Abstract
Protein tyrosine phosphatases (PTPs) have long been thought to play a role in tumor suppression due to their ability to antagonize the growth promoting protein tyrosine kinases. Recently, a candidate tumor suppressor from 10q23, termed P-TEN, was isolated, and sequence homology was demonstrated with members of the PTP family, as well as the cytoskeletal protein tensin. Here we show that recombinant P-TEN dephosphorylated protein and peptide substrates phosphorylated on serine, threonine, and tyrosine residues, indicating that P-TEN is a dual-specificity phosphatase. In addition, P-TEN exhibited a high degree of substrate specificity, showing selectivity for extremely acidic substrates in vitro. Furthermore, we demonstrate that mutations in P-TEN, identified from primary tumors, tumor cells lines, and a patient with Bannayan–Zonana syndrome, resulted in the ablation of phosphatase activity, demonstrating that enzymatic activity of P-TEN is necessary for its ability to function as a tumor suppressor.
Keywords: cancer, tyrosine phosphorylation/signal transduction/protein tyrosine phosphatase
A variety of techniques have been used to identify genes involved in the etiology of cancer. From these studies, a surprisingly large number of protein tyrosine kinases (PTKs) have been implicated in carcinogenesis (1). The PTKs are activated by amplification, deletion, or mutation of important negative regulatory domains, or by genetic rearrangements that result in the production of activated fusion proteins (1, 2). Further support for the importance of tyrosine phosphorylation in oncogenesis comes from the finding that expression of v-crk, a small adaptor protein that does not contain intrinsic PTK activity, results in an increase in the levels of cellular phosphotyrosine and cellular transformation (3). The critical role that tyrosine phosphorylation plays in oncogenesis has led to the suggestion that many protein tyrosine phosphatases (PTPs) would act as tumor suppressors. Although PTPs have been linked to the inhibition of cell proliferation, there had been no clear cut examples of these enzymes functioning as tumor suppressors.
P-TEN, a candidate tumor-suppressor gene identified on chromosome 10, also known as MMAC1, shares homology with the PTP family, as well as with the cytoskeletal protein tensin (4, 5). P-TEN was isolated from a locus on chromosome 10, 10q22-23, which is deleted in a large number of tumors, especially glioblastomas (4, 5). Subsequently, it was shown that P-TEN is deleted or mutated in a significant fraction of glioblastomas and prostate tumors (4, 5). Importantly, germ-line mutations in P-TEN give rise to Cowden disease, which is typified by the formation of multiple, benign tumors and an increased susceptibility to malignant cancers. Mutations in P-TEN were also found in patients suffering from disorders similar to Cowden disease, such as Lhermitte–Duclos disease, which has additional pathologies, including ataxia, macrocephaly, and dysplastic cerebellar gangliocytomatosis (6, 7). The detection of germ-line mutations in four of five Cowden/Lhermitte–Duclos kindreds verified that P-TEN functions as a tumor suppressor and also suggests that P-TEN plays a role in the proper development and formation of certain tissues (8). Recent data also suggest that germ-line mutations of P-TEN are also responsible for Bannayan–Zonana syndrome (C.E., unpublished data), an autosomal dominant disorder, that in addition to the mental retardation, macrocephaly, and thyroid disease shared with Cowden disease, is characterized by lipomatosis, speckled penis, and an early onset of the neoplastic disease (9).
All PTPs contain the catalytic signature motif HCXXGXXRS/T (10). The cysteine residue in this motif is absolutely required for catalysis, as it acts as a nucleophile to attack the phosphorous atom in the phosphate moeity of its substrate, forming a thiol-phosphate intermediate (11). Mutation of this cysteine to serine or alanine results in the complete loss of phosphatase activity (12). The dual-specificity phosphatases, which catalyze the hydrolysis of phospho-seryl, -threonyl, and -tyrosyl residues, also contain the canonical PTP catalytic motif (13). The crystal structure of PTP1B, the prototypic PTP, reveals that the stringent amino acid selectivity of the PTPs is determined, in part, by the location of the catalytic cysteine at the base of a cleft (11, 14). The depth of the cleft (9 Å) matches the length of a phosphotyrosine residue and the shorter phospho-seryl and -threonyl residues are not able to reach the catalytic cysteine. In addition, the deep cleft is lined with hydrophobic residues that help stabilize the interaction with the hydrophobic tyrosine moiety (12). In contrast to PTP1B, the phosphate-binding loop in the dual-specificity phosphatase VHR is at the base of a much shallower cleft, which can accommodate all three phosphorylated hydroxyl amino acids (15). Regardless of the depth of the catalytic pocket, both classes of enzymes proceed through similar steps of catalysis. The initial nucleophilic attack of the cysteine residue results in the formation of an enzyme–substrate complex. This complex is disrupted by the catalytic acid (Asp-181 in PTP1B and Asp-92 in VHR) which protonates the phenolic oxygen of the tyrosyl group, releasing the dephosphorylated substrate from the complex. The phosphorous atom remains associated with the active site cysteine as a thiol–phosphate. In the case of the PTPs, the active enzyme is regenerated by the hydrolysis of the thiol–phosphate bond by a water molecule, possibly activated by the same aspartate residue (11, 14, 15). The efficiency of this reaction is illustrated by the finding that PTP1B can undergo ≈2,000 catalytic cycles per minute (12).
Recent data indicate that the PTPs exhibit a great deal of substrate specificity in vivo and are not simply unregulated antagonists of the signals mediated by the PTKs (12, 16). In fact, PTPs can exert both positive and negative effects on signaling pathways, indicating that they do not simply function as “off switches” (17, 18). In spite of the large number of PTKs that have been shown to play a role in oncogenesis (1), there were no examples of PTPs that function as classical tumor suppressors. Although P-TEN has been identified as a tumor suppressor and displays some of the structural features of the PTP family, its phosphatase activity has not been characterized. We have expressed P-TEN in Escherichia coli as a glutathione S-transferase (GST) fusion protein and demonstrated that P-TEN is catalytically active. Purified GST–P-TEN showed a strong preference for only the most acidic substrates. Furthermore, P-TEN was able to dephosphorylate serine, threonine, and tyrosine residues, establishing it as a dual-specificity phosphatase. Finally, we demonstrated that the phosphatase activity of P-TEN is necessary for its ability to function as a tumor suppressor, because a variety of point mutations, derived from tumor samples or Bannayan–Zonana syndrome, ablated P-TEN activity.
MATERIALS AND METHODS
Expression and Purification of GST–P-TEN.
A full-length P-TEN cDNA was generated by ligating the NotI–BglII fragment from EST264611 with the BglII–EcoRI fragment of EST365465 into pBluescript digested with NotI–EcoRI. The resulting full-length P-TEN cDNA was amplified by PCR using pfu polymerase (Stratagene) and primers that add a 5′ BamHI site (5′-CGCGGATCCATGACAGCCATCATCAAAGAGATCGTTAGC) and a 3′ EcoRI site (5′-CGCGAATTCTCAGACTTTTGTAATTTGTGTATGC). The resulting fragment was subcloned into pGEX2T (Pharmacia), and the sequence was verified by automated sequencing. Expression of P-TEN was induced in 500 ml of mid-log phase bacteria (A600 = 0.600) by the addition of isopropyl β-d-thiogalactoside to 200 μM. The culture was shifted to room temperature and expression was allowed to proceed for 12 h. The bacteria were harvested by centrifugation, the supernatant was removed, and the bacterial pellet was frozen at −80°C. The frozen pellets were resuspended in 5 ml of ice cold 20 mM Tris, 150 mM NaCl, and 5 mM EDTA (pH 8.0) supplemented with lysozyme (1 mg/ml), aprotinin (5 μg/ml), leupeptin (5 μg/ml), and benzamidine (1 mM) and incubated on ice for 15 min. The bacteria were lysed by sonicating three times for 1 min each with a Branson model 450 sonifier, power setting 4, 70% duty cycle. The lysate was cleared by centrifugation at 30,000 × g for 10 min, diluted with an equal volume of HBS (50 mM Hepes/150 mM NaCl, pH 7.4). Glutathione-Sepharose 4B (300 μl) was added, and the resulting slurry was incubated at 4°C on a rocking platform for 1–2 h. The glutathione-Sepharose was washed five times, each with 10 ml of ice cold HBS, and the washed fusion proteins were eluted with a solution containing 20 mM glutathione, 50 mM Hepes, and 30% glycerol (pH 8.0). Protein concentrations were determined by the method of Bradford, using BSA as a standard, and the integrity of the fusion proteins were verified by SDS/PAGE. Mutations identified from tumor samples and cell lines [refs. 4 and 5; and L. Hedrick (Johns Hopkins University) personal communication] or identified from Cowden disease (8) and Bannayan–Zonana (C.E., unpublished data) were introduced into pGEX2T–P-TEN using the Quickchange mutagenesis kit as described by the manufacturer (Stratagene). For all mutations, the entire P-TEN ORF was sequenced to confirm that no other mutations had been introduced.
Substrate Preparation.
All tyrosine-phosphorylated substrates were phosphorylated with the cytoplasmic fragment of the β subunit of the insulin receptor kinase (β-IRK) and purified as described (19). Serine-phosphorylated substrates were phosphorylated with recombinant protein kinase A (New England Biolabs) or with recombinant casein kinase II (a gift from D. Litchfield, University of Western Ontario) in a reaction mixture consisting of 50 mM Hepes (pH 7.2), 10 mM MgCl2, 2 mM DTT, 2 mM ATP, and 1 mCi of [γ-32P]ATP (1 Ci = 37 GBq) in a total volume of 1 ml. Casein was used at a concentration of 10 mg/ml, myelin basic protein (MBP) at 4 mg/ml, and the peptides RRRDDDSDDD (DSD) and RRREEETEEE (ETE) were used at a concentration of 0.5 mg/ml. Protein substrates were precipitated by the addition of ammonium sulfate to 80%, incubated on ice for 30 min, and harvested by centrifugation. The precipitated proteins were washed three times with 80% ammonium sulfate and then resuspended in 500 μl of 1 M Hepes (pH 7.5). The solubilized proteins were dialyzed against several changes of 50 mM imidizole (pH 7.2). Peptide substrates were purified using a Sep-pak C18 reverse phase cartridge (Waters) as described (20). The purified peptides were lyophilized to dryness and resuspended in 50 mM imidizole (pH 7.2). Random copolymers of glutamate and tyrosine, with a 4:1 ratio of glutamate to tyrosine (polyGlu4Tyr1) or with a 1:1 ratio (polyGlu1Tyr1), were purchased from Sigma and phosphorylated with β-IRK at a final polymer concentration of 1 mg/ml and purified, as described, using Sep-pak C18 reverse phase chromatography. Phosphorylated ERK2 was produced in E. coli by coexpression of activated MEK and was a gift from D. Barford (Oxford University).
Phosphatase Assays.
The standard phosphatase assay contained 10 μM substrate, 50 mM Hepes (pH 7.0), 10 mM MgCl2, and 10 mM DTT. The reaction was initiated by the addition of enzyme, typically 1–2 μg, to prewarmed (30°C) substrate mix, resulting in a final volume of 60 μl. The reactions were allowed to proceed at 30°C for the indicated times and stopped by the addition of a suspension of activated charcoal in 900 mM HCl, 90 mM NaPPi, and 2 mM NaPi (21). Dephosphorylation of ERK2 was performed essentially as described for the radioactive substrates; at the indicated times duplicate aliquots were removed and stopped by the addition of 5× Laemmli sample buffer and processed for immunoblot analysis as described (20). Immunoblots were probed with a 1:5,000 dilution of anti-mitogen-activated protein (MAP) kinase ascites or with a mixture of anti-phosphotyrosine antibodies [1:2,000 dilution of G-104 and G-2-98 ascites (22)] and were developed with ECL reagents (Amersham) in conjunction with an horseradish peroxidase-labeled rat anti-mouse κ-chain antibody (Zymed).
RESULTS
Although P-TEN displays structural features of members of the PTP family of enzymes an important, and essential, step toward understanding the function of P-TEN is the characterization of its enzymatic activity. To test whether P-TEN encodes an active phosphatase, we expressed it as a fusion protein in E. coli. As a control, we assayed all substrates with a mutant of P-TEN in which the essential cysteine from the signature motif had been mutated to serine (P-TENC124S). This mutant would be expected to be catalytically inactive, and ensures that any activity seen was the result of P-TEN and not of a copurifying bacterial phosphatase.
PTP Activity of P-TEN.
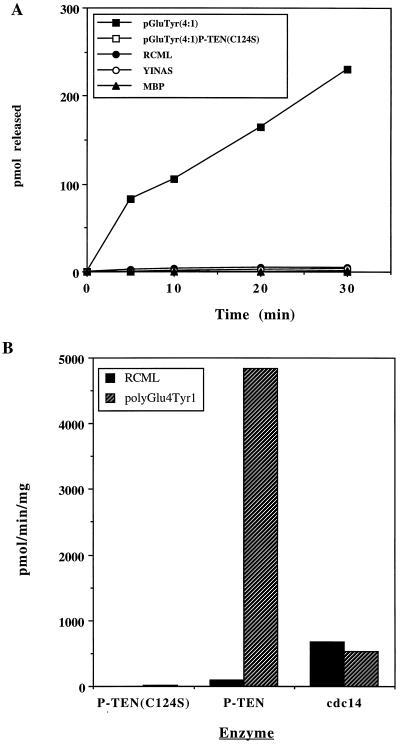
Initially, we assayed P-TEN against a number of tyrosine-phosphorylated proteins and peptides, including reduced carboxyamidomethylated and maleylated lysozyme (RCML), MBP, polyGlu4Tyr1, and polyGlu1Tyr1, as well as the peptide EDNDYINASL. The activity of P-TEN toward the classical substrates RCML and MBP was weak (<80 pmol/min per mg). However, P-TEN, exhibited robust phosphatase activity when polyGlu4Tyr1 was used as a substrate (4,840 pmol/min per mg) (Table 1 and Fig. 1A). Significantly, addition of unphosphorylated polyGlu4Tyr1 to reactions using RCML as a substrate did not result in an increase in activity toward RCML (data not shown), indicating that P-TEN was not activated by the polyanionic character of polyGlu4Tyr1. In fact, inclusion of unphosphorylated polyGlu4Tyr1 in reactions containing RCML resulted in the inhibition of the already limited dephosphorylation of RCML, suggesting that even unphosphorylated polyGlu4Tyr1 was capable of binding to P-TEN, displacing the more weakly interacting RCML. We also observed that P-TEN dephosporylated polyGlu1Tyr1. When the stoichiometries of phosphorylation were normalized, no significant differences were detected in the rates of dephosphorylation of polyGlu4Tyr1 and polyGlu1Tyr1, suggesting that the acidic character of these substrates is an important determinant of substrate recognition. P-TEN, however, exhibited reduced activity when assayed with the acidic peptide EDNDYINASL (Table 1), suggesting that the mere presence of acidic residues was not sufficient to ensure that a peptide would be a substrate. cdc14, a dual-specificity phosphatase that is closely related to P-TEN (8), does not discriminate between RCML and polyGlu4Tyr1 (Fig. 1B), indicating that polyGlu4Tyr1 is not a universal substrate for the dual-specificity phosphatases, and that the substrate specificity of P-TEN is likely to be unique feature determined by structural motifs separate from the catalytic motif. Caution should be exercised when comparing the activity of P-TEN to that of other dual-specificity phosphatases because of the inherent substrate specificity exhibited by these enzymes.
Table 1.
Activity of P-TEN measured with tyrosine, serine, and threonine phosphorylated substrates
| Substrate | Activity, pmol/min per mg |
|---|---|
| Phosphotyrosyl substrates | |
| polyGlu4Tyr1 | 4,840 ± 140 |
| RCML | 88 ± 6.6 |
| EDNDYINASL peptide | 51 ± 4.0 |
| MBP | 21 ± 1.2 |
| Phospho-seryl and -threonyl substrates | |
| DSD peptide | 210 ± 4.2 |
| ETE peptide | 161 ± 1.8 |
| Casein (casein kinase II) | 14.2 ± 1.2 |
| Casein (protein kinase A) | 27.5 ± 2.2 |
| MBP | 11.3 ± 0.6 |
P-TEN activity was measured with the indicated substrates in triplicate. The activity is expressed as the mean pmol phosphate released per min per mg enzyme ± SD.
Figure 1.
Tyrosine phosphatase activity of purified P-TEN. (A) P-TEN was tested for protein phosphatase activity using the indicated tyrosine-phosphorylated substrates. Activity is expressed as pmol of phosphate released. A catalytically inactive mutant of P-TEN (P-TENC124S) was included as a control to rule out the possibility of contaminating bacterial phosphatases. (B) Comparison of P-TEN and cdc14 activities. P-TEN and cdc14 were assayed as above with RCML or polyGlu4Tyr1, and the activity is expressed as pmol of phosphate released per min per mg.
Dual-Specificity Phosphatase Activity of P-TEN.
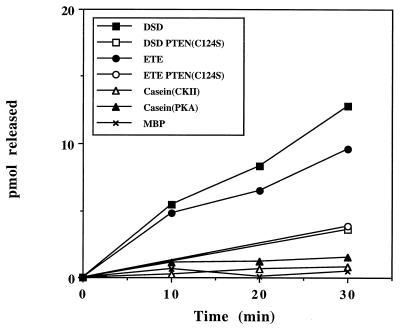
To test whether P-TEN falls into the class of dual-specificity phosphatases, we assayed activity using a number of proteins and peptides phosphorylated on serine and threonine residues. Similar to the findings with tyrosine-phosphorylated substrates, P-TEN dephosphorylated serine/threonine residues in substrates that had a preponderance of acidic residues. Specifically, P-TEN dephosphorylated two peptide substrates (DSD and ETE) with the highest efficiency (Table 1 and Fig. 2). Furthermore, P-TEN showed specificity even among acidic serine/threonine substrates, exhibiting a reduced activity when casein, phosphorylated by casein kinase II or protein kinase A, was used as substrate (Table 1 and Fig. 2). As might be anticipated in light of these properties, P-TEN exhibited almost undetectable activity when assayed with polybasic substrates, such as MBP or Kemptide (LRRASLG) (Fig. 2 and data not shown). The finding that P-TEN dephosphorylated two peptide substrates of casein kinase II also indicates that the inability of P-TEN to dephosphorylate EDNDYINASL does not simply reflect its inability to dephosphorylate small peptide substrates. Although many dual-specificity phosphatases show a preference for tyrosine residues, it is possible that the reduction in the activity of P-TEN toward serine/threonine substrates may be the result of these residues being located in a suboptimal substrate backbone.
Figure 2.
Dual-specificity phosphatase activity of purified P-TEN. P-TEN was tested for protein phosphatase activity using the indicated serine/threonine-phosphorylated substrates. Activity is expressed as pmol of phosphate released. A catalytically inactive mutant of P-TEN (P-TENC124S) was included to control for contaminating bacterial phosphatases. Casein was phosphorylated, as indicated, with casein kinase II (CKII) or protein kinase A (PKA).
Dephosphorylation of ERK2.
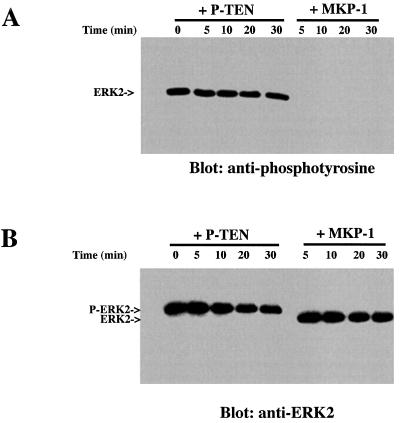
A number of dual-specificity phosphatases show a preference for members of the MAP kinase family (13, 23, 24). Therefore, we tested whether P-TEN could dephosphorylate ERK2. We could not detect dephosphorylation of ERK2 by P-TEN, either by changes in anti-phosphotyrosine antibody reactivity or by changes in the electrophoretic mobility of ERK2. In contrast, MAP kinase phosphatase 1 (MKP-1) quickly and completely dephosphorylated ERK2, as shown by the removal of phosphotyrosine from ERK2 and an increase in its electrophoretic mobility (Fig. 3 A and B, respectively).
Figure 3.
P-TEN does not dephosphorylate ERK2. Phosphorylated ERK2 was incubated with P-TEN or MKP-1 for the indicated times and ERK2 assayed for residual phosphotyrosine by immunoblotting (A) with an anti-phosphotyrosine antibody or (B) by immunoblotting with anti-MAP kinase antibodies to visualize the change in electrophoretic mobility.
Effects of Point Mutations on the Activity of P-TEN.
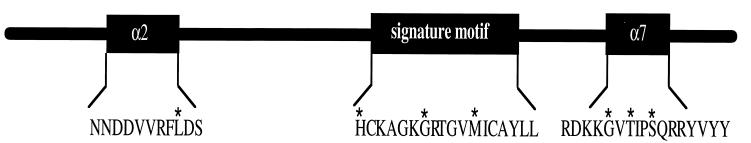
To test whether the activity of P-TEN was altered during tumorigenesis, we assessed the effect on activity of a variety of point mutations that occur in P-TEN isolated from tumor specimens. Many of these mutations occur in or near the catalytic motif of P-TEN, including His-123 → Tyr (H123Y), Gly-129 → Arg (G129R) or Gly-129 → Glu (G129E), and Met-134 → Leu (M134L). In addition, several mutations were found that occurred outside the conserved catalytic motif, including Leu-57 → Trp (L57W) and a cluster of mutations C terminal to the catalytic loop [Gly-165 → Arg (G165R), Thr-167 → Pro (T167P), and Ser-170 → Arg (S170R)]. The exact positions of these mutations in P-TEN are indicated in Fig. 4. We produced GST fusion proteins in which P-TEN was mutated at each of these positions to mimic the mutant alleles, and the phosphatase activity of the resulting recombinant proteins was measured using polyGlu4Tyr1 as a substrate (Fig. 5). With the exceptions of the M134L and the G129E mutation, all the mutations tested resulted in a dramatic decrease in the activity of P-TEN (Fig. 5). These data indicate that the catalytic activity of P-TEN has been disrupted in the majority of these tumors.
Figure 4.
Location of P-TEN mutations. A diagram of P-TEN showing the locations of the point mutations, indicated by an ∗, that were tested in this study. In addition, the predicted structural motifs (see text) in which these mutations lie is also indicated.
Figure 5.
Disruption of P-TEN activity by point mutations found in tumor samples. The indicated point mutations were introduced into recombinant P-TEN and their effects on phosphatase activity were determined. Assays were performed with polyGlu4Tyr1 for 15 min. Activity is expressed as pmol of phosphate liberated per min per mg of P-TEN. Assays were performed in triplicate and are expressed as the mean ± SD. The catalytically inactive mutant of P-TEN (P-TENC124S) was included as control to rule out the possibility of contaminating bacterial phosphatases.
The activity exhibited by the G129E mutation was surprising, especially in light of the finding that mutation of this same residue to arginine (G129R) resulted in a significant reduction of P-TEN activity. Other potential effects of this mutation on P-TEN, such as changes in protein stability in the complex environment of the mammalian cell or changes in substrate specificity, have not been addressed. On the other hand, the essentially wild-type activity exhibited by the M134L mutation was not totally unexpected because many other dual-specificity phosphatases contain a leucine residue at this position (25).
DISCUSSION
The most common chromosomal deletion in glioblastoma occurs around 10q22-23, suggesting the presence of a tumor suppressor at this locus (26). Mapping of the deletions at 10q22-23, as well as representational difference analysis (27), led to the identification of P-TEN as the tumor suppressor residing at this locus (4, 5). P-TEN shares sequence homology with the cytoskeletal protein tensin and with the family of PTPs (4, 5). A substantial fraction of all glioblastoma samples tested have either deleted or mutated P-TEN alleles. In addition, P-TEN has been shown to be disrupted in a large number of breast and prostate tumor samples (4, 5). Importantly, germ-line mutations in P-TEN give rise to a variety of genetic disorders, most notably Cowden disease, a disorder characterized by the formation of multiple benign tumors (hamartomas), as well as an increased susceptibility to malignant cancers of the breast and thyroid (28), confirming that P-TEN functions similarly to classical tumor-suppressor genes (8). Significantly, many of the mutations isolated from tumor samples, as well as from Cowden disease and Bannayan–Zonana syndrome, were predicted to disrupt the phosphatase domain of P-TEN. These studies, however, did not address whether P-TEN is a functional phosphatase or whether these mutations abrogate activity.
To address these issues, we measured the enzymatic activity of recombinant P-TEN. Initially, we were frustrated by the low activity of the fusion proteins when assayed with proteins that are commonly used as PTP substrates, such as RCML or MBP. Recently, another group has also reported weak phosphatase activity when RCML was used as a substrate (29). However, we demonstrate that P-TEN exhibits robust phosphatase activity when assayed with a random copolymer of glutamate and tyrosine, polyGlu4Tyr1, suggesting that it exhibits an unusual substrate specificity. P-TEN did not efficiently dephosphorylate the acidic peptide EDNDYINASL, suggesting that the presence of acidic residues may not be sufficient to create an optimal substrate and that higher order structures, such as those found in protein substrates, may be required.
On the basis of the presence of the “AYLL/M” motif found in many dual-specificity phosphatases, as well as the absence of some of the sequence motifs found in PTPs, particularly those that contribute to creating a deep, substrate-binding cleft (30), one would predict that P-TEN is a dual-specificity phosphatase. P-TEN was shown to exhibit activity against serine/threonine, as well as tyrosine-phosphorylated proteins (Table 1), verifying that it is a dual-specificity phosphatase. Like the tyrosine-phosphorylated substrates, P-TEN dephosphorylated only the most acidic serine/threonine-phosphorylated substrates tested. Even the best serine/threonine substrates, the ETE and DSD peptides, were not as efficiently dephosphorylated as polyGlu4Tyr1 (Table 1). Casein, phosphorylated by casein kinase II, was dephosphorylated poorly by P-TEN (Table 1 and Fig. 2), suggesting that P-TEN may require multiple acidic residues positioned both N and C terminally to the phosphorylated residue. It is currently unclear if the apparent amino acid selectivity of P-TEN is an intrinsic characteristic or if it is caused by differences in the affinity of P-TEN for polyGlu4Tyr1 and the ETE or DSD peptides, and may reflect a preference for protein over peptide substrates.
Mutations that occur in P-TEN during tumorigenesis fall into three large classes: (i) genomic deletions encompassing all or most of P-TEN, (ii) frameshift mutations resulting in the production of truncated P-TEN proteins, and (iii) point mutations resulting in the substitution of one amino acid for another (4, 5). We have chosen to introduce a variety of the point mutations found in tumor samples into P-TEN, because they are less likely than the frameshift deletions to cause unpredictable changes in the conserved secondary structure found in all PTPs (30). Comparisons of P-TEN with other phosphatases, whose crystal structures have been solved, aids in predicting how these point mutations might disrupt P-TEN activity. For example, the mutation of His-123 → Tyr (H123Y) and Gly-129 → Arg, two residues located in the catalytic motif, resulted in the complete loss of phosphatase activity. One would have predicted that the H123Y mutation, detected in an endometrial cancer, would be catastrophic to activity because of the critical importance of this histidine in correctly orienting the catalytic cysteine so it can act as a nucleophile (11). Although, Gly-129 is not as highly conserved as other residues found in the catalytic motif, the substitution of the large, positively charged side chain of arginine, a mutation found in a glioblastoma cell line, for the much smaller glycine, is likely to have a deleterious effect on the overall structure of the phosphate-binding loop and is likely to impede the binding of phosphate. In contrast, a mutation that was identified in a prostate tumor cell line, in which Met-134 was changed to Leu (M134L), had no effect on P-TEN activity. However, among the dual-specificity phosphatases, leucine is commonly found at this position (25), suggesting that the M134L allele may represent a naturally occurring polymorphism rather than a mutation.
Several other mutations that reside outside of the catalytic motif were also tested for their effect on P-TEN activity. A point mutation discovered in a glioblastoma sample that changes Leu-57 → Trp (L57W) also eliminated P-TEN phosphatase activity. This amino acid is located in a conserved α-helix (α2), which is found in both PTP1B and VHR. Although not directly involved in catalysis, this helix helps form the overall secondary structure of the enzyme (14, 15). Similarly, mutation of residues in this helix of LAR, a receptor PTP, also resulted in a significant loss of phosphatase activity (31). A second cluster of point mutations was discovered in the last conserved structural motif found in most PTPs and dual-specificity phosphatases, which also is an α-helix (14, 15). In YopH, a PTP isolated from the causative agent of bubonic plague, a hydrophobic residue in this α-helix is important for coordinating the water molecule necessary for regenerating the active enzyme (15). All three mutations found in this region—Gly-165 → Arg (found in a glioblastoma), Thr-167 → Pro (found in a breast cancer), and Ser-70 → Arg (found in a patient with a Bannayan–Zonana syndrome)—resulted in the loss of P-TEN activity. The G165R and the S170R mutations would result in substitution of relatively small, uncharged amino acids for a much larger positively charged residue, potentially disrupting important interactions between this α-helix and surrounding structures. Moreover, the substitution of Thr-167 → Pro is also likely to disrupt these interactions by interrupting the proper folding of this α-helix. These data indicate that this conserved helix (α7 in YopH and VHR) is a required motif in the dual-specificity phosphatases.
All of the mutations tested were derived from tumor samples that, with two exceptions, resulted in the complete loss of, or greatly reduced, enzymatic activity of P-TEN, even when the mutations resided well outside of the conserved catalytic motif. These observations indicate that the inhibition of enzymatic activity was required for the progression of these cells to a cancerous state.
M134L was one of two mutations found in which activity was not lost. However, leucine is commonly found at this position in other dual-specificity phosphatases (25) and the wild-type allele was not lost in the tumor from which this mutation derived. Hence, M134L is likely to be a naturally occurring polymorphism.
We were surprised by the finding that the G129E mutation, a mutation found in the germ line of patients suffering from Cowden disease, did not disrupt P-TEN activity, whereas a mutation of this same residue to arginine (found in a glioblastoma) ablated P-TEN activity. Although we were unable to detect a change in the activity of P-TEN carrying the G129E mutation, it is likely that this mutation effects the ability of P-TEN to function in vivo, as this mutation has been isolated from two independent Cowden disease kindreds but not from unaffected patients (8). Moreover, deletions of the wild-type allele were found in the tumors that develop in this Cowden disease kindred (8). Although the G129E mutation does not effect activity when measured in vitro using artificial substrates, this mutation may interfere with the ability of P-TEN to dephosphorylate its physiological targets. Furthermore, this mutation may decrease the stability or half-life of P-TEN in the complex environment of the mammalian cell. A full understanding of the effects of this mutation on P-TEN will require the identification of its physiological substrates, regulators and perhaps even the elucidation of the structural changes elicited by this amino acid substitution.
Even though the sample size is small, there does appear to be a correlation between the severity in the disruption of P-TEN activity and the pathology of the disease. In contrast to the G129E mutation, the S170R mutation exhibited a significant reduction in activity and was isolated from a patient suffering from a severe disorder, Bannayan–Zonana, that shares many features with Cowden disease, but also manifests additional pathologies, including early onset of the disease at birth (9).
To understand how P-TEN functions as tumor suppressor, it will be necessary to identify its physiological substrates. Most dual-specificity phosphatases dephosphorylate and inactivate the MAP kinases (13, 23, 24), and moreover, the MAP kinases are found to be hyperphosphorylated in breast cancer (32), a cancer that frequently contains P-TEN mutations or deletions (8). Therefore, we tested if P-TEN could dephosphorylate the MAP kinase ERK2. However, our data indicate that P-TEN was incapable of dephosphorylating ERK2 in vitro, strongly suggesting that the MAP kinases are not regulated by P-TEN in vivo. Thus, the physiological substrates of P-TEN are still unknown. The unique substrate specificity of P-TEN, however, should aid in identifying its in vivo substrates. Importantly, the requirement for acidic residues may be satisfied by the presence of phosphorylated residues, suggesting that multiply phosphorylated proteins may serve as P-TEN substrates.
Acknowledgments
We thank Drs. D. Barford, K. Lerea, and H. Charbonneau for providing reagents and Dr. L. Hedrick for providing information on P-TEN mutations in endometrial cancer. We are grateful to M. Daddario for expert technical support and to T. Tiganis, S.H. Zhang and especially to H. Charbonneau for helpful discussions. We also thank Drs. E. Fischer and B. Vogelstein for critical reading of the manuscript. This work was supported by grants to N.K.T. from the National Institutes of Health (CA53840 and GM 55989). M.P.M. was supported by a National Cancer Institute training grant (5T32 CA09311-18). C.E. is the Lawrence and Susan Marx Investigator in human cancer genetics. M.H.W. is an American Cancer Society Research Professor and is supported by the U.S. Department of the Army (DAMD-17-94-14247), the National Cancer Institute (5R35 CA39829), Amplicon Corporation, and the “1 in 9” breast cancer organization.
ABBREVIATIONS
- PTP
protein tyrosine phosphatase
- PTK
protein tyrosine kinase
- GST
glutathione S-transferase
- MBP
myelin basic protein
- MAP
mitogen-activated protein
- RCML
reduced carboxyamidomethylated and maleylated lysozyme
References
- 1.Rosen N. In: Molecular Basis of Cancer. Mendelsohn V, Howley P M, Israel M A, Liota L A, editors. Philadelphia: Saunders; 1995. pp. 105–140. [Google Scholar]
- 2.Gauwerky C E, Croce C M. In: Molecular Basis of Cancer. Mendelsohn V, Howley P M, Israel M A, Liota L A, editors. Philadelphia: Saunders; 1995. pp. 18–37. [Google Scholar]
- 3.Barker K, Hanafusa H. Mol Cell Biol. 1990;10:3813–3817. doi: 10.1128/mcb.10.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steck P A, Perhouse M A, Jasser S A, Yung W K A, Lin H, Ligon A H, Lauren A L, Baumgard M L, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng D H F, Tavtigan S V. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang S, Puc J, Milliaresis C, Rodgers L, McCombie R, Bigner S H, Giovanella B C, Ittman M, Tycko B, Hibshoosh H, Wigler M H, Parsons R. Science. 1997;275:1943–1946. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 6.Eng C, Murday V, Seal S, Mohammed S, Hodgson S V, Chaudray M A, Fentiman I S, Ponder B A J, Eeles R A. J Med Genet. 1994;31:458–461. doi: 10.1136/jmg.31.6.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eng C. J Genet Counsel. 1997;6:181–191. doi: 10.1023/A:1025664119494. [DOI] [PubMed] [Google Scholar]
- 8.Liaw D, Marsh D J, Li J, Dahia P L M, Wang S I, Zheng Z, Bose S, Call K M, Tsou H C, Peacocke M, Eng C, Parsons R. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 9.Grolin R J, Cohen M M, Condon L M, Burke B A. Am J Med Genet. 1992;44:307–314. doi: 10.1002/ajmg.1320440309. [DOI] [PubMed] [Google Scholar]
- 10.Charbonneau H, Tonks N K. Annu Rev Cell Biol. 1992;8:463–493. doi: 10.1146/annurev.cb.08.110192.002335. [DOI] [PubMed] [Google Scholar]
- 11.Barford D, Flint A J, Tonks N K. Science. 1994;263:1397–1404. [PubMed] [Google Scholar]
- 12.Flint A J, Tiganis T, Barford D, Tonks N K. Proc Natl Acad Sci USA. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun H, Charles C H, Lau L F, Tonks N K. Cell. 1993;75:487–493. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- 14.Stuckey J A, Schubert H, Fauman E B, Zhang Z-Y, Dixon J E, Saper M A. Nature (London) 1994;370:571–575. doi: 10.1038/370571a0. [DOI] [PubMed] [Google Scholar]
- 15.Yuvaniyama J, Denu J M, Dixon J E, Saper M A. Science. 1996;272:1328–1331. doi: 10.1126/science.272.5266.1328. [DOI] [PubMed] [Google Scholar]
- 16.Garton A J, Flint A J, Tonks N K. Mol Cell Biol. 1996;16:6408–6418. doi: 10.1128/mcb.16.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hertog J, Tracy S, Hunter T. EMBO J. 1994;13:3020–3032. doi: 10.1002/j.1460-2075.1994.tb06601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diamond R H, Cressman D E, Laz T M, Abrams C S, Taub R. Mol Cell Biol. 1994;14:3752–3762. doi: 10.1128/mcb.14.6.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flint A J, Gebbink M F G B, Franza B R, Hill D E, Tonks N K. EMBO J. 1993;12:1937–1946. doi: 10.1002/j.1460-2075.1993.tb05843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myers M P, Murphy M B, Landreth G E. Mol Cell Biol. 1994;14:6954–6961. doi: 10.1128/mcb.14.10.6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S-H, Eckberg W R, Yang Q, Samatar A A, Tonks N K. J Biol Chem. 1995;270:20067–20072. doi: 10.1074/jbc.270.34.20067. [DOI] [PubMed] [Google Scholar]
- 22.Garton A J, Flint A J, Tonks N K. Mol Cell Biol. 1996;16:6408–6418. doi: 10.1128/mcb.16.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwak S P, Hakes D J, Martell K J, Dixon J E. J Biol Chem. 1994;269:3569–3604. [PubMed] [Google Scholar]
- 24.Mourey R J, Vega Q C, Campbell J S, Wenderoth M P, Hauschka S D, Krebs E G, Dixon J E. J Cell Biol. 1996;271:3795–3802. doi: 10.1074/jbc.271.7.3795. [DOI] [PubMed] [Google Scholar]
- 25.Barford D. Curr Opin Struct Biol. 1995;5:728–34. doi: 10.1016/0959-440x(95)80004-2. [DOI] [PubMed] [Google Scholar]
- 26.Fults D, Pedone C A, Thomas G A, White R. Cancer Res. 1990;50:5784–5789. [PubMed] [Google Scholar]
- 27.Lisitsyn N A, L N M, Dalbagni G, Barker P, Sanchez C A, Gnarra J, Linehan W M, Reid B J, Wigler M H. Proc Natl Acad Sci USA. 1995;92:151–155. doi: 10.1073/pnas.92.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mallory S B. Dermatol Clin. 1995;13:27–31. [PubMed] [Google Scholar]
- 29.Li D-M, Sun H. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 30.Barford D, Jia Z, Tonks N K. Nature Struct Biol. 1995;2:1043–1053. doi: 10.1038/nsb1295-1043. [DOI] [PubMed] [Google Scholar]
- 31.Streuli M, Krueger N X, Thai T, Tang M, Saito H. EMBO J. 1990;9:2399–407. doi: 10.1002/j.1460-2075.1990.tb07415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sivaraman V S, Wang H, Nuovo C J, Malbon C C. J Clin Invest. 1997;99:1478–1483. doi: 10.1172/JCI119309. [DOI] [PMC free article] [PubMed] [Google Scholar]