Abstract
Chorismate mutase acts at the first branchpoint of aromatic amino acid biosynthesis and catalyzes the conversion of chorismate to prephenate. Comparison of the x-ray structures of allosteric chorismate mutase from the yeast Saccharomyces cerevisiae with Escherichia coli chorismate mutase/prephenate dehydratase suggested conserved active sites between both enzymes. We have replaced all critical amino acid residues, Arg-16, Arg-157, Lys-168, Glu-198, Thr-242, and Glu-246, of yeast chorismate mutase by aliphatic amino acid residues. The resulting enzymes exhibit the necessity of these residues for catalytic function and provide evidence of their localization at the active site. Unlike some bacterial enzymes, yeast chorismate mutase has highest activity at acidic pH values. Replacement of Glu-246 in the yeast chorismate mutase by glutamine changes the pH optimum for activity of the enzyme from a narrow to a broad pH range. These data suggest that Glu-246 in the catalytic center must be protonated for maximum catalysis and restricts optimal activity of the enzyme to low pH.
Keywords: Claisen rearrangement, site-directed mutagenesis
Chorismate mutase (chorismate pyruvatemutase, EC 5.4.99.5) catalyzes the isomerization of chorismate to prephenate within the tyrosine and phenylalanine biosynthetic pathway. This isomerization occurs through a Claisen rearrangement and is the single example of an enzyme-catalyzed pericyclic reaction. The reaction takes place spontaneously and is enhanced by a factor of 106 by enzymatic catalysis (1). Studies of the bifunctional chorismate mutase-prephenate dehydrogenase of Escherichia coli (T protein) indicate that ionizing groups are important for the mutase reaction (2). The activity of the yeast enzyme shows a narrow pH profile with maximum activity preferentially at acidic pH (3). In contrast, bacterial chorismate mutases as e.g., chorismate mutase-prephenate dehydratase of Escherichia coli (P protein) and chorismate mutase of Salmonella typhimurium have highest catalytic activities at alkaline pH (4, 5). The monofunctional chorismate mutase of Bacillus subtilis has a shallow pH profile with highest mutase activity between pH 5 and 9 (6).
The reaction mechanism of chorismate mutase has been investigated by different approaches such as stereochemical studies applying molecular orbital calculations (1, 7) and by the examination of isotope effects using labeled substrate (8, 9). The data indicate that both the uncatalyzed and catalyzed reactions proceed through a transition state with chairlike geometry. Different models for the mechanism of the enzymatic catalysis have been suggested, including nucleophilic attack of an active side residue or conformational restriction of the substrate by binding to the active site (9, 10). In solution 10–20% of chorismate exists as the energetically less favored pseudodiaxial form in dynamic equilibrium with the pseudodiequatorial form (11). It is assumed that the reaction is accelerated by binding of this active conformer to the enzyme, which locks the substrate via a series of electrostatic and hydrogen bonding and van der Waals interactions in the requisite chair conformation for rearrangement (12–16). Furthermore, the transition state might be stabilized by electrostatic interactions with active site residues (10, 17). In an alternative mechanism an acidic residue is proposed to protonate the ether oxygen O7 of the substrate chorismate (9). This protonation could catalyze the formation of the enol of pyruvate and an cyclohexadienyl cation probably stabilized by the enzyme (9).
The x-ray structures of monofunctional chorismate mutases from B. subtilis (13, 18), from a fragment of the bifunctional chorismate mutase-prephenate dehydratase of E. coli (P protein) harboring the chorismate mutase domain (15), and from the catalytic antibody 1F7 raised against a bicyclic hapten designed as a structural analog of the reaction’s putative transition-state (14), could be obtained in complex with the endo-oxabicyclic transition state analogue. The structures for the activated and inhibited state of Saccharomyces cerevisiae chorismate mutase also have been described (19, 20). Yeast chorismate mutase is a monofunctional enzyme with a molecular mass of 60,000 Da and is composed of two identical subunits (3). The activity of yeast chorismate mutase is regulated by the allosteric binding of the crosspathway activator tryptophan and the feedback inhibitor tyrosine, respectively (3). The almost all-helical structure of the enzyme is completely different from the structure of the B. subtilis enzyme, which consists mainly of β-sheet elements. Moreover, no sequence similarities exist. Low sequence homology exists between the yeast enzyme and E. coli chorismate mutase (P protein), yet similarity in folds of the two structures has been described (19, 21). Computer graphics modeling located a putative active site cavity with similarity to the active site of the E. coli enzyme (P protein) (21). Four residues (Arg-16, Arg-157, Lys-168, and Glu-198) of yeast chorismate mutase are conserved among the two enzymes of yeast and E. coli. Asn-194, Thr-242, and Glu-246 of yeast chorismate mutase in E. coli are replaced by aspartate, serine, and glutamine, respectively. The proposed binding of a transition state-like inhibitor to the active site of the yeast enzyme is shown in Fig. 1 (21). This location of the active site has very recently been confirmed in an x-ray diffraction study still in progress. The corresponding residues in the active site of E. coli chorismate mutase are given in brackets.
Figure 1.
Proposed active site of the allosteric chorismate mutase from yeast. The end-oxabicyclic transition state analogue could be modeled into a region with high similarity to the active site of E. coli chorismate mutase-prephenate dehydratase (P protein) (21). Residues of the yeast chorismate mutase are indicated. Notations in brackets give the corresponding residues in the enzyme of E. coli.
Here, several amino acid residues of the proposed active site of yeast chorismate mutase have been replaced by site-directed mutagenesis and demonstrated to be part of the catalytic center. The enzymes were expressed in yeast with recombinant DNA technology. The chorismate mutases were purified to homogeneity and kinetic data determined. We identified Glu-246, an active site residue to be responsible for the narrow pH optimum curve of yeast chorismate mutase.
MATERIALS AND METHODS
Yeast Strains, Media, Plasmids, and Transformation.
All yeast strains used are derivatives of the S. cerevisiae laboratory strain X2180–1A (Mata, gal2, SUC2, mal, and CUP1) and X2180–1B (Matα, gal2, SUC2, mal, and CUP1). Derivatives of plasmid pME605 (22) were used for overexpression of chorismate mutase enzymes in strain RH1242 (Matα, aro7, and leu2–2). Plasmid pME781 was constructed by inserting a blunt-ended XhoI/BamHI fragment from pGEM7Zf+ (Promega) carrying the ARO7 gene as EcoRI fragment into the XbaI/HindIII-digested and flushed pJDB207 (23). The 5′ or 3′ terminal portions of the gene were replaced by a PCR-mutagenized (24) NdeI/XbaI or XbaI/BamHI fragment, respectively. The NdeI site has been introduced to the vector by site-directed mutagenesis at the ATG start codon of the ARO7 ORF. Plasmid YEp352 containing the different ARO7 alleles was used for expression of unfunctional chorismate mutases in yeast strain RH2185 by selection on the URA3 marker. RH2185 (Mata, suc2δ9, ura3–52, leu2–3, leu2–112, his4–519, and aro7::LEU2) was constructed by disruption of the ARO7 locus in RH1405 (Mata, suc2δ9, ura3–52, leu2–3, leu2–112, and his4–519), replacing most of the coding sequence with a functional LEU2 gene. Transformation was carried out by the LiOAc method (25). Minimal vitamins minimal medium for the cultivation of yeast was described earlier (26). Strains requiring amino acids were supplemented with 30 mg/liter l-tyrosine or 50 mg/liter l-phenylalanine.
Purification of Yeast Chorismate Mutase.
Yeast cells were grown at 30°C in 10-liter rotatory fermentors under aeration. Cells were harvested in mid-log phase at an OD546 of 4–6, washed twice with 50 mM K-phosphate buffer, and stored in 1 ml buffer/g wet cells at −20°C containing protease inhibitors [0.1 mM phenylmethylsulfonyl fluoride (PMSF), 0.2 mM EDTA, and 1 mM DTT]. For purification, 80–110 g of cell paste was thawed and run three times through a French pressure cell (18,000 psi). Cell debris was pelletted by centrifugation at 30,000 × g for 20 min. The enzyme was purified by ammonium sulfate precipitation, hydrophobic interaction chromatography on ethylamino-Sepharose, anion exchange chromatography on MonoQ, and gel filtration on Superdex200 according to the procedure described by Schmidheini et al. (3). Additionally, PMSF was added to the equilibration buffer for the ethylamino-Sepharose column, dialysis was used to desalt protein extracts, and chorismate mutase was applied to a second MonoQ column (HR 5/5). Chorismate mutase was detected by SDS/PAGE (27), enzyme assays, and immunoblotting with rabbit antibody raised against purified yeast chorismate mutase and a second antibody with alkaline phosphatase activity. Measurements of protein concentrations were performed by using the Bradford assay calibrated with BSA (28).
Enzyme Assays (29).
A stop assay as described previously was used for measuring enzymatic activity of yeast chorismate mutase. The assay was standardized by keeping enzymatic reactions at 30°C and equilibrating the spectrophotometer cell to the same temperature. Reaction volumes of 250 μl containing 100 mM Tris at pH 7.6, 2 mM EDTA, 20 mM DTT, optionally 0.1 mM tyrosine or 0.01 mM tryptophan, chorismate mutase enzyme, and chorismate in a range from 0.25 to 10 mM were used. The reaction was started by adding a mix of all ingredients to the prewarmed chorismate solution. Reaction was stopped by adding 250 μl of 1 M HCl. After an incubation time of 10 min, 4 ml of 1 M NaOH were added, and OD320 was measured against H2O. Blanks of increasing chorismate concentrations without enzyme were prepared, and their absorbances were subtracted from optical densities measured for enzyme activities. A calibration curve was measured with different known phenylpyruvate concentrations that had been treated in the same way as the enzyme reactions. The molecular extinction coefficient at 30°C was determined as 13,095 M−1⋅cm−1. The collected data were transformed to international units (μmol/min) per mg enzyme. The maximum velocity Vmax, the Hill coefficient nH, and the substrate concentration at half-maximal velocity S0.5 or Km were determined using a computer program applying the Quasi-Newton method (Davidon–Fletcher–Powell algorithm) to fit optimal curves to the data (30). For substrate saturation curves data were fitted either to the Michaelis–Menten equation [v = Vmax [S] (Km + [S])−1] or to the Hill equation [v = Vmax [S]n ([S]n + S′)−1], where S′(1/n) = S0.5. Eadie–Hofstee plots (v [S]−1 vs. v) were drawn to decide to which equation a set of kinetic data had to be applied. Enzyme kinetics without cooperativity result in a linear curve, whereas even small degrees of cooperativity result in concave curvatures of the kinetic data (31). Hill plots (log(v (Vmax − v)−1) vs. log [S]) were used to calculate Hill coefficients. The resulting Vmax values were transformed to catalytic constants [kcat = Vmax Mr E0−1 (60s)−1; substrate turnover per enzyme dimer]. The numerical values turned out to be identical due to the use of 60,000 as molecular weight for chorismate mutase. Determination of catalytic activity of chorismate mutases expressed from plasmid Yep352 in strain RH2185 was performed with crude extracts, which were prepared from 500 ml of minimal vitamins medium following the method used for enzyme purification. Additionally, crude extracts were desalted over PD-10 columns. For determination of the pH optima, a universal buffer solution with a pH range of 2.6–11.8 containing 30 mM citric acid, 30 mM KH2PO4, 30 mM H3BO3, 30 mM diethylbarbituric acid, and different concentrations of NaOH was used.
RESULTS
Analysis of Active Site Residues of Yeast Chorismate Mutase.
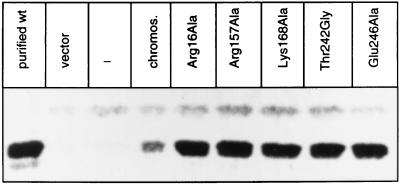
The residues that were proposed to compose the active site of yeast chorismate mutase are based on a comparison between the x-ray structures of yeast and E. coli chorismate mutase. We have analyzed these residues by a mutational study. Arg-16, Arg-157, Lys-168, and Glu-246 were exchanged for alanine and residues Glu-198 and Thr-242 for glycine, respectively. The resulting mutant alleles of the ARO7 gene were named ARG16ALA, ARG157ALA, LYS168ALA, GLU198GLY, THR242GLY, and GLU246ALA. Arg-16, Arg-157, Lys-168, and Glu-198 are conserved between yeast and E. coli. Glu-246 is a glutamine at the corresponding position of E. coli chorismate mutase (P protein), and Thr-242 is a serine (Fig. 1). When expressed in yeast, ARG16ALA, ARG157ALA, LYS168ALA, GLU246ALA, and THR242GLY were not able to complement the auxotrophy of an aro7 null mutation, whereas the GLU198GLY allele complemented the aro7 phenotype. Western blots of cell extracts of yeast strain RH2185 harboring the different ARO7 alleles on the 2-μ plasmid YEp352 reveal that all unfunctional ARO7 alleles are expressed in similar amounts as stable proteins with a molecular weight that corresponds to purified wild-type chorismate mutase (Fig. 2). However, cell extracts of strains expressing alleles ARG16ALA, ARG157ALA, LYS168ALA, GLU246ALA and THR242GLY showed no detectable chorismate mutase activity at a fixed substrate concentration of 1 mM chorismate even if excess amounts of protein extracts were used. Thus, residues Arg-16, Arg-157, Lys-168, Glu-246, and Thr-242 are essential for chorismate mutase activity and are likely to compose the active site of the enzyme. Presumably hydrogen bonds between these residues and the substrate might play a critical role in catalysis (Fig. 1).
Figure 2.
The chorismate mutases with amino acid replacements Arg16Ala, Arg157Ala, Lys168Ala, Thr242Gly, and Glu246Ala, which exhibit no in vitro activity are properly expressed in yeast. The immunoblot shows 20 μg of crude extracts of yeast strain RH2185 harboring the different ARO7 alleles on the high copy number plasmid YEp352. The protein was hybridized with polyclonal rabbit antibody raised against purified chorismate mutase.
Comparison of Active Site Residues Not Conserved Between Yeast and E. coli.
Thr-242, Asn-194, and Glu-246 of yeast chorismate mutase are replaced by a serine, an aspartate, and a glutamine residue at the corresponding positions in the E. coli P protein, respectively (Fig. 1). Thr-242 in yeast and Ser-84 in E. coli, respectively, are proposed to bind to the ring carboxylate of chorismate by their hydroxyl groups and might have a similar function in the catalytic process of both enzymes. Asn-194 is supposed to interact with the substrate by its backbone NH group. The sidechain of Glu-246 presumably interacts with the ether oxygen of chorismate, and the replacement to glutamine in E. coli manifests the most basic difference between the active sites of these two enzymes. Therefore, Asn-194 and Glu-246 of yeast chorismate mutase were replaced by aspartate and glutamine, respectively, to match the E. coli sequence. Both of these ARO7 alleles are able to complement an aro7 mutation when expressed in yeast. In addition, both ASN194ASP and GLU246GLN are stably expressed in yeast and the crude extracts show considerable chorismate mutase activity (data not shown). Therefore, we decided to purify these proteins and determine the catalytic parameters.
Replacements Glu198Gly and Glu246Gln Have Strong Effects on Chorismate Mutase, Whereas Asn194Asp Resulted in Minor Changes.
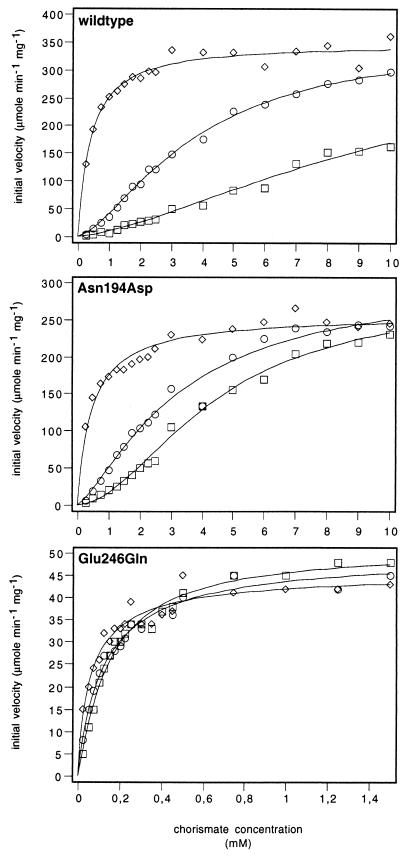
Asn194Asp, Glu198Gly, and Glu246Gln proteins that were purified to homogeneity and compared with wild-type enzyme exhibited no difference in the elution behavior on the different FPLC columns or in their stability during purification. These observations indicate that no serious damage on the overall enzyme structure occurred by the amino acid replacements. Kinetic properties of the purified enzymes were then determined and compared with wild-type enzyme (Fig. 3; Table 1). The substrate saturation curves of the purified enzymes were determined in the absence of effectors, in the presence of the inhibitor tyrosine (100 μM), and in the presence of the activator tryptophan (10 μM) (Fig. 3). Data were fitted to equations describing Michaelis–Menten-type hyperbolic saturation or Monod–Wyman–Changeux-type cooperative saturation. The kinetic parameters are given in Table 1. The unliganded and the tyrosine-inhibited wild-type chorismate mutases show cooperativity with respect to chorismate. Addition of tryptophan lead to Michaelis–Menten-type kinetics. Regulation by the effectors tyrosine and tryptophan is expressed by an increase or decrease in substrate affinity, respectively (Table 1).
Figure 3.
Substrate saturation plots of wild-type and mutant chorismate mutases. The enzymes were assayed with 10 μM tryptophan (◊), without effectors (○), or in the presence of 100 μM tyrosine (□). The data were fitted to functions describing cooperative or Michaelis–Menten-type saturation.
Table 1.
Kinetic parameters of wild-type and mutant chorismate mutases
| Protein | Value of protein with amino acid replacement
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inhibited (100 μM tyrosine)
|
Unliganded
|
Activated (10 μM tryptophan)
|
||||||||||
| kcat, s−1 | Km, S0.5, mM | nH | kcat/Km, mM−1⋅s−1 | kcat, s−1 | Km, S0.5, mM | nH | kcat/Km, mM−1⋅s−1 | kcat, s−1 | Km, S0.5, mM | nH | kcat/Km, mM−1⋅s−1 | |
| Wild type | 387 | 11.8* | 1.4 | 32.8 | 360 | 3.8 | 1.6 | 94.7 | 348 | 0.4 | 1.1† | 870.0 |
| Asn194Asp | 282 | 4.4 | 1.77 | 64.1 | 305 | 3.3 | 1.4 | 92.4 | 256 | 0.5 | 1.0† | 512.0 |
| Glu246Gln | 52 | 0.15 | 1.1† | 346.7 | 50 | 0.13 | 0.92† | 384.6 | 46 | 0.06 | 0.85† | 766.7 |
Values for kcat, Km, S0.5 were determined by fitting initial velocity data to equations describing hyperbolic or cooperative saturation, respectively. Hill coefficients (nH) were calculated from Hill plots by linear regression.
Values resulted in uncertainty intervals >10% from the fitting procedure.
Linear Eadie–Hofstee plots indicated hyperbolic saturation.
Purified Glu198Gly chorismate mutase exhibits a dramatic drop in in vitro chorismate mutase activity. Measurement of chorismate mutase activity of this mutant enzyme was only possible when tryptophan was added to the reaction. Yet, chorismate concentrations up to 10 mM were not sufficient for saturation indicating markedly increased Km values (data not shown). These data suggest that hydrogen bonding to the C4 hydroxyl group of chorismate is critical for enzymatic catalysis (Fig. 1).
Replacement of Asn-194 by Asp does not cause significant alterations on the catalytic and regulatory properties of yeast chorismate mutase. Turnover numbers of about 300 were found, which does not differ markedly from the wild-type enzyme (Table 1). Asn194Asp is still regulated by both effectors, tyrosine and tryptophan. The Km value is somewhat lower than that of wild-type chorismate mutase and cannot be shifted to a high value by addition of tyrosine as found for wild-type chorismate mutase.
Replacement of Glu-246 by Gln results in a profound effect on catalytic activity of yeast chorismate mutase. The mutant enzyme shows a large increase in affinity toward the substrate chorismate. The Km value of 0.4 mM found for activated wild-type chorismate mutase is further reduced to approximately 0.1 mM in the Glu246Gln mutant enzyme. This value is not markedly changed upon addition of tyrosine and tryptophan, respectively, and the Glu246Gln enzyme does not show marked response toward the regulatory amino acids. Although Glu246Gln exhibits a 7-fold decrease in catalytic activity compared with the wild-type enzyme, catalytic efficiency is elevated due to its highly decreased Km value. This suggests that a glutamate in yeast chorismate mutase in close proximity to the ether oxygen is required for strong regulation.
Effect of pH on Enzyme Activities.
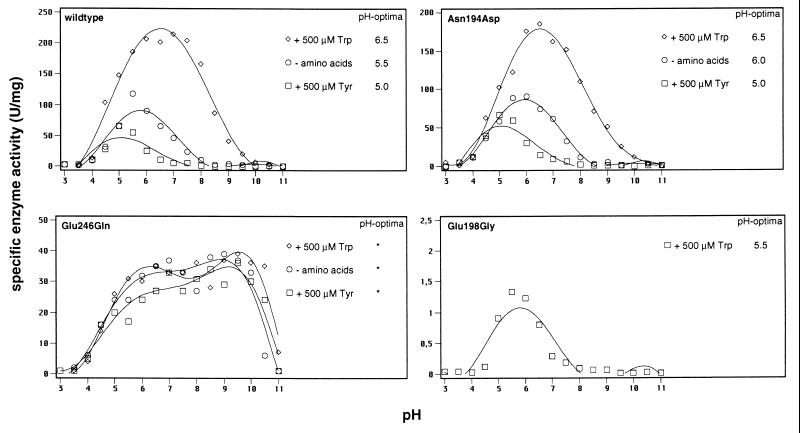
Activity of wild-type yeast chorismate mutase shows strong pH-dependency with highest activities at acidic pH (Fig. 4). Addition of the inhibitor tyrosine further lowers the optimum pH. In contrast, tryptophan increases the pH value where highest activity is measured. Because our mutations altered the overall charge of yeast chorismate mutase, Asn194Asp, Glu198Gly, and Glu246Gln were tested for the dependency of enzymatic activity on pH. The specific enzyme activities of these mutants were determined over a pH range of 2.5–11 in the absence of amino acids and in the presence of tyrosine and tryptophan (Fig. 4).
Figure 4.
pH optima for the wild-type and mutant chorismate mutases under different effector conditions. The optima are given on the right side. Glu198Gly was assayed only with tryptophan because specific activity could not be determined for the nonactivated state.
Activity of Glu198Gly chorismate mutase was only detectable in the presence of the activator tryptophan. Glu198Gly displays highest activity at acidic pH similar to wild-type chorismate mutase. Compared with the tryptophan-activated wild-type enzyme, the optimum value of Glu198Gly is shifted to a slightly lower pH.
Dependency of activity for Asn194Asp chorismate mutase on pH is similar to that observed for wild-type chorismate mutase under all effector conditions, and almost identical values for the pH optima of both enzymes were found (Fig. 4).
To our surprise, activity of Glu246Gln chorismate mutase does not respond to variations of the pH over a broad range. Catalytic activity of the enzyme is observed over a pH range between 4 and 10 with no well defined optimum pH. Measurements at subsaturating chorismate concentrations of 100 μM resulted in the same wide region of optimum pH for this enzyme (data not shown). We therefore conclude that a single amino acid substitution at the active site of yeast chorismate mutase changes the well defined pH optimum curve to enzymatic activity over a broad range resulting in a shallow pH profile for chorismate mutase.
DISCUSSION
The regulation of cytoplasmic pH is necessary for every living cell. Almost every physiochemical and biochemical reaction is influenced by the hydrogen concentration. Usually a narrow range of optimum pH values limits the activity and/or stability of most proteins. Many enzymes are irreversibly denatured at extreme pH values. Because marked fluctuations of the pH often occur in biological systems, pH homeostasis has to compensate for changes in the environment. The intracellular pH value of yeast is kept at a value of between pH 5 and 6 (32). Optimum pH for activity of yeast chorismate mutase between 5.0 and 6.5 coincides with the findings for intracellular pH values and the cellular biochemistry here seems to be well adapted to intracellular conditions. Although enzymes normally contain several ionizing groups, the variation of initial velocity with pH might result in a bell-shaped curve because it is the first group to protonate or deprotonate as the pH is moved up or down from the optimum. The activity of yeast chorismate mutase was found to be highly pH-dependent, rising dramatically under acidic conditions. We have identified Glu-246 of yeast chorismate mutase as active site residue that is responsible for the narrow pH optimum of the enzyme. The Glu246Gln enzyme closely resembles B. subtilis chorismate mutase in that it shows no preferential pH optimum over a broad range (6). In B. subtilis no equivalent residue to Glu-246 was found in proximity to the ether oxygen. In E. coli P protein a glutamine interacts with the ether oxygen of the substrate, and the enzyme shows optimum pH at alkaline conditions. It appears that Glu-246 in yeast chorismate mutase has to be protonated for functionality of the enzyme and plays a critical role in catalyzing the regulated rearrangement of chorismate to prephenate. In comparison to glutamate, glutamine for chemical reasons is expected to form a weaker hydrogen bond, which could provide an explanation for the reduction in catalytic activity. Also a more active role might be attributed to Glu-246. This residue could be involved in protonation of the ether oxygen of chorismate catalyzing breakage of the C-O bond. In any case, wild-type yeast chorismate mutase would require an undissociated carboxylic group of Glu-246 either to form a hydrogen bond to the ether oxygen and stabilize partial negative charge or to protonate the ether oxygen. Kinetic studies of the chorismate mutase/prephenate dehydrogenase from E. coli (T protein) also proposed protonation of the bridge oxygen of the substrate by a protonated ionizable group from the enzyme (9). Guilford et al. (9) concluded that their data were consistent with a reaction pathway in which the rate-limiting step could be the cleavage of the chorismate ether bond. At low substrate concentration, the rate limiting transition state occurs before the transition state of the chemical rearrangement (8). At saturating conditions, dissociation of prephenate is rate limiting (6). The existence of a glutamate in the active site of yeast chorismate mutase might indicate a similar mechanism where cleavage of the C-O ether bond is also facilitated by protonation of the ether oxygen. Contrarily, studies of the monofunctional enzyme of B. subtilis favor a mechanism that is exclusively mediated by binding of the substrate to the active site. Here no functional residue is specifically involved in a catalytic step, although there may well be electrostatic and van der Waals influences on the conformer of the substrate that is bound (13). Conservation in the distribution of charged amino acids between the yeast and the bacterial enzymes of E. coli (P protein) and B. subtilis and functionality of the Glu246Gln yeast enzyme indicate that all enzymes support the rearrangement of chorismate by binding of the active conformer and providing the optimal structure and electrostatic environment for the transition state of the rearrangement. The uncatalyzed reaction has been shown to be strongly dependent on the polarity of the solvent (11), indicating polarity of the transition state. The active site of yeast chorismate mutase, with a negative charge at the apex and three positive groups at the other three vertices, might be electrostatically appropriate for stabilizing the polarity of the transition state. Several interactions between the enzyme and chorismate are necessary to overcome the energy penalty for the chorismate mutase reaction, possibly by freezing internal rotations of the enolpyruvyl side chain, thus allowing the reaction to proceed. Glu-246 in yeast and a glutamate in proximity to the ether oxygen in the chorismate mutase of E. coli (T protein) may additionally function as ionizable groups, facilitating protonation of negative charge developing on the ether oxygen. (We assume here that the chorismate mutase regions of the E. coli P and T proteins have similar three-dimensional structures in view of a sequence identity of 26%.)
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie and by Grant GM 06920 from the National Institutes of Health.
References
- 1.Andrews P R, Smith G D, Young I G. Biochemistry. 1973;12:3492–3498. doi: 10.1021/bi00742a022. [DOI] [PubMed] [Google Scholar]
- 2.Turnbull J, Cleland W W, Morrison J F. Biochemistry. 1991;30:7777–7782. doi: 10.1021/bi00245a016. [DOI] [PubMed] [Google Scholar]
- 3.Schmidheini T, Mösch H-U, Evans J N S, Braus G. Biochemistry. 1990;29:3660–3668. doi: 10.1021/bi00467a011. [DOI] [PubMed] [Google Scholar]
- 4.Schmit J C, Zalkin H. Biochemistry. 1969;8:174–181. doi: 10.1021/bi00829a025. [DOI] [PubMed] [Google Scholar]
- 5.Dopheide T A A, Crewther P, Davidson B E. J Biol Chem. 1972;247:4447–4452. [PubMed] [Google Scholar]
- 6.Gray J V, Eren D, Knowles J R. Biochemistry. 1990;29:8872–8878. doi: 10.1021/bi00489a051. [DOI] [PubMed] [Google Scholar]
- 7.Sogo S G, Widlanski T S, Hoare J H, Grimshaw C E, Berchthold G A, Knowles J R. J Am Chem Soc. 1984;106:2701–2703. [Google Scholar]
- 8.Addadi L, Jaffe E K, Knowles J R. Biochemistry. 1983;22:4494–4501. doi: 10.1021/bi00288a022. [DOI] [PubMed] [Google Scholar]
- 9.Guilford W J, Copley S D, Knowles J R. J Am Chem Soc. 1987;109:5013–5019. [Google Scholar]
- 10.Gray J V, Knowles J R. Biochemistry. 1994;33:9953–9959. doi: 10.1021/bi00199a018. [DOI] [PubMed] [Google Scholar]
- 11.Copley S D, Knowles J R. J Am Chem Soc. 1987;109:5008–5013. [Google Scholar]
- 12.Görisch H. Biochemistry. 1978;17:3700–3705. doi: 10.1021/bi00611a004. [DOI] [PubMed] [Google Scholar]
- 13.Chook Y M, Gray J V, Ke H, Lipscomb W N. J Mol Biol. 1994;240:476–500. doi: 10.1006/jmbi.1994.1462. [DOI] [PubMed] [Google Scholar]
- 14.Haynes M R, Stura E A, Hilvert D, Wilson I A. Science. 1994;263:646–652. doi: 10.1126/science.8303271. [DOI] [PubMed] [Google Scholar]
- 15.Lee A Y, Karplus P A, Ganem B, Clardy J. J Am Chem Soc. 1995;117:3627–3628. [Google Scholar]
- 16.Lee A Y, Steward J D, Clardy J, Ganem B. Chem Biol. 1995;2:195–203. doi: 10.1016/1074-5521(95)90269-4. [DOI] [PubMed] [Google Scholar]
- 17.Galopin C C, Zhang S, Wilson D B, Ganem B. Tetrahedron Lett. 1996;37:8675–8678. [Google Scholar]
- 18.Chook Y M, Ke H, Lipscomb W N. Proc Natl Acad Sci USA. 1993;90:8600–8603. doi: 10.1073/pnas.90.18.8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue Y, Lipscomb W N, Graf R, Schnappauf G, Braus G. Proc Natl Acad Sci USA. 1994;91:10814–10818. doi: 10.1073/pnas.91.23.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sträter N, Hakånsson K, Schnappauf G, Braus G, Lipscomb W N. Proc Natl Acad Sci USA. 1996;93:3330–3334. doi: 10.1073/pnas.93.8.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue Y, Lipscomb W N. Proc Natl Acad Sci USA. 1995;92:10595–10598. doi: 10.1073/pnas.92.23.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graf R, Dubaquié Y, Braus G H. J Bacteriol. 1995;177:1645–1648. doi: 10.1128/jb.177.6.1645-1648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beggs J D. Nature (London) 1978;275:104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- 24.Giebel L B, Spritz R A. Nucleic Acids Res. 1990;18:4947. doi: 10.1093/nar/18.16.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito H, Jukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miozzari G, Niederberger P, Hütter R. J Bacteriol. 1978;134:48–59. doi: 10.1128/jb.134.1.48-59.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lämmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 29.Schmidheini T, Sperisen P, Paravicini G, Hütter R, Braus G. J Bacteriol. 1989;171:1245–1253. doi: 10.1128/jb.171.3.1245-1253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fletcher R, Powell M J D. Computer J. 1963;6:163–168. [Google Scholar]
- 31.Newell J O, Schachman H K. Biophys Chem. 1990;37:183–196. doi: 10.1016/0301-4622(90)88018-n. [DOI] [PubMed] [Google Scholar]
- 32.Cimprich P, Slavik J, Kotyk A. FEMS Microbiol Lett. 1995;130:245–252. doi: 10.1111/j.1574-6968.1995.tb07727.x. [DOI] [PubMed] [Google Scholar]