Summary
Intrinsic transcription termination by bacterial RNA polymerase (RNAP) occurs at sequences coding for a G:C-rich RNA hairpin followed by a 7–9-nt U-rich tract. We used single-molecule optical trapping techniques to investigate the mechanism by which elements from three representative terminators (his, t500, tR2) destabilize the elongation complex (EC). For his and tR2 terminators, loads exerted to bias DNA translocation did not affect termination efficiency (TE). However, the force-dependent kinetics of release at the t500 terminator and the force-dependent TE of a mutant imply a forward translocation mechanism for this terminator. Tension applied to isolated U-tracts induced transcript release in a manner consistent with shearing of the RNA:DNA hybrid. We deduce that different mechanisms of EC dissociation, involving hypertranslocation or RNA:DNA shearing, operate at terminators with different U-tract sequences. Tension applied to RNA at full terminators suggests that closure of the final 2-3 hairpin bases supplies the energy that destabilizes the hybrid, and that competing nascent RNA structures modulate TE by slowing terminator hairpin formation. We propose a quantitative model based on energetics that successfully predicts the behavior for all three terminators and several mutant variants.
Introduction
In carrying out transcription, RNA polymerase must satisfy opposing constraints. On one hand, the enzyme must form a highly stable EC that can move processively along DNA over distances of kilobases and more. On the other hand, the EC must become labile enough to dissociate with high efficiency upon receipt of a signal to terminate. In prokaryotes, about 50% of transcripts are terminated by specific DNA sequences—intrinsic terminators—that encode for RNA consisting of a G:C-rich hairpin followed by a 7–9-nt U-rich tract (Lesnik et al., 2001; Nudler and Gottesman, 2002). Terminators may be situated at the ends of coding regions, to halt transcription and prevent read-through into adjacent genes, or they may lie in regulatory regions upstream of a gene, where the efficiency of termination, which affects the transcription of downstream sequences, is linked to metabolic signals (Merino and Yanofsky, 2005; Platt, 1981; Yanofsky, 1981). Although the regulation of termination is vital to cellular function and development, fundamental questions remain regarding the molecular mechanism by which terminator elements, specifically the RNA hairpin and U-rich tract, cause RNAP to dissociate from the DNA template and release the nascent transcript.
Three models have been proposed to explain the role of the hairpin in the termination process. The forward translocation model proposes that the formation of the hairpin drives RNAP forward along the DNA template without concomitant transcript elongation, placing the polymerase in a ‘hypertranslocated’ state, shortening the RNA:DNA hybrid by up to several base pairs, and thereby destabilizing the EC (Santangelo and Roberts, 2004; Yarnell and Roberts, 1999). The RNA pullout or shearing model proposes that the hairpin extracts the nascent RNA by shearing the RNA:DNA hybrid without a need for forward translocation (Macdonald et al., 1993; Toulokhonov and Landick, 2003). The allosteric model proposes that the terminator hairpin induces a structural alteration in the RNAP active-site cleft that facilitates melting of the RNA:DNA hybrid again without requiring RNAP translocation (Gusarov and Nudler, 1999; Komissarova et al., 2002; Platt, 1981; Toulokhonov et al., 2001).
The role played by the U-rich sequence adjacent to the hairpin has also been of interest. Both bulk and single-molecule transcription assays suggest that a U-rich RNA:DNA hybrid may induce a transcriptional pause immediately downstream of the hairpin-encoding sequence (Gusarov and Nudler, 1999; Shundrovsky et al., 2004; Yarnell and Roberts, 1999), causing the U-tract to function as a kinetic trap that prevents RNAP from moving beyond the termination site before the hairpin forms and destabilizes the TEC. A further function for the U-tract was suggested by data from thermal denaturation studies of rU:dA duplexes in solution, which indicate that a U-rich RNA:DNA hybrid is significantly less stable than structurally similar rA:dT duplexes (Martin and Tinoco, 1980). This property may facilitate transcript release by lowering the stability of the RNA:DNA hybrid inside RNAP, in effect acting as a ‘slippery’ sequence. However, structural investigations and a recent single-molecule study suggest that the RNA:DNA hybrid is stabilized by extensive protein contacts within the RNAP active-site cleft (Dalal et al., 2006; Gnatt et al., 2001; Korzheva et al., 2000; Korzheva et al., 1998; Vassylyev et al., 2007a; Vassylyev et al., 2007b). The energy conferred by interactions between the hybrid and the protein may therefore offset any destabilizing effects of a weak rU:dA hybrid. Only a direct determination of hybrid stability in the context of RNAP can unambiguously determine the contribution of a U-rich tract towards destabilization of the TEC.
To investigate the mechanism by which the hairpin and U-rich sequence combine to produce termination, as well as to differentiate among candidate models, we employed a high-resolution optical trapping assay that allows force to be exerted either on the DNA template or on the RNA transcript relative to RNAP (Abbondanzieri et al., 2005; Dalal et al., 2006; Neuman et al., 2003), and studied three representative terminator sequences that display a range of TEs and U-tract interruptions (his, t500, and tR2; Figure 1A). To test the forward translocation model, we exerted assisting and hindering loads along the DNA. The application of external load selectively perturbs the free energy landscape for reaction steps that involve a component of enzyme motion along the DNA (Neuman et al., 2003). We observed no systematic variation in termination efficiency for two of the three terminators across the full range of loads, implying that no significant displacement of DNA with respect to the enzyme (~0.5 bp or greater) is involved in transcript release. Hypertranslocation may therefore not represent a general feature of termination, as previously conjectured (Santangelo and Roberts, 2004). However, for the specific case of the t500 terminator, which exhibits high efficiency, load was found to modulate the kinetics of a pre-termination delay, rather than the termination efficiency. No corresponding effect was observed for the other terminators studied, which lack such a delay. Furthermore, t500 terminators carrying a mutation in the hairpin stem that lowers the TE showed clear load dependence, consistent with forward translocation in this terminator (only). To probe the stability of the RNA:DNA hybrid within the enzyme, we applied forces between the 5′ end of RNA and RNAP during the transcription of U-rich tracts derived from each of the three terminators. Our results imply that the application of sufficiently high loads directly to the RNA can recapitulate the action of the terminator hairpin in extracting the RNA, leading to transcript release. Taken together, our results suggest that termination occurs through alternative mechanisms of shearing or forward translocation, depending on the composition of the U-rich tract.
Figure 1. Intrinsic Terminators and Single-Molecule DNA-pulling Assay.
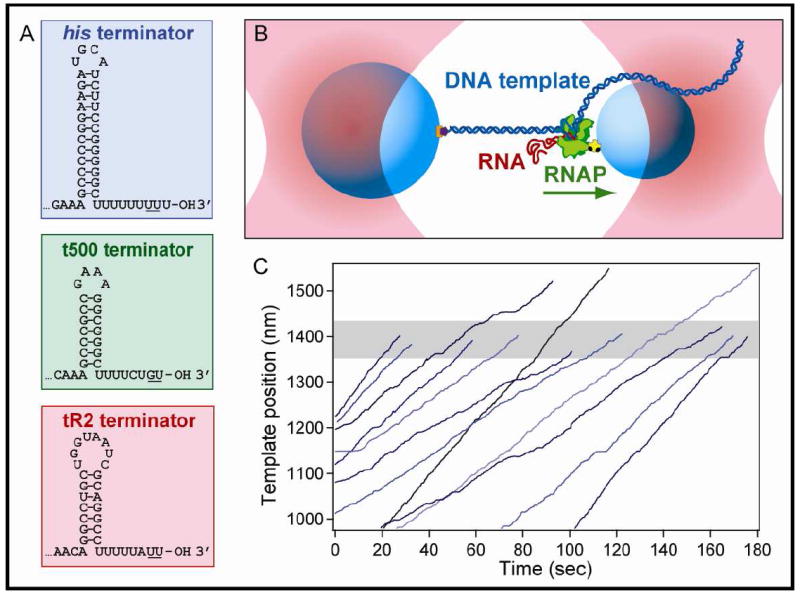
(A) Sequences and expected secondary structure for the three intrinsic hairpin terminators and U-tracts used in this study. Underlined bases indicate the transcript termination positions.
(B) Experimental geometry for the DNA-pulling dumbbell assay (not to scale). Two polystyrene beads (light blue) are held in two separate optical traps (pink). RNAP (green) is attached to the smaller of the two beads via a biotin-avidin linkage (yellow and black), and the DNA template (dark blue) is attached to the larger bead via a digoxigenin-antidigoxigenin linkage (purple and orange). The RNA transcript (red) emerges co-transcriptionally from RNAP and is untethered. The direction of transcription is shown (green arrow); in this particular orientation of the DNA template, the applied tension assists RNAP translocation.
(C) Twelve representative records of RNAP elongation on the his terminator template under 18 pN of assisting load. Nine records terminated within ±40 nm of the expected position of the terminator (shaded gray area), whereas three ran through the terminator position.
By applying force to the 5′ end of the RNA as RNAP transcribed complete terminator sequences, we mapped TE as a function of load, which was found to be biphasic. At high loads, beyond those sufficient to unfold the terminator hairpin, TE increased with load in manner consistent with hybrid shearing. However, for moderate loads, sufficient only to perturb the closure of the base of the hairpin stem, TE also increased, in this case, as force was reduced. Surprisingly, TE for moderate loads could surpass the unloaded efficiency. This result can be explained by the role of force in suppressing the formation of secondary structure that otherwise competes with terminator hairpin formation. The behavior of TE over the full range of loads for all three terminators could be understood by a quantitative model involving an energetic competition between termination and elongation, with the barrier to termination being modulated both by the formation of the terminal bases of the hairpin stem and the energetic stability of the RNA:DNA hybrid. The model also successfully accounts for the TE measured in three additional terminators bearing specific mutations designed to test its central predictions.
Results
Termination Efficiency is Not Generally Sensitive to Force on DNA
We measured termination efficiency as a function of force along the DNA for three well-studied intrinsic terminators (Figure 1A): the his terminator (a variant of the sequence in the his operon leader region of S. typhimurium with a G-insertion in the hairpin loop) (Yin et al., 1999), the t500 terminator (a mutant construct derived from the late gene terminator sequence of bacteriophage 82) (Yarnell and Roberts, 1999), and the tR2 terminator (a terminator sequence from the bacteriophage λ rightward operon). All three terminators encode the canonical G:C-rich stem-loop hairpin element followed by an 7-9 nt U-rich tract. However, each terminator is characterized by a different intrinsic efficiency, has a different stem or loop length, and varies in the number and placement of interruptions in the U-rich tract.
Transcription was assayed using the dumbbell geometry, where one end of a template DNA is attached via a digoxigenin-antibody linkage to a polystyrene bead held in one optical trap, and the elongating RNAP molecule is attached through a biotin-avidin linkage to another bead held in a second trap (Figure 1B) (Shaevitz et al., 2003). In this geometry, applied force affects translocation of RNAP along DNA, but not folding of nascent RNA. Relative motions of the two beads, recorded by position-sensitive photodetectors, permit direct measurement of transcriptional progress under controlled loads. Single-molecule DNA-pulling records for templates carrying the his terminator display elongation followed by sequence-specific template release (rupture of the dumbbell tether), corresponding to the termination event, during which DNA dissociates from the RNAP (Figure 1C). The properties of the elongation phase correspond closely to previous single-molecule studies of RNAP, with records showing periods of active movement, during which RNAP transcribes at a rate of ~10-20 bp/s, interrupted by frequent transcriptional pausing (Adelman et al., 2002; Dalal et al., 2006; Davenport et al., 2000; Herbert et al., 2006; Neuman et al., 2003). Termination is expected to occur at a DNA tether contour length of ~1400 nm in our construct, with small deviations in the observed position arising from 5-10% variations in the bead diameter (Dalal et al., 2006). Therefore, all records showing tether rupture within a region corresponding to ±40 nm of the expected position were scored as termination events (NT); those that elongated beyond this region were scored as read-through events (NRT). The TE was operationally defined as the ratio of ECs that dissociated to the total number that reached the terminator position: TE = NT/(NT + NRT). An minor population of rupture events was observed at low frequency prior to the terminator region, which we attributed to occasional breakage of the digoxigenin-antibody linkage (Dalal et al., 2006): this population was discarded from subsequent analysis.
The TE at the his terminator was unchanged by both assisting and hindering loads applied to the DNA template, and the interpolated value at F = 0 matched the efficiency measured independently in a bulk biochemical assay (77%) (Figure 2A). Furthermore, the TE for a deletion of the his terminator, missing half the hairpin stem sequence and therefore encoding only the 9-nt U-tract, was also independent of load, and corresponded closely to the low release level found in bulk solution (<6%). This result indicates that forces applied to the DNA cannot induce termination at a perfect U-tract sequence (Figure 2A). The TE was also measured as a function of force along DNA for both tR2 and t500 terminators, which have interrupted U-tracts (Figure 1A). Here again, the TE was independent of load placed on the DNA, and measured values matched those found in bulk solution (30% and 98%, respectively) (Figure 2A,B).
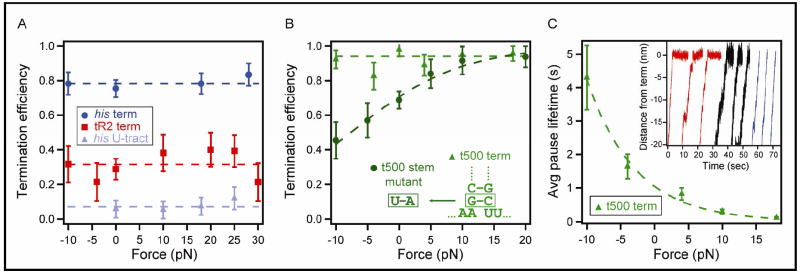
Figure 2. TE and Kinetics as a Function of Force Applied to the DNA.
Termination efficiency and terminal dwell lifetimes (mean ± SEM) are plotted as functions of the load applied between the DNA template and the RNAP; positive force values correspond to loads assisting RNAP translocation. (A) TE for the his terminator (dark blue circles), tR2 terminator (red squares), and his U-tract (light blue triangles). Fits of each data set to a constant are displayed (dotted lines): 78 ± 3% (his), 31 ± 3% (tR2), and 7 ± 3% (his U-tract).
(B) TE for the t500 terminator (light green triangles) and t500 stem mutant (dark green circles). The force-dependence of the t500 stem mutant terminator was fit by Equations 2,3 (dark green dotted line), yielding a distance parameter of 0.49 ± 0.13 nm (~1.4 bp). The inset shows substitutions made to the base of the hairpin stem for the t500 mutant.
(C) Average terminal dwell times at the t500 terminator (light green triangles). Data were fit to Equation 1 (light green dotted line) giving a force-dependent distance parameter of 0.52 ± 0.10 nm. Inset: Three groups of three representative, aligned records that terminated at the t500 sequence under loads of -10 pN (red), -4 pN (black), and +10 pN (blue).
Kinetics of t500 and TE for a t500 Mutant are Force-dependent
Although the fraction of terminating RNAP molecules remained nearly invariant under load for all terminators, the kinetics of EC dissociation at the t500 terminator exhibited significant force dependence (Figure 2C). At large hindering loads (10 pN), we observed a delay at the site of the terminator, with an average lifetime of ~4 s prior to dissociation. This delay, which we call a “terminal dwell,” persisted at lower hindering loads (~2 s at 4 pN), but became insignificant for assisting loads (Figure 2C, inset). Such a dwell does not constitute a transcriptional pause, by definition, because it was not followed by continued elongation. Although a terminal dwell has previously been reported for the his terminator under unloaded conditions (Yin et al., 1999), no effect of load on the kinetics was found for either the his or tR2 terminators (data not shown), and no corresponding terminal dwell was apparent.
The effect of force on the lifetime of the terminal dwell (τterm) was fit by a single-barrier, Boltzmann-type expression:
| (1) |
where F is the applied force, δrelease corresponds to the characteristic distance moved along DNA prior to release, τ0 is the lifetime of the terminal dwell at F = 0, and c is a constant average lifetime for any force-independent steps that occur prior to release. The fit yielded c ≈ 0 s, τ0 = 1.1 ± 0.2 s, and δrelease = 0.52 ± 0.10 nm, a distance consistent with RNAP forward translocation by ~1.5 bp to reach the transition state on the path to transcript release (assuming 0.34 nm/bp for dsDNA). To account for the load-independence of TE, this translocation event would have to occur after an essentially irreversible commitment step toward termination.
The effect of load on the kinetics of the t500 terminator led us to measure the force dependence of termination efficiency in a mutant t500 terminator with a weaker A:U pair positioned at the base of the hairpin stem, replacing G:C (Figure 2B). Because the efficiency of the unmodified t500 terminator is already near 100%, an effect of assisting load in raising the TE may not be otherwise apparent, even if hypertranslocaton occurred. The t500 mutant, however, has a reduced TE of ~70% at F = 0, and therefore supplies a more sensitive probe. It exhibited force-dependence that was fit by a two-state Boltzmann relation (Figure 2B):
| (2) |
| (3) |
where Ecommit is the energy difference between the barrier to elongation and the barrier to termination at F = 0 and δcommit corresponds to the characteristic distance moved along DNA in reaching the transition to commitment to termination. The fit yielded δcommit = 0.49 ± 0.13 nm (~1.4 bp), a distance roughly consistent with the 2-4 bp hypertranslocated displacement at the t500 terminator inferred by a previous termination study, which positioned a series of roadblocks to inhibit such movements (Santangelo and Roberts, 2004). Just as for the unmodified t500 terminator, a terminal dwell was observed for the t500 mutant when a large hindering load was exerted along the DNA (data not shown).
Loads on the RNA Lead to Specific Transcript Release at U-tracts
We next assessed the destabilizing effects of isolated U-rich tracts, derived from the three terminators, in the context of actively transcribing RNAP molecules. In each case, the template region coding for the upstream half of each terminator hairpin sequence was deleted to preclude stem formation, leaving an unpaired sequence in the nascent RNA followed by a downstream U-rich element. In this experimental arrangement, force was exerted between RNAP and the 5′ end of the nascent transcript (rather than the template DNA), and the efficiency of transcript release, as assayed by tether rupture, was measured. The RNA was placed under controlled loads using a modification of the single-molecule RNA-pulling geometry described by (Dalal et al., 2006). To form an RNA tether, a stalled EC with a 29 nt RNA transcript was attached to an avidin-coated bead, and the 5’ end of the nascent RNA was hybridized to a 25 nt complementary single-stranded overhang of a ~3-kb dsDNA ‘handle’ molecule. This handle was attached at its distal end to a second bead, either by a digoxigenin-antibody linkage, or, to sustain larger forces, by a biotin-avidin linkage (Figure 3B). In this assay geometry, applied force can affect RNA folding, movement of the nascent RNA relative to RNAP, movement of the RNA:DNA hybrid relative to RNAP, or some combination of these. With reference to transcriptional motion, we note that pulling the 5′ end of RNA away from RNAP tends to promote forward— that is, transcriptionally downstream—motion of the polymerase molecule along DNA. Thus, the effect of applying force on the RNA is similar to that of applying an assisting load that pulls the upstream end of the DNA away from RNAP (Dalal et al., 2006).
Figure 3. Single-Molecule RNA-pulling Transcription Assay.
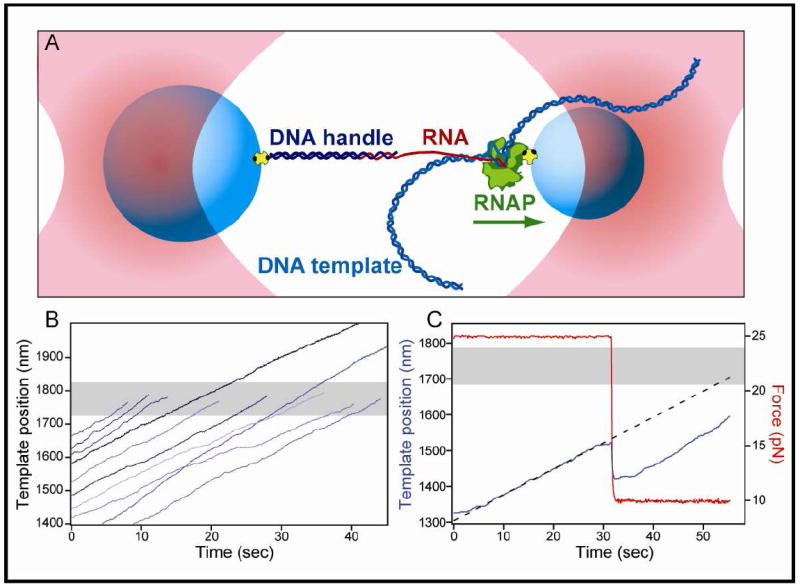
(A) Experimental geometry for the RNA-pulling dumbbell assay. The nascent RNA (red) is hybridized to a 25-nt overhang of a 3 kb DNA handle (dark blue), which is attached at its distal end to a polystyrene bead (light blue) through a biotin-avidin linkage (yellow and black).
(B) Ten representative records of RNAP elongation on the his U-tract template under 30 pN assisting load. Eight records terminated within ±50 nm of the expected position of the his U-tract (shaded gray area), whereas two ran through this region.
(C) Record of RNA extension (blue) on the his U-tract template at both high and low loads (red). Initially, elongation took place under 25 pN load along the nascent transcript. At 32 sec, the force was dropped to 10 pN, causing a decrease in the apparent tether extension. Under 10 pN of load, the record shows that RNAP continues transcription at very nearly the same rate at that observed at 25 pN. At 55 sec, the tether ruptured. Using the average rate of elongation at 25 pN and extrapolating from the extension of the tether immediately prior to the drop in force, we can accurately estimate the template position at which the RNA was released (dotted line). In this record, the extrapolated termination location occurred within ±50 nm of the expected position of the his U-tract (shaded gray area).
Load-dependent, sequence-specific transcript release was found to occur at the U-tracts for all three terminators; representative records for the his terminator at 30 pN are shown (Figure 3B). Forces greater than ~18 pN are sufficient to remove secondary structure from the nascent RNA (Dalal et al., 2006). At such high forces, the tether extension at which the RNAP reached the terminator site was determined by modeling the elasticity of the system as two elements in series: (1) a dsDNA of fixed length, corresponding to the handle (modeled as a worm-like chain, WLC) (Odijk, 1995), followed by (2) a ssRNA, corresponding to the transcript (modeled as a freely-jointed chain, FJC) (Smith et al., 1992), as described (Dalal et al., 2006). To compute the RNA tether length for forces below 18 pN, the nascent transcript was initially extended under high force (20-25 pN) to determine the initial template position and extension rate. The load was subsequently reduced into the measurement range when RNAP had reached a position ~150 nm upstream of the terminator U-tract. Any reduction in force below ~18 pN causes the apparent extension to decrease, due to the formation of secondary structure in ssRNA (Liphardt et al., 2001). However, by extrapolating the tether length, based on the average rate of extension prior to the reduction in force, we could compute the template position as a function of time (Figure 3C). If termination failed to occur, the force was later increased to 20-25 pN to re-measure the tether length unambiguously and confirm that RNAP had run through the U-tract position.
For each terminator U-tract, the TE increased with load on the RNA (Figure 4A-C), in accordance with a two-state Boltzmann relation (Equation 2). Because single-stranded nucleic acids are significantly more compliant than double-stranded forms (for forces beyond ~5 pN), a portion of the work performed by the trap goes into changing the length of the emerging transcript as it transitions from the RNA:DNA hybrid to ssRNA. This confers load dependence to the energy required to stretch the RNA thus liberated, as well as to the extension of the RNA released from the enzyme. Consequently, the expression for ΔEtotal becomes:
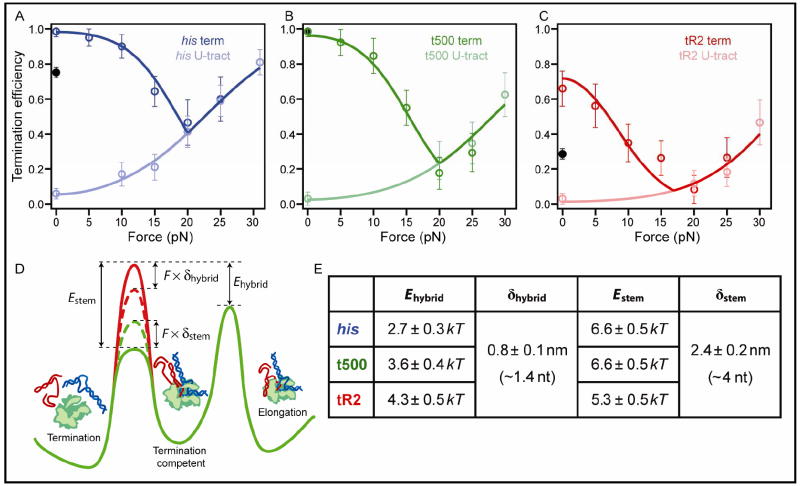
Figure 4. Force Applied Along RNA Affects TE and an Energetic Model of Termination.
(A)-(C) Termination efficiency (mean ± SEM) plotted as a function of the load applied between the nascent RNA and RNAP. (A) The complete his terminator (dark blue) and associated U-tract alone (light blue), (B) the complete t500 terminator (dark green) and associated U-tract alone (light green), (C) the complete tR2 terminator (dark red) and associated U-tract alone (light red). For each complete terminator, two values for TE at F = 0 are plotted: the TE determined in bulk solution using a gel-based assay (black filled circles) and the TE measured in the same fashion, but in the presence of a complementary oligo that prevents secondary structure from forming in the upstream region (the endogenous TE, shown by the open, colored circle at F = 0). Solid light-colored lines show global fits to Equations 2 & 4, solid dark-colored lines show fits to Equations 2 & 5. (D) An energy landscape representation of the quantitative model for termination. The TE is determined by the difference, ΔEtotal, between energy barriers to elongation (right) and termination (left). The portion of the landscape depicted in red represents the barrier to termination for terminator U-tracts, which can be modulated by force on the RNA, lowering it by ~F ×·δhybrid (dotted red line; the exact expression for modulation of this barrier includes corrections for the mechanical properties of ssRNA, not shown here; see Supplemental Materials and Equation 4) The inclusion of a hairpin stem lowers the termination barrier by Estem (solid green line). Force on RNA biases against folding of the terminator hairpin, raising the barrier to termination by ~F ×·δstem (dotted green line). The overall TE is related to ΔEtotal through Equation 2. (E) Table of global best-fit parameters, obtained by fitting the TE for all three terminators and associated U-tracts at all forces to Equation 2 & 5.
| (4) |
where Ehybrid represents the energy difference between the barrier to termination and the barrier to elongation at F = 0, δhybrid is a distance parameter representing the contour length of the liberated ssRNA, xssRNA is the force-dependent extension of the liberated ssRNA, and Whybrid is the energy required to stretch the liberated ssRNA to this extension, assuming a worm-like chain model (Seol et al., 2004) (Figure 4D and Figure S3 in Supplemental Materials). A global fit to all three terminators with a common distance parameter, δhybrid, but individual hybrid energies, Ehybrid, (4 parameters) yielded δhybrid = 0.8 ± 0.1 nm, which corresponds to ~1.4 bp of motion along ssRNA between the barriers to elongation and termination, assuming a rise per base of 0.59 nm (Woodside et al., 2006a). Values for Ehybrid are summarized in Figure 4E.
Force Biases Folding of the Hairpin
To probe the mechanism by which the complete terminator destabilizes the EC, we next measured the TE for each of the three full terminator sequences as functions of load exerted on the RNA. At forces greater than the hairpin unfolding force, the efficiency of transcript release was indistinguishable from that measured for the corresponding terminator U-tracts alone, as anticipated (Figure 4). Numerical estimates for the hairpin unfolding force, FU, for each of the hairpins under our buffer conditions were computed (Supplemental Materials). The behavior of the complete his terminator above and below this transition point (FU ~ 20 pN) is shown (Figure 4A); similar curves were obtained for the t500 and tR2 terminators (FU ~ 22 and 15 pN, respectively) (Figures 4B,C).
In all three cases, for loads below the hairpin unfolding transition, we observed a monotonic increase in the TE as force was further reduced. Furthermore, for the his and tR2 terminators, the TE at the lowest loads (F = 5-10 pN) rose to a level significantly greater than the unloaded value measured independently in bulk solution in the identical buffers (Figures 4A, 4C). Tension on the U-tract alone cannot be responsible for such a large increase in TE at low forces, because our results with isolated his and tR2 U-tracts indicate that such forces are capable of stimulating only a negligible fraction of the overall transcript release (<6 % at 5 pN). Instead, we attributed the dramatic increase in TE to a force-mediated suppression of secondary structure in the transcript. Although weak forces applied to the RNA are incapable of unfolding an energetically stable terminator hairpin, they nevertheless disfavor the co-transcriptional formation of smaller secondary structures (or short hairpins with large loops) (Woodside et al., 2006b).
To test this hypothesis, we measured the TE in bulk solution (F = 0) for all three terminators in the presence of a synthetic oligonucleotide (oligo) complementary to the 56-bp region of the transcript immediately upstream of the terminator hairpin sequence. In the presence of 100 μM oligo, the efficiency for the his terminator increased from 77% to beyond 98% (Figure 4A). This result suggests that in an unloaded situation, termination at the his terminator invariably occurs once the hairpin has an opportunity to fold properly. For tR2, the addition of 100 μM oligo increased the TE from 30% to 50%, which grew even further for increasing concentrations until saturating at ~65% for ≥0 .5 mM oligo (Figure 4C and Figure S1C in Supplemental Materials). This dose-dependent effect is not believed to be a consequence of oligo-stimulated transcript release (Santangelo and Roberts, 2004), because another oligo, complementary to a different 56-bp transcript region located farther upstream from the terminator hairpin, did not affect the TE when present at the same concentration level (data not shown). Moreover, oligo-stimulated transcript release has only been observed under limiting NTP conditions, whereas our experiments were conducted at saturating levels (1 mM NTPs).
To model the dependence of the TE with load for forces below FU, we introduced a second term to the expression for ΔEtotal (Equation 4) that accounts for the transition state energy and distance change associated with hairpin folding:
| (5) |
where Estem represents the change in the energy barrier to termination corresponding to the final pairing of the nucleotides located at the base of the hairpin stem, δstem is the contour length over which these same terminal bases fold, xssRNA is the force-dependent extension of the unfolded ssRNA, and Wstem represents the work required to stretch the unfolded ssRNA to this extension (Figure S3 in Supplemental Materials). In the absence of load on the RNA, the barrier against termination is reduced by an energetic contribution arising from the folding of the terminal base pairs in the hairpin stem, Estem. At intermediate forces, hairpin closure is impeded by the applied load, increasing the barrier to termination and lowering the TE. For sufficiently large forces, however, the terminator hairpin becomes completely unfolded, so the corresponding energetic term reduces to zero, leading to termination directly through force-stimulated release, as previously described. All together, these effects explain the biphasic shapes of TE-load curves. We globally fit the curves for all three terminators using Equation 5 (Figure 4E); the model includes 7 parameters and captures the behavior across the accessible force range. The distance parameter for hairpin closure, taken to be the same for all terminators, was δstem = 2.4 ± 0.2 nm, equivalent to a contour length of ~4 nt of ssRNA. The his and t500 terminators were constrained to have the same energy for folding (Estem) because the terminal bases of their hairpin stems are identical (Figure 1A), whereas the corresponding energy for tR2 was allowed to float.
Discussion
Termination occurs by multiple pathways: hypertranslocation and shearing
We found that hindering the forward progress of RNAP using force applied to the DNA had no measurable effect, either upon the TE or upon the release kinetics, at the his and tR2 terminators. This finding immediately places a constraint on the reaction pathway leading to transcript release. Upon encountering an intrinsic terminator, RNAP either reads through the sequence, continuing normal elongation, or branches off the elongation pathway, releasing the transcript and template DNA. The insensitivity of termination to force at this branch point therefore implies an irreversible “commitment” step that does not involve additional motion of the enzyme along the DNA. However, if the distance to the transition state for termination (measured along the DNA) were sufficiently small, e.g., the distance subtended by single base pair or less, then we might be unable to modulate the energetic barrier to termination sufficiently to observe a change in TE, even if forward translocation did take place. The forward translocation model proposed a 3-4 bp step, but evidence suggests that this distance could be variable, and may be as small as 1-2 bp (Santangelo and Roberts, 2004). In these experiments, we were able to modulate force on the DNA from 25 pN (assisting load) to -10 pN (hindering load) before either linkage rupture or irreversible backtracking of RNAP occurred, respectively. Over the span of a single dsDNA base pair (0.34 nm), these forces correspond to a modulation of the energy barrier to termination by -2.1–0.8 kBT. The “endogenous” termination efficiency for the his and t500 terminators, operationally defined here as the unloaded TE measured in bulk solution with oligo-mediated suppression of upstream secondary structures, is very nearly 100% (Figure 4A & B). This high value implies that a large hindering force would be required to produce any measurable effect on termination, given a sufficiently small distance parameter. These data alone, therefore, cannot exclude the possibility that we failed to detect a limited amount of forward translocation (on the order of 1 bp or less), but we found no evidence for any larger motions.
Additional lines of evidence argue against even small amounts of forward translocation for the his and tR2 terminators. The endogenous TE for the tR2 terminator is reduced compared to either of the two others, reaching only ~65% (Figure 4C). We found that we could remove the effect of competing secondary structure on tR2—and therefore any need for suppressing oligos—by mutating the 8 bp tR2 terminator loop (which is unstructured) to a smaller, more stable tetraloop (Tuerk et al., 1988). The tR2-tetraloop terminator yielded the same TE in the presence and absence of complementary oligos (~55%). In a forward translocation scenario, the efficiency of this terminator ought to be strongly modulated by the application of load on the DNA over our experimental range. However, we did not observe any significant change in TE under either assisting (20 pN) or hindering (-10 pN) loads (Figure S2B in Supplemental Materials). We also tested a mutant of the his terminator for evidence of forward translocation. Although the endogenous TE for his is ~100%, changing the closing basepair at the base of the hairpin stem from G:C to A:U reduces the TE to ~90% (Figure 5), allowing a more sensitive measurement of load dependence over the accessible force range. The TE of this his mutant was also found to be independent of force applied to the DNA (Figure S2C in Supplemental Materials). Thus, we conclude that forward translocation without nucleotide addition is not a step in termination at the his or tR2 terminators.
Figure 5. Model Prediction for TE of Mutant Terminators.
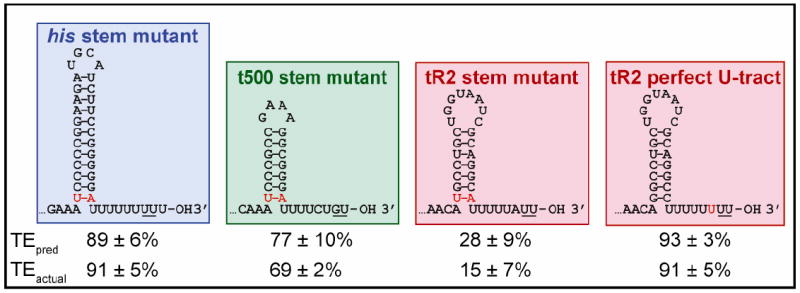
Sequences and expected secondary structures for 4 mutant terminator constructs based on his, t500, and tR2 (mutated bases are shown in red). Model predictions (TEpred) for the endogenous TE were calculated from Equation 2 & 5 at F =0, using previously determined values for Ehybrid (Figure 4) and computing Estem from MFOLD predictions for the energy difference in pairing nucleotides at the base of the hairpin stem between the mutant and unmodified terminators. Experimental values (TEactual) for the endogenous TE were measured in solution in the presence of saturating concentrations of complementary oligos designed to block competing upstream secondary structure.
Nonetheless, our results suggest that forward translocation does occur at the t500 terminator, much as previously proposed (Santangelo and Roberts, 2004). Although the TE for t500 was independent of load on DNA (Figure 2B), that observation is attributable to its high endogenous TE and an inability to modulate the termination energy barrier over the accessible force range. When we measured the force-dependence for the t500 hairpin stem mutant with a reduced endogenous TE of ~70%, we observed significant force-dependence, with a distance parameter, δcommit = 0.49 ± 0.13 nm (1.4 bp), corresponding to forward translocation along DNA during the termination commitment step. This displacement is very nearly the same as the distance to the transition state along RNA prior to termination, δhybrid = 1.4 nt, obtained by global fits (Figure 4), suggesting that RNA and DNA may move with respect with RNAP, but remain in register during the t500 termination commitment step. We also observed a load-dependent increase in the lifetime of a pre-termination delay for the t500 terminator, indicating that additional RNAP translocation may be required for transcript release subsequent to the initial commitment step. The characteristic distance for this additional displacement after commitment but prior to release is δrelease = 0.52 ± 0.10 nm (1.5 bp). Together, the magnitudes of δcommit plus δrelease (2.9 bp) are consistent with the 2-4 bp forward translocation movement postulated for the t500 terminator by (Santangelo and Roberts, 2004).
The sensitivity of t500 transcript release kinetics to load, and the force-dependence of the t500 stem mutant, contrast sharply with the comparative insensitivity of both the his and tR2 terminators and mutants, suggesting that hypertranslocation may not be a general phenomena: different mechanisms may hold for different intrinsic terminators, depending on the composition of their U-tracts. For his and tR2 terminators, where the U-tract consists of a perfect stretch of uridines or has a single adenosine interruption at the 6th hybrid position, respectively, the transcript may simply slide past the DNA (i.e., hybrid ‘shear’) during release, precluding any need for hypertranslocation (Toulokhonov and Landick, 2003). Since during shearing, the RNA moves independently of the remainder of the EC at weak hybrid positions, any dependence on forces placed on the DNA would be eliminated. Conversely, the t500 terminator U-tract, which carries two interruptions, may require forward translocation to fully release the transcript. In support of this notion, previous biochemical investigations reported that the efficiencies of terminators with U-tracts containing large numbers of non-uridine interruptions are dependent on the energetic stability of downstream DNA (Reynolds and Chamberlin, 1992), consistent with a forward translocation mechanism, where such downstream base pairs must be broken prior to transcript release. However, the efficiency at a terminator with a perfect U-tract was found to be independent of the downstream DNA sequence, consistent with a mechanism that does not require forward translocation. Further studies of terminators with different U-tracts should help determine whether a need for hypertranslocation is dictated by the particular sequence of the RNA:DNA hybrid at termination, and what hybrid positions may be critical to maintaining stability.
U-tract Lowers Mechanical Stability of the Hybrid
We observed sequence-specific transcript release at all three terminator U-tracts with loads applied to the nascent RNA. In the absence of RNAP, 8-9 bp U-tracts are not predicted to form stable RNA:DNA duplexes at room temperature. The fact that significant loads placed on the RNA are required to stimulate transcript release from the EC therefore reflects significant stabilization of such hybrids inside RNAP. The force-dependent release was fit by a single distance parameter, δhybrid = 0.8 ± 0.1 nm, consistent with a 1.4 nt displacement of ssRNA relative to the EC. Had the RNA had remained in register with the DNA during the release event, we would have expected a dependence on the force applied to the DNA as well, contrary to observation. However, the expected distance parameter would be smaller, because a displacement by one base of dsDNA moves RNAP just 0.34 nm (Abbondanzieri et al., 2005), lengthening the dumbbell DNA tether by this same amount. The smaller distance produced by a single-base translocation in a DNA-pulling assay (compared to a RNA-pulling assay) decreases the energetic effect of force applied to DNA by roughly 50% relative to loads on the RNA. However, when 25 pN of load was applied to DNA (equivalent to applying ~14 pN in the RNA-pulling assay, for which an increase in TE was observed), we did not detect releases beyond background levels at the his U-tract (Figure 2A and Figure S2D in Supplemental Materials). These observations suggest that RNA moves independently of DNA as it is extracted from the EC.
A mechanism consistent with our results is that the rate-limiting step for transcript release at the his and tR2 terminators involves a small, ~1 nt shearing motion, during which the transcript moves out-of-register with the DNA template, shortening the hybrid by 1 bp. In this model, the transition state to hybrid shearing constitutes the highest energy barrier in the pathway of termination, and leads to blockage of further transcriptional elongation by removing the 3′ end of the RNA transcript from the RNAP active site.
An alternative termination mechanism, equally consistent with our measured value of δhybrid, postulates that RNAP commits to termination when several upstream bases (i.e., bases distal from the active site) of the RNA:DNA hybrid melt (unpair) without shearing. This mechanism finds some support from previous studies, which found that shortening the upstream end of the hybrid by several base pairs significantly destabilizes the TEC (Gusarov and Nudler, 1999; Komissarova et al., 2002). If 2-3 upstream RNA hybrid nucleotides were to unpair from the template, the released bases would subsequently stretch, causing the interphosphate distance to increase from that of dsDNA (~0.34 nm) to that of ssRNA (~0.4-0.6 nm, depending upon the force applied), leading to a distance change roughly consistent with our measured value for δhybrid The melting mechanism involves only the 2-3 most upstream bases of the hybrid, however, and therefore—unlike the shearing mechanism—it cannot readily explain the range of load-dependent stabilities (Ehybrid) observed for the different U-tracts, because the terminators studied here all contain identical rU:dA bases at the remaining downstream positions. A hybrid shearing mechanism induced by terminator hairpin closure is therefore the most parsimonious explanation for our current findings, but alternative models are not excluded. However, any such models would have to be shown to be quantitatively, as well as qualitatively, consistent with the data here for termination efficiencies under load.
The perfect U-tract of the his terminator had the lowest measured stability, 2.7 ± 0.3 kBT, and was also the easiest to extract by force. It is computed to form the weakest duplex: -4 kBT based on nearest-neighbor interaction energies (Sugimoto et al., 1995). Furthermore, shearing across a perfect U-tract by one base maintains the canonical base-pair interactions within the core of the hybrid, reducing the energetic cost of such a motion. For the interrupted t500 and tR2 U-tracts, however, Ehybrid did not correlate well with thermodynamic stability of the duplex. This result is not unexpected because the energy required to remove RNA from the enzyme is not merely a function of this stability, but also of the extensive energetic contacts made between the duplex and the protein. The tR2 U-tract, which has a single adenosine interruption, had the largest Ehybrid (4.3 ± 0.5 kBT) but a computed stability of -5 kBT, similar to that of the his U-tract. The t500 U-tract had an intermediate Ehybrid (3.6 ± 0.4 kBT), but the greatest duplex stability, -10 kBT. We note, however, that hypertranslocation at the t500 terminator shortens the hybrid prior to shearing, thereby avoiding the energetic cost of breaking the two stable G:C pairs interrupting the U-tract. This hypertranslocation lowers the energetic cost of RNA extraction compared to a pure shearing mechanism.
Generalized Attenuation and a Quantitative Model for Termination
We sought to measure the energy of terminator hairpin formation, Estem, which is postulated to aid in overcoming the energy barrier against RNA:DNA hybrid disruption, Ehybrid, ultimately leading to transcript release. To do so, we exerted forces on the RNA as RNAP transcribed complete hairpin terminator sequences (his, t500, and tR2) and measured the change in TE with load (Figure 4).
For forces beyond the hairpin unfolding force, FU, the efficiency of transcript release for each of the three terminators was indistinguishable from that observed for the corresponding U-tract alone under identical loads. This suggests that mechanical denaturation of the terminator hairpin under force is equivalent to a mutagenic deletion of the hairpin stem, and that transcript release at higher forces is a direct consequence of mechanical disruption of the RNA:DNA duplex, presumably through RNA shearing (for his and tR2) or forward translocation (for t500), as discussed.
For forces below FU, the TE increased monotonically with reduction in the applied load, rising to match the endogenous TE at F = 0. The maximal value attained by the TE was greater than that measured in bulk solution by traditional means, in the absence of any suppression of competing secondary structure by complementary oligos. The important role played by upstream RNA secondary structure has been underscored in extensive studies of transcriptional attenuation, where the nascent transcript forms one of two alternative hairpin structures that determine whether RNAP continues or terminates transcription (Yanofsky, 2000). In our studies, the three terminator sequences were inserted into a region of the rpoB gene devoid of known regulatory elements that might form large secondary structures and thereby block terminator formation. However, even in the absence of any specific ‘anti-terminator’ sequences (such as those found in the attenuator regions of the trp and his operons), we observed a large effect of upstream sequence on terminator efficiency. Our results imply a general mode of transcriptional attenuation, in which non-specific secondary structure forms in the upstream RNA with sufficient frequency and stability to limit the formation of terminator hairpins. This generalized attenuation mechanism supplies an alternative explanation for why an inverse correlation exists between elongation rate and TE (McDowell et al., 1994), and why pausing at the U-tract may be important for termination (Gusarov and Nudler, 1999). Assuming that the formation of upstream secondary structure—which is synthesized first—is favored kinetically, but not thermo-dynamically, then either a reduction in elongation rate or pausing at a U-tract would facilitate greater equilibration among competing structures in the RNA, favoring formation of the more stable terminator hairpin, and thereby increasing the efficiency. An immediate implication of this kinetic competition is that the efficiency achieved by a given terminator is not merely an intrinsic property of the sequence of the hairpin and U-tract, but depends also on the context of the upstream sequence in a given gene.
The dependence of TE on force below FU could be fit by a characteristic distance parameter, δstem =2.4 ± 0.2 nm, equivalent to a contour-length change of ~4 nt of ssRNA, and consistent with the displacement necessary to pair several nucleotides at the base of the hairpin stem. The global fit to our data also yielded two folding energies, Estem: one for the his and t500 terminators, and the other for the tR2 terminator (Figure 4E). The folding energy for t500 and his is slightly higher than for tR2, (6.6 vs. 5.3 kBT), and the difference (~1.3 kBT) is consistent with the difference in energy for pairing the last few bases of the hairpin stems, as predicted by MFOLD (Zuker, 2003). Taken together, these findings suggest that formation of the final 3-4 bp in the hairpin stem provides the energy necessary for hybrid disruption. The importance of these bases in termination is further supported by bulk measurements of TE for mutated tR2 terminators. Mutations that weaken the base of the hairpin stem strongly reduce the TE, whereas hairpin-stabilizing mutations that do not change the base of the stem have no discernable effect, in general agreement with other mutational studies of the tR2 terminator (data not shown)(Cheng et al., 1991; Wilson and von Hippel, 1995).
Using Equations 2 and 5 at F = 0 in conjunction with a value for the energy associated with the final base pairs of the hairpin stem returned by MFOLD, we can predict the TE for several terminator variants. We mutated the terminal G:C pair in each hairpin stem to a weaker U:A pair (Figure 5) and measured the endogenous TE (i.e., the bulk TE in the presence of a complementary oligo). To estimate Estem for the variant, we subtracted the change in thermodynamic energy for the formation of the final four bases produced by the mutation, computed by MFOLD, from the Estem value of the original terminator. We used the same value for Ehybrid because the duplex-forming sequences for the terminator and its variant are identical. The predictions were in excellent agreement with the measured bulk TE values (Figure 5). We were also able to predict the TE for a chimeric construct in which the tR2 hairpin was paired with the perfect U-tract of the his terminator.
Taken all together, our findings paint a consistent picture of the intrinsic termination pathway (Figure 6). Upon reaching a termination sequence, secondary structure formed in the upstream RNA can prevent the terminator hairpin from folding in some fraction of cases, causing RNAP to read directly through the terminator. The misfolded fraction is governed by the exact sequence of upstream RNA, as well as by the composition and length of the terminator hairpin (Supplemental Materials). Hairpin misfolding is likely to represent the major determinant of efficiency for a number of terminators, including his and tR2. For those cases where the terminator hairpin folds properly, the TE will be governed by a balance between the energetic barriers to termination and elongation, which is largely controlled by the sequence of the U-rich tract. Pairing of the terminal bases in the hairpin stem then provides a second energetic component that biases the outcome by lowering the barrier against termination, stabilizing a transition-state intermediate that likely involves shearing the RNA out of register with the template DNA (for his and tR2) or forward translocation (for t500), or both. We suggest that forward translocation is likely to occur whenever such motion incurs a smaller energetic penalty than simply shearing the hybrid. In the case of the t500 terminator, a second translocation motion of 1.5 bp is subsequently required for complete EC dissociation. The ability of the simple model presented here to explain multiple aspects of termination, including its overall efficiency, the effects of load on DNA and RNA, and the consequences of several mutations, suggests that the termination process is predominantly governed by energetic considerations that relate to hairpin closure and hybrid stability. Such energetic considerations do not preclude a further role for enzyme allostery, however, particularly in triggering the release of DNA and RNA, which take place concomitant with the cessation of elongation. The modularity of these energetic components—one moderately stabilizing component, determined by the U-tract hybrid, and one destabilizing component, supplied by hairpin stem closure—allows Nature to tune the termination efficiency over a wide range.
Figure 6. Model of the Termination Pathway.
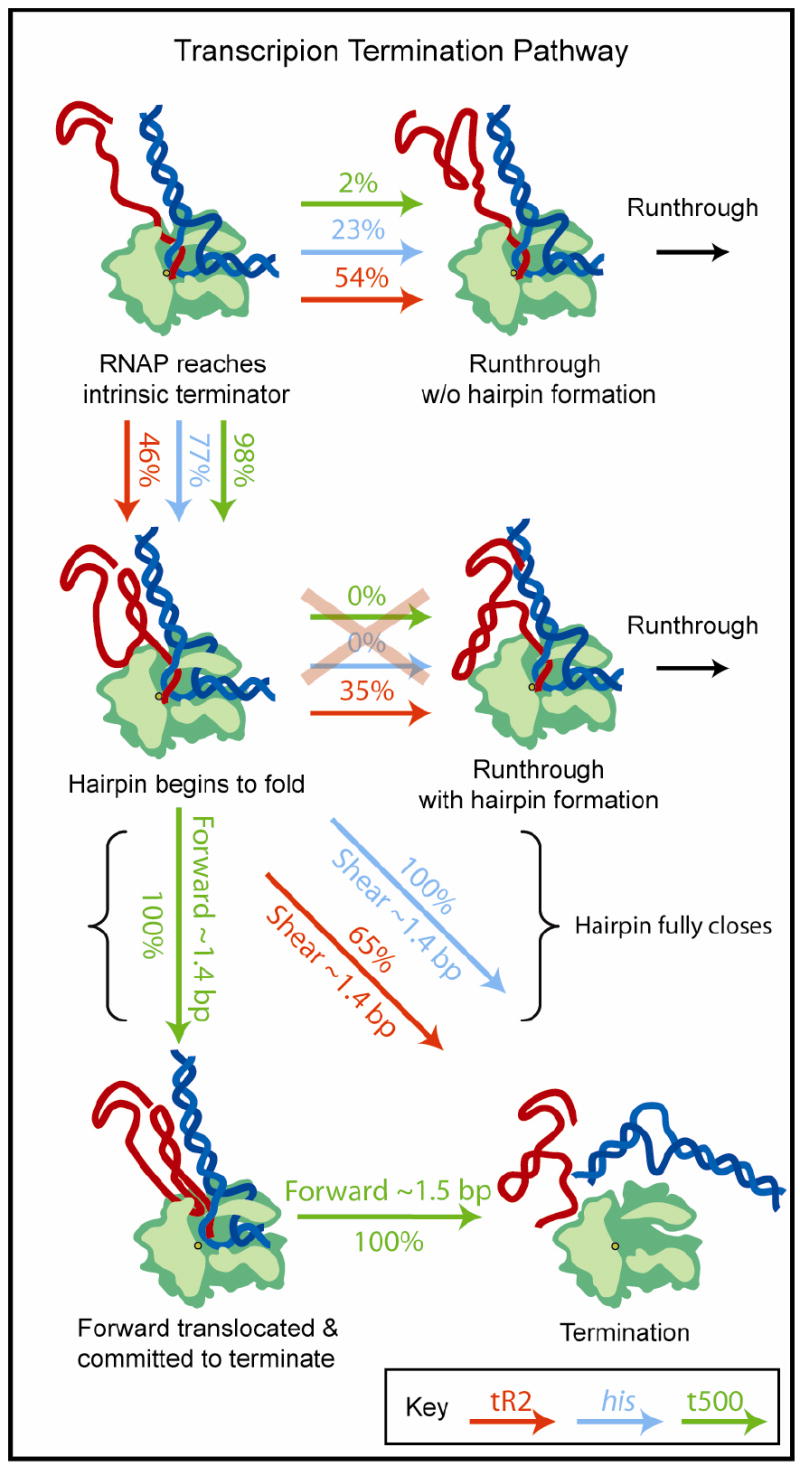
The pathways to termination or transcriptional run-through for the his, t500, and tR2 terminators. Upon reaching an intrinsic terminator, a fraction of terminators hairpins fail to fold due to kinetic competition with upstream secondary structure, resulting in run-through. For the fraction that fold, the subsequent decision to terminate is set by a competition between the energy barriers for termination and elongation (Figure 4D). The his and t500 terminators are energetically biased to terminate with high probability once the hairpin folds, whereas the tR2 terminator terminates only ~65% of the time. Once the decision to terminate is made, the his and tR2 terminators terminate without forward translocation, likely by a shearing motion of the RNA:DNA hybrid through ~1.4 bp. At the t500 terminator, RNAP first forward translocates by ~1.4 bp along DNA to reach a state that is committed to termination (the 3’ end of the transcript has been removed from the RNAP active-site, blocking further elongation). The existence of this committed intermediate leads to a terminal dwell prior to termination. A second translocation step of ~1.5 bp is required for complete complex dissociation.
Experimental Procedures
Experimental Assays
Sample preparation for the DNA pulling assays were performed as described (Abbondanzieri et al., 2005). For the RNA pulling assays, the nascent RNA was placed under tension using a 3057 bp DNA handle with a 25 nt ssDNA overhang complementary to the nascent RNA of the stalled TECs. The DNA handle was created by autosticky PCR of the M13mp18 plasmid, as described previously (Dalal et al., 2006; Lang et al., 2004). This handle was attached to a polystyrene bead, either through a digoxygenin-antibody linkage, or via a biotin-avidin linkage. To achieve the dual biotin-avidin attachment chemistry, stalled TECs were incubated at a near-stoichiometric ratio with 730 nm avidin-coated beads, whereas the sticky DNA handle (used to bind the nascent RNA) was incubated at 100-fold molar excess with 600 nm beads for one hour at room temperature. After this initial incubation, the beads mixed with handle DNA were washed, combined with the beads previously mixed with stalled TECs, and incubated again for 90 min. For some experiments, the bead sizes incubated with the handle and TECs were reversed to improve surface interactions between RNAP dumbbells and the coverglass surface.
An oxygen-scavenger system, consisting of 40 U/mL glucose oxidase (Calbiochem), 185 U/mL catalase (Sigma), and 8.3 mg/mL glucose (Sigma) in transcription buffer (50 mM HEPES pH 8.0, 130 mM KCl, 4 mM MgCl2, 0.1 mM EDTA, 0.1 mM DTT) was used to reduce damage to the TEC from oxygen radicals created by the optical trap (Neuman et al., 1999). To remove RNase contamination, glucose oxidase was FPLC-purified (GE Healthcare, ÄKTA) using a Superdex 200 10/300 GL size-exclusion column (GE Healthcare), and the concentration measured on a UV/Vis spectrometer (PerkinElmer, Lambda 40). The transcription buffer and oxygen-scavenger components were tested for RNase activity using the Ambion RNaseAlert™ Lab Test Kit. An RNase inhibitor (Superase·In, Ambion) was added at a concentration of 0.3 U/μL to further protect against RNase cleavage.
Data collection and Analysis
Data were collected using the apparatus described previously (Abbondanzieri et al., 2005). Force was applied either through an active feedback method (Abbondanzieri et al., 2005), or by using a passive, all-optical force clamp (Greenleaf et al., 2005). We estimate the uncertainty in force due to calibration errors and bead size variations at 10-20%. Position data were acquired at 2 kHz using custom software (Labview), filtered at 1 kHz by an 8 pole low-pass Bessel filter, and analyzed using custom software (Igor Pro). Tether lengths for the DNA-pulling assay were calculated using methods described in (Abbondanzieri et al., 2005); tether lengths for the RNA-pulling assay were calculated using methods described in (Dalal et al., 2006).
Termination events in the single-molecule assay were signaled by tether rupture within ±40 nm of the expected terminator position for the DNA-pulling assay, and ±50 nm of the expected position for the RNA-pulling assay, because RNA-pulling records exhibit more noise, due to the larger compliance of ssRNA (Dalal et al., 2006). Statistical uncertainties in Figures 2A,B and 4A-C were calculated from , where N is the number of records. The transcription records displayed in Figures 1C and 3B,C were additionally smoothed using a 100-point boxcar filter.
Bulk Termination Assays
All terminator sequences were inserted into a site in the rpoB gene following the T7A1 promoter. Bulk measurements of TE were carried out in the presence of 1 mM NTPs and allowed to proceed for 20-25 min in the same transcription buffer used for single-molecule assays. For complementary oligonucleotide disruption experiments, a 56 nt oligo was diluted in transcription buffer and added prior to the addition of NTPs. For all templates, terminated transcripts were separated from run-through transcripts by more than 2.3 kb, producing two clearly resolvable bands. The TE was determined by quantifying the intensity of the termination and run-through bands using image analysis routines (Igor Pro).
Supplementary Material
Supplemental data include text, experimental procedures, and three figures and can be found with this article online at www.cell.com.
Acknowledgments
The authors thank Jeff Gelles for illuminating discussions, Peter Anthony and Kirsten Frieda for critical reading of the manuscript, Jennifer Chen and Kristina Herbert for help with sample preparation, and the Block Lab for intellectual stimulation. MHL acknowledges the support of an NSF graduate research fellowship. SMB and RL acknowledge NIH grants GM057035 and GM38660 for support, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K, La Porta A, Santangelo TJ, Lis JT, Roberts JW, Wang MD. Single molecule analysis of RNA polymerase elongation reveals uniform kinetic behavior. Proc Natl Acad Sci U S A. 2002;99:13538–13543. doi: 10.1073/pnas.212358999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SW, Lynch EC, Leason KR, Court DL, Shapiro BA, Friedman DI. Functional importance of sequence in the stem-loop of a transcription terminator. Science. 1991;254:1205–1207. doi: 10.1126/science.1835546. [DOI] [PubMed] [Google Scholar]
- Dalal RV, Larson MH, Neuman KC, Gelles J, Landick R, Block SM. Pulling on the nascent RNA during transcription does not alter kinetics of elongation or ubiquitous pausing. Mol Cell. 2006;23:231–239. doi: 10.1016/j.molcel.2006.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport RJ, Wuite GJ, Landick R, Bustamante C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- Greenleaf WJ, Woodside MT, Abbondanzieri EA, Block SM. Passive all-optical force clamp for high-resolution laser trapping. Phys Rev Lett. 2005;95:208102. doi: 10.1103/PhysRevLett.95.208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, Block SM. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komissarova N, Becker J, Solter S, Kireeva M, Kashlev M. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol Cell. 2002;10:1151–1162. doi: 10.1016/s1097-2765(02)00738-4. [DOI] [PubMed] [Google Scholar]
- Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst SA. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- Korzheva N, Mustaev A, Nudler E, Nikiforov V, Goldfarb A. Mechanistic model of the elongation complex of Escherichia coli RNA polymerase. Cold Spring Harb Symp Quant Biol. 1998;63:337–345. doi: 10.1101/sqb.1998.63.337. [DOI] [PubMed] [Google Scholar]
- Lang MJ, Fordyce PM, Engh AM, Neuman KC, Block SM. Simultaneous, coincident optical trapping and single-molecule fluorescence. Nat Methods. 2004;1:133–139. doi: 10.1038/nmeth714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnik EA, Sampath R, Levene HB, Henderson TJ, McNeil JA, Ecker DJ. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 2001;29:3583–3594. doi: 10.1093/nar/29.17.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- Macdonald LE, Zhou Y, McAllister WT. Termination and slippage by bacteriophage T7 RNA polymerase. J Mol Biol. 1993;232:1030–1047. doi: 10.1006/jmbi.1993.1458. [DOI] [PubMed] [Google Scholar]
- Martin FH, Tinoco I., Jr DNA-RNA hybrid duplexes containing oligo(dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res. 1980;8:2295–2299. doi: 10.1093/nar/8.10.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JC, Roberts JW, Jin DJ, Gross C. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science. 1994;266:822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- Merino E, Yanofsky C. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet. 2005;21:260–264. doi: 10.1016/j.tig.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- Neuman KC, Chadd EH, Liou GF, Bergman K, Block SM. Characterization of photodamage to escherichia coli in optical traps. Biophys J. 1999;77:2856–2863. doi: 10.1016/S0006-3495(99)77117-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E, Gottesman ME. Transcription termination and anti-termination in E. coli. Genes Cells. 2002;7:755–768. doi: 10.1046/j.1365-2443.2002.00563.x. [DOI] [PubMed] [Google Scholar]
- Odijk T. Stiff chains and filaments under tension. Macromolecules. 1995;28:7016–7018. [Google Scholar]
- Platt T. Termination of transcription and its regulation in the tryptophan operon of E. coli. Cell. 1981;24:10–23. doi: 10.1016/0092-8674(81)90496-7. [DOI] [PubMed] [Google Scholar]
- Reynolds R, Chamberlin MJ. Parameters affecting transcription termination by Escherichia coli RNA. II. Construction and analysis of hybrid terminators. J Mol Biol. 1992;224:53–63. doi: 10.1016/0022-2836(92)90575-5. [DOI] [PubMed] [Google Scholar]
- Santangelo TJ, Roberts JW. Forward translocation is the natural pathway of RNA release at an intrinsic terminator. Mol Cell. 2004;14:117–126. doi: 10.1016/s1097-2765(04)00154-6. [DOI] [PubMed] [Google Scholar]
- Seol Y, Skinner GM, Visscher K. Elastic properties of a single-stranded charged homopolymeric ribonucleotide. Phys Rev Lett. 2004;93:118102. doi: 10.1103/PhysRevLett.93.118102. [DOI] [PubMed] [Google Scholar]
- Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shundrovsky A, Santangelo TJ, Roberts JW, Wang MD. A single-molecule technique to study sequence-dependent transcription pausing. Biophys J. 2004;87:3945–3953. doi: 10.1529/biophysj.104.044081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SB, Finzi L, Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol Cell. 2003;12:1125–1136. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gauss P, Thermes C, Groebe DR, Gayle M, Guild N, Stormo G, d’Aubenton-Carafa Y, Uhlenbeck OC, Tinoco I, Jr, et al. CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988;85:1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007a;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007b;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- Wilson KS, von Hippel PH. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc Natl Acad Sci U S A. 1995;92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodside MT, Anthony PC, Behnke-Parks WM, Larizadeh K, Herschlag D, Block SM. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006a;314:1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodside MT, Behnke-Parks WM, Larizadeh K, Travers K, Herschlag D, Block SM. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc Natl Acad Sci U S A. 2006b;103:6190–6195. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981;289:751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Transcription attenuation: once viewed as a novel regulatory strategy. J Bacteriol. 2000;182:1–8. doi: 10.1128/jb.182.1.1-8.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnell WS, Roberts JW. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- Yin H, Artsimovitch I, Landick R, Gelles J. Nonequilibrium mechanism of transcription termination from observations of single RNA polymerase molecules. Proc Natl Acad Sci U S A. 1999;96:13124–13129. doi: 10.1073/pnas.96.23.13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data include text, experimental procedures, and three figures and can be found with this article online at www.cell.com.