Abstract
Behaviors, morphologies, and genetic loci directly involved in reproduction have been increasingly shown to be polymorphic within populations. Explaining how such variants are maintained by selection is crucial to understanding the genetic basis of fertility differences, but direct tests of how alleles at reproductive loci affect fertility are rare. In the sea urchin genus Echinometra, the protein bindin mediates sperm attachment to eggs, evolves quickly, and is polymorphic within species. Eggs exposed to experimental sperm mixtures show strong discrimination on the basis of the males’ bindin genotype. Different females produce eggs that nonrandomly select sperm from different males, showing that variable egg–sperm interactions determine fertility. Eggs select sperm with a bindin genotype similar to their own, suggesting strong linkage between female choice and male trait loci. These experiments demonstrate that alleles at a single locus can have a strong effect on fertilization and that reproductive loci may retain functional polymorphisms through epistatic interactions between male and female traits. They also suggest that positive selection at gamete recognition loci like bindin involves strong selection within species on mate choice interactions.
Genes directly involved in reproduction are tightly associated with organismal fitness, and the simple evolutionary expectation is that selection should act quickly to weed out all but the best variant at these loci (1, 2). Despite this, there is a surprising degree of natural polymorphism in reproductive tactics and in reproductive genes within many species. Polymorphic mating tactics or mate preferences exist in insects, fish, birds, lizards, and amphibians (3–8). Among insects, accessory gland proteins are passed to females during insemination, and different alleles are associated with differential sperm success (1). The ability to displace sperm from a prior mating varies widely among males in insects, crustacea, molluscs, and mammals (9–12) and appears to be under genetic control (13). In humans, alleles that reduce fertility are generally treated as genetic diseases, but some variants associated with reduced reproduction are widespread (14). Is it possible that such variation is maintained by selection? Or are alleles at reproductive loci always subject to powerful selective sweeps (2) in which the best allele quickly comes to dominate?
It has been difficult to answer in detail how selection operates at most polymorphic loci involved in mating strategies because the genetic determinants of these traits are generally unknown (15). In some cases, however, adult interactions during mating are simplified and may rely on fewer gene products, especially in many terrestrial plants and marine taxa in which gamete recognition is pivotal (16). Study of molecular evolution of genes involved in gamete recognition has shown that positive selection for amino acid divergence occurs between species (17–21), but whether this selective regimen also operates within species is unknown.
In sea urchins, external fertilization is mediated by attachment to the egg of the sperm protein bindin (18, 22, 23). In the genera Echinometra and Strongylocentrotus, bindin varies greatly between species and, like many proteins involved in gamete recognition (24) or other reproductive interactions (25, 26), evolves by positive selection for amino acid divergence (21, 27). In species for which positive selection has been reported, bindin is also highly polymorphic, with multiple allelic polymorphisms that are generated by amino acid replacements or coding region insertion/deletions (21, 28). Although high replacement/silent site variation in these alleles suggests that they are under selection (21), there have been no direct tests of functional differences among bindin alleles. One alternative is that bindin allele differences are functionally neutral and that the marked bindin polymorphism is a reflection of overall high genetic variation in the sea urchin nuclear genome (29). Understanding functional differences among bindin alleles may clarify the mechanisms underlying positive selection on gamete recognition loci and allow characterization of selection rules governing evolution of mating system polymorphisms.
Materials and Methods
Characterization of Bindin Alleles in Adults.
Adults used in this study were collected from Pidi Reef, Guam, and Oahu, HI. Only Hawaiian animals were used in crosses. These were shipped to Boston and maintained in recirculating sea water tables until use. Adult genotypes were obtained by PCR of tube foot DNA isolated from stabled animals. PCR was conducted with the error-correcting enzyme mix rTth (Perkin–Elmer) by using primers listed in ref. 21. PCR products from the first 105 codons of the mature bindin gene [showing the strongest signal of positive selection (21)] were cloned into plasmid vectors. Between 6 and 10 positive clones were sequenced for each individual to obtain diploid gene sequences for each adult used as a parent in this study. Phylogenetic trees from aligned allele sequences were generated by using parsimony (paup 3.1.1). Gaps were treated as single characters, weighted five times as much as nucleotide substitutions. Sequences from the sister species to Echinometra mathaei, E. sp. nov. A, were used as outgroups.
Crosses.
Gametes were obtained by minimal KCl injection. Eggs were washed three times in filtered, artificial sea water. Dry sperm collected at the gonopores were diluted ≈1,000-fold before use. Sperm from different males were diluted, and concentrations were measured by using a Coulter cell counter. Sperm were mixed in approximately equal concentrations and aliquoted into separate culture dishes. Eggs from different females were added to each dish, and fertilization was allowed to proceed for 30 min. All crosses were done within 10 min of gamete dilution. Crosses with the same sperm mixtures were started within 1 min of one another. Fertilized eggs were washed in filtered sea water and allowed to develop for 48 h at room temperature, at which time larvae were at an early pluteus stage. Eggs that did not show at least 95% fertilization were discarded.
Characterization of Bindin Alleles in Larvae.
Single larvae were collected in 5 μl of filtered sea water, 5 μl of 4 mg/ml proteinase K was added, and the mixture was incubated at 70°C for 15 min and then 95°C for 5 min. The most variable section of the mature bindin coding region was amplified from 0.5 μl of the DNA extraction from each individual larva tested. Bindin allele clusters A and B (Fig. 1) can be positively identified with DdeI or ApoI restriction digestions of the associated PCR products, respectively. PCR product from each larva was digested with DdeI or ApoI, and the restriction fragment length polymorphism patterns were visualized on ethidium bromide-stained 3% agarose gels. In each cross, individual genotypes of 40–70 larvae were recorded.
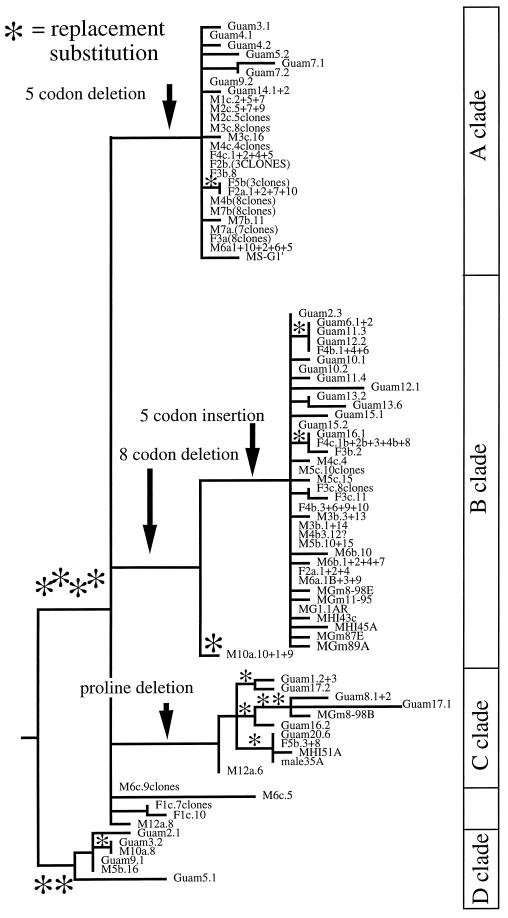
Figure 1.
Allele genealogy for bindins in the sea urchin E. mathaei. Sequences from the first 105 codons of the mature bindin coding region show multicodon insertions and deletions that define major clades. Alleles not labeled “Guam” are from Hawaiian individuals. The tree is rooted to the sister species E. sp. nov. A and is a minimum length, strict consensus phylogram (paup 3.1.1). All polymorphic substitutions and insertions/deletions have a consistency index of 1.0. Polymorphic amino acid changes are denoted as asterisks.
Sperm Utilization Analysis.
The percentage of larvae sired by each male was inferred from the restriction fragment length polymorphism patterns of the 40–70 larvae assayed. These percentages were compared with random expectation by a χ2 test of the observed number of offspring of each male versus the number expected if sperm use was random. The expectations were based on the measured concentrations of sperm from each male used in each cross. Sperm fertilization probabilities corrected for differences in sperm density were calculated as SR/(SR + 1) where SR = (L1/L2)∗(S2/S1). L1 and L2 are the number of larvae sired by males 1 and 2, respectively. S1 and S2 are the sperm concentrations from the two males.
Results
Allele Genealogies.
Among 87 bindin sequences from 53 individuals of the tropical sea urchin E. mathaei, alleles fall into four main classes that differ enormously in the 105-codon region of the protein under positive selection (Fig. 1). Allele clusters A and B are the most common, appear to be the most recently derived, and occur in both Hawaii and Guam. Clades C and D are more basal and make up ≈30–35% of the alleles in each population.
The major clades show strong amino acid differentiation but few silent substitutions. Clades A and B differ by two separate deletions (of 5 and 8 codons, respectively, at positions 307–321 and 244–273) plus an insertion (5 codons, positions 94–108). These two clades differ at 18 of 105 aligned amino acid positions because of the insertion or deletion of groups of repeated amino acid motifs (Fig. 2). Of these 18 differences, 11 involve charged or polar residues, and clade B sequences have two more negatively charged amino acids than clade A sequences. There are no silent substitutions between clades A and B.
Figure 2.
Comparison of the predicted amino acid sequence of the first 105 codons of a representative allele from clades A and B. Vertical tick marks denote identical amino acids.
Within clusters A and B, most sequences have zero or one difference, suggesting they are very recently evolved. Of the three substitutions seen in more than one individual, two are amino acid replacements, and one is silent. Of the 63 individual sequences in these two clades, there were 41 singleton substitutions (not seen in any other individual). This rate of sequence heterogeneity (0.15%) is about that expected from incorporation of errors by rTth polymerase (30).
Clade C is defined by a deletion of a proline and a single silent substitution. Within this clade, all five phylogenetically informative nucleotide substitutions are replacement changes (Fig. 1, asterisks). Clades A, B, and C have diverged significantly from one another but cluster together in a clade defined by four substitutions, all of which are amino acid replacements. Finally, clade D is defined by two amino acid replacements and includes a single phylogenetically informative substitution, which is also a replacement.
In total, there are 15 polymorphic amino acid replacements and four polymorphic insertions/deletions that occur in >1 individual (Fig. 1). By contrast, there are only two silent polymorphisms that occur in more than one individual. Ignoring the insertions, the ratio of replacement to silent polymorphisms is slightly higher than neutral expectations (P = 0.04), consistent with the action of positive selection on divergence of alleles within species.
Functional Differences Among Alleles.
To test whether bindin alleles in clades A and B are functionally different, sperm competition/egg choice experiments were conducted in which sperm from males of known bindin genotype were mixed and used to fertilize eggs of individual females. The resulting 48-h larvae were individually subjected to PCR and restriction digestion to identify their sires and to test paternity against expectations based on concentrations of sperm from each male in the original mixtures.
These experiments show that eggs are often fertilized nonrandomly by sperm from different males and that males have different fertilization characteristics based on their bindin genotypes (Table 1). In 35 of 57 experiments (n = 3,363 larvae), there was nonrandom sperm use. Sperm choice was more apparent in experiments with homozygote males; when eggs were given the choice of sperm from AA and BB males, nonrandom fertilization was seen in 80% of the crosses (Table 1). Differential fertilization patterns were often strong, with up to 95% of larvae resulting from one of the males. The probability of fertilization averaged 76% (±3% SEM) for the winning male and 24% for the loser. Adjusted for slight differences in sperm density, these results show that the winning male is three times more likely to fertilize a particular egg than the losing male.
Table 1.
Sperm utilization patterns from experiments with sperm mixtures
| Homozygote vs. homozygote males
| |||||
|---|---|---|---|---|---|
| AA vs. BB sperm | AA wins* | BB wins | Ties† | No. of females | No. of males‡ |
| AA females | 12 | 0 | 2 | 7 | 7, 5 |
| BB females | 1 | 5 | 1 | 4 | 5, 5 |
| AB females | 1 | 1 | 2 | 3 | 4, 3 |
| Homozygote vs. homozygote males
| |||||
|---|---|---|---|---|---|
| AA (or BB) vs. AB sperm | Homozygote wins | Heterozygote wins | Tie | No. of females | No. of males‡ |
| AA females | 2 | 5 | 8 | 4 | 7, 4 |
| BB females | 1 | 3 | 2 | 2 | 3, 4 |
| AB females | 0 | 4 | 7 | 4 | 7, 4 |
Winners show significantly higher fertilization (χ2, P < 0.02). Average number of larvae screened per cross = 59.
† No statistically significant dominance.
‡ Number of different males used in experimental mixtures. Listed as male 1 and male 2.
Further examination of these results revealed that the female bindin genotype is a good predictor of egg choice. In particular, AA females (n = 7) tended to choose AA males over BB males (12 of 14 tests). Likewise, BB females (n = 4) tended to choose BB males (5 of 7 tests, Fig. 3). Females chose sperm from the male with which they shared the largest number of bindin alleles in 28 of 32 experiments showing nonrandom sperm use (Table 2). An additional 21 tests were ties, and in 4 tests, both males were equally similar. Random sperm use patterns tended to be associated with crosses involving heterozygotes. For example, females that did not choose among sperm tended to have heterozygous bindin genotypes. Of the 22 trials in Table 1 that showed no strong choice, 9 involved AB females. Most of the rest of the ties (10 of 13) involved heterozygote males. Overall, 60% of crosses with AB females and 53% of crosses with AB males showed no choice.
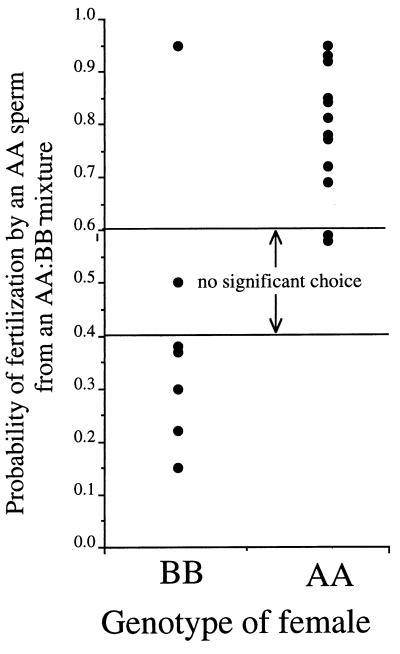
Figure 3.
Relationship between fertilization and female genotype. Females with BB genotypes tended to be fertilized best by sperm from BB males, whereas AA females tended to use sperm of AA males. Points represent separate experiments from which 50–70 larvae were typed. Horizontal lines represent 95% confidence limits for detecting significantly nonrandom sperm choice in an experiment with an average number of typed larvae. Sample sizes: experiments with AA females = 7, 6, 5 (females, AA males, BB males). BB females = 4, 4, 4.
Table 2.
Sperm use patterns depend on the similarity of male and female bindin genotypes
| Female genotype | Winning male
|
Ties | |
|---|---|---|---|
| Most similar | Least similar | ||
| AA | 16 | 3 | 11 |
| AB | 4 | 0 | 8 |
| BB | 8 | 1 | 2 |
In each cross, the male that shared the largest number of bindin alleles with a female is considered the most similar. Values represent the number of crosses in which most similar or least similar males showed significantly enhanced fertilization. Females with different bindin genotypes consistently tended to use sperm from males with the greatest bindin similarity.
Two lines of evidence suggest that strong differences between use of sperm from different males do not merely represent the success of more vigorous sperm. First, eggs from different females do not all choose the same sperm. Experiments in which sperm mixtures were used to fertilize eggs of different females showed that some eggs accepted sperm from the first male more easily, whereas others chose sperm from second male. For example, among 13 pairs of males used to fertilize eggs of 2–4 females with different genotypes, each male of the pair was the winner in at least one cross. The second line of evidence is that eggs from the same female tended to make consistent choices based on the bindin genotype of the males in separate experiments. Female choice was considered consistent if eggs from the same female chose males with the same bindin genotype. Overall consistency in choices was 93% among five females, each tested with three to four sets of AA:BB males.
Discussion
These experiments show that different bindin alleles confer different fertilization properties in free spawning sea urchins. Although other aspects of individual history like nutritional condition and readiness to spawn may affect fertilization ability, variable choice among females with different genotypes and high consistency within females suggests that fertilization differences reflect male bindin genotype, not some other aspect of male genotype or physiology. Moreover, fertilization differences result from the interaction of egg and sperm traits that are under the control of both female and male genotypes. There is no universal “best bindin.” Instead, a bindin allele has a functional advantage only in the context of a particular female genotype. Because different bindin alleles function best with different mating partners, fertility variation within populations is not necessarily eliminated by rapid selective sweeps. Such complex effects of alleles at multiple loci are thought to stabilize other complex polymorphisms, like the apo E polymorphism in humans linked to cholesterol metabolism (31), and may be a common feature in reproductive polymorphisms as well.
In sea urchins, bindin is not known to be expressed in females (22). Thus, the association of female bindin genotype with sperm choice pattern must be caused by expression of independent loci, the products of which interact at gamete surfaces with bindin. The presumptive female loci are currently undescibed but must be polymorphic as well. Nonrandom fertilization associated with the bindin polymorphism may reflect the linkage disequilibrium between male and female loci that is predicted by models of assortative mating (32). Mating system genes in fungi are known to be tightly linked (33), but the gene responsible for the urchin sperm receptor is not well characterized (34), and direct linkage tests are currently impossible in sea urchins. An alternative might be that bindin is expressed very slightly on egg surfaces and interacts with sperm bindin in a fashion analogous to the peptide pheromone of the ciliate Euplotes raikovi (35).
In Echinometra mathaei, individuals with similar bindin genotype fertilize best together (Table 1) and represent potential mating guilds within populations, where mating guilds are defined as groups of individuals that reproduce more successfully within the group than between groups (20). Existence of mating guilds is predicted by some models of mating system evolution based on interaction of male and female loci (32) and is seen as intraspecific assortative mating patterns in a number of taxa (36). However, in many such models, polymorphism generated by mating guilds is unstable, and genetic hitchhiking among loci in linkage disequilibrium results in fixation of one set of alleles (37, 38).
What mechanisms could explain persistence of functional fertilization differences in E. mathaei? In the case of the bindin-egg receptor system, at least three possible mechanisms exist for the maintenance of multilocus polymorphism. Heterozygote advantage may operate in bindin and would slow the loss of alleles (39). Although heterozygote males are frequently involved in ties, they are more successful than homozygote males in ≈40% of crosses completed to date (Table 1). Mild heterozygote advantage may account for the maintenance of polymorphism and the fact that there is only a slight excess of homozygote bindin genotypes within populations (43% instead of the 37% predicted under Hardy–Weinberg assumptions; n = 137, P = 0.07). Heterozygote advantage at other loci under positive selection like MHC or the s-locus of plants is known to enhance polymorphism but also results in transspecies allele clades (26, 40–42). Such sharing of allele clades among species is not observed in bindin and is not predicted for loci thought to be intrinsic to reproductive isolation among species (16). However, it is possible that only certain bindin heterozygotes are at a reproductive advantage and that general models of balancing selection (39), in which all alleles are equally advantageous when they are in a heterozygous state, do not apply. Further experiments are needed to understand the mating rules involving males that are heterozygous at the bindin locus.
Alternatively, egg choice of sperm may be nontransitive with females preferring male A over B, B over C, but C over A. Such nontransitivities result in a rock-scissors-paper game and can enhance mate choice diversity and genetic variation for mating tactics (8). A third alternative is that male and female mating strategies are in conflict (43). When sperm densities are high, selection may favor eggs with rare receptor mutations that slow sperm entry to limit polyspermy (20). Selection on males to overcome this barrier may in turn lead to selection for mutant bindins. This molecular coevolution could generate positive selection of gamete recognition proteins (20), monophyly of species at recognition loci, and intraspecific polymorphism.
Positive selection for amino acid variation has been observed in a number of other loci expressed on gamete surfaces in mammals, molluscs, and plants (18, 44, 45) and is a common feature of bindin sequences in sympatric sea urchins (21, 28). To date, however, the underlying mechanisms responsible for positive selection are poorly understood. Metz et al. (46) showed that the positive selection signal was absent in allopatric species of the sea urchin genus Arbacia, suggesting that selection within a species was not enough to “rachet along the differentiation of bindin” (p. 194). If this were also true in the broadly sympatric species of Echinometra (47), then polymorphisms of bindin would be predicted to be neutral, as they may be in the genus Arbacia. However, the results reported here on functional differences among bindin alleles show that selection within species of Echinometra is likely to be an important component of rapid evolution at this locus.
Among the 10 species of sea urchins in which bindin has been studied in detail, all show amino acid polymorphism except Arbacia dufresnei (Table 3). Interestingly, species showing positive selection for bindin tend to have polymorphisms for insertions and deletions similar to those that define functional differences in E. mathaei bindin alleles (Fig. 1). These associations suggest that functional differentiation of bindins is not strictly because of amino acid substitution and that positive selection may operate on length variation as well.
Table 3.
Examples of bindin amino acid polymorphism in sea urchins
| Species | Sequence* | No. of sequences | No. of alleles | No. of variable amino acids | No. of variable indels | Positive selection? | Ref. |
|---|---|---|---|---|---|---|---|
| Echinometra mathaei | 5′ | 85 | 15 | 16 | 4 | Y | This study |
| Echinometra oblonga | 5′ | 16 | 9 | 11 | 1 | Y | 21 |
| Echinometra sp. A | 5′ | 8 | 6 | 5 | 0 | Y | 21 |
| Strongylocentrotus pallidus | FL | 10 | 7 | 4 | 6 | Y | 48 |
| Strongylocentrotus droebachiensis | FL | 13 | 5 | 19 | 5 | Y | 48 |
| Strongylocentrotus franciscanus | 5′ | 134 | 14 | 9 | 0 | N | 49 |
| Arbacia lixula | FL | 7 | 5 | 4 | 0 | N | 46 |
| Arbacia punctulata | FL | 4 | 3 | 3 | 0 | N | 46 |
| Arbacia incisa | FL | 5 | 3 | 2 | 0 | N | 46 |
| Arbacia dufresnei | FL | 4 | 1 | 0 | 0 | N | 46 |
*Sequences compared are either the coding region 5′ of the intron (5′) or the complete coding region of the mature bindin gene (FL).
Other reproductive loci, like mating type loci in the basidiomycetous fungus Coprinus cinereus, are also known to be polymorphic (50), but this contrasts strongly with the low polymorphism in reproductive genes like abalone lysin and 18-kDa proteins (51), and in mating type loci among strains of Chlamydomonas (33). Rapid, species-wide selective sweeps have been proposed as the mechanism generating high rates of amino acid evolution and low polymorphism in lysin and the 18-kDa protein. If this is true, then periodically, a new amino acid substitution must confer such strong reproductive benefits that a new allele quickly comes to dominate widespread coastal populations. The dynamics of selection on these alleles and subsequent geographic spread is speculative because so few polymorphisms exist in current populations. For bindin, selection on individual alleles and rapid geographic spread are clearly observed. Allele clades A and B have recently evolved but have come to dominate E. mathaei across a large part of its range in the Indo-West Pacific. Although it is possible that we are witnessing a selective sweep in progress, the higher fertilization by AA males with AA females and higher fertilization by BB males with BB females may provide a mechanism to help prevent a sweep to fixation of any one allele type.
Eggs have often been thought of as passive acceptors of sperm (15), and fertilization has been characterized as a race among males (9). This view has been increasingly challenged as the genes involved in reproduction have become more widely understood (52). The current results show that positive selection on male gamete interaction loci is associated with strong differences among alleles in fertilization properties, but that both male and female loci are intrinsic to fertilization choice patterns. Because different bindin alleles are favored in interactions with eggs of different females, strong polymorphisms at loci intrinsic to reproductive success may be maintained without rapid selective sweeps. In this case, variation at female loci can help stabilize polymorphisms and allow the persistence of multiple, functionally different alleles at loci directly linked to fitness (2, 32). This population level sorting depends on both male and female genotype and may be the basis for some observed variation in reproductive function between individuals.
Acknowledgments
David Plouffe provided technical assistance. The manuscript was improved by discussions with P. Barber, S. Cohen, T. Duda, D. Haig, M. Hare, K. Ingram, K. Shaw, and two anonymous reviewers. This work was supported by the National Science Foundation.
Footnotes
References
- 1.Clark A G, Aguadé M, Prout T, Harshman L G, Langley C H. Genetics. 1995;139:189–201. doi: 10.1093/genetics/139.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prout T, Clark A G. Genetics. 1996;144:401–408. doi: 10.1093/genetics/144.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomaz D, Beall E, Burke T. Proc R Soc London Ser B. 1997;264:219–226. [Google Scholar]
- 4.Yamana A. Behav Process. 1998;43:239–249. doi: 10.1016/s0376-6357(98)00018-7. [DOI] [PubMed] [Google Scholar]
- 5.Vinnedge B, Verrell P. Anim Behav. 1998;56:443–448. doi: 10.1006/anbe.1998.0776. [DOI] [PubMed] [Google Scholar]
- 6.Erbelding D C, Schroeder J H, Schartl M, Nanda I, Schmid M, Epplen J T. Behav Genet. 1994;24:95–101. doi: 10.1007/BF01067933. [DOI] [PubMed] [Google Scholar]
- 7.Lanctot R B, Weatherhead P, Kempenaers B, Scribner K. Anim Behav. 1998;56:419–432. doi: 10.1006/anbe.1998.0841. [DOI] [PubMed] [Google Scholar]
- 8.Sinervo B, Lively C M. Nature (London) 1996;380:240–243. [Google Scholar]
- 9.Birkhead T R. Evolution. 1998;52:1212–1218. doi: 10.1111/j.1558-5646.1998.tb01848.x. [DOI] [PubMed] [Google Scholar]
- 10.Cooper G, Miller P L, Holland P W. Proc R Soc London Ser B. 1996;263:1343–1349. doi: 10.1098/rspb.1996.0197. [DOI] [PubMed] [Google Scholar]
- 11.Baur B. Behav Ecol Sociobiol. 1994;35:413–421. [Google Scholar]
- 12.Urbani N, Sainte-Marie B, Sevigny J M, Zadworny D, Kuhnlein U. Can J Fish Aquat Sci. 1998;55:1104–1113. doi: 10.1046/j.1365-294x.1998.00306.x. [DOI] [PubMed] [Google Scholar]
- 13.Lewis S, Austad S. Am Nat. 1990;135:351–359. [Google Scholar]
- 14.Georgiou I, Konstanelli M, Syrrou M, Messinis I, Lolis D E. Hum Reprod. 1997;12:1430–1433. doi: 10.1093/humrep/12.7.1430. [DOI] [PubMed] [Google Scholar]
- 15.Eberhard W. Female Control: Sexual Selection by Cryptic Female Choice. Princeton: Princeton Univ. Press; 1996. p. 501. [Google Scholar]
- 16.Palumbi S R. Trends Ecol Evol. 1992;7:114–118. doi: 10.1016/0169-5347(92)90144-Z. [DOI] [PubMed] [Google Scholar]
- 17.Shaw A, McRee D E, Vacquier V D, Stout C D. Science. 1993;262:1864–1867. doi: 10.1126/science.8266073. [DOI] [PubMed] [Google Scholar]
- 18.Vacquier V D, Swanson W J, Hellberg M. Dev Growth Differ. 1995;37:1–10. doi: 10.1046/j.1440-169X.1995.00001.x. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y-H, Ota T, Vacquier V D. Mol Biol Evol. 1995;12:231–238. doi: 10.1093/oxfordjournals.molbev.a040200. [DOI] [PubMed] [Google Scholar]
- 20.Palumbi S R. In: Endless Forms: Species and Speciation. Howard D, Berlocher S, editors. New York: Oxford Univ. Press; 1998. pp. 271–278. [Google Scholar]
- 21.Metz E C, Palumbi S R. Mol Biol Evol. 1996;13:391–406. doi: 10.1093/oxfordjournals.molbev.a025598. [DOI] [PubMed] [Google Scholar]
- 22.Minor J, Gao B, Davidson E. In: The Molecular Biology of Fertilization. Schatten H, Schatten G, editors. San Diego: Academic; 1989. pp. 73–88. [Google Scholar]
- 23.Vacquier V D, Moy G W. Proc Natl Acad Sci USA. 1977;74:2456–2460. doi: 10.1073/pnas.74.6.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vacquier V D, Lee Y-H. Zygote. 1993;1:181–196. doi: 10.1017/s0967199400001465. [DOI] [PubMed] [Google Scholar]
- 25.Aguadé M. Genetics. 1998;150:1079–1089. doi: 10.1093/genetics/150.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark A G, Kao T-H. Proc Natl Acad Sci USA. 1991;88:9823–9827. doi: 10.1073/pnas.88.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metz E C, Yanagimachi H, Palumbi S R. In: Biology of Echinodermata. Yanagisawa T, Yasumasu I, Oguro C, Suzuki N, Motokawa T, editors. Rotterdam: Balkema; 1991. pp. 131–138. [Google Scholar]
- 28.Biermann C. Mol Biol Evol. 1998;15:1761–1771. doi: 10.1093/oxfordjournals.molbev.a025902. [DOI] [PubMed] [Google Scholar]
- 29.Grula J W, Hall T J, Hunt J A, Giugini D, Graham J, Davidson E H, Britten R J. Evolution. 1982;36:665–676. doi: 10.1111/j.1558-5646.1982.tb05434.x. [DOI] [PubMed] [Google Scholar]
- 30.France S, Tachino N, Duda T F, Shleser R A, Palumbi S R. Mar Biotech. 1999;1:261–268. doi: 10.1007/pl00011775. [DOI] [PubMed] [Google Scholar]
- 31.Jarvik G P. Ann Epidemiol. 1997;7:357–362. doi: 10.1016/s1047-2797(97)00028-8. [DOI] [PubMed] [Google Scholar]
- 32.Wu C-I. Evolution. 1986;39:66–82. doi: 10.1111/j.1558-5646.1985.tb04080.x. [DOI] [PubMed] [Google Scholar]
- 33.Ferris P J, Pavlovic C, Fabry S, Goodenough U W. Proc Natl Acad Sci USA. 1997;94:8634–8639. doi: 10.1073/pnas.94.16.8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mauk R, Jaworski N, Kamei D, Glabe C. Dev Biol. 1997;184:31–37. doi: 10.1006/dbio.1997.8512. [DOI] [PubMed] [Google Scholar]
- 35.Miceli C, LaTerza A, Bradshaw R A, Luporini P. Proc Natl Acad Sci USA. 1992;89:1988–1992. doi: 10.1073/pnas.89.5.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marin I. J Evol Biol. 1994;7:303–314. [Google Scholar]
- 37.Payne R J H, Krakauer D C. Evolution. 1997;51:1–9. doi: 10.1111/j.1558-5646.1997.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 38.Turner G, Burrows M T. Proc R Soc London Ser B. 1995;260:287–292. [Google Scholar]
- 39.Takahata N. Proc Natl Acad Sci USA. 1990;87:2419–2423. doi: 10.1073/pnas.87.7.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes A L, Nei M. Nature (London) 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 41.Ioerger I R, Clarke A G, Kao T-H. Proc Natl Acad Sci USA. 1990;87:9732–9735. doi: 10.1073/pnas.87.24.9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richman A D, Kohn J R. Trends Ecol Evol. 1996;11:497–502. doi: 10.1016/s0169-5347(96)10051-3. [DOI] [PubMed] [Google Scholar]
- 43.Holland B, Rice W. Evolution. 1998;52:1–7. doi: 10.1111/j.1558-5646.1998.tb05132.x. [DOI] [PubMed] [Google Scholar]
- 44.Vacquier V D, Carner K R, Stout C D. Proc Natl Acad Sci USA. 1990;87:5792–5796. doi: 10.1073/pnas.87.15.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richman A D, Uyenoyama M, Kohn J R. Science. 1996;273:1212–1216. doi: 10.1126/science.273.5279.1212. [DOI] [PubMed] [Google Scholar]
- 46.Metz E C, Gomez G G, Vacquier V D. Mol Biol Evol. 1998;15:185–195. doi: 10.1093/oxfordjournals.molbev.a025914. [DOI] [PubMed] [Google Scholar]
- 47.Palumbi S R. J Exp Mar Biol Ecol. 1996;203:75–92. [Google Scholar]
- 48.Biermann C. Ph.D. dissertation. Stony Brook: State Univ. of New York; 1997. p. 233. [Google Scholar]
- 49.Debenham P. Ph.D. dissertation. Santa Barbara: University of California; 1997. p. 215. [Google Scholar]
- 50.May G, Chevanton L L, Pukkila P J. Genetics. 1991;128:529–538. doi: 10.1093/genetics/128.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Metz E C, Robles S R, Vacquier V D. Proc Natl Acad Sci USA. 1998;95:10676–10681. doi: 10.1073/pnas.95.18.10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brevis I A, Moore H D M. Hum Reprod. 1997;12S:156–165. [PubMed] [Google Scholar]