki et al. (2), who showed that the stability of a phasin trimer bound to PHB granules was better than that of a PhaP1Reu monomer.
Very recently, the Aeromonas hydrophila phasin PhaPAhy (13 kDa) was crystallized, and its structure was investigated by using X-ray analysis (59). The authors found a tetrameric structure for this phasin, which is not homologous to PhaP1Reu and therefore not a phasin sensu stricto. A tetramer of PhaPAhy having a molecular mass of about 52 kDa is almost the same size as the PhaP1Reu trimer (60 kDa). A molecular mass of a PHB-binding unit of about 50 to 60 kDa is probably optimal to provide a layer at the surface of PHB granules.
From the revealed structure and size of the phasin protein, it was calculated that about 6,130 PhaP1Reu homotrimers or 18,390 individual PhaP1Reu molecules are necessary to completely cover the entire surface of a PHB granule with a diameter of 250 nm. Assuming that a single cell possesses about 20 such PHB granules in the late accumulation phase and that the average protein content of a single cell is about 0.155 pg (27), it can be calculated that PhaP1Reu accounts for about 7.8% of the total cellular protein. This is in good agreement with some previous estimates (50) and observations (56), although in other studies workers concluded that the phasin covers only 27 to 54% of the granule surface (52).
Surface coating of nanoparticles is an important part of nanoparticle synthesis. PHB granules are of interest especially for medical applications, as they are biodegradable and nontoxic. PHB forms water-insoluble intracellular inclusions (granules) that are accumulated as carbon and energy storage compounds in a large variety of prokaryotes. For optimized generation of functionalized nanoparticles based on phasin fusion proteins, knowledge about the binding of phasins to the granules should be helpful. It seems to be impossible to recognize a specific binding motif of phasins by in silico analyses. All four phasins of R. eutropha lack a highly conserved domain, but they have homologous hydrophobic regions. In addition, phasins occurring in other PHB-accumulating bacteria, like Azotobacter vinelandii, also do not have highly conserved amino acid residues.
Acknowledgments
This study was supported in part by a grant provided by the Deutsche Forschungsgemeinschaft (grant Ste 386/6-4) and in part by the European Commission (contract 509012, NEST).
Footnotes
Published ahead of print on 25 January 2008.
REFERENCES
- 1.Anderson, A. J., and E. A. Dawes. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54450-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banki, M. R., T. U. Gerngross, and D. W. Wood. 2005. Novel and economical purification of recombinant proteins: intein-mediated protein purification using in vivo polyhydroxybutyrate (PHB) matrix association. Protein Sci. 141387-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnard, G. C., J. D. McCool, D. W. Wood, and T. U. Gerngross. 2005. Integrated recombinant protein expression and purification platform based on Ralstonia eutropha. Appl. Environ. Microbiol. 715735-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 71513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deleage, G., C. Blanchet, and C. Geourjon. 1997. Protein structure prediction. Implications for the biologist. Biochimie 79681-686. [DOI] [PubMed] [Google Scholar]
- 7.Friedrich, B., C. Hogrefe, and H. G. Schlegel. 1981. Naturally occurring genetic transfer of hydrogen-oxidizing ability between strains of Alcaligenes eutrophus. J. Bacteriol. 147198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griebel, R., Z. Smith, and J. M. Merrick. 1968. Metabolism of poly-β-hydroxybutyrate. I. Purification composition and properties of native poly-β-hydroxybutyrate granules from Bacillus megaterium. Biochemistry 73676-3681. [DOI] [PubMed] [Google Scholar]
- 9.Guermeur, Y., C. Geourjon, P. Gallinari, and G. Deleage. 1999. Improved performance in protein secondary structure prediction by inhomogeneous score combination. Bioinformatics 15413-421. [DOI] [PubMed] [Google Scholar]
- 10.Guinier, A., and G. Fournet. 1955. Small angle scattering of X-ray. Wiley, New York, NY.
- 11.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 12.Hänisch, J., M. Wältermann, H. Robenek, and A. Steinbüchel. 2006. The Ralstonia eutropha H16 phasin PhaP1 is targeted to intracellular triacylglycerol inclusions in Rhodococcus opacus PD630 and Mycobacterium smegmatis mc2155 and provides an anchor to target other proteins. Microbiology 1523271-3280. [DOI] [PubMed] [Google Scholar]
- 13.Hänisch, J., M. Wältermann, H. Robenek, and A. Steinbüchel. 2006. Eukaryotic lipid body proteins in oleogenous actinomycetes and their targeting to intracellular triacylglycerol inclusions: impacts on model of lipid body biogenesis. Appl. Environ. Microbiol. 726743-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoppert, M., and F. Mayer. 1999. Principles of macromolecular organization and cell function in bacteria and archaea. Cell. Biochem. Biophysiol. 31247-284. [DOI] [PubMed] [Google Scholar]
- 15.Kataoka, M., Y. Hagihara, K. Mihara, and Y. Goto. 1993. Molten globule of cytochrome c studied by the small angle X-ray scattering. J. Mol. Biol. 229591-596. [DOI] [PubMed] [Google Scholar]
- 16.Kataoka, M., I. Nishii, T. Fujisawa, T. Ueki, F. Tokunaga, and Y. Goto. 1995. Structural characterization of molten globule and native states of apomyoglobin by solution X-ray scattering. J. Mol. Biol. 249215-228. [DOI] [PubMed] [Google Scholar]
- 17.King, R. D., and M. J. E. Sternberg. 1996. Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci. 52298-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koch, M., P. Vachette, and D. Svergun. 2003. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q. Rev. Biophys. 36147-227. [DOI] [PubMed] [Google Scholar]
- 19.Kovach, M. E, P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166175-176. [DOI] [PubMed] [Google Scholar]
- 20.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157105-132. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lee, S. J., J. P. Park, T. J. Park, S. Y. Lee, S. Lee, and J. K. Park. 2005. Selective immobilization of fusion proteins on poly(hydroxyalkanoate) microbeads. Anal. Chem. 775755-5759. [DOI] [PubMed] [Google Scholar]
- 23.Liebergesell, M., B. Schmidt, and A. Steinbüchel. 1992. Isolation and identification of granule-associated proteins relevant for poly(3-hydroxybutyric acid) biosynthesis in Chromatium vinosum D. FEMS Microbiol. Lett. 99227-232. [DOI] [PubMed] [Google Scholar]
- 24.Maehara, A., S. Ueda, H. Nakano, and T. Yamane. 1999. Analyses of a polyhydroxyalkanoic acid granule-associated 16 kilodalton protein and its putative regulator in the pha locus of Paracoccus denitrificans. J. Bacteriol. 1812914-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acids from microorganisms. J. Mol. Biol. 3208-218. [Google Scholar]
- 26.Moldes, C., P. Garcia, J. L. Garcia, and M. A. Prieto. 2004. In vivo immobilization of fusion proteins on bioplastics by the novel tag BioF. Appl. Environ. Microbiol. 703205-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neidhardt, C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates Inc., Sunderland, MA.
- 28.Occhipinti, E., P. Martelli, F. Spinozzi, F. Corsi, C. Formantici, L. Molteni, H. Amenitsch, P. Mariani, P. Tortora, and R. Casadio. 2003. 3D structure of Sulfolobus solfataricus carboxypeptidase developed by molecular modeling is confirmed by site-directed mutagenesis and small-angle X-ray scattering. Biophys. J. 851165-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohura, T., K. I. Kasuya, and Y. Doi. 1999. Cloning and characterization of the polyhydroxybutyrate depolymerase gene of Pseudomonas stutzeri and analysis of the function of substrate-binding domains. Appl. Environ. Microbiol. 65189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park, T. J., J. P. Park, S. J. Lee, H. J. Hong, and S. Y. Lee. 2006. Polyhydroxyalkanoate chip for the specific immobilization of recombinant proteins and its applications in immunodiagnostics. Biotechnol. Bioproc. Eng. 11173-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez, J., P. Vachette, D. Russo, M. Desmadril, and D. Durand. 2001. Heat-induced unfolding of neocarzinostatin, a small all-β protein investigated by small-angle X-ray scattering. J. Mol. Biol. 308721-743. [DOI] [PubMed] [Google Scholar]
- 32.Peters, V., and B. H. A. Rehm. 2005. In vivo monitoring of PHA granule formation using GFP-labeled PHA synthase. FEMS Microbiol. Lett. 24893-100. [DOI] [PubMed] [Google Scholar]
- 33.Peters, V., and B. H. A. Rehm. 2006. In vivo enzyme immobilization by use of engineered polyhydroxyalkanoate synthase. Appl. Environ. Microbiol. 721777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pieper-Fürst, U., M. H. Madkour, F. Mayer, and A. Steinbüchel. 1994. Purification and characterization of a 14-kDa protein that is bound to the surface of polyhydroxyalkanoic acid granules in Rhodococcus ruber. J. Bacteriol. 1764328-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pieper-Fürst, U., M. H. Madkour, F. Mayer, and A. Steinbüchel. 1995. Identification of the region of a 14-kDa protein of Rhodococcus ruber that is responsible for the binding of this phasin to polyhydroxyalkanoic acid granules. J. Bacteriol. 1772513-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pohlmann, A., W. F. Fricke, F. Reinecke, B. Kusian, H. Liesegang, R. Cramm, T. Eitinger, C. Ewering, M. Pötter, E. Schwartz, A. Strittmatter, I. Voβ, G. Gottschalk, A. Steinbüchel, B. Friedrich, and B. Bowien. 2006. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat. Biotechnol. 241257-1262. [DOI] [PubMed] [Google Scholar]
- 37.Pollack, L., M. W. Tate, N. C. Darnton, J. B. Knight, S. M. Gruner, W. A. Eaton, and R. H. Austin. 1999. Compactness of the denatured state of a fast-folding protein measured by submillisecond small angle X-ray scattering. Proc. Natl. Acad. Sci. USA 9610115-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pötter, M., M. H. Madkour, F. Mayer, and A. Steinbüchel. 2002. Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology 1482413-2426. [DOI] [PubMed] [Google Scholar]
- 39.Pötter, M., H. Müller, F. Reinecke, R. Wieczorek, F. Fricke, B. Bowien, B. Friedrich, and A. Steinbüchel. 2004. The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. Microbiology 1502301-2311. [DOI] [PubMed] [Google Scholar]
- 40.Pötter, M., H. Müller, and A. Steinbüchel. 2005. Influence of homologous phasins (PhaP) on PHA accumulation and regulation of their expression by the transcription repressor PhaR in Ralstonia eutropha H16. Microbiology 151825-833. [DOI] [PubMed] [Google Scholar]
- 41.Pötter, M., and A. Steinbüchel. 2005. Poly(3-hydroxybutyrate) granule-associated proteins: impacts on poly(3-hydroxybutyrate) synthesis and degradation. Biomacromolecules 6552-560. [DOI] [PubMed] [Google Scholar]
- 42.Pötter, M., and A. Steinbüchel. 2006. Biogenesis and structure of polyhydroxyalkanoate granules, p. 109-136. In J. M. Shively (ed.), Inclusions in prokaryotes. Microbiology monographs, vol. 1. Springer, Heidelberg, Germany. [Google Scholar]
- 43.Rost, B., and C. Sander. 1993. Prediction of protein secondary structure at better than 70 percent accuracy. J. Mol. Biol. 232584-599. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 45.Schlegel, H. G., G. Gottschalk, and V. Bartha. 1961. Formation and utilization of poly-β-hydroxybutyric acid by knallgas bacteria (Hydrogenomonas). Nature 29463-465. [DOI] [PubMed] [Google Scholar]
- 46.Schlegel, H. G., H. Kaltwasser, and G. Gottschalk. 1961. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch. Mikrobiol. 38209-222.13747777 [Google Scholar]
- 47.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1784-791. [Google Scholar]
- 48.Spinozzi, F., F. Carsughi, and P. Mariani. 1998. Particle shape reconstruction by small-angle scattering: integration of group theory and maximum entropy to multipole expansion method. J. Chem. Phys. 10910148-10158. [Google Scholar]
- 49.Steinbüchel, A., and H. G. Schlegel. 1989. Excretion of pyruvate by mutants of Alcaligenes eutrophus, which are impaired in the accumulation of poly(β-hydroxybutyric acid) (PHB), under conditions permissive for synthesis of PHB. Appl. Microbiol. Biotechnol. 31168-175. [Google Scholar]
- 50.Steinbüchel, A., K. Aerts, W. Babel, C. Föllner, M. Liebergesell, M. H. Madkour, F. Mayer, U. Pieper-Fürst, A. Pries, H. E. Valentin, and R. Wieczorek. 1995. Considerations on the structure and biochemistry of bacterial polyhydroxyalkanoic acid inclusions. Can. J. Microbiol. 41(Suppl. 1)94-105. [DOI] [PubMed] [Google Scholar]
- 51.Tessmer, N., S. König, R. Reichelt, M. Pötter, and A. Steinbüchel. 2007. Heat shock protein HspA and other proteins mimicking the function of phasins sensu stricto in recombinant strains of Escherichia coli accumulating polythioesters or polyhydroxyalkanoates. Microbiology 153366-374. [DOI] [PubMed] [Google Scholar]
- 52.Tian, J., A. He, A. G. Lawrence, P. Liu, N. Watson, A. J. Sinskey, and J. Stubbe. 2005. Analysis of transient polyhydroxybutyrate production in Wautersia eutropha H16 by quantitative Western analysis and transmission electron microscopy. J. Bacteriol. 1873825-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 764350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trewhella, J. 1997. Insights into biomolecular function from small-angle scattering. Curr. Opin. Struct. Biol. 7702-708. [DOI] [PubMed] [Google Scholar]
- 55.Reference deleted.
- 56.Wieczorek, R., A. Pries, A. Steinbüchel, and F. Mayer. 1995. Analysis of a 24-kDa protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J. Bacteriol. 1772425-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williamson, D. H., and J. F. Wilkinson. 1958. The isolation and estimation of the poly-β-hydroxybutyrate inclusions of Bacillus species. J. Gen. Microbiol. 19198-209. [DOI] [PubMed] [Google Scholar]
- 58.York, G. M., J. Stubbe, and A. J. Sinskey. 2002. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J. Bacteriol. 18459-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao, M., Z. Li, W. Zheng, Z. Lou, and G.-Q. Chen. 2006. Crystallization and initial X-ray analysis of polyhydroxyalkanoate granule-associated protein from Aeromonas hydrophila. Acta Crystallogr. Sect. F 62814-819. [DOI] [PMC free article] [PubMed] [Google Scholar]